Abstract
Meat quality is a key factor determining the economic viability of the broiler industry, particularly in native broiler breeds. Skeletal muscles contain a mixture of muscle fibers, each possessing unique physicochemical properties; the composition of myofiber types within these muscles is closely linked to meat quality. However, comprehension of the regulatory mechanisms governing this trait remains limited. Therefore, we conducted a genome-wide association study (GWAS) with a population of 400 yellow-feather broilers to explore genetic variations associated with myofiber-type composition at the genomic level. Whole-genome resequencing was employed to detect genetic variations and immunohistochemistry was used for muscle fiber typing in the sartorius muscle. We identified 1 and 18 single-nucleotide polymorphisms (SNPs) significantly and potentially associated with the proportion of slow muscle fibers, respectively, and 1 and 12 SNPs significantly and potentially associated with the area proportion of slow muscle fibers, respectively. We annotated several candidate genes, including DMD, KLF7, CREB1, EFCAB11, GADD45A, GSTT1, and GSTT1L, which are related to myofiber type composition. We also demonstrated that myofiber composition traits exhibit low-to-medium heritability, indicating potential for enhancement through genetic selection. These findings provide a crucial reference for further studies on the regulatory mechanisms of poultry meat quality and for advancing the breeding of superior-quality broiler chickens.
Keywords: Genome-wide association study, Myofiber type composition, Poultry meat quality, Broiler breeding
Introduction
As the growth rate and market share of modern broiler chickens continue to rise, a decline in meat quality presents a significant challenge for industrial development. Myofibers are the basic units that constitute muscle tissue (Soglia et al., 2021). In chickens, myofibers are classified into two primary types: red myofibers (type I/IIA), which are characterized by oxidative metabolism and slow contraction, and white myofibers (type IIB), known for glycolytic metabolism and rapid contraction. The proportion of these myofiber types within the muscle substantially influences meat quality, as well as impacts factors such as pH, water-holding capacity, and shear force (Ismail and Joo, 2017; Jeong et al., 2017; Huo et al., 2022; Park et al., 2022). With advances in genomic selection technologies (Liu et al., 2023; Chen et al., 2024), comprehending the regulatory mechanisms that control myofiber composition is vital for expediting the genetic enhancement of meat quality in broiler breeding programs.
Myofiber composition results from complex biological processes and is influenced by various factors, including nutrition, environment, and genetics (Li et al., 2007; Chaillou, 2018; Mo et al., 2023). Despite numerous transcriptome studies and overexpression or knockdown experiments identifying candidate genes associated with chicken myofiber types, such as PPARGC1A, CSRP3, SIX1, and SOX6, the regulatory mechanisms remain largely unexplored (Shu et al., 2017; Ma et al., 2018; Liu et al., 2020; Liu et al., 2022; Shan et al., 2023). With the rapid advancement of high-density single-nucleotide polymorphism (SNP) arrays and high-throughput sequencing technologies, genome-wide association studies (GWAS)have emerged as a powerful approach for investigating the regulatory mechanisms of complex traits and diseases in livestock (Tan et al., 2023). This methodology is extensively applied in studies focused on traits such as growth (Zhang et al., 2021), meat quality (Munyaneza et al., 2022), disease resistance (Psifidi et al., 2016), and egg production (Gao et al., 2022) in chickens. However, owing to challenges in myofiber type identification, only a few researchers have pursued GWAS on myofiber traits in humans and pigs (Guo et al., 2020; Semenova et al., 2022).
In our study, we employed whole-genome sequencing for a GWAS to analyze myofiber composition traits in a population of yellow-feather broiler breeders. We aimed to enhance the understanding of the regulatory mechanisms that affect meat quality, thereby facilitating the acceleration of broiler breeding improvements.
Material and methods
Ethics statement
The protocol for the animal study was reviewed and approved by the Animal Care and Use Committee of the Jiangsu Institute of Poultry Science (Approval ID: S202210).
Chicken population
Four hundred roosters from a breeding population of yellow-feather broiler chickens from Jiangsu Lihua Livestock Co., Ltd. were included in this study. These chickens were raised at the Jintan Breeding Farm in Jiangsu Province (Changzhou, 31°43′25.5″ N, 119°34′57.3″ E, 7.0 m above sea level) from July 2022 to September 2022, during the autumn months when outdoor temperatures ranged from 30–35°C. The breed, designated as “Huashan No. 2 Broiler,” is a fast-growing chicken collaboratively developed by Lihua Animal Husbandry Co., Ltd. and the Jiangsu Institute of Poultry Science. This breed has undergone over ten generations of selection for growth rate and feed conversion efficiency, enabling it to reach a marketable age of 60 days with an approximate weight of 2.5 kg. During the study, all chickens had unrestricted access to feed and water under uniformly recommended nutritional and environmental conditions. Blood samples were collected from all individuals and preserved at −20°C for subsequent analyses.
Muscle sampling and carcass traits measurement
Chicken leg muscles typically comprise a mixture of fast- and slow-twitch fibers. However, different regions within the same muscle, such as the deep and superficial layers, may vary in fiber-type composition. The sartorius, the longest skeletal muscle, is responsible for functions such as knee joint movement. In this population, the width and thickness of this muscle are approximately 1 cm and 0.5 cm, respectively, meeting sectioning requirements without trimming. Therefore, we selected the sartorius muscle as the research material to minimize the sampling errors.
At 60 days, chickens were euthanized; the right sartorius muscle of each bird was fixed in polyformaldehyde for myofiber-type analysis. Key performance traits, including live body weight, carcass weight, dressed percentage, and leg muscle weight and percentage, were measured and recorded for each individual.
Myofiber type determination
The sartorius muscles from all experimental birds were subjected to immunohistochemical staining, as described previously (Weng et al., 2022), to identify slow myofibers. Anti-Slow Myosin Skeletal Heavy chain antibody (MYH7B, 1:4000, ab11083, Abcam) used for this staining resulted in darkly stained slow myofibers. Accordingly, the proportion of slow myofibers (SM%) and the percentage area of slow myofibers (SMA%) were quantified with Image-Pro Plus (Media Cybernetics, Rockville, MD, USA) using the following formulas:
and
For each individual, a subset of at least 500 myofibers was analyzed from each of the 388 chickens processed.
Correlation analysis between myofiber and carcass traits
R software was used to analyze the correlation between SM%, SMA%%, and carcass traits to explore associations with myofiber composition.
Whole-genome resequencing
Genomic DNA was isolated from the blood samples using the conventional phenol-chloroform method. DNA quantity and quality were determined using a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). A DNA library was constructed, and whole-genome sequencing (WGS) was performed according to standard BGISEQ procedures. Genomic DNA from 388 roosters with characterized myofiber composition phenotypes were sequenced using the DNBSEQ T7 platform (BGI, Shenzhen, China), generating 150 bp paired-end reads at a sequencing depth of 10 × .
Quality control and variant calling
Raw sequencing reads were processed with fastp software (Chen, 2023) to remove adapters and low-quality sequences. Clean reads were mapped to the chicken reference genome GRcg7b using the BWA-MEM algorithm with its default settings (Li and Durbin, 2009). Format conversion and sequence sorting were performed using Picard 3.1 (http://broadinstitute.github.io/picard/) and SAMtools 1.18 (Li et al., 2009). SNPs were detected using GATK V3.8 (McKenna et al., 2010). To isolate high-quality SNPs for GWAS, stringent filtering criteria recommended by the GATK guidelines were implemented for all variants with Quality by Depth < 2.0, Fisher Strand > 60.0, root mean square Mapping Quality < 40.0, MQRankSum < −12.5, HaplotypeScore > 13.0, and ReadPosRankSum < −8.0. SNPs that passed these filters were further screened based on quality control thresholds: minor allele frequency >0.05 and missing rate <0.02. Individuals with genotype call rates below 0.8 were excluded. Following quality control, 388 individuals and 10,266,594 SNPs were retained for further analyses.
Population structure analysis
Principal component analysis (PCA) was performed to assess the population structure among 388 chickens using the PCA program within the PLINK v1.9 software package (Purcell et al., 2007).
Genome-wide association studies
A univariate mixed linear model was used for the GWAS analysis of myofiber composition traits using GEMMA (Zhou and Stephens, 2012). The statistical model employed is as follows:
Where y denotes the phenotypic values of 388 individuals, W is the fixed effects matrix including the first three principal components from PCA, α is the fixed effects vector, x represents the genotype vector, β is the effect vector for the SNPs, u is a vector of random effects with a covariance structure that refers to the n-dimensional multivariate normal distribution , indicates the ratio of genetic variance to residual variance (the random effects vector () and the residual vector (), Additionally represents the residual variance, where K is the relatedness matrix calculated based on SNPs using GEMMA, and is the identity matrix.
Linkage disequilibrium (LD) pruning was performed with parameters set to a 50 SNP window size, 10 SNP step size, and an r2 threshold of 0.2, resulting in 432,529 independent SNP markers. The genome-wide significance level was established using a 5% Bonferroni correction applied to the number of estimated independent SNPs, leading to a significance threshold of 0.05/432,529 = 1.16e-07 [-log10 (P-value) =6.94], and the threshold for suggestive significance was set at 1/432,529 = 2.31e-06 [−log10(P-value) =5.64]. The CMplot package in R was used to produce Manhattan and quantile–quantile (Q–Q) plots of the analyzed traits.
SNP annotation
Based on the chicken reference genome GRcg7b, significant and potentially associated SNPs were annotated, and neighboring genes were identified using the Variant Effect Predictor (VEP) online tool (https://asia.ensembl.org/Tools/VEP). A selection criterion of 100 Kb upstream and downstream was used to screen neighboring genes for SNPs.
Genetic parameters estimation
The restricted maximum likelihood (REML) method was used to estimate genetic parameters using GCTA software version 1.93 (Yang et al., 2011). The SNP-based heritability (h2) was derived from the genetic relationship matrix (GRM) across individual pairs. Genetic correlations across pairs of traits were evaluated using the bivariate REML option.
To identify specific chromosomes harboring significant SNPs linked to particular traits, the total genomic variance was partitioned by chromosome. For each chromosome, individual GRMs were calculated from the SNPs using the GCTA software. The variance attributed to each chromosome for every GRM was calculated independently using the linear model:
Where Y represents the vector of phenotypes, β is the vector denoting fixed effects and covariates, accompanied by its incidence matrix X, n represents the number of distinct SNP subsets utilized in the partitioning, gG denotes the vector of random additive genetic effects attributed to the aggregated SNP information, and e represents a vector of random residuals.
Results
Phenotypic characterization
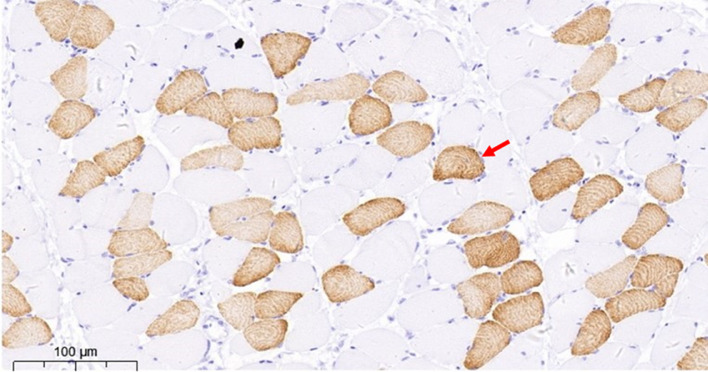
The immunohistochemical results of the sartorius muscle are illustrated in Fig. 1, where darker regions indicate slow myofiber staining with the antibody. Table 1 presents the descriptive statistics for myofiber composition and carcass traits in the experimental population of 388 roosters. The mean values of SM% and SMA% were 27.39% and 21.99%, respectively. The coefficients of variation for SMA% were significantly higher than those for SM%, indicating a higher degree of genetic variability associated with SMA% in the sampled population. Table 2 presents the correlation analysis between the two myofiber phenotypes and five carcass traits, showing a weak correlation between SM%, SMA%, and carcass traits.
Fig. 1.
Assessment of muscle fiber types using immunohistochemistry. Arrows indicate slow muscle fibers that appear dark owing to antibody staining (magnifications 20 ×). Bar = 100μm.
Table 1.
Descriptive statistics for myofiber composition and carcass traits.
| Traits | N_records1 | Mean | SD2 | Maximum | Minimum | CV3(%) |
|---|---|---|---|---|---|---|
| Proportion of slow myofiber numbers (SM%) | 388 | 27.93 | 5.69 | 43.36 | 12.10 | 20.38 |
| Percentage area of slow myofibers (SMA%) | 388 | 22.34 | 6.72 | 43.66 | 8.50 | 30.08 |
| Live body weight (g) | 388 | 2537.85 | 186.62 | 3082 | 1714 | 7.35 |
| Carcass weight (g) | 388 | 2351.62 | 187.68 | 2920 | 1630 | 7.98 |
| Dressed percentage (%) | 388 | 92.67 | 3.05 | 76.41 | 96.96 | 3.29 |
| Leg muscle weight (g) | 388 | 192.18 | 28.43 | 283.70 | 20.30 | 14.79 |
| Percentage of leg muscle (%) | 388 | 10.75 | 1.30 | 13.95 | 1.16 | 12.06 |
N_records: number of chickens with phenotypic data.
SD: standard deviation.
CV: coefficient of variation.
Table 2.
Correlation analysis between myofiber composition and carcass traits.
| Live body weight | Carcass weight | Dressed percentage | Leg muscle weight | Percentage of leg muscle | |
|---|---|---|---|---|---|
| Proportion of slow myofiber numbers (SM%) | −0.0083 | 0.0795 | 0.2099 | 0.2304 | 0.1242 |
| Percentage area of slow myofibers (SMA%) | −0.0069 | 0.0586 | 0.1520 | 0.1542 | 0.0631 |
Summary of genome sequencing and SNP calling
Following whole-genome sequencing conducted according to standard BGISEQ procedures, 4.25 Tb of raw data were generated from 388 individuals, achieving an average depth of 10.17 × . The filtering process yielded 4.24 Tb of high-quality sequencing data, with an average mapping rate of 98.73% when aligned to the reference chicken genome. The average GC content was 45.65%. After SNP calling using GATK, 16,411,878 SNPs were obtained for subsequent analysis.
Population structure analysis
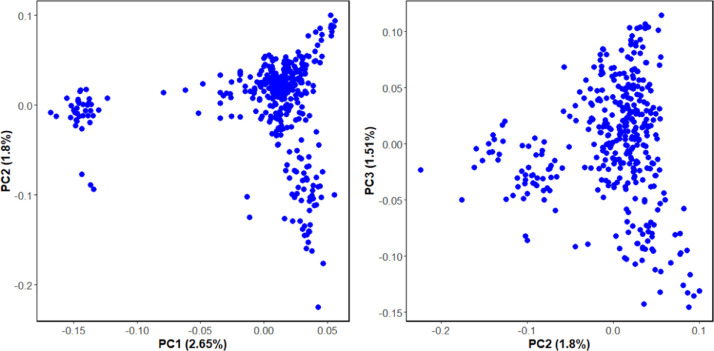
Fig. 2 displays the results of the population structure analysis, indicating slight stratification in the population of 388 individuals. We incorporated the top three principal components as fixed effects in the statistical model to mitigate the impact of population structure on GWAS results, thereby minimizing the risk of false positives in the association analysis and enhancing the reliability of our findings.
Fig. 2.
Principal components analysis (PCA) of chicken populations (388 individuals). PC1, PC2, PC3 explain the percentages of 2.65%, 1.8% and 1.51%, respectively.
Genome-wide association study of myofiber composition traits
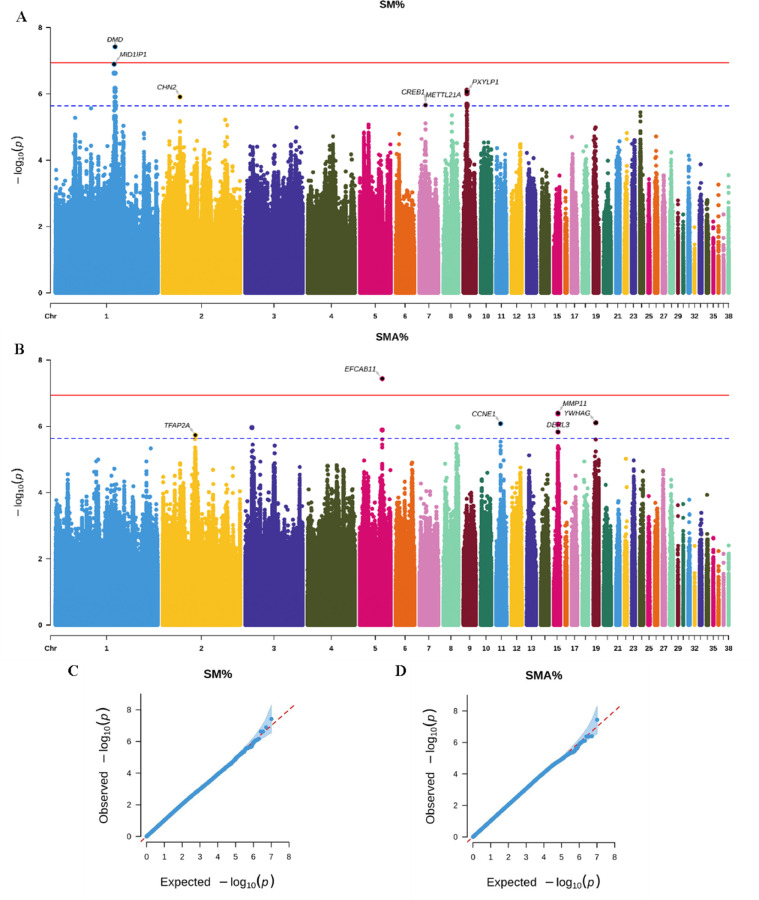
After filtering for SNPs, a GWAS was conducted on 10,266,594 SNPs to identify genomic regions associated with SM% and SMA%. Manhattan and quantile–quantile (Q-Q) plots for each trait are depicted in Fig. 3. The analysis helped identify two SNPs with genome-wide significant associations with myofiber traits at a P-value threshold of < 1.16E-7, and 30 SNPs at a suggestively significant level of P-value < 2.31E-6. The inflation coefficients calculated for SM% and SMA% were 1.022 and 1.024, respectively, indicating minimal bias from population stratification. Moreover, 68 genes located proximal to the significant SNPs, within a 100 kb window upstream and downstream, were suggested as potential candidate genes affecting these traits. As detailed in Fig. 3A and Table 3, an SNP site significantly associated with the SM% trait was identified on chromosome 1 within DMD. Eighteen loci with potential associations with this trait were observed across chromosomes 1, 2, 7, and 9 near 13 genes, including MID1IP1, GK2, CHN2, CREB1, and KLF7.
Fig. 3.
Genome-wide association study (GWAS) results for myofiber type composition traits. (A) Manhattan plot for the number of slow myofibers (SM%). (B) Manhattan plot for the area percentage of slow myofibers (SMA%). (C) Quantile–quantile plot for SM%. (D) Quantile–quantile plot for SMA%. The red solid line denotes the 5% Bonferroni genome-wide significance threshold (P = 1.16e-07), and the blue dotted line represents the suggestive significance threshold (P = 2.31e-06). Highlighted areas indicate the nearest genes within 100 kb upstream and downstream of significant loci.
Table 3.
Annotations of 5% Bonferroni genome-wide significant and suggestive-level SNPs associated with slow myofiber numbers (SM%).
| SNP1 | Position2 | Alt/Ref3 | P-value | Location | Candidate/Surrounding gene (100 kb)4 |
|---|---|---|---|---|---|
| rs736451963 | 1:114556280 | T/A | 3.80E-8 | intron | DMD |
| rs314441718 | 1:112778555 | C/T | 2.37E-7 | intron | MID1IP1 |
| rs314806781 | 1:112778583 | A/G | 1.28E-7 | intron | MID1IP1 |
| rs317158254 | 1:114561947 | C/T | 1.24E-6 | intron | DMD |
| rs15392642 | 1:114562726 | C/T | 8.43E-7 | intron | DMD |
| rs312575899 | 1:114562804 | T/C | 6.83E-7 | intron | DMD |
| rs15392647 | 1:114566874 | A/G | 2.28E-6 | intron | DMD |
| rs313949350 | 1:114584695 | T/C | 1.78E-6 | intron | DMD |
| rs738093891 | 1:114594712 | G/A | 2.39E-7 | intron | DMD |
| rs13668302 | 2:33529776 | A/G | 1.24E-6 | intron | CHN2 |
| 7:11802820 | A/G | 2.19E-6 | downstream | METTL21A | |
| 7:11802820 | A/G | 2.19E-6 | downstream | CREB1 | |
| rs739533409 | 9:6475388 | T/C | 2.06E-6 | intron | PXYLP1 |
| rs733424755 | 9:6475392 | C/T | 2.30E-6 | intron | PXYLP1 |
| rs738453647 | 9:6477588 | T/C | 9.97E-7 | intron | PXYLP1 |
| rs313304726 | 9:6477645 | A/G | 2.30E-6 | intron | PXYLP1 |
| rs737019788 | 9:6478235 | A/G | 8.62E-7 | intron | PXYLP1 |
| 9:6479187 | T/C | 7.75E-7 | intron | ENSGALG00010011362 |
SNP: rs ID of the locus
Position: Physical location of the SNP (chromosome: position).
Alt/Ref: Alternative allele and reference allele.
Candidate/Surrounding gene (100 kb): Genes annotated in the 100 kb region upstream and downstream of the SNP
A significant SNP associated with the SMA% was identified within the genomic regions of EFCAB11, TDP1, KCNK13, and FOXN3 on chromosome 5 (Fig. 3B and Table 4). Moreover, 12 SNPs of suggestive significance were located on chromosomes 2, 3, 5, 8, 11, 15, and 19, in the vicinity of genes such as GCNT2, TFAP2A, SERBP1, GADD45A, CCNE1, GSTT1, GSTT1L, TMEM120A.
Table 4.
Annotations of 5% Bonferroni genome-wide significant and suggestive-level SNPs associated with area percentage of slow myofibers (SMA%).
| SNP1 | Position2 | Alt/Ref3 | P-value | Location | Candidate/Surrounding gene (100 kb)4 |
|---|---|---|---|---|---|
| rs314708444 | 5:43140218 | G/A | 3.64E-8 | downstream | EFCAB11 |
| 2:63027523 | C/T | 1.85E-6 | intron | TFAP2A | |
| 3:12873236 | A/C | 1.10E-6 | intron | ENSGALG00010019286 | |
| rs317587148 | 5:43140058 | G/A | 1.29E-6 | downstream | EFCAB11 |
| rs15940870 | 8:28402606 | C/G | 1.05E-6 | regulatory region | ENSGALR00000432893 |
| 11:8108098 | C/T | 8.31E-7 | downstream | CCNE1 | |
| rs738833579 | 15:7987459 | A/G | 8.74E-7 | intron | MMP11 |
| 15:7988165 | T/C | 4.02E-7 | intron | MMP11 | |
| 15:7988199 | C/A | 4.20E-7 | intron | MMP11 | |
| 15:7988202 | G/A | 4.20E-7 | intron | MMP11 | |
| 15:8019698 | T/C | 1.48E-6 | upstream | DERL3 | |
| rs431888656 | 19:4735002 | C/T | 7.81E-7 | intron | YWHAG |
SNP: rs ID of the locus
Position: Physical location of the SNP (chromosome: position).
Alt/Ref: Alternative allele and reference allele.
Candidate/Surrounding gene (100 kb): Genes annotated in the 100 kb region upstream and downstream of the SNP
Genetic parameter estimation
Heritability, genetic correlation, and phenotypic correlation among the myofiber composition traits were calculated using the REML method with GCTA. The estimated heritability values for SM% and SMA were 0.09 and 0.21, respectively. A strong positive genetic correlation (0.42) and phenotypic correlation (0.95) were observed between SM% and SMA%.
Discussion
Myofiber composition plays a critical role in determining meat quality; for instance, a high content of slow-twitch myofibers enhances juiciness and flavor (Joo et al., 2013). Compared with the progress in mammalian research, the molecular mechanisms underlying myofiber characteristics in chickens remain largely unexplored. The complexity and time-consuming nature of obtaining myofiber phenotypes have limited studies on the relationship between quantitative trait loci (QTLs) or SNPs and these traits (Guo et al., 2020). To our knowledge, this is the first GWAS conducted on myofiber types in chickens.
While chicken breast muscles primarily consist of fast-twitch fibers, leg muscles contain a mix of fast- and slow-twitch fibers (Liu et al., 2020). The chicken sartorius muscle, a slender muscle in the leg, was selected to ensure consistent sampling locations for myofiber type determination (Dziedzic et al., 2014). We employed an immunofluorescence method utilizing antibodies specific to slow-twitch myosin heavy chains to enhance the efficiency and accuracy of myofiber type identification, yielding phenotypic data for sartorius muscles of 388 chickens (Huo et al., 2022).
In our study, the genomic heritability of SM% and SMA% was estimated at 0.09 and 0.21, respectively, which is similar to the results observed in pigs (Yan et al., 2024). These low-to-moderate heritability estimates indicate the potential for genetic improvements in myofiber composition. Our GWAS helped identify 32 SNPs significantly or potentially associated with myofiber traits in yellow-feather broilers.
Our analysis revealed a region on chromosome 1 closely associated with SM%, wherein the most significant SNP is located within the intron region of DMD. DMD, which encodes dystrophin, is crucial for maintaining muscle cell structure and function (Fortunato et al., 2021); mutations in this gene can lead to Duchenne muscular dystrophy, a condition characterized by muscle degeneration and the transition of myofibers from fast to slow type (Webster et al., 1988). This finding suggests that the SNP discovered in our study may regulate myofiber types by affecting DMD gene function. Another SNP associated with slow muscle fiber proportion was identified near KLF7. The Kruppel-like factor (KLF) gene family is a group of transcription factors containing highly conserved zinc-finger motifs, which play crucial roles in cell proliferation and differentiation (Pearson et al., 2008). Notably, KLF15 regulates the expression of slow myosin heavy chains in C2C12 myotubes through NFATc1 (Wang et al., 2014). KLF7 and KLF15 are highly expressed in chicken muscle tissues, with expression levels in fast-twitch muscle tissues being higher than those in slow-twitch muscles (Ling et al., 2023). Although no direct studies link KLF7 to muscle fiber types, the association identified in our study suggests it is a vital candidate gene.
Another noteworthy SNP related to muscle fiber proportion was found near CREB1, an essential transcription factor involved in muscle metabolism and myofiber-type regulation (Berdeaux and Hutchins, 2019). SIRT6 can reportedly activate CREB1, facilitating the transition to slow-twitch fibers (Song et al., 2022). CREB1 also regulates key genes like SOX6 and PPARGC1A involved in muscle fiber-type transition (Mäkelä et al., 2016). This evidence further confirms the role of CREB1 in affecting the distribution of muscle fiber types in chicken muscles.
We identified a significant SNP associated with the ratio of slow muscle fiber area at 43040218 bp on chromosome 5, located 1 kb downstream of EFCAB11. This gene encodes a protein belonging to the EF-hand calcium-binding protein family, which is crucial in various intracellular signaling pathways, including calcium-regulated myofiber type transformation (Chen et al., 2015). The calcium pathway plays a crucial role in regulating the transformation of myofiber types (Mu et al., 2007), suggesting that this gene is a new candidate gene for trait distribution of chicken myofiber types. Another SNP potentially related to myofiber types was also found near GADD45A on chromosome 8. This gene is a member of the GADD45 family, whose proteins are closely related to the cellular response to various environmental stresses. The expression level of GADD45A in muscles is associated with muscle weakness in humans, and its overexpression can lead to a reduction in glycolytic muscle fibers and an increase in oxidative fibers in mice (Marcotte et al., 2023). SNP mutations in this gene are related to meat quality traits in pigs (Cho et al., 2015), and the gene is associated with stress-induced declines in the meat quality of goats (Naldurtiker et al., 2022).
Furthermore, our study focused on the relationship between GSTT1, GSTT1L, and the proportion of myofiber area. GSTT1 and GSTT1L are members of the glutathione S-transferase (GSTs) family, a group of proteins primarily responsible for the detoxification of oxidative stress and chemical toxins in the body. The functions of GSTT1 and GSTT1L in meat quality regulation have been preliminarily confirmed; notably, reduced GSTT1 expression and hypermethylation in the promoter region of GSTT1L are among the reasons for changes in meat quality during the later growth stages of chickens (Zhang et al., 2017, 2023). Thus, these genes can be considered candidate genes that affect myofiber type composition and provide a reference for marker-assisted selection in yellow-feather chickens. However, the underlying mechanisms require further investigation.
Conclusions
Our genome-wide association analysis using whole-genome resequencing data from 400 yellow-feather broilers has provided significant insights into the molecular regulatory mechanisms of myofiber-type composition traits. We identified 32 SNPs significantly or potentially related to myofiber composition and discovered that DMD, KLF7, CREB1, EFCAB11, GADD45A, GSTT1, and GSTT1L may serve as candidate genes influencing myofiber types. These findings offer substantial theoretical support for elucidating the molecular regulatory basis of chicken meat quality traits.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by the STI2030—Major Projects (2023ZD04064), Natural Science Foundation of China (32472900), Natural Science Foundation of Jiangsu Province (BK20211121 and BK20230729), “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS [2021]107), and Chinese Agricultural Research System (CARS-41).
References
- Berdeaux R., Hutchins C. Anabolic and pro-metabolic functions of CREB-CRTC in skeletal muscle: advantages and obstacles for type 2 diabetes and cancer cachexia. Front. Endocrinol. (Lausanne) 2019;10:535. doi: 10.3389/fendo.2019.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou T. Skeletal muscle fiber type in hypoxia: adaptation to high-altitude exposure and under conditions of pathological hypoxia. Front. Physiol. 2018;9:1450. doi: 10.3389/fphys.2018.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta. 2023;2:e107. doi: 10.1002/imt2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo Y., Ge F., Gao H., Zhou J., Wu X., Qian C., Wang Z., Wang Z., Zhu B., Xu L., Gao X., Zhang L., Gao H., Li J. Developing a liquid capture chip to accelerate the genetic progress of cattle. Anim. Res. One Health. 2024;2:204–216. [Google Scholar]
- Chen W., Wang M., Zhang Z., Tang H., Zuo X., Meng X., Xiong M., Zhou F., Liang B., Dai F., Fang J., Gao J., Zhu J., Zhu Y., Wan H., Wang M., Chan S., Sun L. Replication the association of 2q32.2-q32.3 and 14q32.11 with hepatocellular carcinoma. Gene. 2015;561:63–67. doi: 10.1016/j.gene.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Cho E.S., Lee K.T., Choi J.W., Jeon H.J., Lee S.W., Cho Y.M., Kim T.H. Novel SNPs in the growth arrest and DNA damage-inducible protein 45 alpha gene (GADD45A) associated with meat quality traits in Berkshire pigs. Genet. Mol. Res. 2015;14:8581–8588. doi: 10.4238/2015.July.31.6. [DOI] [PubMed] [Google Scholar]
- Dziedzic D., Bogacka U., Ciszek B. Anatomy of sartorius muscle. Folia Morphol. (Warsz) 2014;73:359–362. doi: 10.5603/FM.2014.0037. [DOI] [PubMed] [Google Scholar]
- Fortunato F., Farnè M., Ferlini A. The DMD gene and therapeutic approaches to restore dystrophin. Neuromuscul. Disord. 2021;31:1013–1020. doi: 10.1016/j.nmd.2021.08.004. [DOI] [PubMed] [Google Scholar]
- Gao J., Xu W., Zeng T., Tian Y., Wu C., Liu S., Zhao Y., Zhou S., Lin X., Cao H., Lu L. Genome-wide association study of egg-laying traits and egg quality in LingKun chickens. Front. Vet. Sci. 2022;9 doi: 10.3389/fvets.2022.877739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Gao J., Yang B., Yan G., Xiao S., Zhang Z., Huang L. A whole genome sequence association study of muscle fiber traits in a White Duroc × Erhualian F2 resource population. Asian-Australas. J. Anim. Sci. 2020;33:704–711. doi: 10.5713/ajas.18.0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo W., Weng K., Li Y., Zhang Y., Zhang Y., Xu Q., Chen G. Comparison of muscle fiber characteristics and glycolytic potential between slow- and fast-growing broilers. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2021.101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail I., Joo S.T. Poultry meat quality in relation to muscle growth and muscle fiber characteristics. Korean J. Food Sci. Anim. Resour. 2017;37:873–883. doi: 10.5851/kosfa.2017.37.6.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.Y., Jeong T.C., Yang H.S., Kim G.D. Multivariate analysis of muscle fiber characteristics, intramuscular fat content, and fatty acid composition in porcine longissimus thoracis muscle. Livest. Sci. 2017;202:13–20. [Google Scholar]
- Joo S.T., Kim G.D., Hwang Y.H., Ryu Y.C. Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci. 2013;95:828–836. doi: 10.1016/j.meatsci.2013.04.044. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yuan L., Yang X., Ni Y., Xia D., Barth S., Grossmann R., Zhao R.Q. Effect of early feed restriction on myofibre types and expression of growth-related genes in the gastrocnemius muscle of crossbred broiler chickens. Br. J. Nutr. 2007;98:310–319. doi: 10.1017/S0007114507699383. [DOI] [PubMed] [Google Scholar]
- Ling X., Wang Q., Zhang J., Zhang G. Genome-wide analysis of the KLF gene family in chicken: characterization and expression profile. Animals (Basel) 2023;13:1429. doi: 10.3390/ani13091429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shan Y., Tu Y., Zhang M., Ji G., Ju X., Shi S., Fan C., Li Y., Shu J. Designing and evaluating a cost-effective single nucleotide polymorphism liquid array for Chinese native chickens. Anim. Res. One Health. 2023;1:168–179. [Google Scholar]
- Liu Y., Zhang M., Shan Y., Ji G., Ju X., Tu Y., Sheng Z., Xie J., Zou J., Shu J. miRNA-mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. Sci. Rep. 2020;10:10619. doi: 10.1038/s41598-020-67482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.F., Zhang M., Shan Y.J., Pang L.C., Ji G.G., Ju X.J., Tu Y.J., Shi S.Y., Bai H., Zou J.M., Shu J.T. Transcriptome sequencing analysis of the role of miR-499-5p and SOX6 in chicken skeletal myofiber specification. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Cai B., Jiang L., Abdalla B.A., Li Z., Nie Q., Zhang X. lncRNA-Six1 is a target of miR-1611 that functions as a ceRNA to regulate Six1 protein expression and fiber type switching in chicken myogenesis. Cells. 2018;7:243. doi: 10.3390/cells7120243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä J., Tselykh T.V., Kukkonen J.P., Eriksson O., Korhonen L.T., Lindholm D. Peroxisome proliferator-activated receptor-γ (PPARγ) agonist is neuroprotective and stimulates PGC-1α expression and CREB phosphorylation in human dopaminergic neurons. Neuropharmacology. 2016;102:266–275. doi: 10.1016/j.neuropharm.2015.11.020. [DOI] [PubMed] [Google Scholar]
- Marcotte G.R., Miller M.J., Kunz H.E., Ryan Z.C., Strub M.D., Vanderboom P.M., Heppelmann C.J., Chau S., Von Ruff Z.D., Kilroe S.P., McKeen A.T., Dierdorff J.M., Stern J.I., Nath K.A., Grueter C.E., Lira V.A., Judge A.R., Rasmussen B.B., Nair K.S., Lanza I.R., Ebert S.M., Adams C.M. GADD45A is a mediator of mitochondrial loss, atrophy, and weakness in skeletal muscle. JCI Insight. 2023;8:22. doi: 10.1172/jci.insight.171772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M., Zhang Z., Wang X., Shen W., Zhang L., Lin S. Molecular mechanisms underlying the impact of muscle fiber types on meat quality in livestock and poultry. Front. Vet. Sci. 2023;10 doi: 10.3389/fvets.2023.1284551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X., Brown L.D., Liu Y., Schneider M.F. Roles of the calcineurin and CaMK signaling pathways in fast-to-slow fiber type transformation of cultured adult mouse skeletal muscle fibers. Physiol. Genom. 2007;30:300–312. doi: 10.1152/physiolgenomics.00286.2006. [DOI] [PubMed] [Google Scholar]
- Munyaneza J.P., Kim E.T., Kim M. Genome-wide association studies of meat quality traits in chickens: a review. Korean J. Agric. Sci. 2022;49:407–420. [Google Scholar]
- Naldurtiker A., Batchu P., Kouakou B., Terrill T.H., Shaik A., Kannan G. RNA-seq exploration of the influence of stress on meat quality in Spanish goats. Sci. Rep. 2022;12:20573. doi: 10.1038/s41598-022-23269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Song S., Cheng H., Im C., Jung E.Y., Moon S.S., Choi J., Hur S.J., Joo S.T., Kim G.D. Comparison of meat quality and muscle fiber characteristics between porcine skeletal muscles with different architectures. Food Sci. Anim. Resour. 2022;42:874–888. doi: 10.5851/kosfa.2022.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R., Fleetwood J., Eaton S., Crossley M., Bao S. Krüppel-like transcription factors: a functional family. Int. J. Biochem. Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Psifidi A., Banos G., Matika O., Desta T.T., Bettridge J., Hume D.A., Dessie T., Christley R., Wigley P., Hanotte O., Kaiser P. Genome-wide association studies of immune, disease, and production traits in indigenous chicken ecotypes. Genet. Sel. Evol. 2016;48:74. doi: 10.1186/s12711-016-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E.A., Zempo H., Miyamoto-Mikami E., Kumagai H., Larin A.K., Sultanov R.I., Babalyan K.A., Zhelankin A.V., Tobina T., Shiose K., Kakigi R., Tsuzuki T., Ichinoseki-Sekine N., Kobayashi H., Naito H., Burniston J., Generozov E.V., Fuku N., Ahmetov I.I. Genome-wide association study identifies CDKN1A as a novel locus associated with muscle fiber composition. Cells. 2022;11:3910. doi: 10.3390/cells11233910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y.J., Ji G.G., Ming Z., Liu Y.F., Tu Y.J., Ju X.J., Shu J.T., Zou J.M. Use of transcriptome sequencing to explore the effect of CSRP3 on chicken myoblasts. J. Integr. Agric. 2023;22:1159–1171. [Google Scholar]
- Shu J.T., Xiao Q., Shan Y.J., Zhang M., Tu Y.J., Ji G.G., Sheng Z.W., Zou J.M. Oxidative and glycolytic skeletal muscles show marked differences in gene expression profile in Chinese Qingyuan partridge chickens. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soglia F., Petracci M., Davoli R., Zappaterra M. A critical review of the mechanisms involved in the occurrence of growth-related abnormalities affecting broiler chicken breast muscles. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.Y., Han C.Y., Moon Y.J., Lee J.H., Bae E.J., Park B.H. Sirt6 reprograms myofibers to oxidative type through CREB-dependent Sox6 suppression. Nat. Commun. 2022;13:1808. doi: 10.1038/s41467-022-29472-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., He Z., Fahey A.G., Zhao G., Liu R., Wen J. Research progress and applications of genome-wide association study in farm animals. Anim. Res. One Health. 2023;1:56–77. [Google Scholar]
- Wang J., Chen T., Feng F., Wei H., Pang W., Yang G., Shen Q.W. KLF15 regulates slow myosin heavy chain expression through NFATc1 in C2C12 myotubes. Biochem. Biophys. Res. Commun. 2014;446:1231–1236. doi: 10.1016/j.bbrc.2014.03.091. [DOI] [PubMed] [Google Scholar]
- Webster C., Silberstein L., Hays A.P., Blau H.M. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988;52:503–513. doi: 10.1016/0092-8674(88)90463-1. [DOI] [PubMed] [Google Scholar]
- Weng K., Huo W., Li Y., Zhang Y., Zhang Y., Chen G., Xu Q. Fiber characteristics and meat quality of different muscular tissues from slow- and fast-growing broilers. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2021.101537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Li L., Huang Y., Tang X., Shu Y., Cui D., Yu C., Hu Y., Ma J., Xiao S., Guo Y. Investigation on muscle fiber types and meat quality and estimation of their heritability and correlation coefficients with each other in four pig populations. Anim. Sci. J. 2024;95:e13915. doi: 10.1111/asj.13915. [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen X., Cao J., Chang C., Geng A., Wang H., Chu Q., Yan Z., Zhang Y., Liu H. Proteomic profiling of thigh meat at different ages of chicken for meat quality and development. Foods. 2023;12:2901. doi: 10.3390/foods12152901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yan F.B., Li F., Jiang K.R., Li D.H., Han R.L., Li Z.J., Jiang R.R., Liu X.J., Kang X.T., Sun G.R. Genome-wide DNA methylation profiles reveal novel candidate genes associated with meat quality at different age stages in hens. Sci. Rep. 2017;7:45564. doi: 10.1038/srep45564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Li Y., Wu J., Wang X., Bian C., Tian Y., Sun G., Han R., Liu X., Jiang R., Wang Y., Li G., Li W., Hu X., Kang X. Genome-wide association study reveals the genetic determinism of growth traits in a Gushi-Anka F2 chicken population. Heredity (Edinb.) 2021;126:293–307. doi: 10.1038/s41437-020-00365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]