ABSTRACT
Avian haemosporidian parasites and avian pox virus (APV) are well‐known pathogens for their impact on avian populations, especially in oceanic islands where introduced pathogens show strong virulence for endemic and naïve birds. The Bonin Islands are a group of oceanic islands 1000 km south of Tokyo. Like the Hawaiian Islands, there are many endemic and endangered species as well as introduced species, which have greatly affected the native avian fauna. However, pathogens in wild birds of this archipelago had not been investigated. In this study, we investigated the prevalence of avian haemosporidian parasites and APV among birds and mosquitoes in this unique ecosystem of the Bonin Islands. From 2014 to 2020, 524 birds of 39 species either rescued, deceased, or caught by mist‐netting were sampled. APV‐like lesions were sampled from nine birds. 262 mosquitoes were collected by sweeping nets or CDC traps. All samples were tested via PCR for haemosporidian infection, and lesions were tested for APV.209 birds (39.9%) of 11 species were positive for haemosporidian parasite DNA, and all three parasite genera were detected. Prevalence was particularly high for Plasmodium elongatum (pGRW06) and Prelictum relictum (pGRW04). The former was detected from both resident birds and mosquitoes, suggesting local transmission. An introduced species, the warbling white‐eye ( Zosterops japonicus ), had a particularly high prevalence of pGRW06 (68.3%) and may be a reservoir of this lineage. Both APV and Plasmodium spp. were detected from all APV‐tested birds, suggesting that these two pathogens may be transmitted simultaneously via mosquitoes. The presence of avian haemosporidian parasites and APV was confirmed in the Bonin Islands for the first time. However, the virulence and origin of these pathogens remain unknown, and many bird species are still understudied. Further investigations are required to contribute to the conservation of this unique avifauna.
Keywords: APV, avian haemosporidia, Bonin Islands, mosquito, oceanic islands
Avian haemosporidian parasites and avian pox virus were detected from birds of the Bonin Islands of Japan for the first time, including some that are endemic to the islands. Both pathogens are suggested to be simultaneously transmitted by mosquitoes. Some of the haemosporidian parasite lineages are suggested to have been introduced along with their avian hosts.
1. Introduction
Oceanic islands have isolated history, as they arose from the ocean basin and have therefore never been connected to continental lands. The biotas of the islands are restricted to organisms that were capable of reaching the islands by air or sea, either by passive or active movement. Consequently, unique ecosystems are created through the evolution of endemism (Carlquist 1974; Kawakami and Okochi 2010; Kueffer, Drake, and Fernández‐Palacios 2014). Species of such unique ecosystems are incredibly vulnerable to introduced alien species, which can drastically alter the ecosystem through direct predation, interspecific competition, and the introduction of novel pathogens (Carlquist 1974; Van Riper et al. 1986; Kawakami and Okochi 2010; Kawakami 2019). Avian malaria, caused by Plasmodium parasites, and fowlpox caused by avian poxvirus (APV), are known to have caused population‐level lethal impacts on the avifauna of Hawaii and New Zealand (Warner 1968; Van Riper et al. 1986; Van Riper, Van Riper, and Hansen 2002; Van Riper III 1991; Aruch et al. 2007; Atkinson and Lapointe 2009; Howe et al. 2012; Sijbranda et al. 2016; Samuel et al. 2018). Several native honeycreepers of Hawaii were driven into extinction due to the combination of the two pathogens (Warner 1968; Van Riper et al. 1986; Van Riper, Van Riper, and Hansen 2002). Because endemic species on these oceanic islands had never been exposed to these pathogens, they had not developed immunity against these pathogens and were therefore extremely naïve, resulting in high virulence (Van Riper et al. 1986; Atkinson et al. 2000, 2013; Sorci 2013). Introductions of infected avian populations have been considered to be the primary drivers of such population declines and extinctions (Warner 1968; Van Riper III 1991; Van Riper, Van Riper, and Hansen 2002; Ewen et al. 2012). Both avian malaria parasites and APV are known to be transmitted by mosquitoes (Valkiūnas 2005; Aruch et al. 2007; Yeo et al. 2019), although APV can also be transmitted by other mechanical vectors and by direct contact (Huong et al. 2014; Lee et al. 2017; Yeo et al. 2019). Therefore, the introduction of mosquitoes to Hawaii, where suitable vectors were previously absent, incidentally made transmission within the islands possible (Warner 1968; Van Riper et al. 1986; Van Riper, Van Riper, and Hansen 2002). Novel pathogens via introduced animals can therefore cause devastating impacts on the ecosystems of oceanic islands.
The Bonin Islands are part of the Ogasawara Archipelago, which is located nearly 1000 km south of mainland Tokyo, Japan. The archipelago supports a unique ecosystem involving a wide variety of indigenous species (Momiyama 1930; Shimizu 2003; Kawakami 2008; Kawakami and Okochi 2010) and also serves as an important breeding area for seabirds (Chiba et al. 2007; Kawakami and Okochi 2010). In 2011, the Ogasawara Archipelago was designated as a World Natural Heritage site for its unique ecosystem (UNESCO 2011). Meanwhile, the Bonin grosbeak ( Carpodacus ferreorostris ) and Bonin thrush ( Zoothera terrestris ), which are both extinct species previously endemic to the Bonin Islands, are thought to have been driven into extinction by introduced rats and cats (BirdLife International 2020). Furthermore, major population declines of endemic species have been recorded through direct and indirect effects of introduced species such as flatworms, lizards, plants, and cats. For these reasons, along with other human‐related activities, many island species have decreased in numbers (Tomiyama 1998; Shimizu 2003; Kawakami 2008; Kawakami and Okochi 2010; Sugiura 2016). Endemic species and subspecies, including the Bonin white‐eye ( Apalopteron familiare ) and Bannerman's shearwater ( Puffinus bannermani ), have consequently been designated as national endangered species (Ministry of the Environment 2020). Conservation plans have been organized along with conservational research in order to preserve this vulnerable ecosystem (Takahashi 1973; Shimizu 2003; Kawakami and Higuchi 2003; Toma and Miyagi 2005; Takaoka and Saito 2006; Chiba et al. 2007; Yabe et al. 2009; Kawakami and Okochi 2010; Chiba and Suzuki 2011; Sugita, Kawakami, and Nishiumi 2016; Saitoh et al. 2020). However, few studies have addressed the presence and impact of wildlife diseases on the islands, with only Angiostrongylus cantonensis in invasive rodents, trematodes in fishes, Phellinus noxius in plants, and Salmonella (Tokiwa et al. 2013; Sahashi et al. 2015; Kuramochi 2018; Sumiyama et al. 2020). Bloodsucking arthropod insects, including mosquitoes, blackflies, and biting midges, have been confirmed in the Bonin Islands (Takahashi 1973; Tanaka, Mizusawa, and Saugstad 1979; Wada 1986; Takaoka, Saito, and Suzuki 1999; Toma and Miyagi 2005). The presence of both endemic and introduced avian species, along with possible vector species, creates a similar scenario to other oceanic islands in which avian malaria parasites and APV have had significant impacts. As part of conservational research, we investigated the prevalence of avian haemosporidia and APV in the Bonin Islands.
2. Materials and Methods
2.1. Study Site
Birds and mosquitoes were sampled in the Bonin Islands (Figure 1a). Sampling took place in islands of the Chichijima group (Figure 1b): Chichijima (27°04′N, 142°13′E), Anijima (27°07′N, 142°12′E), Minamijima (27°02′N, 142°10′E), and Higashijima (27°06′N, 142°15′E) and islands of the Hahajima group (Figure 1c): Hahajima (26°40′N, 142°09′E), Mukohjima (26°36′N, 142°07′E), Meijima (26°34′N, 142°13′E), Imotojima (26°33′N, 142°12′E), and Anejima (26°33′N, 142°09′E). These islands are located in the subtropical climate zone. The mean annual temperature of Chichijima is 23°C with a narrow range of 18°C–28°C. The annual precipitation is about 1300 mm, with the most rainfall in May (174 mm) and the least in February (61 mm) (Ministry of the Environment 2010; Japan Meteorological Agency 2020). The archipelago is low‐lying, with the highest elevations on Chichijima and Hahajima being 326 and 462 m, respectively (Government of Japan 2010). Currently, most of the areas are strictly protected, and only parts of Chichijima and Hahajima are inhabited by people (Ministry of the Environment 2010).
FIGURE 1.
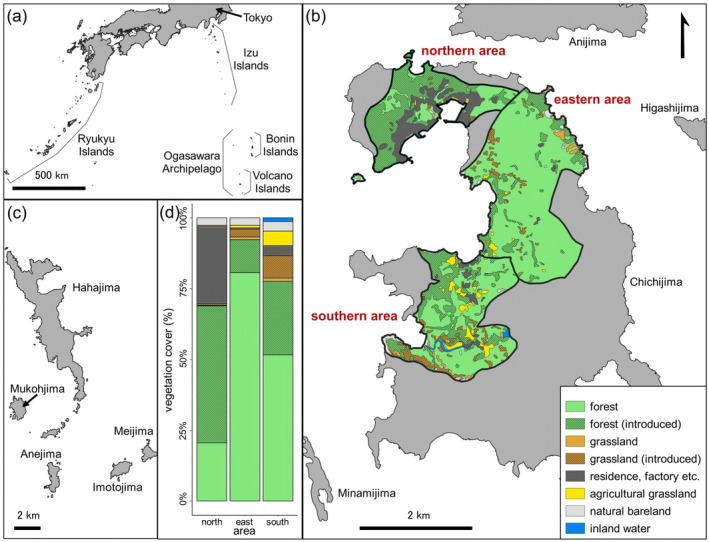
Map of the study area. Location of the Bonin Islands in relation to other areas of Japan (a). Islands of the Chichijima group, with vegetation cover of the three sampling areas (b). Islands of the Hahajima group (c). Vegetation cover proportions of the three sampling areas (d).
Samples of Chichijima were collected in three main areas, classified according to the city they were sampled (Figure 1b). Vegetation cover proportions differ among these areas (Figure 1d), as calculated from vegetation maps using Quantum GIS ver. 3.14 (Biodiversity Center of Japan 2016; National Statistics Center 2018; QGIS Developmental Team 2020). The northern area is the main residential area of Chichijima, surrounded by secondary forests consisting of mainly introduced species such as Casuarina equisetifolia and Leucaena leucocephala communities. The southern area is a smaller residential area, surrounded by agricultural grasslands as well as introduced and natural forests. Human settlement and introduced species have had great impacts on the northern and southern areas. In contrast, the vegetation in the eastern area of sampling is mostly natural and well preserved. The area is mostly covered by Machilus kobu‐Schimetum mertensianae communities, which are currently severely managed for conservation measures. Information on the precise location of collection for many birds could not be obtained, and hence details on the habitat during collection were unknown.
2.2. Sampling Rescued or Deceased Birds
Birds were collected in the Bonin Islands between 2011 and 2019. These include birds that were found dead and rescued birds that died during rehabilitation. 378, two, one, seven, and five bird(s) were found in Chichijima, Anijima, Higashijima, Minamishima, and Hahajima, respectively. The sampling size was particularly large in Chichijima, where the NPO Institute of Boninology actively collected deceased birds. Note that resident bird species were not collected in Higashijima and Minamishima. Additionally, one bird landed on the passenger ship Ogasawara‐maru, which travels between Tokyo and Chichijima. For six birds, a portion of the liver and heart were sampled prior to freezing, and impression smears were prepared from the cut surface of the heart. All other individuals were frozen prior to necropsy. Tissue samples were collected from the liver, lung, heart, or muscle and placed in microtubes containing 70% ethanol directly after necropsy. In eight warbling white‐eyes and one White's thrush ( Zoothera aurea ), APV‐like lesions were seen on the eyelids, nares, legs, and wing joints. These lesions were removed, placed in microtubes, and kept at −20°C until further processes. For one warbling white‐eye, a lesion from the leg was placed in a microtube containing 10% neutral buffered formalin.
In addition, blood from three birds rescued in Chichijima was sampled at the Institute of Boninology in August of 2015. Blood was drawn from the brachial or jugular vein, and blood smears were prepared. The remaining blood was placed in microtubes containing 70% ethanol.
Impression smears and blood smears were fixed with methanol and stained with Hemacolor (Merck KGaA, Darmstadt, Germany). Once dry, the smears were mounted in Eukitt medium (O. Kindler GmbH, Freiburg, Germany). The formalin‐fixed lesion was embedded in paraffin to make thin sections, which were mounted on slides and stained with hematoxylin and eosin.
2.3. Sampling Birds by Mist‐Netting
Resident passerines were captured with mist nets in February 2016, September 2020, and June to July 2021. 38 and 89 birds were captured in Chichijima and the Hahajima group [Hahajima (n = 3), Mukohjima (n = 36), Meijima (n = 11), Imotojima (n = 4), and Anejima (n = 35)]. Blood was obtained from the brachial vein and placed in microtubes containing 70% or 99% ethanol. Blood smears were also prepared for individuals captured in 2016 and 2021. The smears were prepared and stained in the same manner as rescued birds. The birds were released after they were ringed.
2.4. DNA Extraction and Molecular Detection of Avian Haemosporidia From Birds
DNA was extracted from organ tissue and blood samples using the standard phenol‐chloroform method, with tris‐EDTA as the final buffer. DNA concentration was confirmed with Nanodrop One Microvolume UV–Vis Spectrophotometers (Thermo Fisher Scientific, MA, USA) and adjusted to a final concentration of 50 ± 10 ng/μL. A nested PCR targeting the partial mitochondrial cytochrome b (cytb) gene of avian malarial parasites (Plasmodium) and related parasites (Haemoproteus and Leucocytozoon) was performed using the primers HaemNFI/HaemNR3 for the first PCR and HaemF/HaemR2 and HaemFL/HaemR2L for the second PCR (Hellgren, Waldenström, and Bensch 2004). The reaction mixture composition was as follows: 2 mM MgCl2, 0.2 mM deoxynucleotide triphosphate, 10x ExTaq buffer (Mg2+ free; Takara, Ohtsu, Japan), 0.625 U Ex‐Taq (Takara), 0.6 μM each primer, and 50 ng of template DNA, making the final volume 25 μL each. Plasmodium gallinaceum GALLUS01 from an experimentally infected chicken ( Gallus gallus ) and Leucocytozoon sp. OTULEM04 from a rescued Japanese scops‐owl ( Otus semitorques ) were included as positive controls. For a negative control, one sample containing distilled water instead of DNA was also included. Amplification was confirmed on 1.5% agarose gels (Agarose S; Nippon Gene, Tokyo, Japan) containing ethidium bromide (Nacalai Tesque, Kyoto, Japan), which were placed in chambers filled with TAE buffer. One positive control and one negative control were included in every gel. Electrophoresis was performed at 100 V for approximately 20 min. Gels were placed under ultraviolet light to confirm amplification. If any bands were seen in the negative control, all samples in the same gel were retested from the first PCR. Positive bands were cut out of the gel, and DNA was extracted using thermostable β‐agarase (Nippon Gene, Tokyo, Japan).
Additionally, for all samples positive for Plasmodium or Haemoproteus using the above primers, a nested multiplex PCR was carried out in order to detect possible co‐infections (Pacheco et al. 2018). First, PCR targeting a portion of the mitochondrial COX1 gene and the full cytb gene was performed with the primers AE298/AE299. For the second PCR, the primers AE983/AE985 and AE980/AE982 were used to detect Plasmodium and Haemoproteus, respectively. Positive controls consisted of P. gallinaceum GALLUS01 from an experimentally infected chicken and H. larae SPMAG12 from a captive African penguin ( Spheniscus demersus ). Positive bands for Plasmodium were cut out, and DNA was extracted using thermostable β‐agarase (Nippon Gene, Tokyo, Japan). If positive bands for Haemoproteus were detected, an additional PCR was carried out with the primers HaemF/AE982 (Hellgren, Waldenström, and Bensch 2004; Pacheco et al. 2018), and positive bands were cut out and extracted. Reaction mixture compositions and all electrophoresis processes were the same as above. PCR conditions were according to protocol (Pacheco et al. 2018).
2.5. Sampling Mosquitoes
Mosquitoes were collected in Chichijima by sweep netting (March and August 2015) or CDC traps enhanced with yeast‐generated CO2 (July and August 2015) (Tsuda et al. 2009). Sweep netting was carried out in the morning or evening at five, two, and three localities in the northern, eastern, and southern areas, respectively. Two traps each were set in the northern and southern areas. Traps were set in the evening and recovered in the morning. Captured mosquitoes were morphologically identified to a species (Tanaka, Mizusawa, and Saugstad 1979; Tsuda 2019) and then placed in −20°C until further processes.
2.6. DNA Extraction and Molecular Detection of Avian Haemosporidia From Mosquitoes
Mosquitoes were separated into head‐thorax and abdomen under an Olympus SZ61TR stereo microscope (Olympus, Tokyo, Japan). DNA was extracted from each sample individually, using the REDExtract‐N‐Amp Tissue PCR kit (SIGMA, St. Louis, MO, USA). To detect Plasmodium/Haemoproteus parasites, nested PCR targeting the partial cytb gene of avian haemosporidia was performed. We used different primer sets from birds as a result of some trials to more effectively amplify the parasite DNA from mosquitoes. The primers DW2/DW4 were used for the first PCR (Perkins and Schall 2002), followed by a second PCR using HaemNF/HaemNR2 (Waldenström et al. 2004). The composition of the reaction mixture was as follows: 4 mM MgCl2, 0.4 mM deoxynucleotide triphosphate, 10x ExTaq buffer (Mg2+ free; Takara, Ohtsu, Japan), 1 U Ex‐Taq (Takara), 0.4 μM each primer, and 1 μL of template DNA, making the final volume 25 μL each. Cycle conditions were as previously described (Perkins and Schall 2002; Waldenström et al. 2004). Plasmodium gallinaceum GALLUS01 and distilled water were used for the positive and negative controls, respectively. Electrophoresis was carried out in the same method as the avian samples.
2.7. Phylogenetic Analysis of Avian Haemosporidian Lineages
The extracted DNA was sequenced in both directions with the BigDye terminator cycle sequence kit 3.1 (Applied Biosystems, Foster City, CA, USA) and the ABI 3130‐Avant Auto Sequencer (Applied Biosystems). If double‐base callings were detected, the sample was retested on a different day using newly extracted DNA. A sample was considered co‐infected if double‐base callings were seen in both detections. Obtained sequences were assembled and compared with sequences in the GenBank database (Madden 2013) and MalAvi database (Bensch, Hellgren, and Pérez‐Tris 2009). If lineages identical to the positive controls were detected, samples were retested from the first PCR to remove contaminations.
Mr. Bayes version 3.2 was used to construct a Bayesian phylogeny (Ronquist and Huelsenbeck 2003). Morphologically identified lineages of the three haemosporidian genera and molecularly close lineages were included for comparison. Theileria annulata was selected as an outgroup. The Jukes‐Cantor model of substitution was implemented to calculate genetic distances between lineages using MEGA X (Kumar et al. 2018). Using the same software, translated amino acids were also compared between lineages. Using ModelFinder in IQ‐TREE 1.6.12 (Kalyaanamoorthy et al. 2017), the General Time Reversible model with gamma distribution for variable sites and proportion of sites as invariable (GTR+Γ+I) was selected as the best‐fit model under the Bayesian information criterion. Markov chain Monte Carlo (MCMC) sampling was run twice independently for three million generations, with a sampling frequency of every 1000 generations (Ronquist et al. 2012). The first 25% of trees were discarded as “burn‐in,” and the resulting tree was visualized with FigTree 1.4 (Rambaut 2012). A minimum spanning network was generated with PopART 1.7 (Leigh and Bryant 2015) to visualize genetic relationships between the obtained lineages.
2.8. DNA Extraction and Molecular Detection of APV
DNA was extracted from APV‐like lesions using the DNeasy blood & tissue kit (Qiagen, GmbH, Hilden, Germany). A partial sequence of the 4b core protein gene was amplified by PCR using the primers P1x and P2 (Huong et al. 2014). The reaction mixture composition was as the following: 2.5 mM MgCl2, 0.4 mM deoxynucleotide triphosphate, 10× ExTaq buffer (Mg2+ free; Takara, Ohtsu, Japan), 1 U Ex‐Taq (Takara), 0.4 μM each primer, and 50 ng of template DNA, making the final volume 25 μL each. Cycle conditions were according to protocol (Huong et al. 2014). Fowl pox vaccine (Nisseiken Co. Ltd., Tokyo, Japan) was used as a positive control. The negative control contained distilled water instead of DNA. Procedures from electrophoresis to sequence assembly were carried out in the same methods as avian haemosporidian detection. Obtained sequences were compared with sequences in the GenBank database using BLAST (Madden 2013). Genetic distances were calculated by implementing the Jukes‐Cantor model of substitution in MEGA X (Kumar et al. 2018).
2.9. Phylogenetic Analysis of APV Strains
A phylogeny was generated using Mr. Bayes version 3.2 (Ronquist et al. 2012), including sequences from previous studies (Gyuranecz et al. 2013; Banyai et al. 2015; Sarker et al. 2017) and unpublished sequences in the Nihon University College of Bioresource Sciences Laboratory of Biomedical Science database. The transition model with gamma distribution for variable sites and proportion of sites as invariable (TIM+Γ+I) was selected as the best‐fit model under the Bayesian information criterion using ModelFinder implemented in IQ‐TREE 1.6.12 (Kalyaanamoorthy et al. 2017). As TIM is not implemented in Mr. Bayes version 3.2, the substitution model was replaced with General Time Reversible (GTR) model. Markov chain Monte Carlo (MCMC) sampling was run twice independently for three million generations, and samples were taken every 1000 generations. The first 25% of the trees were discarded as a “burn‐in” step. The resulting phylogenetic tree was visualized using FigTree 1.4 (Rambaut 2012).
2.10. Microscopic Examinations
Blood smears, impression smears, and tissue sections were observed under an Olympus BX43 light microscope (Olympus, Tokyo, Japan). Blood smears and impression smears were screened at 400× and 1000× magnification. Detected parasites were identified based on their morphological features (Valkiūnas 2005). The tissue section was observed at 200× and 600× magnification. Photographs were taken using an Olympus IX71 light microscope (Olympus) and software cellSens Standard 1.6 (Olympus).
2.11. Statistical Analysis
Using Fisher's exact test, the prevalence of avian haemosporidia was compared between resident and migratory species. Migratory status was determined based on the Check‐list of Japanese birds, 7th revised edition (Ornithological Society of Japan 2012), as well as field studies on the avifauna of the Bonin Islands (Chiba et al. 2007; Emura 2011; Kawakami et al. 2018). To investigate variables that affect Plasmodium prevalence within the islands, general linear models (GLM) with binomial distribution and logit function were used. Species (warbling white‐eye, blue rock thrush, or White's thrush), age (juvenile or adult), sampling area (north, east, or south), and season of sampling (spring, summer, autumn, or winter) were tested. Only samples from Chichijima were included in the GLM tests. Other resident species were not included due to the small sample size. Furthermore, to avoid potential bias due to parasite lineage variation, only individuals positive for GRW06 were included. For ANOVA calculations, the type II sums of squares were used because of unbalanced data. Tukey's test for multiple comparisons was used for post hoc comparisons. For Cx. boninensis mosquitoes, the effect of sampling area on GRW06 prevalence was tested with Fisher's exact test. All statistical analyses were done using the software R version 3.6.3 (R Core Team 2020), using packages “prevalence,” “car,” and “multcomp” (Hothorn, Bretz, and Westfall 2008; Fox and Weisberg 2011; Devleesschauwer et al. 2014). Statistical significance was determined using the 5% significance level.
3. Results
3.1. Avian Haemosporidia in Birds
Avian haemosporidian DNA was detected from 209 of the 524 examined birds (Table 1). Among resident species, positive individuals were detected from Chichijima, Hahajima, Mukohjima, Meijima, Imotojima, and Anejima (Supporting Information S1). In migratory species, five individuals from Chichijima were positive by PCR. No positive individuals were detected from the remaining three islands (Anijima, Higashijima, Minamijima). The overall prevalence was significantly higher in resident species (53.68%; 95% CI = 48.66%–58.64%) compared to migratory species (3.47%; 95% CI = 1.49%–7.87%) (Fisher's exact test: p < 0.001).
TABLE 1.
Summary of birds sampled in this study, with PCR results of haemosporidian detection.
| Status | Chichijima group | Hahajima group | Total | Lineages a | ||||
|---|---|---|---|---|---|---|---|---|
| Sampled | Infected b | Sampled | Infected b | Sampled | Infected b | |||
| Resident | 290 | 149 (51.38) | 90 | 55 (61.11) | 380 | 204 (53.68) | pGRW04, pGRW06, pCXPIP12, pMONTRI01, hZOSJAP02 c , hZOOLUN01, lZOOAUR01 c , lZOOAUR02 c , lZOOAUR03 c , Leucocytozoon sp. [co‐infected] | |
| Migrant | Migrant breeder | 88 | 0 | 2 | 0 | 90 | 0 | |
| Winter visitor | 34 | 2 (5.88) | 1 | 0 | 35 | 2 (5.71) | pSW2, Plasmodium sp. [co‐infected] | |
| Passage visitor | 4 | 0 | 4 | 0 | ||||
| Irregular or accidental visitor | 14 | 3 (21.43) | 1 | 0 | 15 | 3 (20.00) | pACCBAD01, pGRW06, lANACRE04 c | |
| Sub‐total | 140 | 5 (3.57) | 4 | 0 | 144 | 5 (3.47) | ||
| Total | 430 | 154 (35.81) | 94 | 55 (58.51) | 524 | 209 (39.89) | ||
Lineage names are given according to MalAvi database.
Parentheses: prevalence.
Lineages detected for the first time.
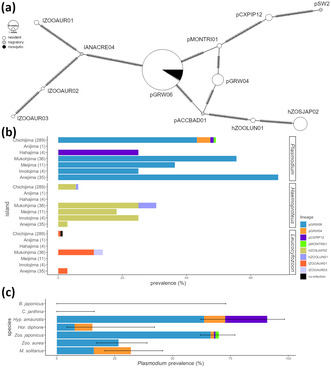
A total of six Plasmodium, two Haemoproteus, and four Leucocytozoon lineages were detected in this study, including five lineages detected for the first time (Table 1, Figure 2, Supporting Information S1). All detected lineages varied from one another by more than 10 bases within the observed genetic region (Figure 2a). Detected Haemoproteus lineages were placed in the clade of parasites belonging to the subgenus Parahaemoproteus (Supporting Information S1). Three birds were co‐infected with parasites of the same genus, and lineages could not be determined. In 27 warbling white‐eyes, the primers HaemF/HaemR2 only detected Plasmodium parasites, but both Plasmodium and Haemoproteus parasites were detected using the genus‐specific primers (Supporting Information S1). For Plasmodium, HaemF/HaemR2 and the genus‐specific primers detected the same lineages (26 pGRW06 and 1 pMONTRI01). The single lineage hZOSJAP02 was detected from all 27 individuals using the Haemoproteus‐specific primers. The only lineage shared between resident and migratory species was pGRW06 of Plasmodium elongatum .
FIGURE 2.
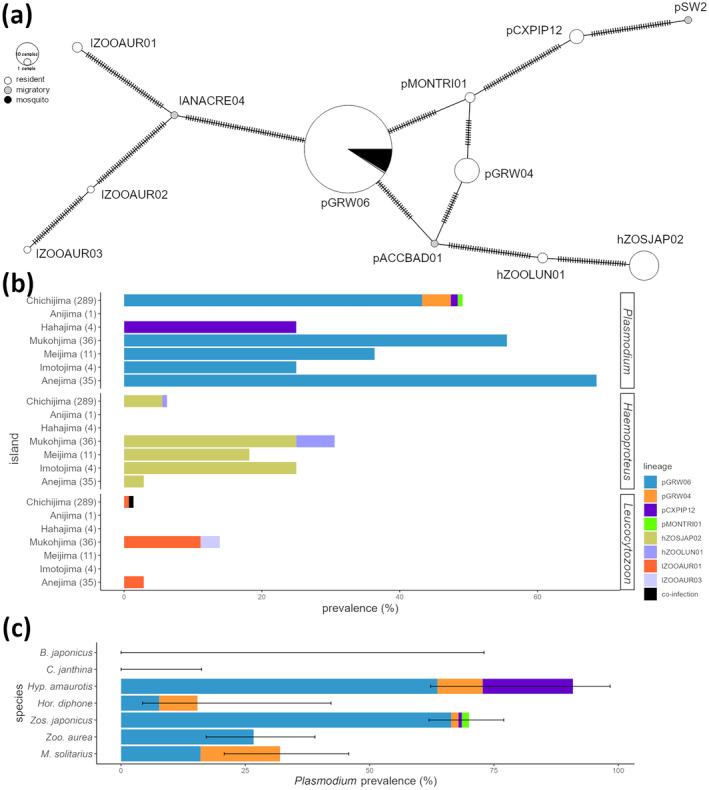
Composition of haemosporidian mitochondrial DNA cytochrome b lineages detected in this study. Minimum spanning network (a), prevalence among resident species by island (b), and Plasmodium prevalence among resident species of Chichijima (c). The colors represent host/vector status (a) or lineages (b, c), as shown in the legends. Islands or species lacking bars show 0% prevalence (i.e., no parasites were detected; b, c). Each hash mark indicates a base mutation (a). Error bars represent ±95% of the confidence interval (c). Note that two species have 0% prevalence but show a long error bar due to the small sample sizes (c).
Nine lineages were detected from resident birds in Chichijima (n = 8), Hahajima (n = 1), Mukojima (n = 5), Meijima (n = 2), Imotojima (n = 2), and Anejima (n = 3). (Figure 2b, Supporting Information S1). Five lineages (pGRW06, pCXPIP12, hZOSJAP02, hZOOLUN01, and lZOOAUR01) were detected from multiple islands, all of which were detected in both the Chichijima and Hahajima groups.
Parasites were detected from six of the eight resident bird species (Table 1). Plasmodium elongatum (pGRW06) was the most prevalent parasite species among all three parasite genera and within Plasmodium lineages (Figure 2b). The second most prevalent Plasmodium species was P. relictum (pGRW04), although detected far less frequently. These two parasite species were detected across multiple resident bird species of Chichijima, although the prevalence greatly differed between host species (Figure 2c, Supporting Information S1). 27 and 10 resident birds were positive for Haemoproteus spp. and Leucocytozoon spp., respectively.
Among the tested individuals, age was not a significant factor regarding pGRW06 prevalence in Chichijima (GLM: LR χ 2 = 1.372, df = 1, p = 0.242), and no interactions were detected. Meanwhile, bird species (GLM: LR χ 2 = 35.153, df = 2, p < 0.001) was a significant factor, with a significant interaction with season (GLM: LR χ 2 = 18.431, df = 6, p = 0.005). No seasonal variations in prevalence were seen in White's thrush (GLM: LR χ 2 = 5.003, df = 3, p = 0.172; GLM (northern area only): LR χ 2 = 3.455, df = 3, p = 0.327) and blue rock thrush (GLM: LR χ 2 = 0.552, df = 3, p = 0.907; GLM (northern area only): LR χ 2 = 0.833, df = 3, p = 0.841), even when compared among only northern samples. Few individuals were sampled in the eastern and southern areas for these two species, and comparing the parasite prevalence among areas was not possible. For the warbling white‐eye, pGRW06 prevalence significantly differed between seasons when all areas were included (GLM: LR χ 2 = 22.22, df = 3, p < 0.001). However, prevalence did not differ among seasons when analyzed separately by area (Table 3). Furthermore, pGRW06 prevalence was higher in the southern area compared to the northern area during the summer and winter, exhibiting a significant difference in prevalence (Tukey's post hoc test for winter: estimate = 2.749, SE = 1.168, z value = 2.353, p < 0.037). The prevalence was considerably higher in the southern area during the autumn as well, although not significantly (Table 3). The prevalence during the winter was higher in the eastern area compared to the northern area, but the small sample size did not allow for accurate comparisons (Tukey's post hoc test: estimate = −19.096, SE = 3584.671, z value = −0.005, p = 1.000).
TABLE 3.
Prevalence of Plasmodium elongatum (pGRW06) in warbling white‐eyes ( Zosterops japonicus ) at each sampling area of Chichijima.
| Season | North | East | South | LR chisq | p | |||
|---|---|---|---|---|---|---|---|---|
| Spring (March to May) | 7/21 | (33.33) a | 0 | 0 | — | — | ||
| Summer (June to August) | 25/43 | (58.14) | 0 | 10/11 | (90.91) | 4.879 | 0.027 | |
| Autumn (September to November) | 7/15 | (46.67) | 0 | 2/2 | (100) | 2.781 | 0.095 | |
| Winter (December to February) | 8/13 | (61.54) | 9/9 | (100) | 25/26 | (96.15) | 10.370 | 0.006 |
| LR chisq | 4.245 | 0.598 | ||||||
| p | 0.236 | 0.742 | ||||||
Note: Significant p values are shown in bold.
Positive/sampled (prevalence by percent).
Of the 100 birds in which smears were obtained, parasites were detected by microscopy from 71 birds (Supporting Information S1). Parasites were not found in the smears of six PCR‐positive individuals. Although some parasites could not be identified to a species, P. elongatum , P. relictum, and H. killangoi were present in many smears. Co‐infections of P. elongatum and H. killangoi were seen in 21 individuals. These microscopy results corresponded to multiplex PCR results, allowing the lineage hZOSJAP02 to be identified as the morphological species H. killangoi (Supporting Information S1). However, note that in two individuals, both P. elongatum and H. killangoi were observed by microscopy, but only P. elongatum was detected molecularly. Similarly, co‐infections of P. elongatum and P. relictum were observed in 2 individuals, but only pGRW06 of P. elongatum was detected by both PCR protocols.
3.2. Avian Haemosporidia in Mosquitoes
In total, 262 mosquitoes of five species were captured (Table 2). Twelve Culex boninensis and one Cx. quinquefasciatus caught by sweep netting were PCR‐positive for P. elongatum (pGRW06). Specifically, pGRW06 was detected from the head‐thorax and abdomen, head‐thorax, and abdomen of five, one, and six unfed Cx. boninensis, respectively, and the total prevalence was 27.27% (95% CI = 16.35%–41.85%). Infected individuals were caught in both March and August. The eastern area had the highest prevalence (31.43%, 95% CI = 18.55%–47.98%), followed by the northern area (20.00%, 95% CI = 3.62%–62.45%). No positive mosquitoes were found in the southern area. The difference between areas was not statistically significant (Fisher's exact test: p = 0.592). Prevalence in Cx. quinquefasciatus was 1.28% (95% CI = 0.23%–6.91%), and pGRW06 was detected from the abdomen of an unfed female caught in the northern area. In addition, Haemoproteus DNA was detected from the abdomen of an unfed Cx. boninensis. The parasite lineage could not be determined due to co‐infection.
TABLE 2.
Mosquito samples collected and tested, including PCR results of haemosporidian detection.
| Method | Species | Northern area | Eastern area | Southern area | Total | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. | positive | No. | Positive | No. | Positive | No. | Positive | ||
| Sweep netting a | Aedes savoryi b | 4/0 | 0 | 1/0 | 0 | 52/0 | 0 | 57/0 | 0 |
| Aedes albopictus | 5/41 | 0 | 1/28 | 0 | 1/1 | 0 | 7/70 | 0 | |
| Culex boninensis b | 1/1 | 1/0 | 0/35 | 0/11 | 3/0 | 0 | 4/36 | 1/11 | |
| Culex quinquefasciatus | 1/1 | 0 | 0/1 | 0 | 1/2 | 0 | |||
| Culex sp. | 1/0 | 0 | 1/0 | 0 | |||||
| Lutzia shinonagai b | 0/1 | 0 | 0/1 | 0 | |||||
| Lutzia sp. | 1/0 | 0 | 1/0 | 0 | |||||
| Sub‐total | 13/43 | 1/0 | 2/65 | 0/11 | 56/1 | 0 | 71/109 | 1/11 | |
| CO2 trap | Aedes albopictus | 2 | 0 | 1 | 0 | 3 | 0 | ||
| Culex boninensis b | 3 | 0 | 1 | 0 | 4 | 0 | |||
| Culex quinquefasciatus | 10 | 1 | 65 | 0 | 75 | 1 | |||
| Sub‐total | 15 | 1 | 67 | 0 | 82 | 1 | |||
| Total | 71 | 2 | 67 | 11 | 124 | 0 | 262 | 13 | |
March/August.
Endemic species.
3.3. APV In Birds
All nine tested individuals were PCR‐positive for APV. Features characteristic of APV infection were also seen in morphological observations (Supporting Information S1). Molecular analysis revealed two novel strains, both placed in clade B by phylogenetic analysis (Figure 3). All eight warbling white‐eyes had the same strain of subclade B4, while the White's thrush strain was placed in subclade B1. By pairwise analysis, a strain detected from a Macqueen's bustard ( Chlamydotis macqueenii ) (divergence: 1.89% compared within 426 bp) and SWPV1 of Shearwaterpox from flesh‐footed shearwaters (Ardenna carneipes) (divergence: 2.84% compared within 570 bp) were closest to the warbling white‐eye strain. The White's thrush strain was closest to a strain from a great tit ( Parus major ) of Germany (divergence: 0.17% compared within 570 bp), although many other strains in subclade B1 were also very close. Avian malaria parasites were detected from all nine APV‐positive individuals. Specifically, pGRW06 was detected from seven warbling white‐eyes and one White's thrush, and pCXPIP12 was detected from one warbling white‐eye.
FIGURE 3.
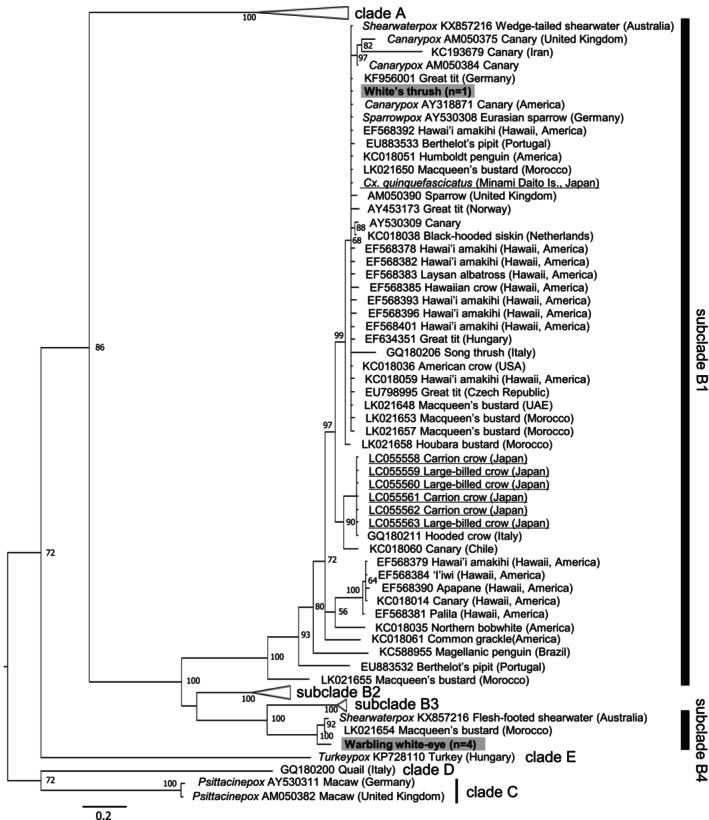
Bayesian phylogenetic analysis of the 4b core protein strains (426 bp) of Avipoxvirus. Posterior probabilities of > 0.60 were indicated. Clade A, subclade B2, and subclade B3 were collapsed. Strains previously detected from Japan are underlined. Dark gray background shows strains detected in this study.
4. Discussion
4.1. Prevalence of Avian Haemosporidia Among Resident Species
Avian haemosporidia was detected from six of the eight resident species tested in this study. Total parasite prevalence and prevalence per lineage widely differed between species. Interestingly, two warbling white‐eyes were co‐infected with P. elongatum and P. relictum , although only P. elongatum GRW06 was detected using two PCR protocols. The difficulty of detecting co‐infections, particularly those of the same genus, has been previously addressed (Bernotiene et al. 2016; Pacheco et al. 2018). Blood smears were obtained for only some individuals, and P. relictum prevalence may possibly be higher than the obtained PCR results.
Among resident bird species of Chichijima, there was a significant difference in P. elongatum (pGRW06) prevalence (Figure 2c). What appeared to be seasonal variations in the warbling white‐eye was due to variations in sampling area, as separate analysis of northern and southern areas revealed no seasonal variations (Table 3). The pGRW06 prevalence in this species was significantly higher in the southern area compared to the northern area. Additionally, although not statistically significant, all individuals from the eastern area were positive for pGRW06. Meanwhile, positive mosquitoes were caught mostly in the eastern area, although the small sample sizes prevented statistical significance. A large proportion of the northern area is residential land, while the eastern area is covered by mostly natural vegetation (Figure 1b,d). The southern area contains a small residential area, but there is a mixture of different vegetation types. Cx. boninensis can be found in various environments but prefers vegetated areas over residential areas, especially areas with vegetation diversity (Maekawa et al. 2021). Warbling white‐eyes in the Bonin Islands utilize a variety of environments, with a preference towards open habitats (Kawakami and Higuchi 2003). Higher parasite prevalence has been recorded in natural habitats (Huang et al. 2015; Ferraguti et al. 2016), in relation to higher vector and host abundance (Bonneaud et al. 2009; Lalubin et al. 2013; Fecchio et al. 2017). The population density of both warbling white‐eyes and Cx. boninensis is thought to be high in the eastern and southern areas, increasing contact between the two and therefore increasing transmission. However, other studies revealed no relationships or inverse relationships between host abundance and parasite prevalence (Fecchio et al. 2017; Van Hoesel et al. 2019). Furthermore, sample sizes were small for some areas and seasons, and re‐analysis using a larger data set would be needed to clarify these results. Differences in prevalence among areas could not be tested in other species, and reasons for the variation in parasite prevalence among host species could not be fully revealed.
4.2. Transmission of Avian Haemosporidia Within the Bonin Islands
A total of nine haemosporidian lineages were detected from resident birds and are therefore thought to be transmitted within the archipelago. The most prevalent lineage, pGRW06, was also detected from Culex boninensis and Cx. quinquefasciatus of Chichijima. In Cx. boninensis, pGRW06 was detected from both the head‐thorax and abdomen of unfed individuals. Considering the life cycle of Plasmodium parasites, detection from the head‐thorax and abdomen would indicate the presence of sporozoites in the salivary gland and oocysts in the mid‐gut, respectively (Valkiūnas 2005). This suggests that Cx. boninensis may be a competent vector species of pGRW06. However, some Plasmodium species undergo abortive sporogonic development in incompetent vectors (Žiegytė 2014; Žiegytė and Valkiūnas 2014), and further studies would be required to prove vector competency. Cx. quinquefasciatus is a competent vector of P. elongatum (pERIRUB01) and may also be competent for pGRW06. However, parasite intensity, environmental conditions, and parasite lineage may affect the competency (Palinauskas et al. 2016). Cx. quinquefasciatus has been experimentally confirmed to be a competent vector of P. relictum (pGRW04) (Lapointe, Goff, and Atkinson 2005; Valkiūnas et al. 2015). Plasmodium sp. CXPIP12 has been molecularly detected from unfed Cx. pipiens (Kim and Tsuda 2010), a closely related species of Cx. quinquefasciatus. While further studies will be necessary to confirm vector competency, our results suggest that Cx. boninensis and Cx. quinquefasciatus are likely to be important vector species of avian malaria in Chichijima. Positive mosquitoes were caught in both March and August, suggesting that if competent, transmission is possible as early as March. The prevalence in mosquitoes (27.27%) was high compared to previous studies ranging from 0.04% to 15.4% (Ferraguti et al. 2024), although reasons are unknown and hope to be investigated in future studies.
Infected resident birds were detected in both the Chichijima group and Hahajima group, suggesting that transmission is possible in both areas. Cx. boninensis and Cx. quinquefasciatus have both been confirmed in Hahajima (Maekawa et al. 2021), suggesting that transmission is possible in Hahajima as well. While five parasite lineages were detected in both island groups, two Plasmodium lineages (pGRW04 and pMONTRI01) were detected from only Chichijima, and lZOOAUR03 was detected from only Mukohjima. Host species of these lineages were investigated in both island groups, and parasite fauna may differ due to limited movement of individuals among the island groups. However, the number of investigated individuals widely differs between islands and island groups. Further sampling may result in a more complete representation of the parasite fauna of each island.
Haemoproteus (subgenus Parahaemoproteus) parasites and Leucocytozoon parasites are generally known to be transmitted by biting midges (Ceratopogonidae) and blackflies (Simuliidae), respectively (Valkiūnas 2005). The two Haemoproteus lineages detected in this study were both placed in the subgenus Parahaemoproteus clade (Figure A1), suggesting that they are likely transmitted by biting midges. Both dipteran groups have been confirmed on Chichijima and Hahajima (Wada 1986; Takaoka, Saito, and Suzuki 1999) and may serve as vectors of these two haemosporidian genera. Future investigations of these dipteran groups would be important to better understand the avian haemosporidian transmission within the Bonin Islands. Abortive Haemoproteus parasites were detected by PCR in Culex mosquitoes for up to 17 days post infection in experimental conditions (Valkiūnas et al. 2013; Žiegytė 2014). In this study, a mixed infection of Haemoproteus parasites was detected from one Cx. boninensis, in which abortive parasites may have been detected by PCR.
4.3. Speculating the Origin of Haemosporidian Parasites in the Bonin Islands
Two and four lineages were detected in only the warbling white‐eye and White's thrush, respectively. Warbling white‐eyes in the Bonin Islands are considered hybrids between Z. j. alani of the Volcanic Islands and Z. j. stejnegeri of the Izu Islands (Momiyama 1930), although a more recent study suggests populations of the Ryukyu Islands may also be involved (Sugita, Kawakami, and Nishiumi 2016). These subspecies were introduced to the Bonin Islands in the early 1900s (Momiyama 1930), and the resultant hybrids are currently one of the dominant species of the Bonin Islands (Kawakami and Okochi 2010; Sugita, Kawakami, and Nishiumi 2016). Meanwhile, White's thrushes naturally established breeding populations in the mid‐1900s during US occupation, due to the introduction of earthworms (Kawakami 2008, 2019). The lineages solely detected from these species were presumably introduced to the islands along with their host species. While molecular investigations of avian haemosporidia have been made in these surrounding areas (Beadell et al. 2004; Murata 2007; Cannell et al. 2013; Zhang et al. 2014; Huang et al. 2015; Inumaru, Murata, and Sato 2017; Tsuda 2017; Eastwood et al. 2019), the Izu Islands and Volcanic Islands have not been investigated thus far. There are only some studies of avian haemosporidia in the Ryukyu Islands (Hagihara et al. 2005; Nagata 2006; Murata 2007; Murata et al. 2008), few of which have molecular information available (Ejiri et al. 2008; Tanaka et al. 2019). Investigations and molecular comparisons of parasites in surrounding island groups may reveal further insights on the parasite fauna of the Bonin Islands.
Plasmodium sp. CXPIP12 was detected from both the warbling white‐eye and brown‐eared bulbul. Like the above‐mentioned lineages, this lineage may have been introduced with warbling white‐eyes. Meanwhile, this lineage was previously detected from brown‐eared bulbuls of mainland Japan and Tsushima, as well as Cx. pipiens of mainland Japan (Kim and Tsuda 2010; Tanigawa et al. 2013; Inumaru, Murata, and Sato 2017). The brown‐eared bulbul population of the Bonin Islands is thought to have originated from populations of the Ryukyu Islands (Sugita, Kawakami, and Nishiumi 2016), and the lineage pCXPIP12 may have been naturally introduced with the host.
The most prevalent lineages, pGRW06 and pGRW04, are two of the most widespread avian malaria lineages (Bensch, Hellgren, and Pérez‐Tris 2009; Ellis et al. 2018). Lineages with wide host ranges and geographical distributions tend to be more successful in new areas. In New Zealand, these two lineages were detected from both introduced and native species but were thought to be introduced along with the introduced host species because they were not detected from the neighboring country Australia while frequently detected in the origin of the introduced host species (Ewen et al. 2012). The fact that introduced species came from a distant area makes such analysis possible. Meanwhile, the introduced warbling white‐eyes originate from neighboring island groups, which is the possible origin of other endemic subspecies of the Bonin Islands (Emura et al. 2013; Sugita, Kawakami, and Nishiumi 2016). The overlap makes speculations on the origins of these widespread lineages difficult, and genetic analyses of other genes may be required (Garcia‐Longoria et al. 2015; Hellgren et al. 2015; Huang et al. 2019). Also, endemic mosquitoes, blackflies, and biting midges have been confirmed on the Bonin Islands (Takahashi 1973; Tanaka, Mizusawa, and Saugstad 1979; Wada 1986; Takaoka, Saito, and Suzuki 1999; Toma and Miyagi 2005), including Cx. boninensis, from which pGRW06 was detected. The availability of possible vector species demonstrates that avian haemosporidia may have been present much earlier than the introduction of alien avian species, and calibrating the divergence of these arthropod vectors would help to understand when transmission became possible within the islands.
4.4. Possibilities on the Introduction of Avian Haemosporidia via Migratory Birds
Collectively, resident birds had a significantly higher parasite prevalence compared to migratory birds (Table 1). A similar case was recently reported, addressing several possible explanations, including parasite host specificity, parasite survival, and host survival (Soares, Latta, and Ricklefs 2020). In our study, one of the primary factors is thought to be the presence of seabirds and waders, which are generally known to have low haemosporidian prevalence (Mendes et al. 2005; Quillfeldt et al. 2010, 2011; Pardal et al. 2014; Clark, Clegg, and Klaassen 2016; Martínez‐De La Puente et al. 2017). Saline environments are not suitable for most vector species, and birds that use these environments are therefore able to minimize exposure to these vectors (Clark, Clegg, and Klaassen 2016). Larvae of some mosquito species, including Aedes savoryi, can tolerate saltwater environments (Dale and Knight 2008; Katano et al. 2010; Tsuda 2019). However, Culex boninensis breeds in freshwater environments (Toma and Miyagi 2005; Dale and Knight 2008; Tsuda 2019; Maekawa et al. 2021) and may therefore have reduced contact with seabirds. Additionally, taxonomic factors related to immunocompetence and host–parasite assemblages have also been proposed to explain the low parasite prevalence in seabirds (Martínez‐Abraín, Esparza, and Oro 2004). Largely due to the location of the islands, most migratory birds in this study were either seabirds or waders, which partially explains the low prevalence of migratory birds.
In addition to the low prevalence in migratory species, only one lineage was shared between migratory and resident species (Figure 2a). Furthermore, all detected lineages were genetically distant from one another. If lineages were regularly traded among resident and migratory species, shared lineages would be expected between the two. The only shared lineage, pGRW06 of P. elongatum , is one of the most widespread Plasmodium lineages (Bensch, Hellgren, and Pérez‐Tris 2009) and has been detected in mainland Japan as well (unpublished data). Meanwhile, the lineage is also the most prevalent lineage on Chichijima. It is therefore unknown whether the migrant was infected in the Bonin Islands or elsewhere. The low sharing of lineages may be a result of host specificity, the lack of proper vector species, or habitual segregations between resident and migratory birds (Beadell et al. 2004; Valkiūnas 2005; Hellgren, Pérez‐Tris, and Bensch 2009; Clark and Clegg 2017; Chahad‐Ehlers et al. 2018; Fecchio et al. 2019). Nonetheless, transmission between migratory and resident species, and therefore establishments of new haemosporidian parasites, is thought to be relatively infrequent in the Bonin Islands. However, if all the necessary conditions are satisfied (i.e., an appropriate vector with access to both an infected migratory bird and an uninfected resident bird), introductions of new parasites may be possible, and careful monitoring will be needed in the future.
4.5. APV In the Bonin Islands
APV was detected from all nine tested individuals. In the tissue section from a warbling white‐eye, characteristic observations confirmed APV infection (Supporting Information S1). Two different strains of APV were detected in this study (Figure 3). Both strains were placed in the canarypox‐like clade B (Gyuranecz et al. 2013). Subclade B1 has the highest host and geographic diversity among all other subclades (Gyuranecz et al. 2013; Le Loc'h et al. 2014; Sarker et al. 2017). Meanwhile, only a few strains of subclade B4 have been detected thus far, but from taxonomically distant species (Le Loc'h et al. 2014; Sarker et al. 2017). APV has generally been considered to be host‐specific to a degree, but is also affected by ecological niche, habitat, and geographical backgrounds (Jarmin et al. 2006; Gyuranecz et al. 2013; Le Loc'h et al. 2014). The two host species in which poxviruses were detected are relatively new species to the Bonin Islands (Kawakami 2008; Sugita, Kawakami, and Nishiumi 2016), as discussed above. However, the relatively relaxed host specificity of APV calls for close monitoring of APV to detect infections of other resident species, as well as shearwaters. The presence of the canarypox‐like clade is particularly alarming in relation to the Bonin greenfinch (Chloris kittlitzi), an endemic species of the Ogasawara Archipelago. This species is a member of the Fringillidae family, which includes canaries (Gill, Donsker, and Rasmussen 2021). Canarypox is highly pathogenic for domestic canaries (Giddens et al. 1971; Shivaprasad et al. 2009), and canarypox‐like strains have been detected from other birds of the Fringillidae family (Warner 1968; Pérez‐Tris et al. 2011; Gyuranecz et al. 2013). The canarypox‐like strains detected in this study may pose an additional threat to this critically endangered species.
APV can be transmitted by mainly two methods: direct contact of broken skin or indirect contact through mechanical vectors (Aruch et al. 2007; Huong et al. 2014; Lee et al. 2017; Yeo et al. 2019), although other methods of transmission have also been recorded (Kleindorfer and Dudaniec 2006). Warbling white‐eyes, along with other Zosterops species, are known to do mutual preening or allopreening (Harrison 1965; Kikkawa and Kakizawa 1981). Such direct contacts between individuals could possibly facilitate the transmission of APV from one individual to another. Alternatively, virions can be transmitted by mechanical vectors, including mosquitoes, biting midges, and mites (Aruch et al. 2007; Huong et al. 2014; Lee et al. 2017; Yeo et al. 2019). All APV‐positive individuals were also positive for avian malaria. In Hawaii, introduced Cx. quinquefasciatus are thought to be the main vector of both avian malaria and APV (Warner 1968; Van Riper et al. 1986; Van Riper, Van Riper, and Hansen 2002; Atkinson and Lapointe 2009; Samuel et al. 2018). Similarly, results from this study suggest that APV is transmitted along with avian malaria through mosquitoes, rather than by direct contact. On the other hand, immune functions related to chronic malaria are also thought to play an important role by making birds more susceptible to pox infections (Samuel et al. 2018). This suggests that individuals infected with avian malaria are more likely to develop severe pox infections. Further monitoring may help in understanding the transmission of APV within the Bonin Islands.
5. Conclusion
In this study, the presence of avian pathogens of avian malaria and fowl pox was confirmed in the Bonin Islands of Japan for the first time. The existence of endemic resident, introduced, and migratory avian species, as well as endemic mosquitoes, depicts a close scenario to that of New Zealand. Detection of both pathogens from the critically endangered Bonin greenfinch was particularly alarming. Meanwhile, it was not possible to fully understand when and where these pathogens came from, and most islands require additional investigations. In the scope of conservation, it would be crucial to investigate the distribution and transmission status of avian haemosporidia at the island level in order to clarify whether conservation actions would involve specific islands, island groups, or the whole archipelago. We were also unable to investigate the endangered and endemic Bonin white‐eye. Furthermore, pathogenicity and possible impacts on the ecosystem of the Bonin Islands are still unknown. Continuous monitoring, including surrounding islands, will be needed to better understand the impact of these pathogens on the ecosystem of the Bonin Islands.
Author Contributions
Mizue Inumaru: data curation (lead), formal analysis (lead), investigation (lead), methodology (lead), resources (lead), validation (lead), visualization (lead), writing – original draft (lead). Rui Kimura: investigation (equal), methodology (equal), validation (equal). Naoko Suzuki: resources (supporting). Hajime Suzuki: resources (supporting). Kazuo Horikoshi: resources (supporting). Isao Nishiumi: resources (supporting), writing – review and editing (supporting). Kazuto Kawakami: resources (equal), writing – review and editing (supporting). Yoshio Tsuda: conceptualization (equal), investigation (supporting), methodology (supporting), supervision (equal), writing – original draft (equal), writing – review and editing (equal). Koichi Murata: conceptualization (supporting), funding acquisition (supporting), writing – review and editing (supporting). Yukita Sato: conceptualization (lead), funding acquisition (lead), project administration (lead), resources (equal), supervision (lead), validation (equal), visualization (equal), writing – original draft (equal), writing – review and editing (lead).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Data S1.
Acknowledgements
First and foremost, we would like to specially acknowledge our co‐author, Dr. Yoshio Tsuda, who passed away prior to the completion of this manuscript. His deep knowledge and passion for mosquitoes and avian haemosporidia constantly guided our study, and he was an essential member of our study group. We deeply thank the islanders of Chichijima who reported the deceased or injured birds to the Institute of Boninology. We also thank Ichiko Kawamukai, Ayako Kawakami, and Kanae Miyamoto for help in molecular detection of avian haemosporidia, and Tetsuyoshi Sakamoto for help in molecular detection of APV. Dr. Shoji Hamao, Dairo Kawaguchi, Kenta Morozumi, Shogo Adachi, and Hitoha Miyagawa helped with mist‐netting in Chichijima and Hahajima. We would like to acknowledge Dr. Hisashi Shibuya and Dr. Hirotaka Kondo for preparing the tissue thin slides and for their helpful insights on histopathological observations. Insightful comments on morphological identifications of haemosporidia were provided by Dr. Gediminas Valkiunas.
Funding: This study was partially supported by the Strategic Research Base Development Program “International Research on the Management of Zoonosis in Globalization and Training for Young Researchers” (project no. S1491007) from the MEXT of Japan and the Grant‐in‐Aid for Scientific Research (KAKENHI nos. 26450484, 19J20367, 21K05961, and 24K09222) from the Japan Society for the Promotion of Science (JSPS). Mist‐netting was conducted as a survey for the conservation management of rare birds by the Forestry Agency of Japan with permission from the Ministry of the Environment of Japan [permit numbers: No. 1603112 (2016), No. 2006031 (2020), No. 21051026 (2021)].
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information S1 of this article, and the obtained lineages were deposited to MalAvi and GenBank (Accession numbers LC765283‐LC765287).
References
- Aruch, S. , Atkinson C. T., Savage A. F., and Lapointe D. A.. 2007. “Prevalence and Distribution of Pox‐Like Lesions, Avian Malaria, and Mosquito Vectors in Kipahulu Valley, Haleakala National Park, Hawai'i, USA.” Journal of Wildlife Diseases 43: 567–575. [DOI] [PubMed] [Google Scholar]
- Atkinson, C. T. , Dusek R. J., Woods K. L., and Iko W. M.. 2000. “Pathogenicity of Avian Malaria in Experimentally‐Infected Hawaii Amakihi.” Journal of Wildlife Diseases 36: 197–204. [DOI] [PubMed] [Google Scholar]
- Atkinson, C. T. , and Lapointe D. A.. 2009. “Introduced Avian Diseases, Climate Change, and the Future of Hawaiian Honeycreepers.” Journal of Avian Medicine and Surgery 23: 53–63. [DOI] [PubMed] [Google Scholar]
- Atkinson, C. T. , Saili K. S., Utzurrum R. B., and Jarvi S. I.. 2013. “Experimental Evidence for Evolved Tolerance to Avian Malaria in a Wild Population of Low Elevation Hawai'i Amakihi ( Hemignathus virens ).” EcoHealth 10: 366–375. [DOI] [PubMed] [Google Scholar]
- Banyai, K. , Dénes B., Farkas S., et al. 2015. “Unique Genomic Organization of a Novel Avipoxvirus Detected in Turkey ( Meleagris gallopavo ).” Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases 35: 221–229. [DOI] [PubMed] [Google Scholar]
- Beadell, J. S. , Gering E., Austin J., et al. 2004. “Prevalence and Differential Host‐Specificity of Two Avian Blood Parasite Genera in the Australo‐Papuan Region.” Molecular Ecology 13: 3829–3844. [DOI] [PubMed] [Google Scholar]
- Bensch, S. , Hellgren O., and Pérez‐Tris J.. 2009. “MalAvi: A Public Database of Malaria Parasites and Related Haemosporidians in Avian Hosts Based on Mitochondrial Cytochrome b Lineages.” Molecular Ecology Resources 9: 1353–1358. [DOI] [PubMed] [Google Scholar]
- Bernotiene, R. , Palinauskas V., Iezhova T., Murauskaite D., and Valkiūnas G.. 2016. “Avian Haemosporidian Parasites (Haemosporida): A Comparative Analysis of Different Polymerase Chain Reaction Assays in Detection of Mixed Infections.” Experimental Parasitology 163: 31–37. [DOI] [PubMed] [Google Scholar]
- Biodiversity Center of Japan . 2016. “The Sixth and Seventh National Surveys on the Natural Environment.”
- BirdLife International . 2020. Threatened Birds of the World. Barcelona and Cambridge, UK: Lynx. [Google Scholar]
- Bonneaud, C. , Sepil I., Mila B., et al. 2009. “The Prevalence of Avian Plasmodium Is Higher in Undisturbed Tropical Forests of Cameroon.” Journal of Tropical Ecology 25: 439–447. [Google Scholar]
- Cannell, B. L. , Krasnec K. V., Campbell K., Jones H. I., Miller R. D., and Stephens N.. 2013. “The Pathology and Pathogenicity of a Novel Haemoproteus Spp. Infection in Wild Little Penguins ( Eudyptula minor ).” Veterinary Parasitology 197: 74–84. [DOI] [PubMed] [Google Scholar]
- Carlquist, S. 1974. Island Biology. New York, NY: Columbia University Press. [Google Scholar]
- Chahad‐Ehlers, S. , Fushita A. T., Lacorte G. A., De Assis P. C. P., and Del Lama S. N.. 2018. “Effects of Habitat Suitability for Vectors, Environmental Factors and Host Characteristics on the Spatial Distribution of the Diversity and Prevalence of Haemosporidians in Waterbirds From Three Brazilian Wetlands.” Parasites & Vectors 11: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, H. , Kawakami K., Suzuki H., and Horikoshi K.. 2007. “The Distribution of Seabirds in the Bonin Islands, Southern Japan.” Journal of the Yamashina Institute 39: 1–17. [Google Scholar]
- Chiba, Y. , and Suzuki T.. 2011. “Breeding Biology of the Ogasawara Buzzard Endemic to the Ogasawara (Bonin) Islands.” Ornithological Science 10: 119–129. [Google Scholar]
- Clark, N. J. , and Clegg S. M.. 2017. “Integrating Phylogenetic and Ecological Distances Reveals New Insights Into Parasite Host Specificity.” Molecular Ecology 26: 3074–3086. [DOI] [PubMed] [Google Scholar]
- Clark, N. J. , Clegg S. M., and Klaassen M.. 2016. “Migration Strategy and Pathogen Risk: Non‐breeding Distribution Drives Malaria Prevalence in Migratory Waders.” Oikos 125: 1358–1368. [Google Scholar]
- Dale, P. E. R. , and Knight J. M.. 2008. “Wetlands and Mosquitoes: A Review.” Wetlands Ecology and Management 16: 255–276. [Google Scholar]
- Devleesschauwer, B. , Torgerson P., Charlier J., et al. 2014. “Prevalence: Tools for Prevalence Assessment Studies.” R Package Version 0.4.0.
- Eastwood, J. R. , Peacock L., Hall M. L., et al. 2019. “Persistent Low Avian Malaria in a Tropical Species Despite High Community Prevalence.” International Journal for Parasitology. Parasites and Wildlife 8: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejiri, H. , Sato Y., Sasaki E., et al. 2008. “Detection of Avian Plasmodium Spp. DNA Sequences From Mosquitoes Captured in Minami Daito Island of Japan.” Journal of Veterinary Medical Science 70: 1205–1210. [DOI] [PubMed] [Google Scholar]
- Ellis, V. A. , Sari E. H. R., Rubenstein D. R., Dickerson R. C., Bensch S., and Ricklefs R. E.. 2018. “The Global Biogeography of Avian Haemosporidian Parasites Is Characterized by Local Diversification and Intercontinental Dispersal.” Parasitology 146: 213–219. [DOI] [PubMed] [Google Scholar]
- Emura, N. 2011. “The Avifauna of Chichijima Island, the Bonin Islands—Comparison With the Record Fifteen Years Ago.” Strix 27: 159–164. [Google Scholar]
- Emura, N. , Ando H., Kawakami K., and Isagi Y.. 2013. “Genetic and Morphological Differences Among Populations of the Japanese Bush‐Warbler (Aves: Sylviidae) on the Ogasawara Islands, Northern Pacific.” Pacific Science 67: 187–196. [Google Scholar]
- Ewen, J. G. , Bensch S., Blackburn T. M., et al. 2012. “Establishment of Exotic Parasites: The Origins and Characteristics of an Avian Malaria Community in an Isolated Island Avifauna.” Ecology Letters 15: 1112–1119. [DOI] [PubMed] [Google Scholar]
- Fecchio, A. , Ellis V. A., Bell J. A., et al. 2017. “Avian Malaria, Ecological Host Traits and Mosquito Abundance in Southeastern Amazonia.” Parasitology 144: 1117–1132. [DOI] [PubMed] [Google Scholar]
- Fecchio, A. , Wells K., Bell J. A., et al. 2019. “Climate Variation Influences Host Specificity in Avian Malaria Parasites.” Ecology Letters 22: 547–557. [DOI] [PubMed] [Google Scholar]
- Ferraguti, M. , Martínez‐De La Puente J., Roiz D., Ruiz S., Soriguer R. C., and Figuerola J.. 2024. “Landscape and Mosquito Community Impact the Avain Plasmodium Infection in Culex pipiens .” iScience 27: 109194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraguti, M. , Martínez‐De La Puente J., Roiz D., Ruiz S., Soriguer R. C., and Figuerola J.. 2016. “Effects of Landscape Anthropization on Mosquito Community Composition and Abundance.” Scientific Reports 6: 29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, J. , and Weisberg S.. 2011. An R Companion to Applied Regression. 3rd ed. Los Angeles, CA: Sage. [Google Scholar]
- Garcia‐Longoria, L. , Hellgren O., Bensch S., De Lope F., and Marzal A.. 2015. “Detecting Transmission Areas of Malaria Parasites in a Migratory Bird Species.” Parasitology 142: 1215–1220. [DOI] [PubMed] [Google Scholar]
- Giddens, W. E. , Swango L. J., Henderson J. D., et al. 1971. “Canary Pox in Sparrows and Canaries (Fringillidae) and in Weavers (Ploceidae).” Veterinary Pathology 8: 260–280. [DOI] [PubMed] [Google Scholar]
- Gill, F. , Donsker D., and Rasmussen P.. 2021. “IOC World Bird List (v11.2).”
- Government of Japan . 2010. “Nomination of the Ogasawara Islands for Inscription on the World Heritage List.”
- Gyuranecz, M. , Foster J. T., Dán Á., et al. 2013. “Worldwide Phylogenetic Relationship of Avian Poxviruses.” Journal of Virology 87: 4938–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara, M. , Amano Y., Nagamine T., Kinjo T., and Murata K.. 2005. “Detection of Haemoproteus Sp. and Microfilaria From the Japanese Black Wood Pigeon ( Columba janthina ) in Okinawa.” Journal of the Japan Veterinary Medical Association 58: 613–616. [Google Scholar]
- Harrison, C. J. O. 1965. “Allopreening as Agonistic Behaviour.” Behaviour 24: 161–208. [Google Scholar]
- Hellgren, O. , Atkinson C. T., Bensch S., et al. 2015. “Global Phylogeography of the Avian Malaria Pathogen Plasmodium relictum Based on MSP1 Allelic Diversity.” Ecography (Cop.) 38: 842–850. [Google Scholar]
- Hellgren, O. , Pérez‐Tris J., and Bensch S.. 2009. “A Jack‐Of‐All‐Trades and Still a Master of Some: Prevalence and Host Range in Avian Malaria and Related Blood Parasites.” Ecology 90: 2840–2849. [DOI] [PubMed] [Google Scholar]
- Hellgren, O. , Waldenström J., and Bensch S.. 2004. “A New PCR Assay for Simultaneous Studies of Leucocytozoon, Plasmodium, and Haemoproteus From Avian Blood.” Journal of Parasitology 90: 797–802. [DOI] [PubMed] [Google Scholar]
- Hothorn, T. , Bretz F., and Westfall P.. 2008. “Simultaneous Inference in General Parametric Models.” Biometrical Journal 50: 346–363. [DOI] [PubMed] [Google Scholar]
- Howe, L. , Castro I. C., Schoener E. R., Hunter S., Barraclough R. K., and Alley M. R.. 2012. “Malaria Parasites (Plasmodium spp.) Infecting Introduced, Native and Endemic New Zealand Birds.” Parasitology Research 110: 913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Dong L., Zhang C., and Zhang Y.. 2015. “Genetic Diversity, Temporal Dynamics, and Host Specificity in Blood Parasites of Passerines in North China.” Parasitology Research 114: 4513–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Rapševičius P., Chapa‐Vargas L., Hellgren O., and Bensch S.. 2019. “Within‐Lineage Divergence of Avian Haemosporidians: A Case Study to Reveal the Origin of a Widespread Haemoproteus Parasite.” Journal of Parasitology 105: 414–422. [PubMed] [Google Scholar]
- Huong, C. T. T. , Murano T., Uno Y., Usui T., and Yamaguchi T.. 2014. “Molecular Detection of Avian Pathogens in Poultry Red Mite (Dermanyssus gallinae) Collected in Chicken Farms.” Journal of Veterinary Medical Science 76: 1583–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inumaru, M. , Murata K., and Sato Y.. 2017. “Prevalence of Avian Haemosporidia Among Injured Wild Birds in Tokyo and Environs, Japan.” International Journal for Parasitology. Parasites and Wildlife 6: 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japan Meteorological Agency . 2020. “Tables of Monthly Climate Statistics”.
- Jarmin, S. , Manvell R., Gough R. E., Laidlaw S. M., and Skinner M. A.. 2006. “Avipoxvirus Phylogenetics: Identification of a PCR Length Polymorphism That Discriminates Between the Two Major Clades.” Journal of General Virology 87: 2191–2201. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy, S. , Minh B. Q., Wong T. K. F., Von Haeseler A., and Jermiin L. S.. 2017. “ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates.” Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katano, R. , Tsuda Y., Saitoh Y., and Kobayashi M.. 2010. “Experimental Studies on the Ecology of Culex Inatomii (Diptera: Culicidae): Effects of Salinity and Temperature on Larval Development and Reproduction of Populations During the Cool Season (Japanese).” Medical Entomology and Zoology 61: 327–333. [Google Scholar]
- Kawakami, K. 2008. “Threats to Indigenous Biota From Introduced Species on the Bonin Islands, Sothern Japan.” Journal of Disaster Research 3: 174–186. [Google Scholar]
- Kawakami, K. 2019. “The History of Anthropogenic Disturbance and Invasive Alien Species Impact on the Indigenous Avifauna of the Ogasawara Islands, Southern Japan.” Japanese Journal of Ornithology 68: 237–262. [Google Scholar]
- Kawakami, K. , Eda M., Izumi H., Horikoshi K., and Suzuki H.. 2018. “Phylogenetic Position of Endangered Puffinus lherminieri bannermani .” Ornithological Science 17: 11–18. [Google Scholar]
- Kawakami, K. , and Higuchi H.. 2003. “Interspecific Interactions Between the Native and Introduced White‐Eyes in the Bonin Islands.” IBIS 145: 583–592. [Google Scholar]
- Kawakami, K. , and Okochi I., eds. 2010. Restoring the Oceanic Island Ecosystem: Impact and Management of Invasive Alien Species in the Bonin Islands. Japan: Springer. [Google Scholar]
- Kikkawa, J. , and Kakizawa R.. 1981. “Agonistic Behaviour of Japanese White‐Eyes Zosterops Japonica on Miyake Island.” Journal of the Yamashina Institute for Ornithology 13: 60–70. [Google Scholar]
- Kim, K. S. , and Tsuda Y.. 2010. “Seasonal Changes in the Feeding Pattern of Culex pipiens Pallens Govern the Transmission Dynamics of Multiple Lineages of Avian Malaria Parasites in Japanese Wild Bird Community.” Molecular Ecology 19: 5545–5554. [DOI] [PubMed] [Google Scholar]
- Kleindorfer, S. , and Dudaniec R.. 2006. “Increasing Prevalence of Avian Poxvirus in Darwin's Finches and Its Effect on Male Pairing Success.” Journal of Avian Biology 37: 69–76. [Google Scholar]
- Kueffer, C. , Drake D. R., and Fernández‐Palacios J. M.. 2014. “Island Biology: Looking Towards the Future.” Biology Letters 10: 20140719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher G., Li M., Knyaz C., and Tamura K.. 2018. “MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms.” Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi, T. 2018. “Digenean Trematode Fauna Parasitic in Fishes From the Ogasawara Islands.” Memoirs of the National Science Museum 52: 95–103. [Google Scholar]
- Lalubin, F. , Delédevant A., Glaizot O., and Christe P.. 2013. “Temporal Changes in Mosquito Abundance ( Culex pipiens ), avian Malaria Prevalence and Lineage Composition.” Parasites & Vectors 6: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe, D. A. , Goff M. L., and Atkinson C. T.. 2005. “Comparative Susceptibility of Introduced Forest‐Dwelling Mosquitoes in Hawai'i to Avian Malaria, Plasmodium relictum .” Journal of Parasitology 91: 843–849. [DOI] [PubMed] [Google Scholar]
- Le Loc'h, G. , Ducatez M. F., Camus‐Bouclainville C., Guérin J.‐L., and Bertagnoli S.. 2014. “Diversity of Avipoxviruses in Captive‐Bred Houbara Bustard.” Veterinary Research 45: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. R. , Koo B.‐S., Kim J.‐T., et al. 2017. “Molecular Edidemiology of Avian Poxvirus in the Oriental Turtle Dove ( Streptopelia orientalis ) and the Biting Midge (Culicoides arakawae) in the Republic of Korea.” Journal of Wildlife Diseases 53: 749–760. [DOI] [PubMed] [Google Scholar]
- Leigh, J. W. , and Bryant D.. 2015. “POPART: Full‐Feature Software for Haplotype Network Construction.” Methods in Ecology and Evolution 6: 1110–1116. [Google Scholar]
- Madden, T. 2013. The BLAST Sequence Analysis Tool. Bethesda: National Center for Biotechnology Information. [Google Scholar]
- Maekawa, Y. , Kimura R., Kasai A., et al. 2021. “Faunal and Genetic Studies of Mosquitoes on Chichi‐Jima and Haha‐Jima, the Ogasawara (Bonin) Islands, Japan.” Medical Entomology and Zoology 72: 237–253. [Google Scholar]
- Martínez‐Abraín, A. , Esparza B., and Oro D.. 2004. “Lack of Blood Parasites in Bird Species: Does Absence of Blood Parasite Vectors Explain It all?” Ardeola 5: 225–232. [Google Scholar]
- Martínez‐De La Puente, J. , Eberhart‐Phillips L. J., Carmona‐Isunza C., et al. 2017. “Extremely Low Plasmodium Prevalence in Wild Plovers and Coursers From Cape Verde and Madagascar.” Malaria Journal 16: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes, L. , Piersma T., Lecoq M., Spaans B., and Ricklefs R. E.. 2005. “Disease‐Limited Distributions?” OIKOS 109: 396–404. [Google Scholar]
- Ministry of the Environment . 2010. “Ogasawara Islands Mangement Plan.”
- Ministry of the Environment . 2020. “The Japanese Red List of Threatened Species, 2020 Version.”
- Momiyama, T. 1930. “On the Birds of Bonin and Iwo‐Islands (Japanese).” Bulletin of the Biogeographical Society of Japan 1: 89–186. [Google Scholar]
- Murata, K. 2007. “Study on Avian Haemosporidian Parasites in Japanese Wild Birds (Japanese).” Journal of Animal Protozooses (Japan) 22: 1–8. [Google Scholar]
- Murata, K. , Nii R., Yui S., et al. 2008. “Avian Haemosporidian Parasites Infection in Wild Birds Inhabiting Minami‐Daito Island of the Northwest Pacific, Japan.” Journal of Veterinary Medical Science 70: 501–503. [DOI] [PubMed] [Google Scholar]
- Nagata, H. 2006. “Reevaluation of the Prevalence of Blood Parasites in Japanese Passerines by Using PCR Based Molecular Diagnosis.” Ornithological Science 5: 105–112. [Google Scholar]
- National Statistics Center . 2018. “Portal Site of Official Statistics of Japan.”
- Ornithological Society of Japan . 2012. Check‐List of Japanese Birds. 7th ed. Shinjuku, Tokyo: Ornithological Society of Japan. [Google Scholar]
- Pacheco, M. A. , Cepeda A. S., Bernotienė R., et al. 2018. “Primers Targeting Mitochondrial Genes of Avian Haemosporidians: PCR Detection and Differential DNA Amplification of Parasites Belonging to Different Genera.” International Journal for Parasitology 48: 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinauskas, V. , Žiegytė R., Iezhova T. A., Ilgūnas M., Bernotienė R., and Valkiūnas G.. 2016. “Description, Molecular Characterisation, Diagnostics and Life Cycle of Plasmodium elongatum (Lineage pERIRUB01), the Virulent Avian Malaria Parasite.” International Journal for Parasitology 46: 697–707. [DOI] [PubMed] [Google Scholar]
- Pardal, S. , Alves J. A., Zé‐Zé L., et al. 2014. “Shorebird Low Spillover Risk of Mosquito‐Borne Pathogens on Iberian Wetlands.” Journal für Ornithologie 155: 549–554. [Google Scholar]
- Pérez‐Tris, J. , Williams R. A. J., Abel‐Fernández E., et al. 2011. “A Multiplex PCR for Detection of Poxvirus and Papillomavirus in Cutaneous Warts From Live Birds and Museum Skins.” Avian Diseases 55: 545–553. [DOI] [PubMed] [Google Scholar]
- Perkins, S. L. , and Schall J. J.. 2002. “A Molecular Phylogeny of Malarial Parasites Recovered From Cytochrome b Gene Sequences.” Journal of Parasitology 88: 972–978. [DOI] [PubMed] [Google Scholar]
- QGIS Developmental Team . 2020. “QGIS Geographic Information System.”
- Quillfeldt, P. , Arriero E., Martínez J., Masello J. F., and Merino S.. 2011. “Prevalence of Blood Parasites in Seabirds—A Review.” Frontiers in Zoology 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillfeldt, P. , Martínez J., Hennicke J., et al. 2010. “Hemosporidian Blood Parasites in Seabirds‐a Comparative Genetic Study of Species From Antarctic to Tropical Habitats.” Naturwissenschaften 97: 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. “R: A Language and Environment for Statistical Computing.”
- Rambaut, A. 2012. “FigTree: Tree Figure Drawing Tool Version 1.4.0.” tree.bio.ed.ac.uk/software/figtree.
- Ronquist, F. , and Huelsenbeck J. P.. 2003. “MrBayes 3: Bayesian Phylogenetic Inference Under Mixed Models.” Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. , Teslenko M., Van Der Mark P., et al. 2012. “MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space.” Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahashi, N. , Akiba M., Ota Y., et al. 2015. “Brown Root Rot Caused by Phellinus Noxius in the Ogasawara (Bonin) Islands, Southern Japan—Current Status of the Disease and Its Host Plants.” Australasian Plant Disease Notes 10: 33. [Google Scholar]
- Saitoh, T. , Kawakami K., Red'kin Y. A., Nishiumi I., Kim C.‐H., and Kryukov A. P.. 2020. “Cryptic Speciation of the Oriental Greenfinch Chloris sinica on Oceanic Islands.” Zoological Science 37: 1–15. [DOI] [PubMed] [Google Scholar]
- Samuel, M. D. , Woodworth B. L., Atkinson C. T., Hart P. J., and LaPointe D. A.. 2018. “The Epidemiology of Avian Pox and Interaction With Avian Malaria in Hawaiian Forest Birds.” Ecological Monographs 88: 621–637. [Google Scholar]
- Sarker, S. , Das S., Lavers J. L., et al. 2017. “Genomic Characterization of Two Novel Pathogenic Avipoxviruses Isolated From Pacific Shearwaters (Ardenna Spp.).” BMC Genomics 18: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, Y. 2003. “The Nature of Ogasawara and Its Conservation.” Global Environmental Research‐English Edition 7: 3–14. [Google Scholar]
- Shivaprasad, H. L. , Kim T., Tripathy D., Woolcock P. R., and Uzal F.. 2009. “Avian Pathology Unusual Pathology of Canary Poxvirus Infection Associated With High Mortality in Young and Adult Breeder Canaries ( Serinus canaria ) Unusual Pathology of Canary Poxvirus Infection Associated With High Mortality in Young and Adult Bre.” Avian Pathology 38: 311–316. [DOI] [PubMed] [Google Scholar]
- Sijbranda, D. C. , Campbell J., Gartrell B. D., and Howe L.. 2016. “Avian Malaria in Introduced, Native and Endemic New Zealand Bird Species in a Mixed Ecosystem.” New Zealand Journal of Ecology 40: 72–79. [Google Scholar]
- Soares, L. , Latta S. C., and Ricklefs R. E.. 2020. “Neotropical Migratory and Resident Birds Occurring in Sympatry During Winter Have Distinct Haemosporidian Parasite Assemblages.” Journal of Biogeography 47: 748–759. [Google Scholar]
- Sorci, G. 2013. “Immunity, Resistance and Tolerance in Bird–Parasite Interactions.” Parasite Immunology 35: 350–361. [DOI] [PubMed] [Google Scholar]
- Sugita, N. , Kawakami K., and Nishiumi I.. 2016. “Origin of Japanese White‐Eyes and Brown‐Eared Bulbuls on the Volcano Islands.” Zoological Science 33: 146–153. [DOI] [PubMed] [Google Scholar]
- Sugiura, S. 2016. “Impacts of Introduced Species on the Biota of an Oceanic Archipelago: The Relative Importance of Competitive and Trophic Interactions.” Ecological Research 31: 155–164. [Google Scholar]
- Sumiyama, D. , Hayashida I., Kanazawa T., Anzai H., and Murata K.. 2020. “Prevalence and Antimicrobial‐Resistance Profiles of Salmonella Spp. Isolated From Green Anoles ( Anolis carolinensis ) Collected on the Haha‐Jima of the Ogasawara Archipelago, Japan.” Journal of Veterinary Medical Science 82: 1558–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S. 1973. “Insects of Medical Importance in Ogasawara (Bonin) Islands (Japanese).” Eisei Dobutsu 24: 143–148. [Google Scholar]
- Takaoka, H. , and Saito K.. 2006. “A New Species of Simulium (Nevermannia) From the Ogasawara (Bonin) Islands, Japan (Diptera: Simuliidae).” Tropical Medicine and Health 34: 155–158. [Google Scholar]
- Takaoka, H. , Saito K., and Suzuki H.. 1999. “Simulium (Nevermannia) Bonninense From the Ogasawara (Bonin) Islands, Japan (Diptera: Simuliidae): Taxonomic Assignment to the Vernum‐Group and Descriptions of Male, Pupa and Mature Larba.” Japanese Journal of Tropical Medicine and Hygiene 27: 189–194. [Google Scholar]
- Tanaka, K. , Mizusawa K., and Saugstad E. S.. 1979. “A Revision of the Adult and Larval Mosquitoes of Japan (Including the Ryukyu Archpelago and the Ogasawara Islands) and Korea (Diptera: Culicidae).” Contributions of the American Entomological Institute 16: 1–987. [Google Scholar]
- Tanaka, K. , Sumiyama D., Kanazawa T., Sato Y., and Murata K.. 2019. “Prevalence and Molecular Phylogeny of Avian Malaria Parasites in Columbiformes in Japan (Japanese).” Japanese Journal of Zoo and Wildlife Medicine 24: 65–71. [Google Scholar]
- Tanigawa, M. , Sato Y., Ejiri H., et al. 2013. “Molecular Identification of Avian Haemosporidia in Wild Birds and Mosquitoes on Tsushima Island, Japan.” Journal of Veterinary Medical Science 75: 319–326. [DOI] [PubMed] [Google Scholar]
- Tokiwa, T. , Hashimoto T., Yabe T., Komatsu N., Akao N., and Ohta N.. 2013. “First Report of Angiostrongylus cantonensis (Nematoda: Angiostrongylidae) Infections in Invasive Rodents From Five Islands of the Ogasawara Archipelago, Japan.” PLoS One 8: e70729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma, T. , and Miyagi I.. 2005. “Notes on Mosquitoes in Chichi‐Jima, Ogasawara Archipelago, Japan and Biology of Culex (Sirivanakarnius) Boninensis (Diptera: Culicidae).” Medical Entomology and Zoology 56: 237–241. [Google Scholar]
- Tomiyama, K. 1998. “Disturbance in Island Ecosystem by Introduced Species in Ogasawara Islands.” Japanese Journal of Ecology 48: 63–72. [Google Scholar]
- Tsuda, Y. 2017. “Review of Recent Studies on Avian Malaria Parasites and Their Vector Mosquitoes (Japanese).” Medical Entomology and Zoology 68: 1–10. [Google Scholar]
- Tsuda, Y. 2019. An Illustrated Book of the Mosquitoes of Japan: Adult Identification, Geographic Distribution and Ecological Note (Japanese). Meguro, Tokyo: Hokuryukan. [Google Scholar]
- Tsuda, Y. , Matsui S., Saito A., et al. 2009. “Ecological Study of Avian Malaria Vectors on the Island of Minami‐Daito, Japan.” Journal of the American Mosquito Control Association 25: 279–284. [DOI] [PubMed] [Google Scholar]
- UNESCO . 2011. “Decisions Adopted by the Word Heritage Committee at Its 35th Session.”
- Valkiūnas, G. 2005. Avian Malaria Parasites and Other Haemosporidia. Boca Raton, FL: CRC Press. [Google Scholar]
- Valkiūnas, G. , Kazlauskienė R., Bernotienė R., Palinauskas V., and Iezhova T. A.. 2013. “Abortive Long‐Lasting Sporogony of Two Haemoproteus Species (Haemosporida, Haemoproteidae) in the Mosquito Ochlerotatus cantans, With Perspectives on Haemosporidian Vector Research.” Parasitology Research 112: 2159–2169. [DOI] [PubMed] [Google Scholar]
- Valkiūnas, G. , Žiegytė R., Palinauskas V., et al. 2015. “Complete Sporogony of Plasmodium relictum (Lineage pGRW4) in Mosquitoes Culex pipiens Pipiens, With Implications on Avian Malaria Epidemiology.” Parasitology Research 114: 3075–3085. [DOI] [PubMed] [Google Scholar]
- Van Hoesel, W. , Marzal A., Magallanes S., Santiago‐Alarcon D., Ibáñez‐Bernal S., and Renner S. C.. 2019. “Management of Ecosystems Alters Vector Dynamics and Haemosporidian Infections.” Scientific Reports 9: 8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Riper, C. I. , Van Riper S. G., Goff M. L., and Laird M.. 1986. “The Epizootiology and Ecological Significance of Malaria in Hawaiian Land Birds.” Ecological Monographs 56: 327–344. [Google Scholar]
- Van Riper, C. I. , Van Riper S. G., and Hansen W. R.. 2002. “Epizootiology and Effect of Avian Pox on Hawaiian Forest Birds.” Auk 119: 929–942. [Google Scholar]
- Van Riper, C., III . 1991. “The Impact of Introduced Vectors and Avian Malaria on Insular Passerifor Bird Populations in Hawaii.” Bulletin of the Society of Vector Ecology 16: 59–83. [Google Scholar]
- Wada, Y. 1986. “Revision of the Claggi Group of the Genus Culicoides Distributed in Japan, With Description of a New Species (Diptera: Ceratopogonidae).” Medical Entomology and Zoology 37: 141–152. [Google Scholar]
- Waldenström, J. , Bensch S., Hasselquist D., and Östman Ö.. 2004. “A New Nested Polymerase Chain Reaction Method Very Efficient in Detecting Plasmodium and Haemoproteus Infections From Avian Blood.” Journal of Parasitology 90: 191–194. [DOI] [PubMed] [Google Scholar]
- Warner, R. E. 1968. “The Role of Introduced Diseases in the Extinction of the Endemic Hawaiian Avifauna.” Condor 70: 101–120. [Google Scholar]
- Yabe, T. , Hashimoto T., Takiguchi M., Aoki M., and Kawakami K.. 2009. “Seabirds in the Stomach Contents of Black Rats Rattus rattus on Higashijima, the Ogasawara (Bonin) Islands, Japan.” Marine Ornithology 37: 293–295. [Google Scholar]
- Yeo, G. , Wang Y., Chong S. M., et al. 2019. “Characterization of Fowlpox Virus in Chickens and Bird‐Biting Mosquitoes: A Molecular Approach to Investigating Avipoxvirus Transmission.” Journal of General Virology 100: 838–850. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Wu Y., Zhang Q., Su D., and Zou F.. 2014. “Prevalence Patterns of Avian Plasmodium and Haemoproteus Parasites and the Influence of Host Relative Abundance in Southern China.” PLoS One 9: e99501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žiegytė, R. 2014. The Experimental Study on Development of Avian Malaria Parasites (Plasmodium) and Haemoproteids (Haemoproteus) in Vectors. Doctoral dissertation. Vilnius: Vilnius University. [Google Scholar]
- Žiegytė, R. , and Valkiūnas G.. 2014. “Recent Advances in Vector Studies of Avian Haemosporidian Parasites.” Ekologija 60: 73–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information S1 of this article, and the obtained lineages were deposited to MalAvi and GenBank (Accession numbers LC765283‐LC765287).


