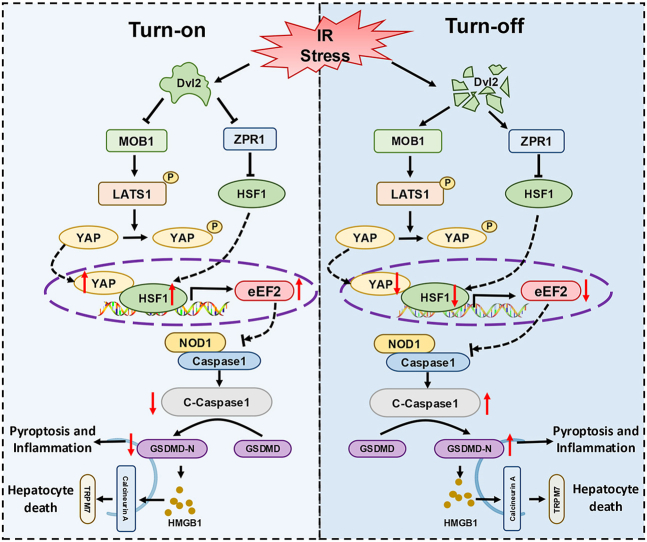
Abstract
Dishevelled 2 (Dvl2) is a key mediator of the Wingless/Wnt signaling pathway that regulates cell proliferation, migration, and immune function. However, little is known about the role of macrophage Dvl2 in modulating NOD1-mediated pyroptosis and hepatocyte death in oxidative stress-induced inflammatory liver injury. In a mouse model of oxidative stress-induced liver inflammation, mice with myeloid-specific Dvl2 knockout (Dvl2M−KO) displayed exacerbated ischemia/reperfusion (IR) stress-induced hepatocellular damage with increased serum ALT levels, oxidative stress, and proinflammatory mediators. Unlike in Dvl2FL/FL controls, Dvl2M−KO enhanced NOD1, caspase-1, GSDMD, and NF-κB activation in liver macrophages after IR. Interestingly, IR stress enhanced YAP colocalized with HSF1 in Dvl2FL/FL macrophages, while macrophage Dvl2 deficiency reduced YAP and HSF1 colocalization in the nucleus under inflammatory conditions. Importantly, Dvl2 deletion diminished nuclear YAP interacted with HSF1 and augmented NOD1/caspase-1 and GSDMD activation in response to inflammatory stimulation. However, Dvl2 activation increased YAP interaction with HSF1 and activated HSF1 target gene eEF2, inhibiting NOD1/caspase-1, GSDMD, and NF-κB activity. Moreover, macrophage eEF2 deletion increased the NOD1-caspase-1 interaction, GSDMD activation, HMGB1 release, and hepatocyte LDH release after macrophage/hepatocyte co-culture. Adoptive transfer of eEF2-expressing macrophages in Dvl2M−KO mice alleviated IR-triggered liver inflammation and hepatocellular damage. Therefore, macrophage Dvl2 deficiency promotes NOD1-mediated pyroptosis and exacerbates IR-induced hepatocellular death by disrupting the YAP-HSF1 axis. eEF2 is crucial for modulating NOD1-driven pyroptosis, inflammatory response, and hepatocyte death. Our findings underscore a novel role of macrophage Dvl2 in modulating liver inflammatory injury and imply the therapeutic potential in organ IRI and transplant recipients.
Keywords: YAP, HSF1, eEF2, GSDMD, TRPM7
Graphical abstract
Highlights
-
•
Macrophage Dvl2 deficiency induces NOD1 and GSDMD activation, exacerbating IR stress-induced liver damage.
-
•
Disruption of macrophage Dvl2 inhibits the YAP and HSF1 signaling pathways in IR-stressed liver.
-
•
YAP acts as a transcriptional coactivator of HSF1 in regulating NOD1-mediated inflammation and pyroptosis in IRI liver.
-
•
Disruption of Dvl2 inhibits the YAP-HSF1 interaction and enhances NOD1 function in macrophages.
-
•
The YAP-HSF1 axis targets eEF2 in response to oxidative stress.
1. Introduction
Hepatic ischemia and reperfusion injury (IRI) is a major cause of hepatic dysfunction and failure following liver transplantation, resection, and hemorrhagic shock [1]. Oxidative stress is considered a key factor in the pathogenesis of liver IR injury by triggering an inflammatory response and cell death signaling [2]. IR stress activates macrophages and releases reactive oxygen species (ROS), which promotes the innate immune response and inflammatory injury in the liver [1,[3], [4], [5]].
Nucleotide-binding oligomerization domain-containing protein 1 (NOD1) is one of the most prominent intracellular Nod-like receptors (NLRs), acting as a crucial sensor of the innate immune system and responsible for detecting different invading pathogens and stress signals arising from tissue injury [6,7]. NOD1 activation can initiate multiple signal transduction mechanisms involved in the stimulation of NF-κB (nuclear factor-κB), mitogen-activated protein kinases (MAPKs), interferon regulatory factors (IRFs), and programmed cell death. NOD1 stimulation recruits immune cells and produces proinflammatory cytokines and chemokines [8]. Activation of NOD1 facilitates macrophage migration and increases immune and inflammatory responses [9], whereas disruption of NOD1 ameliorates tissue inflammatory injury [10]. Oxidative stress induces ROS production and NOD1-dependent inflammation by activating NF-κB [6,11,12]. Moreover, ROS can trigger pyroptosis under oxidative stress conditions [13]. These results suggest that NOD1 signaling could be vital in mediating pyroptotic cell death in response to oxidative stress.
Dishevelled 2 (Dvl2) is a member of the Wingless/Wnt signaling pathway and plays a vital role in developmental biology, nuclear shuttling, and signal transduction [14,15]. Dvl2 functions as a key mediator of the Wnt signaling pathway and modulates cellular processes, including cell proliferation, survival, migration, and differentiation [16]. Previous studies have demonstrated that Dvl2 is associated with multiple Wnt-related signaling pathways such as the Wnt-GSK, the Wnt-calcium, the Wnt-aPKC (atypical Protein Kinase C), and the Wnt-mTOR signaling pathway in tissue and cells [17]. The Dvl2 protein has also been shown to shuttle between the cytoplasm and nucleus, and nuclear localization of Dvl2 is required to regulate gene expression and cell function [18]. Dvl2 reprograms immune cell function by modulating the transcription of immune modulatory genes, while deletion of Dvl2 inhibits mRNA transcription involved in cell proliferation and survival [19]. Moreover, Dvl2 induction depresses NF-κB activation via a tumor necrosis factor α (TNF-α)/tumor necrosis factor receptor 1 (TNFRI)-dependent manner [20]. Although these results indicate the essential role of the Dvl2 on immune regulation and cell function, little is known about the functional role and molecular mechanism of the Dvl2 in controlling NOD1-mediated pyroptosis, inflammatory response, and hepatocyte cell death in IR stress-induced liver injury.
Here, we identified a novel function and regulatory mechanism of macrophage Dvl2 in IR stress-induced liver inflammation and pyroptotic cell death signaling. We demonstrated that macrophage Dvl2 deficiency promotes NOD1-mediated pyroptosis and exacerbates IR-induced hepatocellular death via inhibiting the YAP-HSF1 interaction. Dvl2 activation induces the YAP-HSF1 signaling and activates the HSF1 target gene eEF2, crucial for modulating NOD1-driven pyroptosis, inflammation, and hepatocyte death in IR-stressed livers.
2. Materials and methods
Animals. The floxed Dvl2 (Dvl2FL/FL, B6; 129-Dvl2tm1.1Wds/J), YAP (YAPFL/FL, B6.129P2-YAP1tm1.1Dupa/J), and the mice expressing Cre recombinase under the control of the Lysozyme 2 (Lyz2) promoter (LysM-Cre, B6.129P2-LYZ2tm1(cre)Ifo/J) were obtained from The Jackson Laboratory (Bar Harbor, ME). The Dvl2flox targeting vector was designed to insert a frt-flanked neomycin resistance (neo) cassette followed by a loxP site upstream of exon 2 and a second loxP site of exon 15 distal to the stop codon. Flp-mediated recombination removed the neo cassette. The construct was electroporated into 129P2/OlaHsd-derived E14 embryonic stem (ES) cells. Correctly targeted ES cells were injected into C57BL/6 blastocysts. The resulting chimeric animals were tested for germline transmission. The mice were crossed to mice expressing FLP recombinase to excise the FRT site flanked NEO cassette. Mice that retained the loxP site flanked exons 2–14 were then bred to remove the FLP recombinase allele. Offspring were bred to C57BL/6J mice for at least three generations. To generate myeloid-specific Dvl2 knockout (Dvl2M−KO) mice, a homozygous loxP-flanked Dvl2 mouse was mated with a homozygous Lyz2-Cre mouse to create the F1 mice that were heterozygous for a loxP-flanked Dvl2 allele and heterozygous for the Lyz2-Cre. The F1 mice were then backcrossed to the homozygous loxP-flanked Dvl2 mice, resulting in the generation of Dvl2M−KO (25 % of the offspring), which were homozygous for the loxP-flanked Dvl2 allele and heterozygous for the Lyz2-Cre allele (Fig. S1). The myeloid-specific YAP knockout (YAPM−KO) mice were generated, as described [21]. Mouse genotyping was performed using a standard protocol with primers described in the JAX Genotyping protocols database. The C57BL/6 wild-type (WT) mice were obtained from The Jackson Laboratory. Male mice at 6–8 weeks of age were used in all experiments. This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. Animal protocols were approved by the Institutional Animal Care and Use Committee of The University of California at Los Angeles.
Mouse liver IRI model. We used an established mouse model of warm hepatic ischemia (90min) followed by reperfusion (6h) [21]. Mice were injected with heparin (100U/kg), and an atraumatic clip was used to interrupt the arterial/portal venous blood supply to the cephalad liver lobes. After 90min of ischemia, the clip was removed, and mice were sacrificed at 6h of reperfusion. Some animals were injected via tail vein with eEF2-expressing bone marrow-derived macrophages (BMMs) or control cells (1 × 106 cells in 0.1 ml of PBS/mouse) 24h before ischemia.
Hepatocellular function assay. Serum alanine aminotransferase (sALT) levels, an indicator of hepatocellular injury, were measured by ALT and AST kit (ThermoFisher, Waltham, MA) according to the manufacturer’s instructions.
Histology, immunohistochemistry, and immunofluorescence staining. Liver sections (5-μm) were stained with hematoxylin and eosin (H&E). The severity of IRI was graded using Suzuki’s criteria [22]. Immunofluorescence staining of Dvl2 and GSDMD in Kupffer cells or TRMP7 in hepatocytes were analyzed in the liver sections using primary mouse Dvl2 (Santa Cruz Biotechnology, Santa Cruz, CA) and GSDMD-N Abs (Affinity Bioscience, Pottstown, PA), and rabbit CD68 mAb (ThermoFisher Scientific) or mouse TRPM7 mAb (Santa Cruz Biotechnology) and rabbit HNF4α mAb (Abcam, Fremont, CA). The eEF2 and 4-HNE expression was detected in liver sections by immunohistochemistry staining using primary mouse eEF2 (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit 4-HNE (ThermoFisher Scientific) Abs. The primary rabbit YAP, eEF2 (Cell Signaling Technology, Danvers, MA), rabbit NOD1 (ThermoFisher Scientific), mouse HSF1 and caspase-1 (Santa Cruz Biotechnology) Abs, the secondary AlexFluor488-conjugated AffiniPure donkey anti-rabbit IgG Ab, Cy5-conjugated AffiniPure donkey anti-mouse IgG Ab (Jackson Immunoresearch) were used for staining YAP, HSF1, eEF2, NOD1, and caspase-1 positive cells according to the manufacturer’s instructions. Normal mouse IgG (Santa Cruz Biotechnology) was used as an isotype control. Images for immunofluorescence staining were captured using a fluorescence microscope (Keyence BZ-X810, Osaka, Japan) and analyzed using Image-pro Plus software. Positive cells were counted blindly in 10 HPF/section (x200).
Quantitative RT-PCR analysis. Total RNA was purified from liver tissue or cell cultures using RNeasy Mini Kit (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions. Reverse cDNA transcription was performed using the SuperScript III First-Strand Synthesis System (ThermoFisher Scientific). Quantitative real-time PCR was carried out using the QuantStudio 3 (Applied Biosystems by ThermoFisher Scientific). In a final reaction volume of 25 μl, the following were added: 1 × SuperMix (Platinum SYBR Green qPCR Kit; Invitrogen) cDNA and 10 μM of each primer. Amplification conditions were: 50 °C (2min), 95 °C (5min), followed by 40 cycles of 95 °C (15sec) and 60 °C (30sec). The primer sequences that amplify TNF-α, IL-1β, IL-18, CXCL10, MCP-1, Mob1a, Zpr1, eEF2, Nqo1, Gclc, Gclm, and HPRT were shown in Supplementary Table 1. The target gene expressions were calculated by their ratios to the housekeeping gene HPRT.
Western blot analysis. Protein was extracted from liver tissue or cell cultures with ice-cold protein lysis buffer (50 mM Tris, 150 mM NaCl, 0.1 % sodium dodecyl sulfate, 1 % sodium deoxycholate, 1 % Triton-100). The buffer contains 1 % proteinase and phosphatase inhibitor cocktails (Sigma-Aldrich, St. Louis, MO). Proteins (30 μg/sample) in SDS-loading buffer (50 mM Tris, pH 7.6, 10 % glycerol, 1 % SDS) were subjected to 4–20 % SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membrane (Bio-Rad). The membrane was blocked with 5 % dry milk and 0.1 % Tween 20 (USB, Cleveland, OH). The nuclear and cytosolic fractions were prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher Scientific). Monoclonal rabbit anti-mouse Dvl2, GSDMD, cleaved caspase-1, p-P65, P65, MOB1, p-LATS1. LATS1, p-YAP, YAP, ZPR1, HSF1, NOD1, HMGB1, NLRP3, Tubulin, β-actin, Lamin B2 (Cell Signaling Technology, Danvers, MA), Calcineurin A (ThermoFisher Scientific), TRPM7 (Santa Cruz Biotechnology, Santa Cruz, CA), and GSDMD N-Terminal (Affinity Biosciences) were used. The membranes were incubated with Abs, and then a Western ECL substrate mixture (Bio-Rad) was added for imaging with the iBright FL1000 (ThermoFisher Scientific). Relative quantities of protein were determined by comparing the β-actin expression using iBright image analysis software (ThermoFisher Scientific).
Isolation of primary hepatocytes and liver macrophages. Primary hepatocytes, Kupffer cells, and BMMs from the Dvl2FL/FL, Dvl2M−KO, YAPFL/FL, YAPM−KO or wild-type (WT) mice were isolated as described [3]. In brief, livers were perfused in situ with warmed (37 °C) HBSS solution, followed by a collagenase buffer (collagenase type IV, Sigma-Aldrich). The Perfused livers were dissected and teased through 70-μm nylon mesh cell strainers (BD Biosciences, San Jose, CA). The nonparenchymal cells (NPCs) were separated from hepatocytes by centrifuging at 50×g 2min three times. The NPCs were then suspended in HBSS and layered onto a 50 %/25 % two-step Percoll gradient (Sigma) in a 50-ml conical centrifuge tube and centrifuged at 1800×g at 4 °C for 15min. The liver macrophages (Kupffer cells) in the middle layer were collected and plated to cell culture dishes in DMEM with 10 % FBS, 10 mM HEPES, 2 mM GlutaMax, 100 U/ml penicillin, and 100 μg/ml streptomycin for 15min at 37 °C.
BMM isolation and in vitro transfection. Murine bone-derived macrophages (BMMs) were generated as described [3]. In brief, bone marrow cells were removed from the femurs and tibias of the Dvl2FL/FL, Dvl2M−KO, YAPFL/FL, YAPM−KO, or WT mice and cultured in DMEM supplemented with 10 % FCS and 15 % L929-conditioned medium for seven days. BMMs (1 × 106/well) were transfected with CRISPR/Cas9-Dvl2 activation, CRISPR/Cas9-Dvl2 KO, CRISPR/Cas9-HSF1 KO, CRISPR/Cas9-NOD1 KO, CRISPR/Cas9-eEF2 activation, CRISPR/Cas9-eEF2 KO, or control vector (Santa Cruz Biotechnology) by using Lipofectamine LTX with Plus Reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. After 24–48h, cells were supplemented with LPS (100 ng/ml) for an additional 6h. For lentivirus-mediated gene transfer, BMMs (1 × 106/well) were added with lentivirus-mediated eEF2 (Lv-eEF2) (Santa Cruz Biotechnology) or Lv-GFP control (at a multiplicity of infection 10) (Applied Biological Materials, Richmond, BC, Canada) and incubated at 37 °C overnight. The medium was removed and replaced with fresh medium. After 48h, cells were harvested for in vivo adoptive transfer.
Co-culture of macrophages and primary hepatocytes. Primary hepatocytes were cultured in 6-well plates at 4 × 105 cells per well. After 24h, the 0.4 μm-pore size transwell inserts (Corning) containing 1 × 106 BMMs were placed into the 6-well plate with the initially seeded hepatocytes. The co-cultures were incubated for 24h in the lower chamber.
Reactive oxygen species assay. ROS production in macrophages was measured using the 5-(and-6)-carboxy-2',7'-difluorodihydrofluorescein diacetate (Carboxy-H2DFFDA, ThermoFisher Scientific), as described [3]. In brief, macrophages (2 × 105) were isolated from ischemic livers and cultured on collagen-coated cover slips without or with LPS (100 ng/ml) for 2h at 37 °C. After washing with PBS, cells were incubated with 10 μM of Carboxy-H2DFFDA. The Carboxy-H2DFFDA was converted to a green-fluorescent form when hydrolyzed by intracellular esterase and oxidized in the cells. Cells were fixed with 2 % paraformaldehyde and stained with Hoechst dye. ROS produced by macrophages were analyzed and quantified by fluorescence microscopy. Positive green fluorescent-labeled cells were counted blindly in 10 HPF/section (x200).
RNA-sequencing and bioinformatic analyses. Total RNA was extracted from liver macrophages of IR-challenged Dvl2FL/FL or Dvl2M−KO mice using mirVana™ miRNA ISOlation Kit (Ambion-1561, ThermoFisher Scientific) according to the manufacturer’s protocol. Quality was monitored by NanoDrop ND-2000 (ThermoFisher Scientific) and Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). The samples with RNA Integrity Number (RIN) (7 ≤ RIN values ≥ 8) were subjected to the subsequent analysis. A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Ribosomal RNA was removed by TruSeq Stranded Total RNA with Ribo-Zero Golbin kit (RS-122-2301, Illumina, San Diego, CA), and rRNA free residue was cleaned up by RNAClean XP (A63987, Beckman Coulter, Indianapolis, IN). Sequencing libraries were generated using the rRNA-depleted RNA by NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA) according to the manufacturer’s instructions. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) following manufacturer’s recommendations. After cluster generation, the libraries were sequenced on an Illumina HiSeq 4000 platform, generating 150 bp paired-end reads. Raw fastq files were firstly trimmed to remove adaptors using Trim Galore (v0.5.0) with the following parameters: q 25 -phred33 -length 35 -e 0.1 -stringency 4. Trimmed fastq files were then mapped to the grch38 or grcm39_tran genome utilizing hisat2 (v2.2.0) [23]. Gene expressions were quantified using Cuffdiff (v2.1.1). Differentially expressed genes (DEGs) were analyzed by edgeR R package (v3.18.1) using raw counts. An adjusted P value of less than 0.05 was set as the threshold to define DEGs. KEGG and GO analysis were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; version 6.8) and R package ‘clusterProfiler’ (v3.11.0).
Immunoprecipitation analysis. BMMs treated with CRISPR/Cas9-eEF2 activation, CRISPR/Cas9-eEF2 KO, CRISPR/Cas9-NOD1 KO, or control vector were lysed in NP-40 lysis buffer (50 mM Tris pH7.4, 10 mM EDTA, 150 mM NaCl, 1 % NP-40, ThermoFisher Scientific) containing protease inhibitors. The lysates were incubated with YAP (Cell Signaling Technology) and HSF1 (Santa Cruz Biotechnology), NOD1 (Santa Cruz Biotechnology), and caspase-1 (ThermoFisher Scientific) antibodies or control IgG and protein A/G beads at 4 °C overnight, respectively. After immunoprecipitation, the immunocomplexes were washed with lysis buffer three times and analyzed by standard immunoblot procedures.
Chromatin immunoprecipitation (ChIP). The ChIP analysis was carried out using the ChIP Assay Kit (Abcam). Briefly, BMMs were treated with 1 % formaldehyde for 10 min to cross-link proteins and chromatin. The reaction was stopped by adding 0.125 M glycine for 5 min. Cells were washed with ice-cold PBS and resuspended with ChIP lysis buffer for 10 min. Cell lysates were centrifuged to pellet the nuclei. The cell nuclei were resuspended in nuclei lysis buffer and then subjected to sonication for 15 min. Purified chromatin was analyzed on a 1.5 % agarose gel to analyze DNA fragment size. The sheared chromatin was immunoprecipitated with YAP (Cell Signaling Technology) or HSF1 antibody (Santa Cruz Biotechnology) overnight. As a control, the normal IgG was used as a replacement for YAP or HSF1 antibody. The antibody/chromatin samples were mixed with protein A sepharose beads. Protein-DNA complexes were washed and eluted, followed by a cross-link reversal step, and the resulting DNA was purified. For sequential ChIP, sheared chromatin was first immunoprecipitated with YAP antibody, followed by elution with a second immunoprecipitation using an HSF1 antibody. DNA from each immunoprecipitation reaction was examined by PCR. The primer for the HSF1-responsive region of eEF2 promoter: forward: 5’- CCACCATGATGACGGAAA -3’, reverse: 5’- GTAGATAAGGTGACAAGCATAG -3’.
ChIP-sequencing (ChIP-seq). The ChIP-DNA was amplified to generate a library for sequencing. The workflow consists of fragmentation of whole-genome DNA, end repair to generate blunt ends, A-tailing, adaptor ligation, and PCR amplification. Different adaptors were used for multiplexing samples in one lane. Sequencing was performed on Illumina HiSeq 4000 (Illumina, San Diego, CA) for a single read 50 run at the Technology Center for Genomics & Bioinformatics (TCGB) at UCLA. Data quality check was done on Illumina SAV. Demultiplexing was performed with the Illumina Bcl2fastq2 v 2.17 program. Reads were mapped to mouse mm10 genome using the Bowtie1, and MACS2 was used for the peak calling. ChIPseeker was used for the peak annotation. Genome browser representation files were generated by converting ChIP-seq data to bigWig format. This was done using genomeCoverageBed from bedtools v 2.17.0 to generate a bed file, then UCSC bedGraphToBigWig to convert the bed to bigWig format.
ELISA assay. Cell culture supernatants were harvested for cytokine analysis. ELISA kits were used to measure the IL-1β and HMGB1 (ThermoFisher Scientific) levels according to the manufacturer’s instructions.
LDH activity assay. BMMs (1 × 106) treated with CRISPR/Cas9-eEF2 KO or control vector followed by LPS stimulation were co-cultured with primary hepatocytes (4 × 105/well) for 24 h in the lower chamber. The activity of lactate dehydrogenase (LDH) in the cell culture medium from the lower chamber was measured with a commercial LDH activity assay kit (Stanbio Laboratory, Boerne, TX) according to the manufacturer’s instructions. The Dvl2FL/FL, Dvl2M−KO, YapFL/FL and YaoM−KO mice were subjected to 90min of partial liver warm ischemia, followed by 6h of reperfusion. The serum was collected and diluted. The activity of LDH in the serum was measured with a commercial LDH activity assay kit (Abcam, Fremont, CA) according to the manufacturer’s instructions.
Statistical analysis. Data are expressed as mean ± SD and analyzed using the Permutation t-test and Pearson correlation. Per comparison, two-sided p-values less than 0.05 were considered statistically significant. Multiple group comparisons were made using one-way ANOVA followed by Bonferroni’s post hoc test. When groups showed unequal variances, we applied Welch’s ANOVA to make various group comparisons. All analyses were used by SAS/STAT software, version 9.4.
3. Results
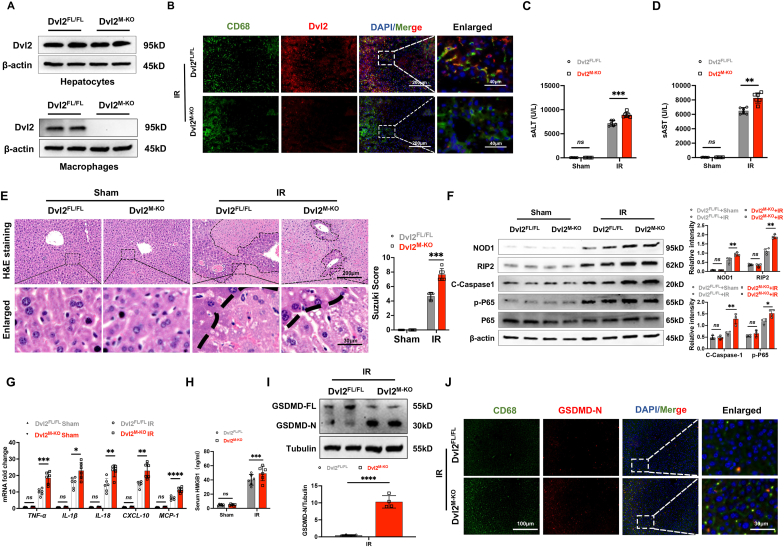
Disruption of myeloid-specific Dvl2 induces NOD1 and GSDMD activation and exacerbates IR stress-induced liver damage. The myeloid-specific Dvl2-deficient (Dvl2M−KO) and Dvl2-proficient (Dvl2FL/FL) mice were subjected to 90min of warm ischemia followed by 6h of reperfusion. The hepatocytes and liver macrophages (Kupffer cells) were isolated from these ischemic livers. Unlike Dvl2FL/FL livers, Dvl2M−KO lacked Dvl2 expression in liver macrophages but not in hepatocytes (Fig. 1A). This was confirmed by immunofluorescence staining, which showed that macrophage did not express Dvl2 in ischemic livers from the Dvl2M−KO mice, compared to the Dvl2FL/FL groups (Fig. 1B). The serum ALT (sALT) and AST (sAST) levels (IU/L) were significantly increased in the Dvl2M−KO compared to Dvl2FL/FL mice at 6h post liver reperfusion (Fig. 1C and D). Using the Suzuki’s histological grading of IR-induced liver damage, the Dvl2M−KO livers displayed severe edema, sinusoidal congestion, and extensive hepatocellular necrosis compared to the Dvl2FL/FL livers, which showed mild to moderate edema, sinusoidal congestion and mild necrosis (Fig. 1E). Dvl2M−KO augmented NOD1, cleaved caspase-1, and NF-κBp65 expression (Fig. 1F), with increased mRNA levels of TNF-α, IL-1β, IL-18, CXCL-10, MCP-1 (Fig. 1G), and HMGB1 release (Fig. 1H) compared to the Dvl2FL/FL controls. Notably, disruption of myeloid-specific Dvl2 induced gasdermin D (GSDMD) activation, evidenced by increased N-terminus of GSDMD (GSDMD-N) expression in ischemic livers from the Dvl2M−KO mice (Fig. 1I). This result was further confirmed by immunofluorescence staining, which showed increased macrophage GSDMD-N expression in Dvl2M−KO livers compared to the Dvl2FL/FL groups (Fig. 1J). Collectively, these data suggest that macrophage Dvl2 deficiency activates NOD1 and GSDMD, which may play a critical role in IR-triggered liver inflammation and pyroptotic cell death.
Fig. 1.
Disruption of myeloid-specific Dvl2 induces NOD1 and GSDMD activation and exacerbates IR stress-induced liver damage. The Dvl2FL/FL and Dvl2M−KO mice were subjected to 90min of partial liver warm ischemia, followed by 6h of reperfusion. (A) The Dvl2 expression was detected in hepatocytes and liver macrophages by Western blot assay. (B) Immunofluorescence staining of macrophage Dvl2 expression in the ischemic livers. Scale bars, 200 μm and 40 μm. (C and D) Liver function was evaluated by serum ALT and AST levels (IU/L) (n = 6 samples/group). (E) Representative histological staining (H&E) of ischemic liver tissue (n = 6 mice/group) and Suzuki’s histological score. Scale bars, 200 μm and 30 μm. (F) The protein expression of NOD1, cleaved caspase-1, p-P65, and P65 was detected by Western blot assay in ischemic livers from the Dvl2FL/FL and Dvl2M−KO mice. The graph shows the quantitation of relative intensity. (G) Quantitative RT-PCR-assisted detection of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 in ischemic livers (n = 6 samples/group). (H) ELISA analysis of HMGB1 levels in the serum samples (n = 6 samples/group). (I) The full-length GSDMD (GSDMD-FL) and N-terminus of GSDMD (GSDMD-N) expression were detected in ischemic livers from the Dvl2FL/FL and Dvl2M−KO mice. (J) Immunofluorescence staining of macrophage GSDMD-N expression in ischemic livers from the Dvl2FL/FL and Dvl2M−KO mice. Scale bars, 200 μm and 40 μm. All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
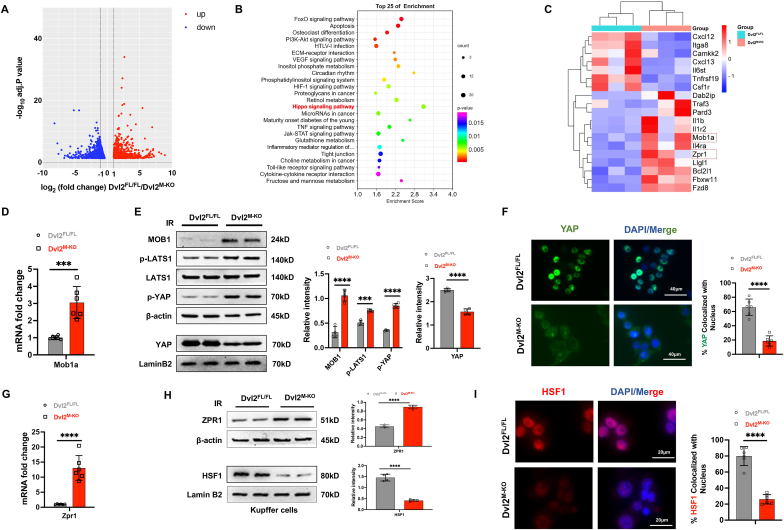
Disruption of macrophage Dvl2 downregulates the YAP and HSF1 signaling pathways in IR-stressed liver. To determine how Dvl2 may regulate NOD1-driven liver inflammation and cell death signaling in liver IRI, the deep RNA-sequencing (RNA-seq) analysis was performed in liver macrophages isolated from the Dvl2FL/FL and Dvl2M−KO mice after liver IRI. We identified 856 genes (red) that were upregulated and 631 genes (green) that were downregulated in Dvl2M−KO macrophages compared to the Dvl2FL/FL groups (Fig. 2A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the downregulated transcripts in Dvl2M−KO macrophages showed enrichment of pathways participating in Hippo-YAP pathway, HSF1 signaling, inflammatory cytokine interaction, and cell death process (Fig. 2B and C). However, the upregulated genes were enriched in pathways involved in the oxidation-reduction process, mitochondrial function, and cell metabolism process (Fig. S2). Notably, Dvl2M−KO increased NOD1 expression (Fig. S5) and the mRNA and protein expression of Mob1 (Fig. 2D, E), a key regulator of the Hippo/YAP pathway [24], leading to augmented LATS1 and YAP phosphorylation, and reduced nuclear YAP expression (Fig. 2E). Moreover, Dvl2M−KO significantly reduced total YAP expression in the cytosolic fraction compared to the Dvl2FL/FL controls (Fig. S9). This result was further confirmed by immunofluorescence staining, which showed decreased localization of YAP in the nucleus in the Dvl2M−KO but not the Dvl2FL/FL groups (Fig. 2F). Interestingly, Dvl2M−KO upregulated the expression of zinc-finger protein 1 (Zpr1) (Fig. 2G), an essential component of HSF1 proteostasis [25]. Disruption of Zpr1 induced rapid and persistent HSF1 activation under cell stress conditions [25]. Consistently, Dvl2M−KO augmented Zpr1 but inhibited HSF1 protein expression (Fig. 2H), with reduced nuclear HSF1 localization (Fig. 2I). To test the crosstalk between Dvl2 and ZPR1 in macrophages after liver IRI, we performed a co-immunoprecipitation analysis. Interestingly, Dvl2 can bind to the ZPR1 in the cytoplasm of macrophages after liver IRI (Fig. S6), suggesting that Dvl2 could directly interact with ZPR1 and regulate ZPR1 and HSF1 activation under cell stress conditions. Moreover, Dvl2M−KO significantly increased serum LDH levels compared to the Dvl2FL/FL control mice after liver IRI (Fig. S8). Collectively, these results suggest that the YAP-HSF1 axis may be crucial for macrophage Dvl2-mediated immune regulation and cell death cascade in liver IRI.
Fig. 2.
Disruption of macrophage Dvl2 downregulates the YAP and HSF1 signaling pathways in IR-stressed liver. Liver macrophages (1 × 106/well) were isolated from the Dvl2FL/FL and Dvl2M−KO mice after liver IRI. Total RNA was extracted and subjected to a deep RNA-sequencing (RNA-seq) analysis. A modified Fisher’s exact test (enrichment score) was used for the functional enrichment analyses. Significantly upregulated or downregulated genes were determined by an adjusted p-value of less than 0.05 was set as the threshold to define DEGs, KEGG, and GO analysis. (A) The log2 fold changes of gene expression in IR-stressed Dvl2M−KO macrophages compared to the Dvl2FL/FL cells. Differentially expressed genes (DEGs) (n = 1487, p < 0.05) in Dvl2M−KO macrophages from the IR-stressed livers are indicated (red, upregulated, n = 856, and green, downregulated, n = 631). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of transcripts differentially expressed in Dvl2M−KO macrophages from the IR-stressed livers. (C) Heat map showing the changed genes in Dvl2M−KO macrophages from the IR-stressed livers. (D) Analysis of Mob1a mRNA levels in Dvl2M−KO or Dvl2FL/FL macrophages from the IR-stressed livers (n = 6 samples/group). (E) Western blot analysis of MOB1, p-LATS1, LATS1, p-YAP, and nuclear YAP protein expression in the indicated groups. The graph shows the quantitation of relative intensity. (F) Immunofluorescence staining of nuclear YAP in Dvl2M−KO or Dvl2FL/FL macrophages from the IR-stressed livers. Scale bars, 40 μm. (G) Analysis of Zpr1 mRNA levels in Dvl2M−KO or Dvl2FL/FL macrophages from the IR-stressed livers (n = 6 samples/group). (H) Western blot analysis of Zpr1 and HSF1 protein expression in the indicated groups. The graph shows the quantitation of relative intensity. (I) Immunofluorescence staining of nuclear HSF1 in Dvl2M−KO or Dvl2FL/FL macrophages in the indicated groups. All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
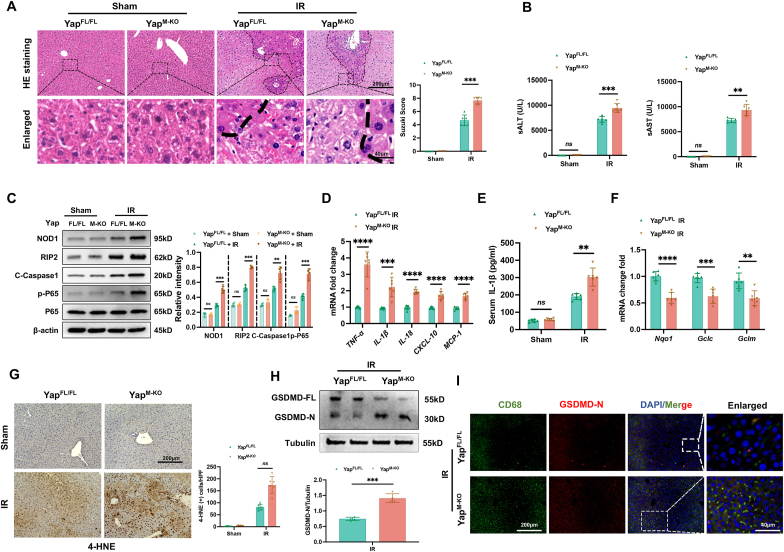
YAP is required to regulate NOD1-mediated liver inflammation and pyroptotic cell death in IR-stressed liver. As myeloid-specific Dvl2 deficiency activated the Hippo/YAP pathway, we then examined whether YAP influenced NOD1 function in IR-stressed livers. Using myeloid-specific YAP knockout mice, we found that YAPM−KO aggravated IR-induced liver damage, evidenced by increased Suzuki’s histological score (Fig. 3A) and sALT and sAST levels compared to the YAPFL/FL controls (Fig. 3B). Unlike YAPFL/FL controls, YAPM−KO augmented NOD1, cleaved caspase-1, and NF-κBp65 expression in ischemic livers (Fig. 3C). The expression of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 (Fig. 3D) with serum levels of IL-1β (Fig. 3E) were significantly increased in the YAPM−KO but not the YAPFL/FL mice. Moreover, increased serum LDH levels was observed from the YAPM−KO but not the YAPFL/FL controls (Fig. S10). YAPM−KO reduced antioxidant Nqo1, Gclc, and Gclm expression (Fig. 3F) with increased the positive cells of 4-hydroxynonenal (4-HNE) (Fig. 3G), a marker of ROS production [26] compared to the YAPFL/FL controls. Strikingly, YAPM−KO induced GSDMD activation as evidenced by increased GSDMD-N expression in ischemic livers (Fig. 3H) and liver macrophages (Fig. 3I). These results suggest the critical role of YAP in modulating NOD1-driven inflammatory response and pyroptotic cell death in IR-stressed livers.
Fig. 3.
YAP is required to regulate NOD1-mediated liver inflammation and pyroptotic cell death in IR-stressed liver. The YAPFL/FL and YAPM−KO mice were subjected to 90min of partial liver warm ischemia, followed by 6h of reperfusion. (A) Representative histological staining (H&E) of ischemic liver tissue (n = 6 mice/group) and Suzuki’s histological score. Scale bars, 200 μm, and 40 μm. (B) Liver function was evaluated by serum ALT and AST levels (IU/L) (n = 6 samples/group). (C) Western-assisted analysis and relative density ratio of NOD1, cleaved caspase-1, p-P65, and P-65 in the YAPFL/FL and YAPM−KO livers after IR stress. (D) Quantitative RT-PCR analysis of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 mRNA levels in ischemic livers (n = 6 samples/group). (E) ELISA analysis of IL-1β levels in the serum samples (n = 6 samples/group). (F) Quantitative RT-PCR analysis of Nqo1, Gclc, and Gclm mRNA levels in ischemic livers (n = 6 samples/group). (G) Immunohistochemistry staining of 4-HNE in ischemic livers (n = 6 mice/group). Quantification of 4-HNE + cells, Scale bars, 200 μm. (H) The expression of the full-length GSDMD (GSDMD-FL) and N-terminus of GSDMD (GSDMD-N) was detected in ischemic livers from the YAPFL/FL and YAPM−KO mice. The graph shows the quantitation of relative intensity. (I) Immunofluorescence staining of macrophage GSDMD-N expression in the indicated groups. Scale bars, 200 μm and 40 μm. All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
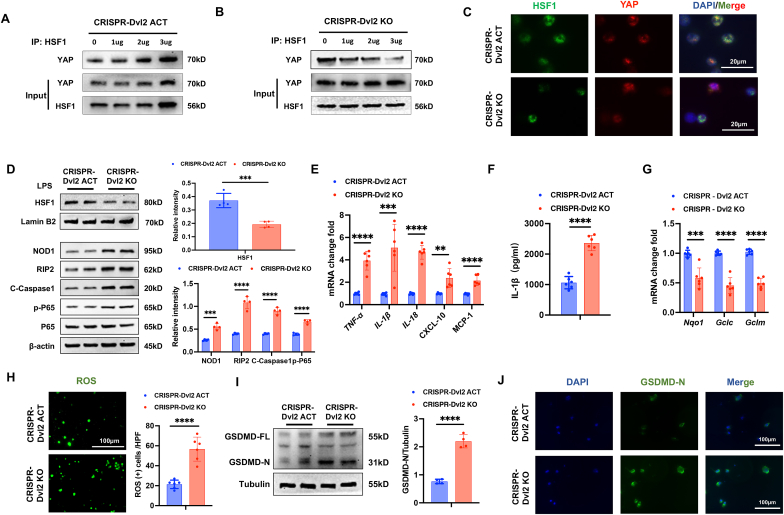
Disruption of Dvl2 inhibits the YAP interaction with HSF1 and enhances NOD1 function in macrophages. As macrophage Dvl2 deficiency inhibited YAP and HSF1 activation in ischemic livers, we next analyzed the crosstalk between YAP pathway and HSF1 signaling in the Dvl2-mediated regulation of NOD1 function in macrophages. Bone marrow-derived macrophages (BMMs) were transfected with CRISPR/Cas9-mediated Dvl2 activation (CRISPR-Dvl2 ACT) or Dvl2 knockout (CRISPR-Dvl2 KO) vector followed by LPS stimulation. Co-immunoprecipitation assays revealed that overexpression of Dvl2 increased YAP bound to endogenous HSF1 in the nucleus of macrophages after LPS stimulation (Fig. 4A). However, deletion of Dvl2 diminished the YAP binding with HSF1 (Fig. 4B). Immunofluorescence staining revealed that increased nuclear YAP and HSF1 colocalization in the CRISPR-Dvl2 ACT but not the CRISPR-Dvl2 KO macrophages (Fig. 4C). Consistently, CRISPR-Dvl2 ACT augmented HSF1 and diminished NOD1, cleaved caspase-1, and NF-κBp65 protein expression while CRISPR-Dvl2 KO reduced HSF1 and increased NOD1, cleaved caspase-1, and NF-κBp65 protein levels in LPS-stimulated macrophages (Fig. 4D). The expression of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 (Fig. 4E) with serum levels of IL-1β (Fig. 4F) were significantly decreased in the CRISPR-Dvl2 ACT but not the CRISPR-Dvl2 KO-treated macrophages. Moreover, CRISPR-Dvl2 ACT-treated macrophages displayed increased antioxidant Nqo1, Gclc, and Gclm levels and decreased ROS production, whereas CRISPR-Dvl2 KO increased ROS production in LPS-stimulation macrophages (Fig. 4G and H). Strikingly, unlike in CRISPR-Dvl2 ACT-treated cells, CRISPR-Dvl2 KO induced GSDMD activation, which showed increased GSDMD-N expression in LPS-stimulated macrophages (Fig. 4I). This result was further confirmed by immunofluorescence staining, which showed increased GSDMD-N expression in Dvl2-deficient macrophages compared to the Dvl2-proficient cells (Fig. 4J). Altogether, these results suggest that induction of Dvl2 activates the YAP-HSF1 signaling and inhibits NOD1 function, whereas deletion of Dvl2 inhibits the YAP-HSF1 activation, enhancing the NOD1-mediated inflammatory response and pyroptotic cell death in macrophages.
Fig. 4.
Disruption of Dvl2 inhibits the YAP interaction with HSF1 and enhances NOD1 function in macrophages. Bone marrow-derived macrophages (BMMs, 1 × 106) were transfected with CRISPR/Cas9-mediated Dvl2 activation (CRISPR-Dvl2 ACT) or knockout vector (CRISPR-Dvl2 KO) (1 μg, 2 μg, and 3 μg, respectively), followed by LPS (100 ng/ml) stimulation for 6h. (A) Immunoprecipitation analysis of YAP and HSF1 in LPS-stimulated macrophages after transfection with CRISPR-Dvl2 ACT vector. (B) Immunoprecipitation analysis of YAP and HSF1 in LPS-stimulated macrophages after transfection with CRISPR-Dvl2 KO vector. (C) Immunofluorescence staining for macrophage HSF1 (green) and YAP (red) colocalization in the nucleus after LPS stimulation. DAPI was used to visualize nuclei (blue). Scale bars, 20 μm. (D) Western blot-assisted analysis and relative density ratio of HSF1, NOD1, cleaved caspase-1, p-P65, and P65 in LPS-stimulated macrophages. (E) Quantitative RT-PCR analysis of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 mRNA levels in the indicated groups (n = 6 samples/group). (F) ELISA analysis of IL-1β levels in the culture supernatant (n = 6 samples/group). (G) Quantitative RT-PCR analysis of Nqo1, Gclc, and Gclm mRNA levels in the indicated groups (n = 6 samples/group). (H) Detection of ROS production by Carboxy-H2DFFDA in the indicated groups (n = 6 samples/group). Quantification of ROS-producing cells (green). Scale bars, 100 μm. (I) The expression of the full-length GSDMD (GSDMD-FL) and N-terminus of GSDMD (GSDMD-N) was detected in the indicated groups. Graph shows the quantitation of relative intensity. (J) Immunofluorescence staining of GSDMD-N expression in the indicated groups. Scale bars, 100 μm. All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
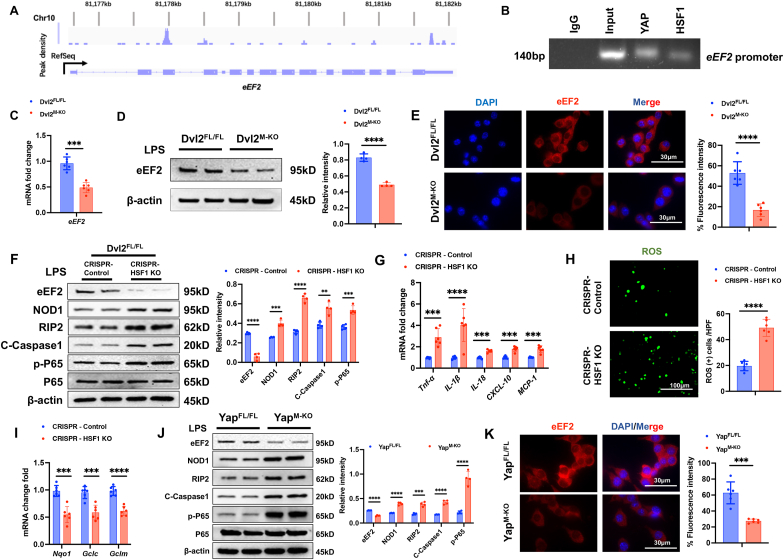
The YAP-HSF1 axis targets eEF2 and regulates NOD1 function in macrophages. To explore the potential mechanism of the YAP-HSF1 axis in modulating NOD1 function in macrophages, we performed HSF1 ChIP coupled to massively parallel sequencing (ChIP-Seq). Indeed, the HSF1 ChIP-seq peaks were identified within the eEF2 gene. One presented the location in the promoter region, and the others were found to be located within the intron/exon of the eEF2 gene (Fig. 5A). To validate the ChIP-seq peak in the eEF2 promoter region, ChIP-PCR was performed using HSF1 and YAP antibodies in macrophages. The primer was designed to detect the HSF1 DNA-binding site in eEF2 promoter by PCR analysis. The sequential ChIPs showed that YAP and HSF1 were bound to the HSF1-binding motif in the HSF1-chromatin complex (Fig. 5B), confirming that YAP and HSF1 are located at the same promoter region of eEF2. Hence, eEF2 is a target gene regulated by the YAP-HSF1 complex. Moreover, Dvl2M−KO diminished eEF2 mRNA levels (Fig. 5C) and protein expression (Fig. 5D) in LPS-stimulated macrophages compared to the Dvl2FL/FL groups. This result was further confirmed by immunofluorescence staining, which showed reduced eEF2 expression in the Dvl2M−KO but not the Dvl2FL/FL macrophages (Fig. 5E). Deletion of HSF1 with a CRISPR-HSF1 KO treatment diminished eEF2 but augmented NOD1, cleaved caspase-1, and NF-κBp65 expression (Fig. 5F), with increased proinflammatory TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 levels (Fig. 5G) and ROS production (Fig. 5H) in macrophages after LPS-stimulation. The expression of antioxidant Nqo1, Gclc, and Gclm was significantly reduced in HSF1-deficient macrophages (Fig. 5I). Consistently, YAP-deficient-macrophages from the YAPM−KO mice displayed reduced eEF2 and augmented NOD1, cleaved caspase-1, and NF-κBp65 expression compared to the YAPFL/FL cells after LPS stimulation (Fig. 5J). Immunofluorescence staining revealed reduced eEF2 expression in the YAPM−KO but not the YAPFL/FL macrophages (Fig. 5K). Collectively, these data indicate that the macrophage YAP-HSF1 axis regulates the HSF1 target gene eEF2, which may play a critical role in regulating NOD1 function during oxidative stress-induced inflammation.
Fig. 5.
The YAP-HSF1 axis targets eEF2 and regulates NOD1 function in macrophages Bone marrow-derived macrophages (BMMs, 1 × 106) were collected and fixed after incubating LPS (100 ng/ml). Following chromatin shearing and HSF1 antibody selection, the precipitated DNA fragments bound by HSF1-containing protein complexes were used for sequencing. (A) Localization of HSF1-binding sites on the mouse eEF2 gene. The 14 exons, 15 introns, 3' untranslated region (UTR), 5’ UTR, and transcription start sites (TSS) of the mouse eEF2 gene on chromosome 10 are shown. (B) ChIP-PCR analysis of HSF1 and YAP binding to the eEF2 promoter. Protein-bound chromatin was prepared from BMMs and immunoprecipitated with HSF1 or YAP antibodies. For sequential ChIP, the protein-bound chromatin was first immunoprecipitated with the HSF1 antibody, followed by elution with a second immunoprecipitation using the YAP antibody. Then, the immunoprecipitated DNA was analyzed using PCR. The normal IgG was used as a negative control. (C) Analysis of eEF2 mRNA levels in LPS-stimulated macrophages from the Dvl2FL/FL and Dvl2M−KO mice (n = 6 samples/group). (D) Western blot analysis and relative density ratio of eEF2 in the indicated groups. (E) Immunofluorescence staining for eEF2 expression in LPS-stimulated macrophages from the Dvl2FL/FL and Dvl2M−KO mice (n = 6 samples/group). Scale bars, 30 μm. (F) Western blot analysis and relative density ratio of eEF2, NOD1, cleaved caspase-1, p-P65, and P65 in LPS-stimulated Dvl2FL/FL macrophages after transfection with CRISPR-HSF1 KO or control vector. (G) qRT-PCR analysis of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 in LPS-stimulated macrophages in the indicated groups (n = 6 samples/group). (H) Detection of ROS production by Carboxy-H2DFFDA in the indicated groups (n = 6 samples/group). Quantification of ROS-producing cells (green). Scale bars, 100 μm. (I) Quantitative RT-PCR analysis of Nqo1, Gclc, and Gclm mRNA levels in the indicated groups (n = 6 samples/group). (J) Western blot analysis and relative density ratio of eEF2, NOD1, cleaved caspase-1, p-P65, and P65 in LPS-stimulated macrophages from the YAPFL/FL and YAPM−KO mice. (K) Immunofluorescence staining for eEF2 expression in LPS-stimulated macrophages from the YAPFL/FL and YAPM−KO mice (n = 6 samples/group). Scale bars, 30 μm. All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
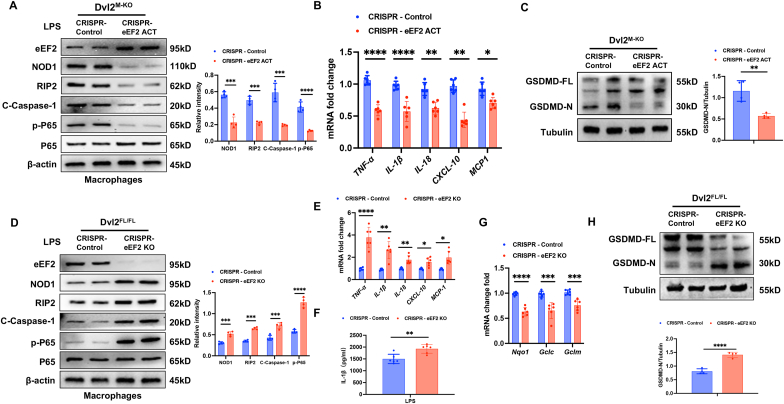
eEF2 is required for the Dvl2-mediated immune regulation of NOD1 function in macrophages. To dissect the role of eEF2 in the Dvl2-mediated immune regulation of NOD1 function in macrophages, BMMs were isolated from the Dvl2FL/FL and Dvl2M−KO mice and transfected with a CRISPR/Cas9-mediated eEF2 activation (CRISPR-eEF2 ACT) or eEF2 knockout (CRISPR-eEF2 KO) vector after LPS stimulation. Indeed, activation of eEF2 with a CRISPR-eEF2 ACT treatment in Dvl2M−KO macrophages reduced NOD1, cleaved caspase-1, and NF-κBp65 expression (Fig. 6A), with decreased mRNA levels of TNF-α, IL-1β, IL-18, CXCL-10, MCP-1 (Fig. 6B). Moreover, CRISPR-eEF2 ACT treatment in Dvl2M−KO macrophages inhibited GSDMD activation, which showed diminished GSDMD-N expression (Fig. 6C) and reduced ROS production (Fig. S11A). However, disruption of eEF2 with a CRISPR-eEF2 KO treatment in Dvl2FL/FL macrophages augmented NOD1, cleaved caspase-1, and NF-κBp65 expression (Fig. 6D), with increased mRNA levels of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 (Fig. 6E) and serum IL-1β levels (Fig. 6F). The expression of antioxidant Nqo1, Gclc, and Gclm was significantly reduced in CRISPR-eEF2 KO-treated Dvl2FL/FL macrophages (Fig. 6G). Notably, CRISPR-eEF2 KO increased GSDMD-N expression in LPS-stimulated Dvl2FL/FL macrophages (Fig. 6H). In one experiment, we also used the Dvl2FL/FL and Dvl2M−KO cells treated with CRISPR-eEF2 ACT. As expected, both Dvl2FL/FL and Dvl2M−KO cells treated with CRISPR-eEF2 ACT displayed reduced levels of TNF-α, IL-1β, IL-18, CXCL-10, MCP-1 (Fig. S11B) and inhibited GSDMD-N expression (Fig. S11C). Collectively, these results suggest that eEF2 is essential for macrophage Dvl2-mediated immune regulation of NOD1 function.
Fig. 6.
eEF2 is required for the Dvl2-mediated immune regulation of NOD1 function in macrophages. (A) Bone marrow-derived macrophages (BMMs) were isolated from Dvl2M−KO mice and transfected with CRISPR/Cas9-mediated eEF2 activation (CRISPR-eEF2 ACT) or control vector followed by 6h of LPS (100 ng/ml) stimulation. Western-assisted analysis of eEF2, NOD1, cleaved caspase-1, p-P65 and P65. (B) qRT-PCR analysis of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 in LPS-stimulated macrophages in the indicated groups (n = 6 samples/group). (C) The expression of the full-length GSDMD (GSDMD-FL) and N-terminus of GSDMD (GSDMD-N) was detected in the indicated groups. The graph shows the quantitation of relative intensity. (D) Bone marrow-derived macrophages (BMMs) were isolated from Dvl2FL/FL mice and transfected with CRISPR/Cas9-mediated eEF2 knockout (CRISPR-eEF2 KO) or control vector followed by 6h of LPS (100 ng/ml) stimulation. Western-assisted analysis of eEF2, NOD1, cleaved caspase-1, p-P65 and P65. (E) qRT-PCR analysis of TNF-α, IL-1β, IL-18, CXCL-10, and MCP-1 in LPS-stimulated macrophages in the indicated groups (n = 6 samples/group). (F) ELISA analysis of IL-1β levels in the culture supernatant (n = 6 samples/group). (G) Quantitative RT-PCR analysis of Nqo1, Gclc, and Gclm mRNA levels in the indicated groups (n = 6 samples/group). (H) The expression of the full-length GSDMD (GSDMD-FL) and N-terminus of GSDMD (GSDMD-N) was detected in the indicated groups. The graph shows the quantitation of relative intensity. All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
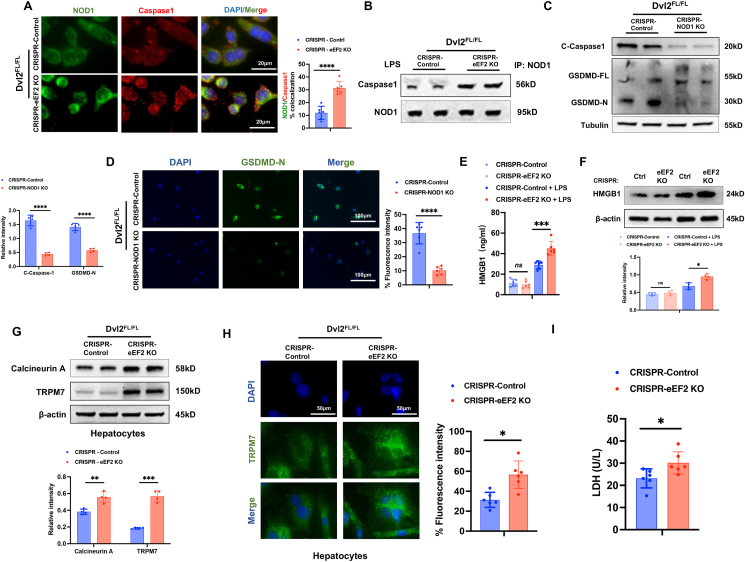
eEF2 is crucial to regulate NOD1-mediated macrophage pyroptosis and hepatocyte death in response to oxidative stress. To elucidate the potential mechanism of eEF2 in regulating NOD1-mediated macrophage pyroptosis and hepatocyte death under cell stress conditions, BMMs from the Dvl2FL/FL mice were transfected with CRISPR-eEF2 KO or control vector followed by LPS stimulation. Interestingly, immunofluorescence staining showed that CRISPR-eEF2 KO increased the colocalization of NOD1 with caspase-1 in LPS-stimulated Dvl2FL/FL macrophages compared to the control vector-treated cells (Fig. 7A). Co-immunoprecipitation assay revealed that increased NOD1 and caspase-1 binding in the CRISPR-eEF2 KO but not the control vector-treated cells (Fig. 7B). Moreover, deletion of NOD1 with a CRISPR/Cas9-mediated NOD1 knockout (CRISPR-NOD1 KO) treatment in Dvl2FL/FL macrophages inhibited caspase-1 and GSDMD activation evidenced by reduced cleaved caspase-1 and GSDMD-N expression in LPS-stimulated macrophages (Fig. 7C). This result was further confirmed by immunofluorescence staining, which showed reduced GSDMD-N expression in macrophages after LPS stimulation (Fig. 7D). Accumulating evidence has demonstrated that oxidative stress induces HMGB1 release, which is vital in activating cell death signaling and liver inflammation [27,28]. Indeed, CRISPR-eEF2 KO increased HMGB1 expression in LPS-stimulated macrophages (Fig. 7E), accompanied by increased HMGB1 release from macrophages (Fig. 7F) compared to the control vector-treated cells. Next, we used a macrophage/hepatocyte co-culture system. BMMs from the Dvl2FL/FL mice were transfected with CRISPR-eEF2 KO or control vector followed by LPS stimulation and then co-cultured with primary hepatocytes. Interestingly, disruption of macrophage eEF2 augmented hepatocyte calcineurin A and TRPM7 expression in response to LPS stimulation after co-culture (Fig. 7G). This result was further confirmed by immunofluorescence staining, which showed that LPS-stimulated CRISPR-eEF2 KO macrophages displayed increased hepatocyte TRPM7 expression after co-culture (Fig. 7H). Consistently, increased LDH release from the CRISPR-eEF2 KO-transfected macrophage/hepatocyte co-culture (Fig. 7I). Collectively, these results indicate that macrophage eEF2 is a critical regulator in activating NOD1-driven pyroptosis and Calcineurin/TRPM7-induced hepatocyte death in response to oxidative stress.
Fig. 7.
eEF2 is crucial to regulate NOD1-mediated macrophage pyroptosis and hepatocyte death in response to oxidative stress. (A) Bone marrow-derived macrophages (BMMs) were isolated from Dvl2FL/FL mice and transfected with CRISPR/Cas9-mediated eEF2 knockout (CRISPR-eEF2 KO) or control vector followed by 6h of LPS (100 ng/ml) stimulation. (A) Immunofluorescence staining for NOD1 (green) and caspase-1 (red) colocalization in LPS-stimulated macrophages (n = 6 samples/group). DAPI was used to visualize nuclei. Scale bars, 20 μm. (B) Immunoprecipitation analysis of caspase-1 and NOD1 in LPS-stimulated macrophages. (C) BMMs were isolated from Dvl2FL/FL mice and transfected with CRISPR/Cas9-mediated NOD1 knockout (CRISPR-NOD1 KO) or control vector followed by 6h of LPS (100 ng/ml) stimulation. Western blot analysis and relative density ratio of cleaved caspase-1, the full-length GSDMD (GSDMD-FL), and N-terminus of GSDMD (GSDMD-N) in LPS-stimulated macrophages. (D) Immunofluorescence staining of GSDMD-N expression in the indicated groups. Scale bars, 100 μm. (E) Western blot analysis and relative density ratio of HMGB1 in the indicated groups. (F) ELISA analysis of supernatant HMGB1 levels in CRISPR-eEF2 KO or control vector-treated BMMs after LPS stimulation (n = 6 samples/group). (G) BMMs from the Dvl2FL/FL mice were transfected with CRISPR/Cas9-mediated eEF2 knockout (CRISPR-eEF2 KO) or control vector followed by 6h of LPS (100 ng/ml) stimulation and then co-cultured with primary hepatocytes for 24h. Western blot analysis and relative density ratio of Calcineurin A and TRPM7 in primary hepatocytes after co-culture with CRISPR-eEF2 KO or control vector macrophages. (H) Immunofluorescence staining for TRPM7 expression in primary hepatocytes (n = 6 samples/group). DAPI was used to visualize nuclei. Scale bars, 50 μm. (I) LDH release from the primary hepatocytes in co-cultures (n = 6 samples/group). All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
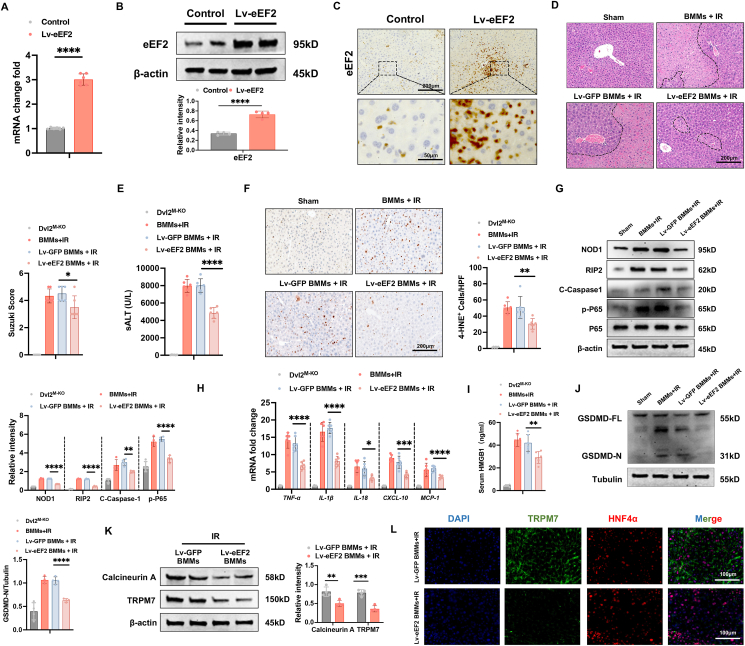
Adoptive transfer of eEF2-expressing macrophages alleviates NOD1-driven inflammation, pyroptosis, and hepatocyte death in IR-stressed livers. Having demonstrated the importance of eEF2 on Dvl2-mediated immune regulation of NOD1 function in macrophages, we then examined whether eEF2 influenced NOD1-driven inflammation, pyroptosis, and Calcineurin/TRPM7-induced hepatocyte death in IR-stressed livers. BMMs were transfected with lentivirus-expressing eEF2 (Lv-eEF2) or GFP control (Lv-GFP). Indeed, the adoptive transfer of Lv-eEF2-treated BMMs markedly increased eEF2 mRNA and protein levels in IR-stressed livers of the Dvl2M−KO mice (Fig. 8A and B). This was further confirmed by immunohistochemistry staining, which showed increased eEF2 expression in Lv-eEF2 BMM-treated livers (Fig. 8C). Lv-eEF2 BMM treatment alleviated IR-induced liver damage in the Dvl2M−KO mice, as evidenced by reduced Suzuki’s histological score (Fig. 8D) and sALT levels (Fig. 8E), compared to the Lv-GFP-treated control cells. Unlike in Lv-GFP-treated controls, Lv-eEF2 BMM treatment decreased the positive cells of 4-HNE (Fig. 8F) in IR-stressed Dvl2M−KO livers. Moreover, Lv-eEF2 BMM treatment reduced the expression of NOD1, cleaved caspase-1, p-P65 (Fig. 8G), with diminished mRNA levels of TNF-α, IL-1β, IL-18, CXCL-10, MCP-1 (Fig. 8H), and HMGB1 levels (Fig. 8I) in ischemic Dvl2M−KO livers. Moreover, the adoptive transfer of Lv-eEF2-treated BMMs inhibited GSDMD-N expression in IR-stressed Dvl2M−KO livers (Fig. 8J). Consistent with this result, Lv-eEF2 BMM treatment diminished Calcineurin A and TRPM7 expression in IR-stressed livers (Fig. 8K). This result was further confirmed by immunofluorescence staining, which showed reduced hepatocyte TRPM7 expression in IR-stressed livers (Fig. 8L). Hence, these results demonstrate the critical role of eEF2 in the macrophage Dvl2-mediated regulation of NOD1-driven inflammation, pyroptosis, and Calcineurin/TRPM7-induced hepatocyte death in IR-triggered liver injury.
Fig. 8.
Adoptive transfer of eEF2-expressing macrophages alleviates NOD1-driven inflammation, pyroptosis, and hepatocyte death in IR-stressed livers. The Dvl2M−KO mice were injected via tail vein with bone marrow-derived macrophages (BMMs, 1 × 106 cells/mouse) transfected with lentiviral-expressing eEF2 (Lv-eEF2) or GFP control (Lv-GFP) 24h before ischemia. (A) Quantitative RT-PCR analysis of eEF2 mRNA levels in IR-stressed livers 24h after tail vein injection (n = 6 samples/group). (B) Western blot analysis and relative density ratio of eEF2 protein expression in ischemic livers after Lv-eEF2 or control BMM treatment. (C) Immunohistochemistry staining of eEF2 in IR-stressed livers 24h after tail vein injection (n = 6 mice/group). Scale bars, 200 μm and 50 μm. (D) Representative histological staining (H&E) of ischemic liver tissue (n = 6 mice/group) and Suzuki’s histological score. Scale bars, 200 μm. (E) sALT levels (IU/L) (n = 6 samples/group). (F) Immunohistochemistry staining of 4-HNE+ cells in IR-stressed livers (n = 6 mice/group). Quantification of 4-HNE+ cells, Scale bars, 200 μm. (G) Western-assisted analysis and relative density ratio of NOD1, p-P65, and P-65 in the indicated groups. (H) qRT-PCR analysis of TNF-α, IL-1β, IL-6, CXCL-10, and MCP-1 in IR-stressed livers (n = 6 samples/group). (I) ELISA analysis of serum HMGB1 levels (n = 6 samples/group). (J) The full-length GSDMD (GSDMD-FL) and N-terminus of GSDMD (GSDMD-N) expression were detected in IR-stressed Dvl2M−KO livers after Lv-eEF2 or control BMM treatment. The graph shows the quantitation of relative intensity. (K) Western-assisted analysis and relative density ratio of Calcineurin A and TRPM7 in the indicated groups. (L) Immunofluorescence staining for TRPM7 expression in hepatocytes from the Dvl2M−KO mice after adoptive transfer of Lv-eEF2 expressing or control BMMs. (n = 6 samples/group). DAPI was used to visualize nuclei. Scale bars, 100 μm. All Western blots represent four experiments, and the data represent the mean ± SD. Statistical analysis was performed using the Permutation t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
4. Discussion
This study is the first to document the key role of macrophage Dvl2 in regulating NOD1-driven inflammation, pyroptosis, and Calcineurin/TRPM7-induced hepatocyte death in IR stress-induced liver injury. Importantly, we show that macrophage Dvl2 controls NOD1 function by regulating the Hippo/YAP pathway and HSF1 activation in IR-stressed livers. First, macrophage Dvl2 deficiency induces NOD1 and GSDMD activation, leading to pyroptosis and increased inflammatory response, exacerbating IR-induced liver damage. Second, disruption of macrophage Dvl2 inhibits the YAP pathway and HSF1 signaling, whereas Dvl2 activation promotes the YAP-HSF1 interaction, diminishing NOD1 function in the IR-stressed liver. Third, the YAP-HSF1 axis activates the HSF1 target gene eEF2, which is critical to regulating NOD1-mediated macrophage pyroptosis, inflammation, and Calcineurin/TRPM7-induced hepatocyte death in IR-stressed livers. Fourth, the adoptive transfer of eEF2-expressing macrophages controls NOD1 activation and alleviates IR-induced liver damage. Our results highlight the importance of the macrophage Dvl2 in regulating NOD1 function in liver IRI.
Pyroptosis is the process of inflammatory cell death, primarily in inflammatory cells, such as macrophages. Excessive pyroptosis induces strong inflammatory responses and severe tissue damage [29]. Pyroptosis requires the cleavage of GSDMD into N- and C-termini by caspase-1. Upon cleavage, the N-terminus of GSDMD (GSDMD-N) forms a transmembrane pore that releases cytokines such as IL-1β, resulting in robust inflammation and cell death. Indeed, as an executor of pyroptosis, DSDMD is critical to trigger immune and inflammatory responses leading to tissue damage and organ dysfunction [30]. Our current study revealed that macrophage Dvl2 deficiency promoted NOD1/caspase-1/GSDMD-mediated pyroptosis, increased IL-1β and HMGB1 release, and exacerbated IR-induced liver damage, demonstrating the crucial role of macrophage Dvl2 in modulating NOD1 function in IR-stressed livers.
The Hippo/YAP pathway is a highly conserved kinase cascade in mammals. The activity of YAP proteins is regulated by its upstream kinases, LATS1/2, which phosphorylates YAP, resulting in cytoplasmic degradation [31]. However, unphosphorylated YAP enters the nucleus and interacts with various transcription factors to regulate gene expression [31]. Previous studies showed that Mob1 is essential for activating LATS1 [24], suggesting that the Mob1-LATS1 complex is a key regulator of YAP activity. Interestingly, our RNA-seq analysis revealed that macrophage Dvl2 deficiency increased mRNA levels of Mob1, which augmented LATS1 and YAP phosphorylation, reducing nuclear YAP expression. Moreover, macrophage Dvl2 deficiency upregulated Zpr1, leading to inhibited HSF1 activation. Thus, we speculate that the YAP-HSF1 axis may be essential in macrophage Dvl2-mediated regulation of NOD1-driven pyroptotic cell death and inflammatory signaling cascade in liver IRI.
The question arises as to how the YAP pathway and HSF1 signaling may selectively affect the NOD1 function in macrophage Dvl2-mediated immune regulation. As a key downstream effector of the Hippo pathway, YAP translocates into the nucleus to activate gene expression [32]. The activation of YAP inhibits immune response, whereas disruption of YAP aggravates tissue inflammatory injury [33]. Our previous studies showed that Hippo signaling regulated innate immunity via a YAP-mediated transcriptional mechanism [21]. Consistent with these findings, we found that disruption of macrophage YAP induced NOD1/caspase-1 activation, with increased proinflammatory mediator and ROS production while inhibiting antioxidant gene expression. Moreover, macrophage YAP deficiency activated GSDMD, leading to macrophage pyroptosis in IR-stressed livers, suggesting that YAP is critical in regulating NOD1-mediated pyroptotic cell death in liver IRI. Notably, YAP was revealed to act as a transcriptional coactivator of HSF1 in macrophage Dvl2-mediated regulation of NOD1 function. Indeed, HSF1 is a transcription factor induced by oxidative stress. HSF1 conferred protection against inflammatory injury and LPS-induced liver damage [34]. Dvl2 induction increased YAP interaction with HSF1 and activated HSF1, inhibiting NOD1/caspase-1 activation and ROS production. Our findings reveal an unexpected role for the macrophage Dvl2 in controlling NOD1 function by activating the YAP-HSF1 axis in IR stress-induced liver injury.
However, how the YAP-HSF1 axis may modulate NOD1-driven macrophage pyroptosis and inflammation response in IR-stressed liver remains unclear. Using the ChIP and ChIP-sequencing approaches, we found that HSF1 was localized on the promoter of eEF2, suggesting that eEF2 is a target gene of HSF1. Indeed, disruption of macrophage HSF1 diminished mRNA levels and protein expression of eEF2 and enhanced NOD1 function, with increased ROS production, proinflammatory mediators, and reduced antioxidant gene expression under cell stress conditions. Moreover, macrophage YAP deletion diminished eEF2 yet enhanced NOD1/caspase-1 activation. Our findings revealed the distinct role of the YAP-HSF1 axis in modulating eEF2 activity in liver IRI.
Of particular interest, eEF2 is a critical regulator in controlling NOD1-mediated pyroptosis and inflammatory response in IR-stressed livers. Indeed, eEF2 is an essential factor for protein synthesis and is required for the modulation of autophagy, cell proliferation, and survival [35]. Phosphorylation of eEF2 by eEF-2 kinase (eEF2K) inhibited eEF2 activity and promoted ER stress-induced cell death [36]. Disruption of eEF2K activated eEF2, which in turn inhibited NF-κB activation and oxidative stress-mediated inflammatory response [37]. These results suggest that eEF2 contributes to the regulation of inflammatory and cell death signaling cascades under cell stress conditions. In line with these findings, we found that activation of macrophage eEF2 expression inhibited NOD1/caspase-1 and NF-κB p65 activation with reduced proinflammatory mediators and ROS production. However, disruption of macrophage eEF2 activated NOD1/caspase-1 and increased proinflammatory mediators while reducing antioxidant gene expression. Strikingly, macrophage eEF2 deficiency promoted GSDMD activation, leading to increased macrophage pyroptosis, indicating the critical role of eEF2 in modulating NOD1-mediated pyroptotic cell death in macrophages.
Another striking finding was that we demonstrated the mechanistic role of eEF2 in modulating NOD1-driven macrophage pyroptosis and hepatocyte cell death under inflammatory conditions. Previous studies showed that caspase-1 activated by NLRP3 cleaved GSDMD and promoted the secretion of IL-1β and IL-18, leading to pyroptotic cell death [38]. As NOD1 is a well-characterized NOD-like Receptors (NLRs) and also contributes to the secretion of IL-1β and IL-18 [39,40], we speculate that the NOD1 could activate caspase-1 to induce pyroptosis upon eEF2 deletion. As expected, macrophage eEF2 deficiency augmented the NOD1-caspase-1 interaction and activated caspase-1, enhancing GSDMD activity and promoting macrophage pyroptosis in response to inflammatory stimulation. Moreover, macrophage eEF2 deletion increased HMGB1 release, a key mediator in driving inflammatory response and tissue damage [28]. HMGB1 can be released from GSDMD-mediated pyroptotic cell death under inflammatory conditions [41]. As eEF2 is a target gene of HSF1 and modulated by Dvl2-mediated YAP-HSF1 axis, our results indicated that Dvl2 could regulate GSDMD-induced HMGB1 activation in liver IRI. Notably, using a macrophage/hepatocyte co-culture system, disruption of macrophage eEF2 induced Calcineurin/TRPM7 activation in hepatocytes. As an ion channel and functional kinase, TRPM7 is regulated by Calcineurin [42], a calcium and calmodulin-dependent serine/threonine protein phosphatase [43]. Activation of Calcineurin increases inward Ca2+ permeation mediated by TRPM7, leading to mitochondrial accumulation of Ca2+ in response to cell stress [44]. Increased mitochondrial Ca2+ induces the mitochondrial permeability transition (MPT) pore, resulting in mitochondria's structural and functional collapse and cell death [45]. In line with these findings, macrophage eEF2 deletion increased hepatocyte TRPM7 expression and LDH release after co-culture. Moreover, in vivo adoptive transfer of eEF2-expressing macrophages alleviated IR-triggered liver damage with depressed NOD1/caspase-1/GSDMD activation, diminished HMGB1 release, and reduced hepatocyte Calcineurin/TRPM7 expression. Thus, our in vitro and in vivo findings reveal the essential role of the macrophage Dvl2-mediated eEF2 in modulating NOD1-driven macrophage pyroptosis and the Calcineurin/TRPM7-induced cell death cascade in IR stress-induced liver inflammatory injury.
It is worth noting that we also examined NLRP3-mediated pyroptosis in our model. Interestingly, Dvl2M−KO only slightly increased NLRP3/caspase-1 and GSDMD-N expression (Fig. S3B) while markedly increased NOD1/cleaved caspase-1 and GSDMD-N (Fig. S3A), suggesting that macrophage Dvl2 deficiency induces GSDMD-mediated pyroptosis mainly by promoting NOD1 activation.
In conclusion, we identify a previously unrecognized role of macrophage Dvl2 on NOD1-mediated pyroptosis, inflammation, and Calcineurin/TRPM7-induced hepatocyte death in IR stress-induced liver injury. Dvl2 controls NOD1 function by activating the YAP-HSF1 signaling, which targets eEF2 and regulates GSDMD-mediated macrophage pyroptosis and Calcineurin/TRPM7-induced hepatocyte death in response to IR stress. By identifying the molecular regulatory mechanism of macrophage Dvl2-mediated regulation of NOD1 function in IR-stressed livers, our findings provide potential therapeutic targets for stress-induced liver inflammation and injury.
CRediT authorship contribution statement
Xiaoye Qu: Writing – review & editing, Validation, Resources, Methodology, Formal analysis, Data curation, Conceptualization. Dongwei Xu: Methodology, Formal analysis, Data curation, Conceptualization. Tao Yang: Writing – review & editing, Supervision, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Yizhu Tian: Software, Resources, Methodology, Investigation, Data curation. Christopher T. King: Visualization, Project administration, Methodology. Xiao Wang: Validation, Resources. Mingwei Sheng: Methodology, Investigation. Yuanbang Lin: Validation, Software. Xiyun Bian: Formal analysis, Data curation. Changyong Li: Visualization, Conceptualization. Longfeng Jiang: Software, Investigation. Qiang Xia: Visualization, Validation, Supervision. Douglas G. Farmer: Supervision, Software, Resources. Bibo Ke: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the NIH grants R01AI139552, P01AI120944, R21AI146742, R21AI112722, and R21AI115133.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103455.
Contributor Information
Tao Yang, Email: dr.taoyang86@gmail.com.
Bibo Ke, Email: bke@mednet.ucla.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Li C., Sheng M., Lin Y., Xu D., Tian Y., Zhan Y., Jiang L., et al. Functional crosstalk between myeloid Foxo1-beta-catenin axis and Hedgehog/Gli1 signaling in oxidative stress response. Cell Death Differ. 2021;28:1705–1719. doi: 10.1038/s41418-020-00695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu L., Xiong X., Wu X., Ye Y., Jian Z., Zhi Z., Gu L. Targeting oxidative stress and inflammation to prevent ischemia-reperfusion injury. Front. Mol. Neurosci. 2020;13:28. doi: 10.3389/fnmol.2020.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yue S., Zhu J., Zhang M., Li C., Zhou X., Zhou M., Ke M., et al. The myeloid heat shock transcription factor 1/beta-catenin axis regulates NLR family, pyrin domain-containing 3 inflammasome activation in mouse liver ischemia/reperfusion injury. Hepatology. 2016;64:1683–1698. doi: 10.1002/hep.28739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu L., Yue S., Jiang L., Li C., Zhu Q., Ke M., Lu H., et al. Myeloid Notch1 deficiency activates the RhoA/ROCK pathway and aggravates hepatocellular damage in mouse ischemic livers. Hepatology. 2018;67:1041–1055. doi: 10.1002/hep.29593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakrabarti S., Visweswariah S.S. Intramacrophage ROS primes the innate immune system via JAK/STAT and toll activation. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keestra-Gounder A.M., Byndloss M.X., Seyffert N., Young B.M., Chavez-Arroyo A., Tsai A.Y., Cevallos S.A., et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532:394–397. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travassos L.H., Carneiro L.A., Girardin S.E., Boneca I.G., Lemos R., Bozza M.T., Domingues R.C., et al. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J. Biol. Chem. 2005;280:36714–36718. doi: 10.1074/jbc.M501649200. [DOI] [PubMed] [Google Scholar]
- 8.Masumoto J., Yang K., Varambally S., Hasegawa M., Tomlins S.A., Qiu S., Fujimoto Y., et al. Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J. Exp. Med. 2006;203:203–213. doi: 10.1084/jem.20051229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motomura Y., Kanno S., Asano K., Tanaka M., Hasegawa Y., Katagiri H., Saito T., et al. Identification of pathogenic cardiac CD11c+ macrophages in nod1-mediated acute coronary arteritis. Arterioscler. Thromb. Vasc. Biol. 2015;35:1423–1433. doi: 10.1161/ATVBAHA.114.304846. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Ramos S., Fernandez-Garcia V., Recalde M., Rodriguez C., Martinez-Gonzalez J., Andres V., Martin-Sanz P., et al. Deletion or inhibition of NOD1 favors plaque stability and attenuates atherothrombosis in advanced atherogenesis (dagger) Cells. 2020;9 doi: 10.3390/cells9092067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pei G., Zyla J., He L., Moura-Alves P., Steinle H., Saikali P., Lozza L., et al. Cellular stress promotes NOD1/2-dependent inflammation via the endogenous metabolite sphingosine-1-phosphate. EMBO J. 2021;40 doi: 10.15252/embj.2020106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A., Singh S., Ahmad S., Gulzar F., Schertzer J.D., Tamrakar A.K. NOD1 activation induces oxidative stress via NOX1/4 in adipocytes. Free Radic. Biol. Med. 2021;162:118–128. doi: 10.1016/j.freeradbiomed.2020.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Shi P., Chen Q., Huang Z., Zou D., Zhang J., Gao X., et al. Mitochondrial ROS promote macrophage pyroptosis by inducing GSDMD oxidation. J. Mol. Cell Biol. 2019;11:1069–1082. doi: 10.1093/jmcb/mjz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallingford J.B., Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 15.Weitzman J.B. Dishevelled nuclear shuttling. J. Biol. 2005;4:1. doi: 10.1186/jbiol21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma M., Castro-Piedras I., Simmons G.E., Jr., Pruitt K. Dishevelled: a masterful conductor of complex Wnt signals. Cell. Signal. 2018;47:52–64. doi: 10.1016/j.cellsig.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao C., Chen Y.G. Dishevelled: the hub of Wnt signaling. Cell. Signal. 2010;22:717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 18.Boligala G.P., Yang M.V., van Wunnik J.C., Pruitt K. Nuclear Dishevelled: an enigmatic role in governing cell fate and Wnt signaling. Biochim. Biophys. Acta Mol. Cell Res. 2022;1869 doi: 10.1016/j.bbamcr.2022.119305. [DOI] [PubMed] [Google Scholar]
- 19.Rasha F., Boligala G.P., Yang M.V., Martinez-Marin D., Castro-Piedras I., Furr K., Snitman A., et al. Dishevelled 2 regulates cancer cell proliferation and T cell mediated immunity in HER2-positive breast cancer. BMC Cancer. 2023;23:172. doi: 10.1186/s12885-023-10647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang F., Cao F., Lu C., He X., Weng L., Sun L. Dvl2 facilitates the coordination of NF-kappaB and Wnt signaling to promote colitis-associated colorectal progression. Cancer Sci. 2022;113:565–575. doi: 10.1111/cas.15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C., Jin Y., Wei S., Sun Y., Jiang L., Zhu Q., Farmer D.G., et al. Hippo signaling controls NLR family pyrin domain containing 3 activation and governs immunoregulation of mesenchymal stem cells in mouse liver injury. Hepatology. 2019;70:1714–1731. doi: 10.1002/hep.30700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki S., Toledo-Pereyra L.H., Rodriguez F.J., Cejalvo D. Neutrophil infiltration as an important factor in liver ischemia and reperfusion injury. Modulating effects of FK506 and cyclosporine. Transplantation. 1993;55:1265–1272. doi: 10.1097/00007890-199306000-00011. [DOI] [PubMed] [Google Scholar]
- 23.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni L., Zheng Y., Hara M., Pan D., Luo X. Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 2015;29:1416–1431. doi: 10.1101/gad.264929.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabbarini I.M., Reif D., McQuown A.J., Nelliat A.R., Prince J., Membreno B.S., Wu C.C., et al. Zinc-finger protein Zpr1 is a bespoke chaperone essential for eEF1A biogenesis. Mol. Cell. 2023;83:252–265. doi: 10.1016/j.molcel.2022.12.012. e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liou G.Y., Storz P. Detecting reactive oxygen species by immunohistochemistry. Methods Mol. Biol. 2015;1292:97–104. doi: 10.1007/978-1-4939-2522-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen R., Kang R., Tang D. The mechanism of HMGB1 secretion and release. Exp. Mol. Med. 2022;54:91–102. doi: 10.1038/s12276-022-00736-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsung A., Sahai R., Tanaka H., Nakao A., Fink M.P., Lotze M.T., Yang H., et al. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J. Exp. Med. 2005;201:1135–1143. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergsbaken T., Fink S.L., Cookson B.T. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasudevan S.O., Behl B., Rathinam V.A. Pyroptosis-induced inflammation and tissue damage. Semin. Immunol. 2023;69 doi: 10.1016/j.smim.2023.101781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saucedo L.J., Edgar B.A. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 32.Hong W., Guan K.L. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramjee V., Li D., Manderfield L.J., Liu F., Engleka K.A., Aghajanian H., Rodell C.B., et al. Epicardial YAP/TAZ orchestrate an immunosuppressive response following myocardial infarction. J. Clin. Invest. 2017;127:899–911. doi: 10.1172/JCI88759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambade A., Catalano D., Lim A., Mandrekar P. Inhibition of heat shock protein (molecular weight 90 kDa) attenuates proinflammatory cytokines and prevents lipopolysaccharide-induced liver injury in mice. Hepatology. 2012;55:1585–1595. doi: 10.1002/hep.24802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kameshima S., Okada M., Yamawaki H. Eukaryotic elongation factor 2 (eEF2) kinase/eEF2 plays protective roles against glucose deprivation-induced cell death in H9c2 cardiomyoblasts. Apoptosis. 2019;24:359–368. doi: 10.1007/s10495-019-01525-z. [DOI] [PubMed] [Google Scholar]
- 36.Boyce M., Py B.F., Ryazanov A.G., Minden J.S., Long K., Ma D., Yuan J. A pharmacoproteomic approach implicates eukaryotic elongation factor 2 kinase in ER stress-induced cell death. Cell Death Differ. 2008;15:589–599. doi: 10.1038/sj.cdd.4402296. [DOI] [PubMed] [Google Scholar]
- 37.Usui T., Okada M., Hara Y., Yamawaki H. Eukaryotic elongation factor 2 kinase regulates the development of hypertension through oxidative stress-dependent vascular inflammation. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H756–H768. doi: 10.1152/ajpheart.00373.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C., Yang T., Xiao J., Xu C., Alippe Y., Sun K., Kanneganti T.D., et al. NLRP3 inflammasome activation triggers gasdermin D-independent inflammation. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abj3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kavathas P.B., Boeras C.M., Mulla M.J., Abrahams V.M. Nod1, but not the ASC inflammasome, contributes to induction of IL-1beta secretion in human trophoblasts after sensing of Chlamydia trachomatis. Mucosal Immunol. 2013;6:235–243. doi: 10.1038/mi.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran L.S., Ying L., D'Costa K., Wray-McCann G., Kerr G., Le L., Allison C.C., et al. NOD1 mediates interleukin-18 processing in epithelial cells responding to Helicobacter pylori infection in mice. Nat. Commun. 2023;14:3804. doi: 10.1038/s41467-023-39487-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volchuk A., Ye A., Chi L., Steinberg B.E., Goldenberg N.M. Indirect regulation of HMGB1 release by gasdermin D. Nat. Commun. 2020;11:4561. doi: 10.1038/s41467-020-18443-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turlova E., Wong R., Xu B., Li F., Du L., Habbous S., Horgen F.D., et al. TRPM7 mediates neuronal cell death upstream of calcium/calmodulin-dependent protein kinase II and calcineurin mechanism in neonatal hypoxic-ischemic brain injury. Transl Stroke Res. 2021;12:164–184. doi: 10.1007/s12975-020-00810-3. [DOI] [PubMed] [Google Scholar]
- 43.Bandyopadhyay J., Lee J., Bandyopadhyay A. Regulation of calcineurin, a calcium/calmodulin-dependent protein phosphatase, in C. elegans. Mol. Cell. 2004;18:10–16. [PubMed] [Google Scholar]
- 44.Duchen M.R. Mitochondria and calcium: from cell signalling to cell death. J. Physiol. 2000;529(Pt 1):57–68. doi: 10.1111/j.1469-7793.2000.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemasters J.J., Theruvath T.P., Zhong Z., Nieminen A.L. Mitochondrial calcium and the permeability transition in cell death. Biochim. Biophys. Acta. 2009;1787:1395–1401. doi: 10.1016/j.bbabio.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.