Abstract
Direct cardiac reprogramming of fibroblasts into induced cardiomyocytes (iCMs) can be achieved by ectopic expression of cardiac transcription factors (TFs) via viral vectors. However, risks like genomic mutations, viral toxicity, and immune response limited its clinical application. Transactivation of endogenous TFs emerges as an alternative approach that may partially mitigate some of the risks. In this study, we utilized a modified CRISPRa/dCas9 strategy to transactivate endogenous reprogramming factors MEF2C, GATA4, and TBX5 (MGT) to induce iCMs from both mouse and human fibroblasts. We identified single-guide RNAs (sgRNAs) targeting promoters and enhancers of the TFs capable of activating various degrees of endogenous gene expression. CRISPRa-mediated Gata4 activation, combined with exogenous expression of Mef2c and Tbx5, successfully converted fibroblasts into iCMs. Despite extensive sgRNA screening, transactivation of Mef2c and Tbx5 via CRISPRa remained less effective, potentially due to de novo epigenetic barriers. While future work and refined technologies are needed to determine whether cardiac reprogramming could be achieved solely through CRISPRa activation of endogenous factors, our findings provide proof of concept that reliance on exogenous TFs for reprogramming can be reduced through CRISPRa-mediated activation of endogenous factors, particularly Gata4, offering a novel strategy to convert scar-forming fibroblasts into iCMs for regenerative purposes.
Keywords: MT: RNA/DNA Editing, guide RNA, CRISPRdCas9 system, endogenous transactivation, direct cardiac reprogramming, MGT
Graphical abstract
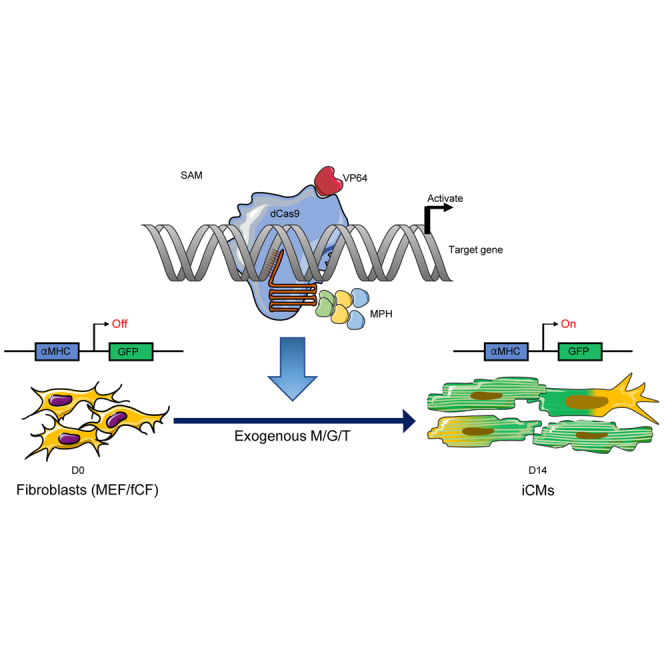
Li and her colleagues demonstrate a CRISPRa-based strategy to activate endogenous cardiac reprogramming factors, reducing reliance on viral vectors. This method successfully converts fibroblasts into induced cardiomyocytes, offering a safer approach for regenerative heart therapies.
Introduction
Acute myocardial infarction (AMI) and subsequent heart failure have remained the leading cause of death worldwide for decades, placing a substantial burden on public health.1 AMI can lead to the loss of nearly 25% of cardiomyocytes within just a few hours,2 which triggers adverse cardiac fibrosis and ventricular remodeling. While effective revascularization therapies such as percutaneous coronary intervention, thrombolytic treatment, and coronary artery bypass grafting have been developed to restore blood flow to the myocardium,3 they do not fully address the fundamental issues of cardiomyocyte loss. Although recent studies have shown that mammalian cardiomyocytes possess some regenerative capacity, the renewal rate is exceedingly slow and insufficient to compensate for the significant loss of cardiomyocytes following AMI.4,5,6,7 Therefore, it is of great importance to explore new strategies to achieve myocardial repair and regeneration.
Direct reprogramming refers to the conversion of somatic cells into another type of mature somatic cell without passing through an intermediate multipotent progenitor or stem cell stage.8 Since 2012, studies have demonstrated the use of viruses to directly reprogram fibroblasts into induced cardiomyocyte-like cells (iCMs) that are phenotypically and functionally similar to cardiomyocytes.9,10,11,12 Forced expression of three cardiac transcription factors (TFs), Mef2c, Gata4, and Tbx5 (MGT), has been shown to transform fibroblasts into functional cardiomyocytes in vitro.9 By using the same TF cocktail in an in vivo mouse model of AMI, approximately 35% of cardiac fibroblasts (CFs) in the border/infarcted zone were successfully converted into iCMs, making a significant breakthrough in efforts to develop novel therapies based on in vivo lineage reprogramming.10 As cardiac reprogramming rapidly advances toward translational application, there is a pressing need to develop reprogramming approaches that faithfully recapitulate cardiomyocyte phenotypes. Therefore, investigating whether and how endogenous activation of cardiac TFs can lead to iCMs is critical to the field.
CRISPR-Cas9 is a powerful, rapidly optimized gene-editing technology that has been adapted for a wide range of applications. By introducing mutations in both endonuclease domains, Cas9 can be engineered into an endonuclease dead form (dCas9) capable of delivering functional protein domains to genomic loci of interest.13,14 When transcriptional activators, such as VP64, are fused to the dCas9 protein, endogenous expression can be efficiently activated by targeting the dCas9 activator to the cis-acting elements of a target gene, a strategy known as CRISPR-based gene activation (CRISPRa).15 Since CRISPRa-based strategies activate endogenous gene expression, they may offer a safer alternative for clinical applications than approaches relying on exogenous expression. Furthermore, the discovery of smaller Cas enzymes has simplified delivery methods, making it possible to package single-guide RNA (sgRNAs) and dCas9 protein into nanoparticles. This advancement holds great potential to mitigate the safety risk associated with viral-based delivery methods and facilitate the translation of direct cardiac reprogramming into clinical application.
In this study, we show that CRISPRa targeting both promoter regions close to the transcription start site (TSS) and enhancer regions distant from the TSS can activate endogenous expression of the cardiac reprogramming factors MGT. We then evaluated the capacity of endogenous MGT expression to reprogram mouse embryonic fibroblasts (MEFs), mouse fresh CFs (fCFs), and human H9-derived fibroblasts (H9F). By combining endogenous Gata4 expression through CRISPRa with exogenous Mef2c and Tbx5 expression from retroviruses, we successfully converted MEFs and H9F into iCMs. Additionally, we screened 124 sgRNAs for Mef2c and 18 sgRNAs for Tbx5 with limited success in transactivating these two genes, likely due to the inaccessibility nature of the chromatin regions at the genomic loci. Nevertheless, the combinatorial approach of Gata4 transactivation with exogenous overexpression of Mef2c and Tbx5 to generate iCMs provides a proof of principle for future refinement of this strategy.
Results
Experimental design
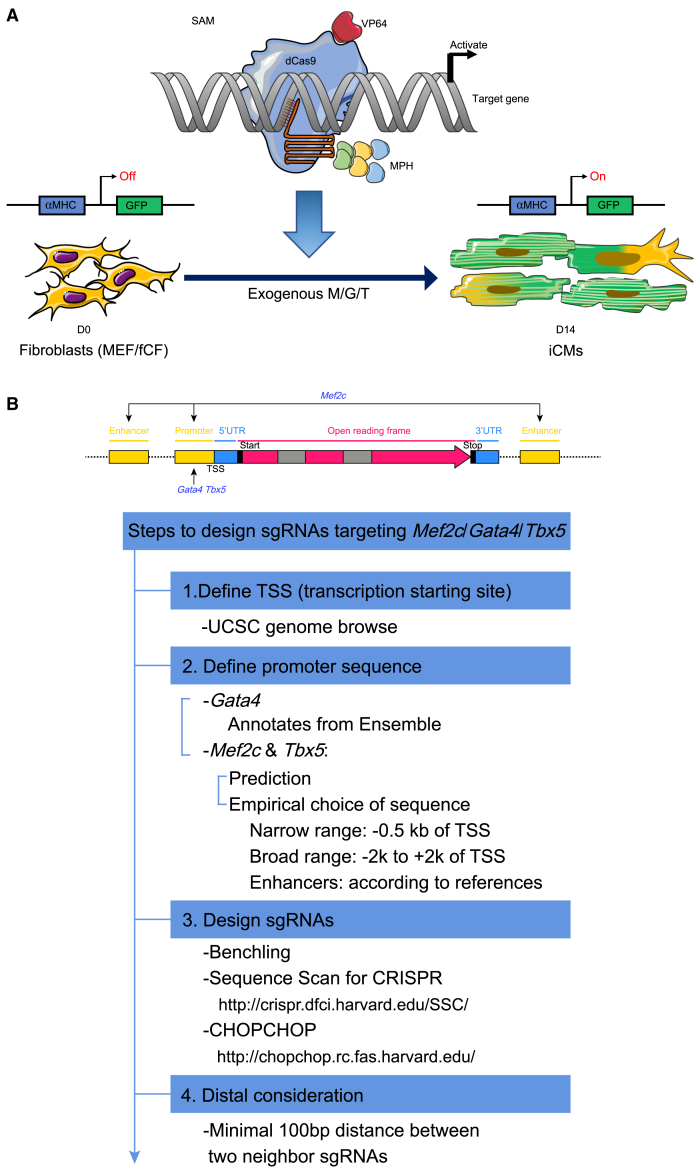
We utilized the second-generation CRISPRa system known as synergistic activation of mediators (SAM),16 which consists of a dCas9-VP64 fusion, a sgRNA containing two MS2 RNA aptamers, and the MCP-p65-hsf1 (MPH) activation helper protein. Using this system to activate endogenous MGT expression, we aimed to leverage SAM CRISPRa to induce cardiac reprogramming (Figure 1A). To begin, we analyzed the structure of the MGT loci to locate their TSSs (narrow range: −0.5 kb of TSS; broad range: −2 kb to +2 kb of TSSs), their promoters, and potential upstream and downstream enhancers. We designed MGT-targeting sgRNAs for these regions using the algorithms Chopchop, Benchling, and Sequence Scan for CRISPR, excluding those with high off-target scores for testing (Figure 1B).
Figure 1.
Graphical representation of cardiac reprogramming by CRISPR-dCas9-based transcriptional activators and sgRNAs design criteria
(A) Schematic of reprogramming mouse embryonic fibroblasts (MEFs) and fresh cardiac fibroblasts (fCFs) into induced cardiomyocytes (iCMs) via SAM system-mediated gene activation. Activation of transcriptional factors, including Gata4, Mef2C, and Tbx5, can reprogram MEFs and fCFs toward iCMs. (B) A graphical workflow for efficient sgRNA design. The process includes defining the TSS, retrieving the promoter sequence, designing sgRNAs, and considering distal regulatory elements for optimal sgRNA performance.
Initial tests of VP64 and SAM activators in MEFs
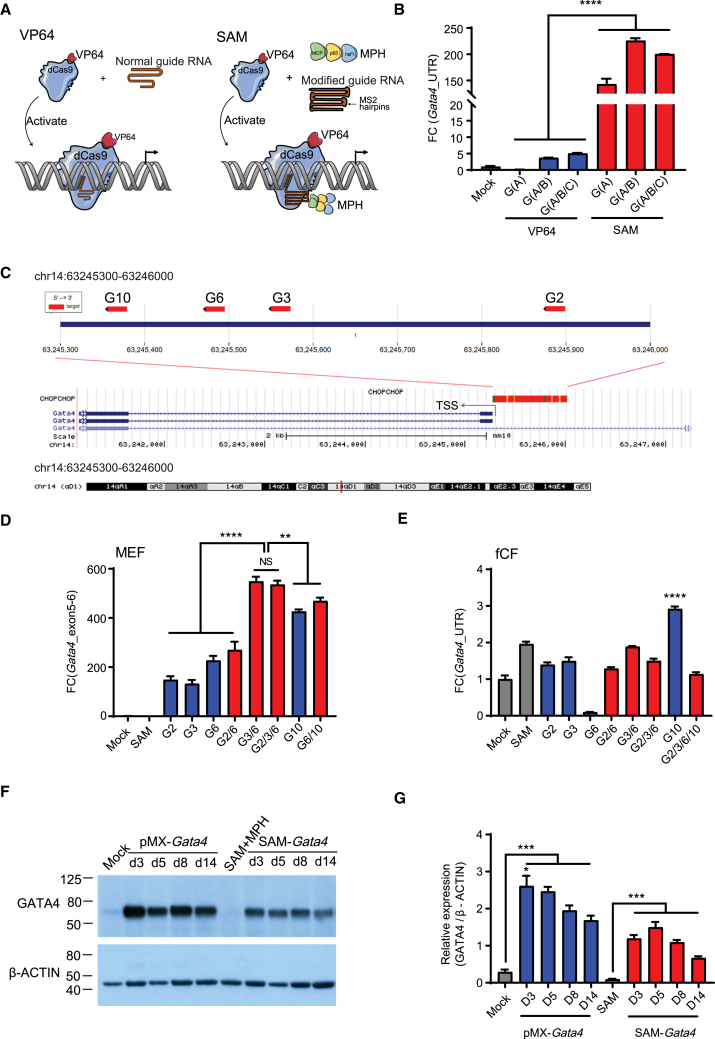
We began by comparing the performance of the first-generation dCas9-VP64 CRISPRa platform with the SAM system by evaluating their ability to activate endogenous Gata4 in MEFs 3 days post-transfection (Figure 2A). The SAM system activated Gata4 expression approximately 150-fold, whereas dCas9-VP64 alone achieved only a 5- to 10-fold activation (Figure 2B). Based on these results, we opted to use the SAM system for the remainder of the study.
Figure 2.
Evaluation of VP64 and SAM systems for Gata4 activation in MEFs in MEFs and fCFs
(A) Schematic of the different systems tested. (B) Comparative analysis of the VP64 and SAM system for Gata4 activation in MEFs. (C) Schematic showing the location of the efficient sgRNA-targeted sites for Gata4 with all four efficient Gata4-sgRNAs (G-sgRNAs) located in −600 to 0 kb of the TSS for the Gata4 gene. (D and E) Transcriptional activation of endogenous Gata4 (Gata4_UTR) in MEFs (D) and fCFs (E) using the SAM system. (F and G) Western blotting analysis of GATA4 protein levels in MEFs with targeted activation (SAM-Gata4) or ectopic overexpression (pMx-Gata4) of Gata4 at various time points post-infection. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Activation of endogenous Gata4 in MEFs and fCFs with dCas9-SAM system
We designed an additional 15 candidate sgRNAs targeting the mouse Gata4 gene (G-sgRNAs) between −1 kbp and +200 bp of the TSS. MEF and fCF were infected with lentiviruses encoding the SAM components and the sgRNAs, and Gata4 transactivation was measured after antibiotic selection. The sequence, location, and activation efficiency of each sgRNA are detailed in Table S1. Compared to mock and non-targeted SAM control, activation of endogenous Gata4 by G-sgRNAs was significantly increased in MEFs, with Gata4-sgRNAs 2, 3, 6, and 10 (G2, G3, G6, and G10) showing the highest activation. These four G-sgRNAs were located between −600 and 0 bp of TSS of Gata4 (Figure 2C).
We then tested whether the co-expression of G2, G3, G6, and G10 in various combinations would further enhance Gata4 transactivation. In MEF, G10 alone mediated the highest activation (284.69 ± 18.10-fold higher than endogenous Gata4 expression), and combining two or three G-sgRNAs significantly boosted the expression level to a 500-fold increase (Figure 2D). In contrast, in fCFs, the activation of endogenous Gata4 by G-sgRNAs was lower, with G10 mediating the highest degree of activation (approximately 3-fold), and combining multiple sgRNAs did not further enhance transactivation (Figure 2E).
To determine whether Gata4 transactivation also leads to increased protein levels, we compared the effect of SAM CRISPRa with G-sgRNAs and the overexpression of Gata4 using retrovirus (pMx-Gata4) by analyzing GATA4 protein levels at different time points post-infection. Both the pMx-Gata4 and G-sgRNA groups significantly increased GATA4 protein levels in MEFs (Figure 2F). A time course analysis of protein expression revealed that exogenous GATA4 expressed by the pMx-Gata4 retroviral vector peaked at day 3, while G-sgRNA-mediated activation of endogenous GATA4 reached a peak at day 5 (Figure 2G).
Activation of endogenous Mef2c in MEFs and fCFs with dCas9-SAM system
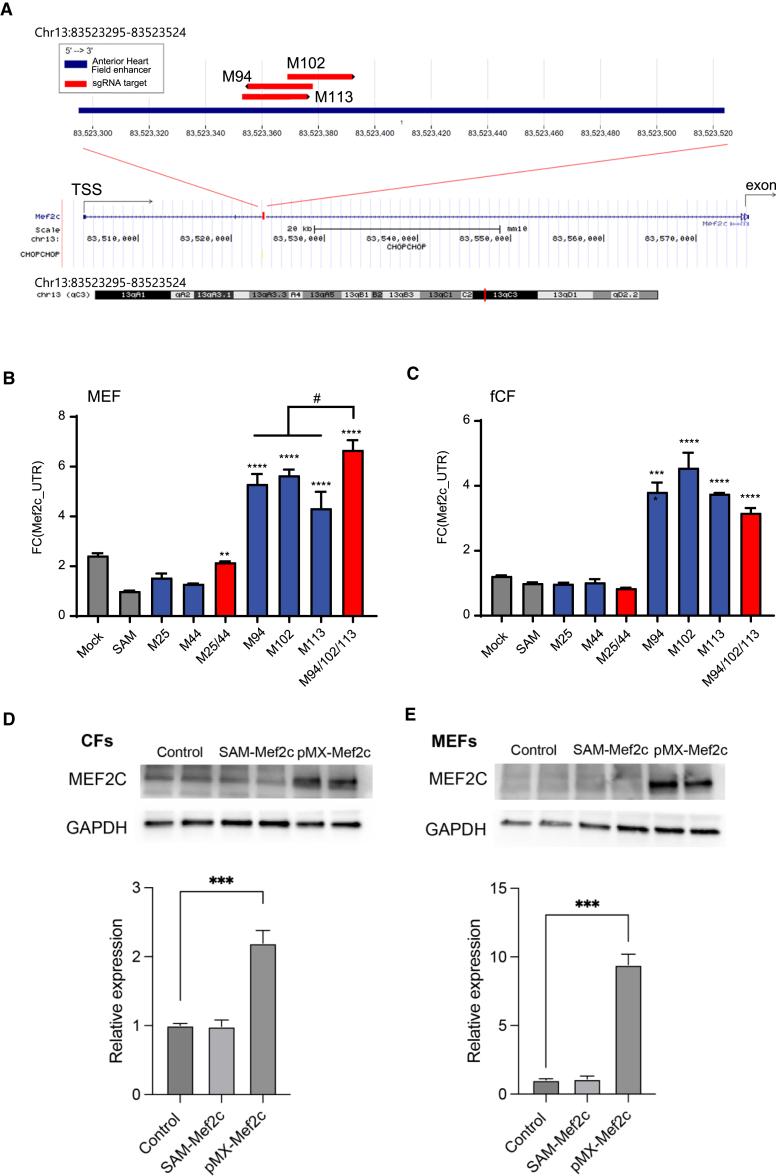
A total of 124 candidate Mef2c sgRNAs (M-sgRNAs) were designed targeting −2 to +2 kbp from the TSS and three enhancers of the Mef2c gene (Table S2). The three enhancers convey muscle, heart field, and endothelial cell-specific expression of Mef2c.17,18,19 The results revealed that compared to the mock and non-targeted SAM controls, most of the M-sgRNAs failed to effectively activate endogenous Mef2c expression (Mef2c_UTR) except for designed Mef2c-sgRNAs 25, 44, 94, 102, and 113 (M25, M44, M94, M102, and M113). We then tested the activation efficiency of these five sgRNAs individually and in combination. M94, M102, and M113, which are targeted to the heart field expression of Mef2c, mediated the highest level of transactivation in MEFs and fCFs (Figure 3A). In MEFs, a single M-sgRNA activated Mef2c expression to 1.60 ± 0.09-fold higher than endogenous Mef2c, and the combined use of two or three M-sgRNAs increased expression by around 6-fold (Figure 3B). In fCFs, a single M-sgRNA activated Mef2c 2.99 ± 0.06-fold. Multiple M-sgRNAs in combination did not further enhance transactivation (Figure 3C). Although the single M-sgRNA activated Mef2c at the mRNA level, SAM-Mef2c failed to activate MEF2C at the protein level in both CFs and MEFs (Figures 3D and 3E), suggesting the inefficacy of the sgRNA. Future work should focus on improving transactivation, not only for Mef2c but also for other reprogramming factors, by utilizing AI-based sgRNA efficiency prediction and leveraging new knowledge about novel enhancer and promoter regions.
Figure 3.
Transcriptional activation of Mef2c in MEFs and fCFs using the SAM system
(A) Schematic of the sgRNA-targeted site for Mef2c, with all three efficient Mef2c-sgRNAs (M-sgRNAs) located within the anterior heart field enhancer of Mef2c gene. (B and C) Transcriptional activation of endogenous Mef2c (Mef2c_UTR) in MEFs (B) and fCFs (C) using the SAM activators. (D and E) Western blotting analysis of MEF2C protein levels in CFs (D) and MEFs (E) with targeted activation (SAM-Mef2c) or ectopic overexpression (pMx-Mef2c). ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
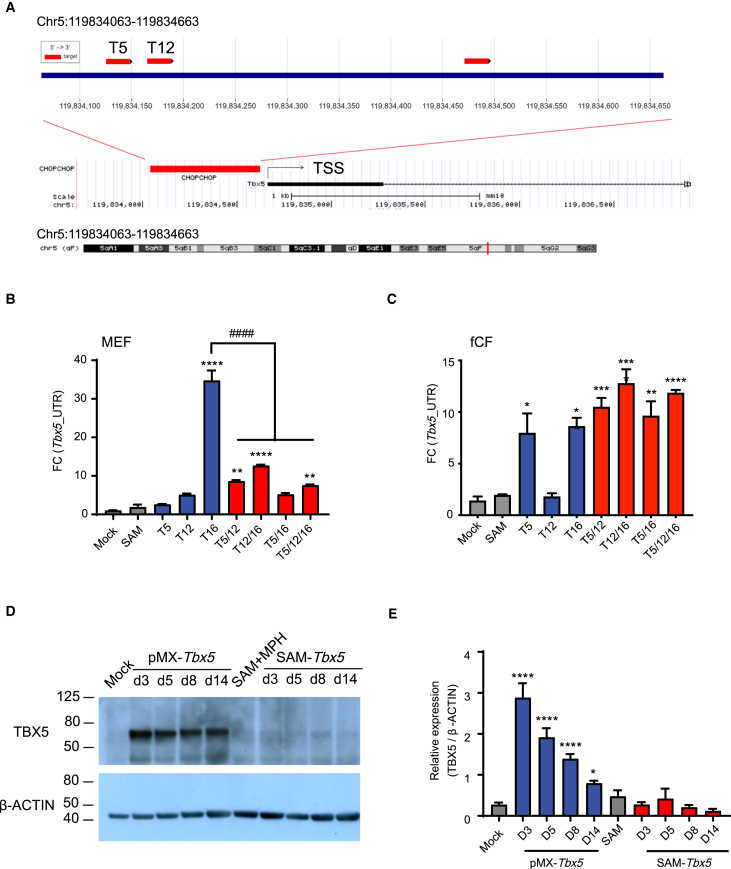
Activation of endogenous Tbx5 in MEFs and fCFs with dCas9-SAM system
We designed 18 candidate Tbx5 sgRNAs (T-sgRNAs) targeting regions spanning −2 to +2 kbp from the TSS (Table S3). Most T-sgRNAs did not effectively activate endogenous Tbx5 expression compared to the mock and non-targeted SAM controls. However, Tbx5-sgRNAs 5, 12, and 16 (T5, T12, and T16), located within the −600- to 0-kbp region of the Tbx5 TSS, showed significant activation (Figure 4A). In MEFs, T16 alone activated Tbx5 expression (Tbx5_UTR) approximately 34.74 ± 4.50-fold. Interestingly, unlike Gata4 and Mef2c CRISPRa, combining multiple T-sgRNAs unexpectedly reduced transactivation compared to T16 alone (Figure 4B). In fCF, T16 and T5 each activated Tbx5 by about 7-fold. No further increase in transactivation was observed with T-sgRNA combinations (Figure 4C). To determine whether Tbx5 transactivation also led to increased protein levels, we compared the effect of SAM CRISPRa with T-sgRNAs to Tbx5 overexpression using the pMx-Tbx5 retrovirus at different time points post-infection. Compared with the mock and non-targeted SAM controls, pMx-Tbx5 significantly increased the expression of TBX5 protein level in MEFs. At the same time, the T-sgRNA group only showed a modest increase in TBX5 protein expression, which did not reach statistical significance (Figures 4D and 4E).
Figure 4.
Transcriptional activation of Tbx5 in MEFs and fCFs using the SAM system
(A) Schematic of sgRNA-targeted sites for Tbx5 with all three efficient Tbx5-sgRNAs (T-sgRNAs) located in −600 to 0 kb of the TSS for the Tbx5 gene. (B and C) Transcriptional activation of endogenous Tbx5 (Tbx5_UTR) in MEFs (B) and fCFs (C) using the SAM system. (D and E) Western blotting analysis of TBX5 protein expression in MEFs following targeted activation (SAM-Tbx5) or ectopic overexpression (pMx-Tbx5) at various time points post-infection. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
OCT1 does not boost sgRNA expression
OCT1 is a widely expressed protein in mammalian cells that recognizes a conserved octamer sequence.20 Previous studies have reported that Oct1 overexpression enhances sgRNA expression by increasing U6 promoter activity.21 To test whether Oct1 can augment the expression of MGT sgRNAs, we used a lentiviral-based system to overexpress Oct1. Although successful overexpression of Oct1 was confirmed (Figure S1A), no enhancement of CRISPRa activity for MGT sgRNAs was observed (Figures S1B–S1G). Thus, Oct1 overexpression was not used in subsequent experiments.
CRISPRa-mediated activation of Gata4 in combination with ectopic retroviral Mef2c and Tbx5 expression induces cardiac reprogramming in MEF
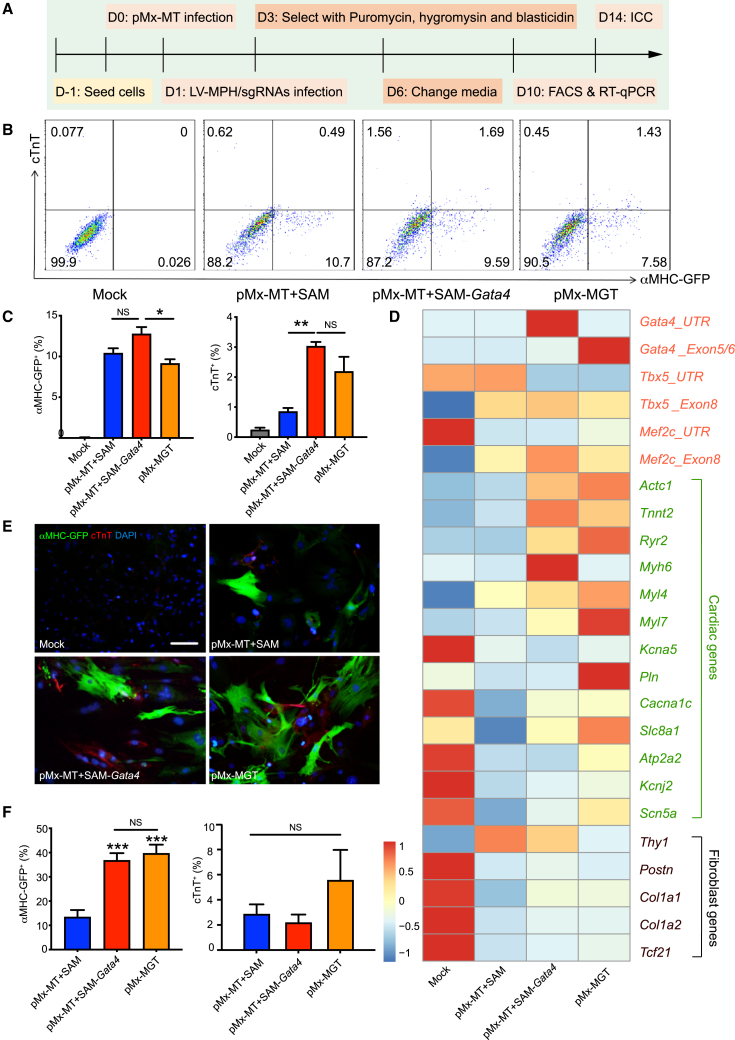
Since the G-sgRNAs demonstrated the highest efficiency in activating their target genes, we evaluated whether CRISPRa-mediated Gata4 expression, in combination with ectopic retroviral expression of Mef2c and Tbx5, could improve reprogramming efficiency compared to pMx-MT or pMx-MGT (Figure 5A). Flow cytometry analysis revealed significant induction of α-major histocompatibility complex (αMHC)-GFP and cardiac troponin T (cTnT) by pMx-MT+SAM, pMx-MT+SAM-Gata4, and pMx-MGT, with the highest efficiency observed in the pMx-MT+SAM-Gata4 group (12.77% αMHC-GFP+ and 3.03% cTnT+; Figures 5B and 5C). Additionally, quantitative real-time PCR (real-time qPCR) was used to measure endogenous and total reprogramming factor expression levels, as well as cardiomyocyte and fibroblast markers, on 8 days post-infection (Figure 5D). Endogenous Gata4 expression was effectively activated by SAM-Gata4, while total Gata4 expression (Gata4_Exo5/6) was the highest in the pMGT group, indicating specific activation of Gata4 by CRISPRa, albeit to a lesser extent than ectopic retroviral Gata4 expression. The expression of sarcomere components and myosin-related genes (Actc1, Tnnt2, Myh6, Myl4, Myl7) was increased in both the pMx-MT+SAM-Gata4 and pMx-MGT groups compared to pMx-MT+SAM. Similarly, the expression of Pln and Slc8a1, which encode key regulators of calcium handling in cardiomyocytes, was highest in the pMx-MT+SAM-Gata4 and pMx-MGT groups. In contrast, fibroblast gene expression (Postn, Col1a1, Col1a2, Tcf21) significantly decreased in all three reprogrammed groups compared to the mock group. Immunofluorescence staining for αMHC-GFP and cTnT confirmed successful cardiac reprogramming in the pMT+SAM-Gata4 and pMx-MGT groups (Figures 4E and 4F). In conclusion, the dCas9-G-sgRNA-mediated activation of endogenous Gata4, when combined with ectopic pMx-MT, can substitute for retroviral Gata4 overexpression to improve the direct cardiac reprogramming of MEFs.
Figure 5.
Reprogramming MEFs into iCMs via SAM-mediated endogenous Gata4 activation and pMx-MT-mediated Mef2c and Tbx5 ectopic overexpression
(A) Schematic of reprogramming MEFs into iCMs using SAM-Gata4 activation and pMx-MT overexpression. D, day; FACS, fluorescence-activated cell sorter; ICC, immunofluorescence staining. (B and C) Flow cytometry analysis of GFP+ and cTnT+ cells (B), with the corresponding statistical graphs (C) showing the percentage of GFP+ or cTnT+ cells in (B). (D) Heatmap displaying the expression levels of transcription factors and cardiac and fibroblast-related marker genes assessed by real-time qPCR on 10 days post-infection in the pMx-MT, pMx-MT+SAM-Gata4, pMx-MGT, and mock groups. (E and F) Immunofluorescence staining of GFP and cTnT on day 14 of reprogramming (E), with the corresponding statistical graph of the percentage of GFP+ or cTnT+ cells (F). Scale bar in (E), 100 μm. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
CRISPRa-mediated activation of endogenous MGT promotes certain cardiac gene expression and suppresses the fibroblast gene program in MEFs and fCFs
After successfully replacing ectopic Gata4 expression with SAM-Gata4 in MEF reprogramming, we investigated whether activating all three reprogramming factors using CRISPRa could induce cardiac reprogramming. Using the best-performing M-sgRNA, G-sgRNA, and T-sgRNA from our screening in MEF, we tested whether SAM-mediated activation of Mef2c, Gata4, and Tbx5 (SAM-MGT) could reprogram both MEFs and fCFs. In MEFs, the proportion of αMHC-GFP+ cells in the SAM-MGT group was significantly higher than that in the mock group (1.86 ± 0.58% vs. 0.31 ± 0.03%, p = 0.02) and the non-targeted SAM group (1.86 ± 0.58% vs. 0.55 ± 0.19%, p = 0.04). However, the proportion of cTnT+ cells did not increase compared to the non-targeted SAM group (Figures S2A and S2B). While SAM-MGT increased both endogenous and total expression of Gata4 and Tbx5, it did not induce Mef2c expression. SAM-MGT also upregulated the expression of some cardiac genes, like Actc1, Tnnt2, and Slc8a1, while downregulating fibroblast markers (Figure S2C). Moreover, immunofluorescence staining showed that a small number of GFP+/cTnT+ cells could be found (Figure S2D). These results demonstrate that CRISPRa-mediated activation of endogenous Gata4 and Tbx5 without activation of Mef2c enhances certain cardiac gene expression and suppresses fibroblast gene program in MEFs, although this effect appears to yield minimal or negligible cardiac reprogramming.
For fCF reprogramming, flow cytometry analysis showed a marginally higher ratio of GFP+ and cTnT+ cells than the mock group. However, this increase was not statistically significant compared to the non-targeted SAM group (Figures S2E and S2F). In contrast to the results observed in MEFs, fCF reprogramming resulted in a substantial increase in the expression of cardiomyocyte structural and ion channel markers in the SAM-MGT group relative to the mock group (Figure S2G). Nevertheless, the overall proportion of GFP+ and cTnT+ cell ratio remained low, as indicated by immunofluorescence staining (Figure S2H). The data suggest that inherent differences in the starting fibroblast populations may result in significantly different reprogramming outcomes independent of the reprogramming approaches employed.
Human cardiac reprogramming using CRISPRa
Next, we initiated the reprogramming of human fibroblasts into iCMs by designing and testing sgRNAs, focusing primarily on potential target sites near the TSS, as proximity to the TSS is strongly associated with sgRNA-mediated gene activation.22 For this purpose, we targeted the TSS of NCBI RefSeq transcripts for each reprogramming factor, as well as those from transcripts with high expression in human ventricular tissue, according to data from the GTEx database.23 In addition to the TSS regions, we sought to identify potential sgRNA target sites with the cardiac-specific enhancers of the reprogramming factors, as cataloged in the VISTA database, which are known to be active during embryogenesis.24 Finally, H3K27Ac enrichments at the MGT loci, as identified in the human cell lines from the ENCODE project, were considered to further optimize our selection of sgRNA target sites.25
We carried out multiple rounds of human sgRNA design, cloning into lentiSAMv2, and testing (Table 1). In each experiment, H9Fs were co-infected with the lentiSAMv2 clone and lentiMPHv2 followed by antibiotic selection and qPCR analysis. We found that the extent of CRISPRa-mediated gene activation varied significantly across sgRNAs and did not consistently correlate with their proximity to the TSS (Figures 6A–6C). Furthermore, siRNAs targeting the same genomic region exhibited considerable variability in gene activation, likely due to the intrinsic biochemical properties of the sgRNAs themselves rather than the targeted genomic regions.26 Through these efforts, we identified two sgRNAs that resulted in a 50- to 70-fold activation of MEF2C compared to the non-targeted control (Figure 6A), while three sgRNAs achieved a 200- to 300-fold activation of GATA4 (Figure 6B). Additionally, 11 sgRNAs were found to induce a 100- to 10,000-fold activation of TBX5 (Figure 6C). These screening results indicate that CRISPRa-mediated activation of human MGT is achievable and represents a promising approach for inducing cardiac reprogramming.
Table 1.
Results of human sgRNA testing
| Gene | sgRNA ID | Target sequence | Experiment 1–replicate 1 | Experiment 2–replicate 1 | Experiment 2–replicate 2 | Experiment 3–replicate 1 | Experiment 3–replicate 2 | Experiment 4–replicate 1 | Experiment 4–replicate 2 | Experiment 5–replicate 1 | Experiment 5–replicate 2 | Average vs. GAPDH | SEM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GATA4 | sgGv1-A | GCCCAGCGGAGG TGTAGCCG |
261.083 | 69.807 | 43.950 | – | – | 234.804 | 111.798 | 701.449 | 544.367 | 281.037 | 94.880 |
| GATA4 | sgGv1-B | TAGCACTTGGGC ATTTTCCG |
21.432 | – | – | – | – | – | – | – | – | 21.432 | NA |
| GATA4 | sgGv1-C | CCTGTGGGAGTC ACGTGCAA |
5.110 | – | – | – | – | – | – | – | – | 5.110 | NA |
| GATA4 | sgGv3-1 | AGGGACCATGTA TCAGAGCTTGG |
– | – | – | – | – | – | – | 314.788 | 174.231 | 244.510 | 70.279 |
| GATA4 | sgGv3-2 | TTGACCTGCGAG GGAGAGAGAGG |
– | – | – | – | – | – | – | 10.909 | 12.211 | 11.560 | 0.651 |
| GATA4 | sgGv3-4 | GCGTGGCTCCTT GACCTGCGAGG |
– | – | – | – | – | – | – | 21.198 | 16.129 | 18.664 | 2.534 |
| GATA4 | sgGv3-5 | TCGCAGGTCAAG GAGCCACGCGG |
– | – | – | – | – | – | – | 24.016 | 107.836 | 65.926 | 41.910 |
| GATA4 | sgGv3-6 | CGCAGGTCAAGG AGCCACGCGGG |
– | – | – | – | – | – | – | 173.596 | 202.758 | 188.177 | 14.581 |
| GATA4 | sgGv4-1 | TGAGGACTGAGT GCCGCGCAGGG |
– | – | – | – | – | – | – | 34.227 | 23.207 | 28.717 | 5.510 |
| GATA4 | sgGv4-2 | GGCGGGCGGCG CAAACTCGGCGG |
– | – | – | – | – | – | – | 1.433 | 0.778 | 1.105 | 0.328 |
| GATA4 | sgGv4-3 | CAGTACTTAAGG CACATCTAAGG |
– | – | – | – | – | – | – | 0.484 | 0.527 | 0.505 | 0.021 |
| GATA4 | sgGv4-4 | AGAGAGGCAGAT GTATAGACAGG |
– | – | – | – | – | – | – | 0.747 | 0.930 | 0.839 | 0.092 |
| GATA4 | sgGv4-5 | ATAAACGGGCCA AAGGTACCTGG |
– | – | – | – | – | – | – | 0.664 | 0.522 | 0.593 | 0.071 |
| GATA4 | sgGv4-6 | ATCTTCAGTCATC AAGGATGGGG |
– | – | – | – | – | – | – | 0.793 | 0.757 | 0.775 | 0.018 |
| GATA4 | sgGv4-7 | ATGTGATGCCCAG AGTCAGTGGG |
– | – | – | – | – | – | – | 0.581 | 0.836 | 0.708 | 0.128 |
| GATA4 | sgGv4-8 | CTTCCTACGATGT TTATGACTGG |
– | – | – | – | – | – | – | 0.495 | 0.562 | 0.528 | 0.034 |
| GATA4 | sgGv4-9 | AATTAAATACATC TACTGAGAGG |
– | – | – | – | – | – | – | 0.612 | 0.495 | 0.553 | 0.058 |
| GATA4 | sgGv4-10 | TCAGCATTTATCC GGAGCGGGGG |
– | – | – | – | – | – | – | 0.758 | 0.911 | 0.835 | 0.077 |
| MEF2C | sgMv1-A | TATCAAGGAAAT AAACTATA |
1.133 | – | – | – | – | – | – | – | – | 1.133 | NA |
| MEF2C | sgMv1-B | ATATATGGTATA TCACAGAC |
0.873 | – | – | – | – | – | – | – | – | 0.873 | NA |
| MEF2C | sgMv1-R1A | TCAAGGAGGTAC AAAAGGAA |
0.872 | – | – | – | – | – | – | – | – | 0.872 | NA |
| MEF2C | sgMv1-R3A | GATGAAATGAAA AACTGGAG |
1.592 | – | – | – | – | – | – | – | – | 1.592 | NA |
| MEF2C | sgMv1-R2A | TCGTGCCAAGGA AAATGGAG |
0.912 | – | – | – | – | – | – | – | – | 0.912 | NA |
| MEF2C | sgMv1-R2B | AAAGTACAGTGG GCTAACAG |
0.694 | – | – | – | – | – | – | – | – | 0.694 | NA |
| MEF2C | sgMv2-9 | CCTCCTCCTCCTC TTCCGAG |
– | – | – | 11.601 | 16.600 | – | – | – | – | 14.101 | 2.500 |
| MEF2C | sgMv2-10 | CCGAGGCCGCTC GGAAGAGG |
– | – | – | 57.353 | 62.174 | 96.684 | 98.050 | 44.579 | 53.283 | 68.687 | 9.373 |
| MEF2C | sgMv2-15 | CCTCGGCGCGCG CGAATGCG |
– | – | – | 0.734 | 0.959 | – | – | – | – | 0.847 | 0.112 |
| MEF2C | sgMv2-17 | CCGCGCATTCGC GCGCGCCG |
– | – | – | 0.672 | 0.708 | – | – | – | – | 0.690 | 0.018 |
| MEF2C | sgMv3-1 | AGCTGTGCAAGTG CTGAAGAAGG |
– | – | – | – | – | 43.275 | 64.776 | – | – | 54.026 | 10.750 |
| MEF2C | sgMv3-3 | ATATATGGTATATC ACAGACAGG |
– | – | – | – | – | 2.427 | 2.701 | – | – | 2.564 | 0.137 |
| MEF2C | sgMv3-4 | GTACCTGTGTATG CTAAGTAGGG |
– | – | – | – | – | 2.849 | 2.696 | – | – | 2.772 | 0.077 |
| MEF2C | sgMv3-6 | GCAGCTAATTCA TTTCTGAAGGG |
– | – | – | – | – | 1.633 | 1.735 | – | – | 1.684 | 0.051 |
| MEF2C | sgMv3-8 | CCATCCTATTTG CATAACGAGGG |
– | – | – | – | – | 1.155 | 1.421 | – | – | 1.288 | 0.133 |
| MEF2C | sgMv3-9 | CCCTCGTTATGC AAATAGGATGG |
– | – | – | – | – | 1.862 | 2.055 | – | – | 1.958 | 0.097 |
| MEF2C | sgMv3-10 | TCCATCCTATTT GCATAACGAGG |
– | – | – | – | – | 1.735 | 2.119 | – | – | 1.927 | 0.192 |
| MEF2C | sgMv3-11 | ACAGACATATCA TGTCTTTGTGG |
– | – | – | – | – | 2.609 | 3.066 | – | – | 2.837 | 0.229 |
| MEF2C | sgMv3-12 | ATTACATACTGG AGATCTCTGGG |
– | – | – | – | – | 2.058 | 2.641 | – | – | 2.350 | 0.291 |
| MEF2C | sgMv3-13 | ATACTGGAGATC TCTGGGTCAGG |
– | – | – | – | – | 4.082 | 3.801 | – | – | 3.941 | 0.140 |
| MEF2C | sgMv3-14 | GAAGAAAAACGG GGACTATGGGG |
– | – | – | – | – | 1.167 | 1.719 | – | – | 1.443 | 0.276 |
| MEF2C | sgMv4-1 | CTGGTCTGGCTT TATTCTGCAGG |
– | – | – | – | – | – | – | 0.859 | 0.786 | 0.823 | 0.037 |
| MEF2C | sgMv4-2 | TGCCATTTTACA TGTTCCAGTGG |
– | – | – | – | – | – | – | 1.075 | 0.841 | 0.958 | 0.117 |
| MEF2C | sgMv4-3 | CACTTATTCAGTA CTCAGCTAGG |
– | – | – | – | – | – | – | 1.424 | 1.281 | 1.353 | 0.071 |
| MEF2C | sgMv4-4 | GGTTTATTGGTAA ACATGTATGG |
– | – | – | – | – | – | – | 1.155 | 1.144 | 1.150 | 0.005 |
| MEF2C | sgMv4-5 | ATATTACTTTTCA TCGAGGGTGG |
– | – | – | – | – | – | – | 0.904 | 0.852 | 0.878 | 0.026 |
| MEF2C | sgMv4-6 | CTACCAAACTCCA CTTTCAGTGG |
– | – | – | – | – | – | – | 1.116 | 1.354 | 1.235 | 0.119 |
| MEF2C | sgMv4-7 | CCAGAGCTAGGT TGTTTATGTGG |
– | – | – | – | – | – | – | 1.067 | 1.229 | 1.148 | 0.081 |
| MEF2C | sgMv4-8 | ATGGTGTGGTCT CACCTTACTGG |
– | – | – | – | – | – | – | 0.940 | 0.962 | 0.951 | 0.011 |
| MEF2C | sgMv4-9 | CTTGTCCAAGGT TCTGCCAAAGG |
– | – | – | – | – | – | – | 1.140 | 1.408 | 1.274 | 0.134 |
| MEF2C | sgMv4-10 | GACAAATACATA CTAATTTGAGG |
– | – | – | – | – | – | – | 1.041 | 1.036 | 1.038 | 0.003 |
| TBX5 | sgTv1-A | CAAATCATTTCA AACGCGGT |
13.794 | – | – | – | – | – | – | – | – | 13.794 | NA |
| TBX5 | sgTv1-B | CTGCTTCTCTGA GCAAGCGG |
20.402 | – | – | – | – | – | – | – | – | 20.402 | NA |
| TBX5 | sgTv1-C | TGAGTGTTCGG GAAATCTAG |
10.990 | – | – | – | – | – | – | – | – | 10.990 | NA |
| TBX5 | sgTv2-12 | GACGTCACGAG TCACGCAAC |
– | – | – | 0.122 | 0.086 | – | – | – | – | 0.104 | 0.018 |
| TBX5 | sgTv2-15 | GCGCTGTCACG TTTGGCTGG |
– | 0.511 | 2.218 | 0.309 | 0.485 | – | – | – | – | 0.881 | 0.448 |
| TBX5 | sgTv2-34 | GCCGCTGCACC TATAGGACT |
– | – | – | 0.141 | 0.113 | – | – | – | – | 0.127 | 0.014 |
| TBX5 | sgTv2-35 | AAACGTGACAG CGCGCGGGC |
– | – | – | 0.751 | 0.594 | – | – | – | – | 0.673 | 0.079 |
| TBX5 | sgTv3-1 | CACCATGGCCG ACGCAGACGAGG |
– | – | – | – | – | 6,851.464 | 4,704.819 | – | – | 5,778.141 | 1,073.323 |
| TBX5 | sgTv3-2 | CTCAGAGCAGA ACCTTGCGCGGG |
– | – | – | – | – | 10,238.454 | 9,834.440 | – | – | 10,036.447 | 202.007 |
| TBX5 | sgTv3-4 | CCTCAGAGCAG AACCTTGCGCGG |
– | – | – | – | – | 13,035.915 | 12,940.372 | 16.275 | 15.789 | 6,502.088 | 3,744.777 |
| TBX5 | sgTv3-5 | TGCTCTGAGGAC AAGAAGCAGGG |
– | – | – | – | – | 1,843.998 | 1,637.516 | – | – | 1,740.757 | 103.241 |
| TBX5 | sgTv3-8 | GCACAATTCTAGT GACAGGAGGG |
– | – | – | – | – | 30,360.732 | 512.313 | – | – | 15,436.522 | 14,924.209 |
| TBX5 | sgTv3-9 | AGTGACAGGAGGGA GCAGTTTGG |
– | – | – | – | – | 381.584 | 418.221 | – | – | 399.903 | 18.318 |
| TBX5 | sgTv3-10 | TGCTGGTAGCTGGA AACTGGGGG |
– | – | – | – | – | 6.473 | 22.619 | – | – | 14.546 | 8.073 |
| TBX5 | sgTv3-11 | GCACCTCCGTTCCC AGCCGAAGG |
– | – | – | – | – | 3,237.790 | 2,387.540 | – | – | 2,812.665 | 425.125 |
| TBX5 | sgTv3-12 | TAGTAACAGTAAT AATTCTAGGG |
– | – | – | – | – | 98.805 | 80.128 | – | – | 89.467 | 9.338 |
| TBX5 | sgTv3-13 | AAAGTAATAGTAA TACTACTAGG |
– | – | – | – | – | 11.146 | 14.097 | – | – | 12.622 | 1.475 |
| TBX5 | sgTv3-14 | ACCCGCGTCAGAC CCGGAGAAGG |
– | – | – | – | – | 49.323 | 50.656 | – | – | 49.990 | 0.666 |
| TBX5 | sgTv3-16 | ATCACGGAGAGCC TGCGCAGGGG |
– | – | – | – | – | 6,474.837 | 6,722.309 | – | – | 6,598.573 | 123.736 |
| TBX5 | sgTv3-17 | TATCACGGAGAGC CTGCGCAGGG |
– | – | – | – | – | 1,998.376 | 2,470.274 | – | – | 2,234.325 | 235.949 |
| TBX5 | sgTv3-18 | TTATCACGGAGAG CCTGCGCAGG |
– | – | – | – | – | 651.057 | 946.434 | – | – | 798.745 | 147.688 |
| TBX5 | sgTv3-19 | GACGGGCAGCTTG CTTATCACGG |
– | – | – | – | – | 2,752.702 | 3,050.598 | – | – | 2,901.650 | 148.948 |
| TBX5 | sgTv3-20 | GAAGTCCAGACAA ATTTGACGGG |
– | – | – | – | – | 121.863 | 170.663 | – | – | 146.263 | 24.400 |
| TBX5 | sgTv3-21 | AGAAGTCCAGACA AATTTGACGG |
– | – | – | – | – | 1.558 | 3.550 | – | – | 2.554 | 0.996 |
| TBX5 | sgTv3-26 | ATTTTTGGAAGCC ACTGGGTAGG |
– | – | – | – | – | 691.631 | 780.797 | – | – | 736.214 | 44.583 |
| TBX5 | sgTv3-28 | GAAATTCTCTAA AGGACACTGGG |
– | – | – | – | – | 14.393 | 13.128 | – | – | 13.761 | 0.633 |
| TBX5 | sgTv3-29 | AGAAATTCTCTAA AGGACACTGG |
– | – | – | – | – | 4.952 | 3.883 | – | – | 4.417 | 0.535 |
| TBX5 | sgTv3-30 | AGAAAACTATTCC TCTTGTGAGG |
– | – | – | – | – | 44.358 | 56.180 | – | – | 50.269 | 5.911 |
| TBX5 | sgTv4-1 | GAGGTCTCTTGCA TAAGGCATGG |
– | – | – | – | – | – | – | 0.448 | 0.465 | 0.456 | 0.009 |
| TBX5 | sgTv4-2 | AACCAGCTAGAGC GGCGCCTCGG |
– | – | – | – | – | – | – | 0.253 | 0.628 | 0.441 | 0.188 |
| TBX5 | sgTv4-3 | CTTGGACCTTTCT CTCCGCAAGG |
– | – | – | – | – | – | – | 1.031 | 0.776 | 0.903 | 0.127 |
| TBX5 | sgTv4-4 | AAATAGAGTGCCT CGTGCCTCGG |
– | – | – | – | – | – | – | 0.290 | 0.514 | 0.402 | 0.112 |
| TBX5 | sgTv4-5 | GTTTTGCAACTCA ACCTAGTGGG |
– | – | – | – | – | – | – | 0.388 | 0.411 | 0.400 | 0.012 |
| TBX5 | sgTv4-6 | AGAATCGAACCCC GAAGCTGGGG |
– | – | – | – | – | – | – | 0.564 | 0.234 | 0.399 | 0.165 |
| TBX5 | sgTv4-7 | CATGCTTCGGAAA GCCCTGGGGG |
– | – | – | – | – | – | – | 2.066 | 1.102 | 1.584 | 0.482 |
| TBX5 | sgTv4-8 | GAAGCTGCCCGCT CTCCCCGCGG |
– | – | – | – | – | – | – | 9.083 | 7.181 | 8.132 | 0.951 |
| TBX5 | sgTv4-9 | TGGGAAACGAAAG CGAAGCTCGG |
– | – | – | – | – | – | – | 7.626 | 9.684 | 8.655 | 1.029 |
| TBX5 | sgTv4-10 | AAACACGAGCCTC GGATTTGGGG |
– | – | – | – | – | – | – | 8.480 | 10.090 | 9.285 | 0.805 |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NA, not applicable.
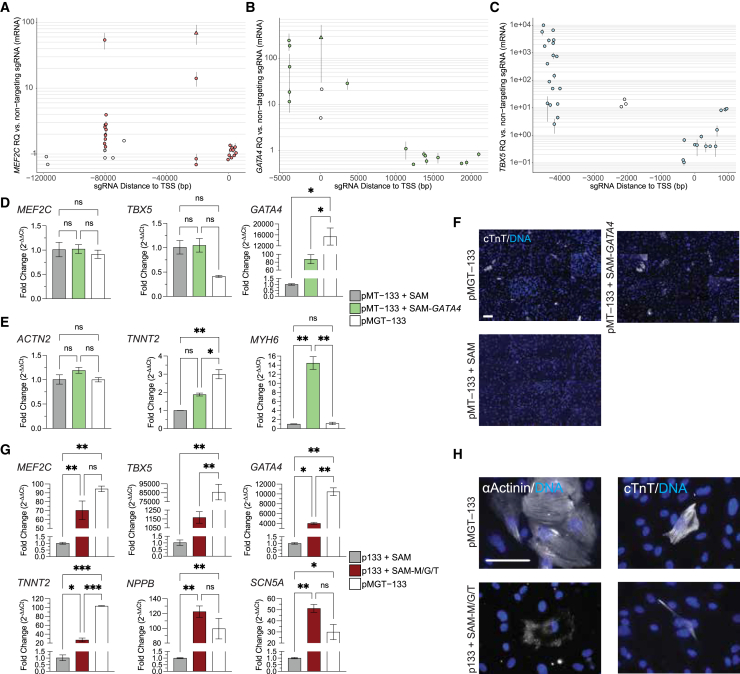
Figure 6.
Reprogramming H9Fs into iCMs via CRISPRa-mediated activation of GATA4, MEF2C, and TBX5
(A–C) Summary of sgRNAs tested for CRISPRa-mediated activation of MEF2C (A), GATA4 (B), and TBX5 (C). Data represent six independent experiments. Point range plots illustrate the activation levels induced by each sgRNA and their relative position to the TSS. Open circles, one replicate; filled circles, two replicates, one experiment; filled triangles, three or more replicates, two or more experiments. (D–F) Replacement of retroviral overexpressing GATA4 with SAM-GATA4. (D) qPCR analysis at day 2. The y axis indicates fold change by 2−ΔΔCt relative to control pMT−133+SAM. n = 2 biological replicates. (E) qPCR analysis at day 10. The y axis indicates fold change by 2−ΔΔCt relative to the pMT−133+SAM control. n = 2 biological replicates. (F) Tiled composite images of ICC staining for cTnT at day 14, with 40× images captured from 16 random fields per well. (G and H) CRISPRa activation of all three reprogramming factors with SAM-MGT (G) qPCR at day 14. The y axis indicates fold change by 2−ΔΔCt relative to the pMT−133+SAM control. n = 2 biological replicates. (H) Representative images of ICC staining for sarcomere components at day 14, acquired at 40× magnification. ns, not significant. Scale bars, 100 μm ∗p < 0.05; ∗∗p < 0.01.
Utilizing the most efficient sgRNA for each gene, we next sought to reprogram fibroblasts into human iCMs with the CRISPRa system. H9Fs were induced to overexpress GATA4 with the CRISPRa system, while MEF2C and TBX5 were overexpressed through retroviral delivery of their respective coding sequences (cDNAs), and miR-133a2 was delivered as a precursor miRNA. The inclusion of miR-133a2 with MGT was based on previous reports indicating that miR133a2 enhances the efficiency of human fibroblast conversion to iCMs.27 To control for the potential effects that miR-133-a2 would have on MGT expression, miR-133a2 was included in all human reprogramming conditions in this study.
We first assessed the expression of MGT following CRISPRa-mediated activation of GATA4. No significant differences in MEF2C or TBX5 expression were observed between non-targeting control (pMT–133+SAM), CRISPRa-mediated GATA4 (pMT–133+SAM-GATA4), and retroviral GATA4 (pMGT–133) groups, suggesting that CRISPRa-mediated activation of GATA4 does not induce the expression of MEF2C or TBX5. However, CRISPRa-mediated GATA4 expression was approximately 80-fold higher than in the non-targeting control, although still significantly lower than that achieved with retroviral GATA4 expression (Figure 6D).
Ten days after initiating reprogramming, the samples with GATA4 CRISPRa exhibited a 14-fold change increase in MYH6 expression compared to both the negative control and retroviral-mediated reprogramming (Figure 6E). Although TNNT2 expression was increased, the increase was not statistically significant compared to the control, and ACTN2 expression levels were similar across samples. By day 14, human iCMs were observed in the sample with GATA4 CRISPRa, as determined by immunofluorescence for cTnT, while no iCMs were detected in the negative control (Figure 6F). Together, these results suggest that CRISPRa-mediated activation of GATA4 substantially contributes to human cardiac reprogramming.
We next attempted to reprogram human fibroblasts into iCMs by CRISPRa-mediated activation of all three MGT factors. H9Fs were infected with separate virus vectors, each encoding a sgRNA targeting MEF2C, GATA4, or TBX5 in the human SAM-MGT system. By day 14, CRISPRa-induced activation reached approximately 70-fold for MEF2C, 1,000-fold for TBX5, and 4,000-fold for GATA4. When compared to retroviral MGT overexpression, CRISPRa-mediated activation of TBX5 and GATA4 was significantly lower, whereas no significant difference was observed for MEF2C expression (Figure 6G). Cardiomyocyte markers TNNT2, NPPB, and SCN5A were significantly upregulated in the CRISPRa-treated samples compared to the negative control (p133+SAM) (Figure 6G). Immunofluorescence staining for cTnT and α-actinin identified a limited number of human iCMs in the CRISPRa samples, although at a low frequency (Figure 6H). Notably, the cTnT signal in the CRISPRa-treated samples was qualitatively weaker, and the morphology of the human iCMs in these samples differed from those produced through retroviral MGT reprogramming. Taken together, these data indicate that a cardiac program in human fibroblasts can be activated by CRISPRa of MGT.
Discussion
In this study, we reprogrammed fibroblasts to iCMs through targeted activation of endogenous reprogramming factors. We utilized the second-generation SAM CRISPRa system as a programmable and transcriptional regulator to activate the expression level of Mef2c, Gata4, and Tbx5 in both mouse and human fibroblasts. Our findings demonstrate that endogenous activation of Gata4 via CRISPRa could effectively substitute for ectopic retroviral Gata4 expression in the reprogramming process.
A comparative study of second-generation CRISPRa platforms, including VPR, SAM, and SunTag, identified SAM as the most effective system.15 In our study, we corroborated this finding, demonstrating that SAM was more effective than the first-generation CRISPRa platform dCAS9-VP64. Notably, sgRNAs targeting Gata4 achieved potent activation in MEFs, and the use of multiplex sgRNAs further enhanced activation. This result aligns with the study of Chavez et al., which showed that the combined use of multiple sgRNAs can synergistically promote target gene expression.15,28 However, in MEFs, combining multiple T-sgRNAs produced a significant antagonistic effect on the transcriptional output compared to singlet T-sgRNAs (Figure 4B). This suggests that while the use of multiple sgRNA may theoretically enhance gene activation, empirical validation is necessary in each specific context. Additionally, we observed that the degree of gene activation varied between cell types, even when using the same sgRNA or sgRNA combination. In MEFs, for instance, several G-sgRNAs activated Gata4 hundreds of times above the endogenous level, whereas in fCFs, the activation was less than 10-fold (Figures 2D and 2E). One potential explanation for this difference is the higher basal Gata4 expression in fCFs compared to MEFs (Figure S3), indicating that CRISPRa-mediated gene activation may be inversely correlated with the basal expression level of the targeted gene, as previously described.29
Various strategies have been proposed for cardiac regeneration and repair, including cytokine administration, skeletal muscle myoblast transplantation, stem cell therapies, and engineered myocardial tissue patches.30 While these approaches have shown promise in animal models, their clinical translation has yielded unsatisfactory outcomes.31 A primary reason for these failures is likely the inability to fully address the permanent loss of cardiomyocytes and reverse the progressive decline in cardiac function. Direct cardiac reprogramming represents a promising alternative for repairing injured hearts.9,10,11,32 However, traditional reprogramming methods, which rely on retroviral infection, pose significant challenges for clinical translation in myocardial regeneration.32
Several alternative approaches have been developed to replace retroviruses-based direct cardiac reprogramming. Mathison et al. constructed a proliferation-deficient-adenoviral GMT expression vector (Ad-GMT) and successfully achieved cardiac reprogramming both in vitro and in vivo.33 Jayawardena et al. identified microRNA (miRNA) combinations, including miRNA-1, -133, -208, and -499, capable of inducing fibroblasts into iCMs.34,35,36 Inspired by the use of small molecules for the generation of induced pluripotent stem cells, Fu et al. developed a small-molecule cocktail CRFVPTZ (C, CHIR99021; R, RepSox; F, Forskolin; V, VPA; P, Parnate; and T, TTNPB) that efficiently reprogrammed MEFs to iCMs.37 These novel strategies represent significant progress toward the clinical translation of direct cardiac reprogramming. Our study provides evidence that cardiac reprogramming could be achieved with CRISPRa, presenting a theoretical opportunity to formulate nanoparticles loaded with SAM-MGT ribonucleoproteins for direct delivery to the border zone of an AMI, thereby circumventing the risks associated with using mutagenic retroviral vectors. The development of fibroblast-specific engineered nanoparticles would further accelerate the clinical translation of CRISPRa-based cardiac reprogramming.
After multiple rounds of empirical testing, we identified sgRNAs capable of activating endogenous MGT expression. However, optimizing our approaches to generate iCM with high molecular and functional fidelity, as well as robust efficiency, remains a significant challenge. Given the importance of MEF2C and TBX5 in cardiac reprogramming, future work may focus on improving the endogenous activation of these factors. Notably, in both human and mouse fibroblasts, the most efficient M-sgRNAs tested were located tens of thousands of base pairs away from the TSS, highlighting the need for a more rational approach to target enhancer elements for the CRISPR strategy. To better understand how to improve reprogramming efficiency, a thorough comparison of the molecular and functional phenotypes of iCMs generated via endogenous activation or ectopic expression of MGT should be undertaken. These future studies will help elucidate the unique mechanism underlying CRISPRa-based cardiac reprogramming, advancing our understanding of the cell fate conversion process, as well as the epigenomic, transcriptomic, and proteomic remodeling involved.
Materials and methods
Mouse lines
Transgenic mice harboring GFP under the control of the αMHC promoter9,10 were used for MEF and fCF preparation. Animal care was performed in accordance with the guidelines established by the University of North Carolina, Chapel Hill.
Generation of MEFs
Embryos from MHC-GFP reporter mice were harvested at embryonic days 12.5–14.5, and their internal organs and head were removed as previously described.16 The body below the head was minced to small pieces and incubated at 37°C for 15 min in 1 mL 0.05% trypsin-EDTA with 100 Kunitz units of DNase I per embryo. After centrifuging, the supernatant was carefully removed and the cell pellet was resuspended in warm MEF medium (DMEM/10% fetal bovine serum [FBS], 50 U/50 μg/mL penicillin/streptomycin, and 1% GlutaMAX supplement [Gibco]) and then plated on a 10-cm dish. The MEF media was changed 24 h after plating. Cells are generally 80%–90% confluent 24 h after plating. MEFs were harvested at 72 h and stored for future use.
Generation of neonatal CFs
Hearts were removed from αMHC-GFP transgenic mice and rinsed thoroughly with chilled PBS to remove blood and other tissues. The hearts were then minced roughly into 1 × 1 mm pieces, transferred to a 50-mL conical tube with 20 mL warm 0.05% trypsin-EDTA, and incubated at 37°C for 20 min. The supernatant was discarded, and the heart pieces were digested with 10 mL warm 0.2% collagenase type II in Hank’s balanced salt solution for 7 min at 37°C, followed by vortexing for 1 min. The supernatant was collected and diluted with 7 mL fibroblast medium. After five rounds of collagenase digestion and collection, a single-cell suspension was obtained by passing the cell solution through a 40-μm cell strainer. The cell suspension was then neutralized with fibroblast media. After centrifuging, the cell pellet was resuspended in 1 mL Red cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA) for 1 min on ice and then resuspended in MACS buffer (Dulbecco’s PBS with 0.5% BSA and 2 mM EDTA) for cell sorting.
H9F derivation and culture
Fibroblasts were previously differentiated from H9 embryonic stem cells and subcultured.34 Cells were expanded, frozen, and stored in liquid nitrogen. A new vial of cells was thawed for each experiment performed, and the cells were passaged at least once prior to seeding for infection. H9Fs were maintained in DMEM 20% FBS 1× non-essential amino acid medium (NEAA) 1× penicillin/streptomycin and kept in an aseptic incubator at 37°C 5% CO2 90% relative humidity.
Magnetic cell sorting
Thy1.2+ CFs were isolated by magnetic-activated cell sorting (MACS; Miltenyi Biotec) according to the manufacturer’s instructions. Briefly, cells (about 1 × 107) were suspended in 90 μL MACS buffer and incubated with 10 μL Thy1.2 biotin anti-mouse CD90.2 antibody (eBioscience, Invitrogen) for 30 min at 4°C. The cell/Thy1.2 biotin anti-mouse CD90.2 antibody/MACS solution was resuspended in 5 mL MACS buffer and centrifuged at 300 × g for 5 min. The supernatant was discarded. The cell/Thy1.2 biotin anti-mouse CD90.2 antibody pellet was then suspended in 90 μL MACS buffer and incubated with 10 μL Anti-Biotin MicroBeads (Miltenyi Biotec) at 4°C for 30 min. The cell/Thy1.2 biotin anti-mouse CD90.2 antibody/Anti-Biotin MicroBeads solution was resuspended in MACS buffer, passed through a 30-μm nylon mesh, and applied to an MACS calibrated LS column. Target cells were flushed out after with two MACS washes and resuspended in explant culture medium for further usage.
Plasmids
Retroviral vectors encoding mouse and human GMT and polycistronic MGT in pMXs-based vectors were described previously.9,10,38,39 To generate the lentiviral vectors encoding GMT targeted sgRNAs, a DNA fragment containing oligonucleotides encoding sgRNAs was synthesized and cloned into SAMv2 vector (Addgene 75112). Lentiviral vector encoding the MS2-P65-HSF1 activator helper complex lentiMPHv2 was also purchased from Addgene (89308).
Cell culture, transfections, and viral transductions
Fibroblasts were maintained in normal or high serum media during viral transduction and subsequently cultured in iCM medium (10% FBS and 90% DMEM/M199 [4:1]) for the duration of the experiment to promote cardiac reprogramming and maturation. PlatE packaging cells (for retrovirus production) and HEK293T cells (for lentivirus production) were maintained in growth media containing DMEM plus 10% FBS, 50 U/50 μg/mL penicillin/streptomycin, 1 μg/mL puromycin (Sigma), and 100 μg/mL of blasticidin S (Life Technologies). All transfections were performed using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s protocol. For positive selection, puromycin (for retrovirus) or blasticidin S and hygromycin (for CRISPRa lentiviruses) were added to transformed cells 3 days after infection. For MEF, fCF, and H9F, concentrations of 2 μg/mL puromycin and 150 μg/mL hygromycin were used for positive selection. For MEF and fCF, a concentration of 100 μg/mL of blasticidin S was used for positive selection, while a concentration of 15 μg/mL blasticidin S was used for positive selection in H9F.
Real-time qPCR
RNA was extracted with Trizol (Invitrogen). First strand cDNAs were synthesized using the Superscript IV first-strand synthesis system (Invitrogen). qRT-PCR with SYBR Green Master Mix was performed using a QuantStudio 6 real-time qPCR system (Applied Biosystems) per the manufacturer’s protocols.
Flow cytometry
At 10 days’ post-infection, the reprogrammed cells were washed with PBS and dissociated with 0.05% trypsin (Life Technologies) for 5 min at 37°C. Cells were washed twice with pre-cold fluorescence-activated cell sorting (FACS) buffer (Dulbecco’s PBS supplemented with 2% FBS and 2 mM EDTA) and subjected to fixation and staining with cell fixation/permeabilization kits (BD Biosciences). Cells were incubated with antibodies at 4°C for 30 min at concentrations recommended by manufacturers. Cells were then suspended with staining buffer (Dulbecco’s PBS with 1% paraformaldehyde) and counted using the BD Accuri C6 flow cytometer. FACS data were analyzed by FlowJo software (Tree Star). The following antibodies were used: mouse anti-troponin T, cardiac isoform (Thermo Scientific, 1:400), rabbit anti-GFP immunoglobulin G (IgG; Invitrogen, 1:500), rabbit anti-α-actinin, (Abcam, ab68167, 1:500), Alexa Fluor 488-conjugated donkey anti-rabbit IgG, and Alexa Fluor 647-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, 1:500).
Immunofluorescence staining
Cells were washed three times with ice-cold PBS and fixed with 4% paraformaldehyde (EMS) at room temperature (RT) for 15 min. For staining, cells were permeabilized with 0.1% Triton/PBS for 30 min and blocked with 5% BSA for 1 h prior to primary antibody incubation at 4°C overnight. Following primary antibody incubation, cells were washed with PBS three times and then secondary antibody was applied for 1 h at RT. Nuclear staining was performed with DAPI in mounting medium Vectashield (Vector Labs). The following primary antibodies were used: cTnT (Thermo Scientific, 1:400), GFP (Invitrogen, 1:500), and α-actinin, (Abcam, ab68167, 1:500). Images were acquired using the EVOS FL Auto Cell Imaging System (Life Technologies).
Western blot
Cells were collected and lysed in 2× SDS loading buffer (Bio-Rad) and subjected to SDS-PAGE. After separation, proteins were transferred to nitrocellulose membranes and probed with the indicated antibodies. The target proteins were detected by chemiluminescence (ECL, Thermo Scientific). The membranes were stripped with stripping buffer (Sigma) and re-probed with antibody against a second protein or β-actin for loading control.
Statistical methods
Statistical analysis was done using GraphPad Prism 8. All data were analyzed with at least three biological replicates and presented as mean ± SEM. Differences between groups were examined for statistical significance using two-way unpaired Student’s t test, a one-way ANOVA, or a two-way ANOVA. A p < 0.05 was regarded as significant.
Data and code availability
All the data related to this study are available within the paper or can be obtained from the authors on request.
Acknowledgments
We thank all the members of the Qian laboratory for helpful discussions and valuable input. This work was supported by American Heart Association (AHA) grant 20EIA35310348 and NIH National Heart, Lung, and Blood Institute (NHLBI) grant R35HL155656 to L.Q. and NIH NHLBI grants R01HL139976 and R01HL139880 and AHA Established Investigator Award 20EIA35320128 to J.L.
Author contributions
P.H. and J.X. conceived the project, performed the experiments, and wrote the manuscript, with contributions from all other authors. Y. Xie, B.K., D.N., Y. Xu, and J.R.H. assisted with experiments and data analysis. S.R. and B.S. assisted with data analysis and edited the manuscript. J.L. edited the manuscript. L.W. and L.Q. supervised the project, provided the funding, and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102390.
Contributor Information
Li Wang, Email: liwang2020@whu.edu.cn.
Li Qian, Email: li_qian@med.unc.edu.
Supplemental information
References
- 1.Benjamin E.J., Muntner P., Alonso A., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Das S.R., et al. Heart disease and stroke statistics—2019 update: A report from the american heart association. Circulation. 2019;139:e56–e528. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 2.Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doenst T., Haverich A., Serruys P., Bonow R.O., Kappetein P., Falk V., Velazquez E., Diegeler A., Sigusch H. Pci and cabg for treating stable coronary artery disease: Jacc review topic of the week. J. Am. Coll. Cardiol. 2019;73:964–976. doi: 10.1016/j.jacc.2018.11.053. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S.L., Szewczykowska M., Jackowska T., Dos Remedios C., et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Kimura W., Xiao F., Canseco D.C., Muralidhar S., Thet S., Zhang H.M., Abderrahman Y., Chen R., Garcia J.A., Shelton J.M., et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature. 2015;523:226–230. doi: 10.1038/nature14582. [DOI] [PubMed] [Google Scholar]
- 6.Nakada Y., Canseco D.C., Thet S., Abdisalaam S., Asaithamby A., Santos C.X., Shah A.M., Zhang H., Faber J.E., Kinter M.T., et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541:222–227. doi: 10.1038/nature20173. [DOI] [PubMed] [Google Scholar]
- 7.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M., Wu T.D., Guerquin-Kern J.L., Lechene C.P., Lee R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J., Du Y., Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., Cao N., Spencer C.I., Nie B., Ma T., Xu T., Zhang Y., Wang X., Srivastava D., Ding S. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014;6:951–960. doi: 10.1016/j.celrep.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., P R Iyer E., Lin S., Kiani S., Guzman C.D., Wiegand D.J., et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W., et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavez A., Tuttle M., Pruitt B.W., Ewen-Campen B., Chari R., Ter-Ovanesyan D., Haque S.J., Cecchi R.J., Kowal E.J.K., Buchthal J., et al. Comparison of Cas9 activators in multiple species. Nat. Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H., et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodou E., Xu S.M., Black B.L. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech. Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- 18.Dodou E., Verzi M.P., Anderson J.P., Xu S.M., Black B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 19.De Val S., Anderson J.P., Heidt A.B., Khiem D., Xu S.M., Black B.L. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev. Biol. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Lin B.R., Natarajan V. Negative regulation of human U6 snRNA promoter by p38 kinase through Oct-1. Gene. 2012;497:200–207. doi: 10.1016/j.gene.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 22.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H., Xiao T., Chen C.H., Li W., Meyer C.A., Wu Q., Wu D., Cong L., Zhang F., Liu J.S., et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y., Liu Z., Welch J.D., Gao X., Wang L., Garbutt T., Keepers B., Ma H., Prins J.F., Shen W., et al. Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming. Cell Stem Cell. 2019;25:149–164.e9. doi: 10.1016/j.stem.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muraoka N., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Isomi M., Nakashima H., Akiyama M., Wada R., Inagawa K., et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black J.B., Adler A.F., Wang H.G., D'Ippolito A.M., Hutchinson H.A., Reddy T.E., Pitt G.S., Leong K.W., Gersbach C.A. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell. 2016;19:406–414. doi: 10.1016/j.stem.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark T., Waller M.A., Loo L., Moreno C.L., Denes C.E., Neely G.G. CRISPR activation screens: navigating technologies and applications. Trends Biotechnol. 2024;42:1017–1034. doi: 10.1016/j.tibtech.2024.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Chen G.H., Xu J., Yang Y.J. Exosomes: promising sacks for treating ischemic heart disease? Am J Physiol Heart Circ Physiol. Am. J. Physiol. Heart Circ. Physiol. 2017;313:H508–H523. doi: 10.1152/ajpheart.00213.2017. [DOI] [PubMed] [Google Scholar]
- 31.Afzal M.R., Samanta A., Shah Z.I., Jeevanantham V., Abdel-Latif A., Zuba-Surma E.K., Dawn B. Adult Bone Marrow Cell Therapy for Ischemic Heart Disease: Evidence and Insights From Randomized Controlled Trials. Circ. Res. 2015;117:558–575. doi: 10.1161/CIRCRESAHA.114.304792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava D., DeWitt N. In Vivo Cellular Reprogramming: The Next Generation. Cell. 2016;166:1386–1396. doi: 10.1016/j.cell.2016.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathison M., Singh V.P., Chiuchiolo M.J., Sanagasetti D., Mao Y., Patel V.B., Yang J., Kaminsky S.M., Crystal R.G., Rosengart T.K. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: Implications for clinical myocardial regeneration. J. Thorac. Cardiovasc. Surg. 2017;153:329–339.e3. doi: 10.1016/j.jtcvs.2016.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jayawardena T.M., Finch E.A., Zhang L., Zhang H., Hodgkinson C.P., Pratt R.E., Rosenberg P.B., Mirotsou M., Dzau V.J. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015;116:418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayawardena T., Mirotsou M., Dzau V.J. Direct reprogramming of cardiac fibroblasts to cardiomyocytes using microRNAs. Methods Mol. Biol. 2014;1150:263–272. doi: 10.1007/978-1-4939-0512-6_18. [DOI] [PubMed] [Google Scholar]
- 37.Fu Y., Huang C., Xu X., Gu H., Ye Y., Jiang C., Qiu Z., Xie X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015;25:1013–1024. doi: 10.1038/cr.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Liu Z., Yin C., Asfour H., Chen O., Li Y., Bursac N., Liu J., Qian L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015;116:237–244. doi: 10.1161/CIRCRESAHA.116.305547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Huang P., Near D., Ravi K., Xu Y., Liu J., Qian L. Isoform specific effects of Mef2C during direct cardiac reprogramming. Cells. 2020;9:268. doi: 10.3390/cells9020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data related to this study are available within the paper or can be obtained from the authors on request.