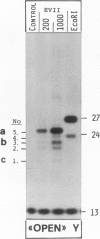
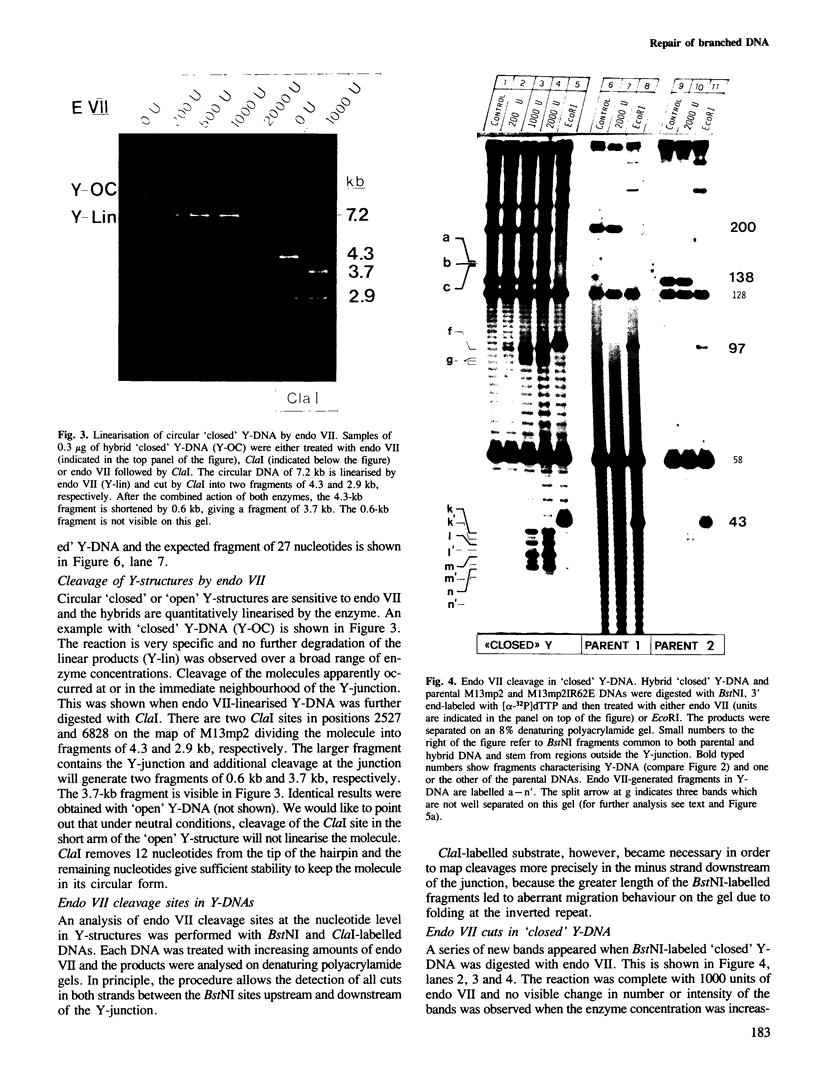
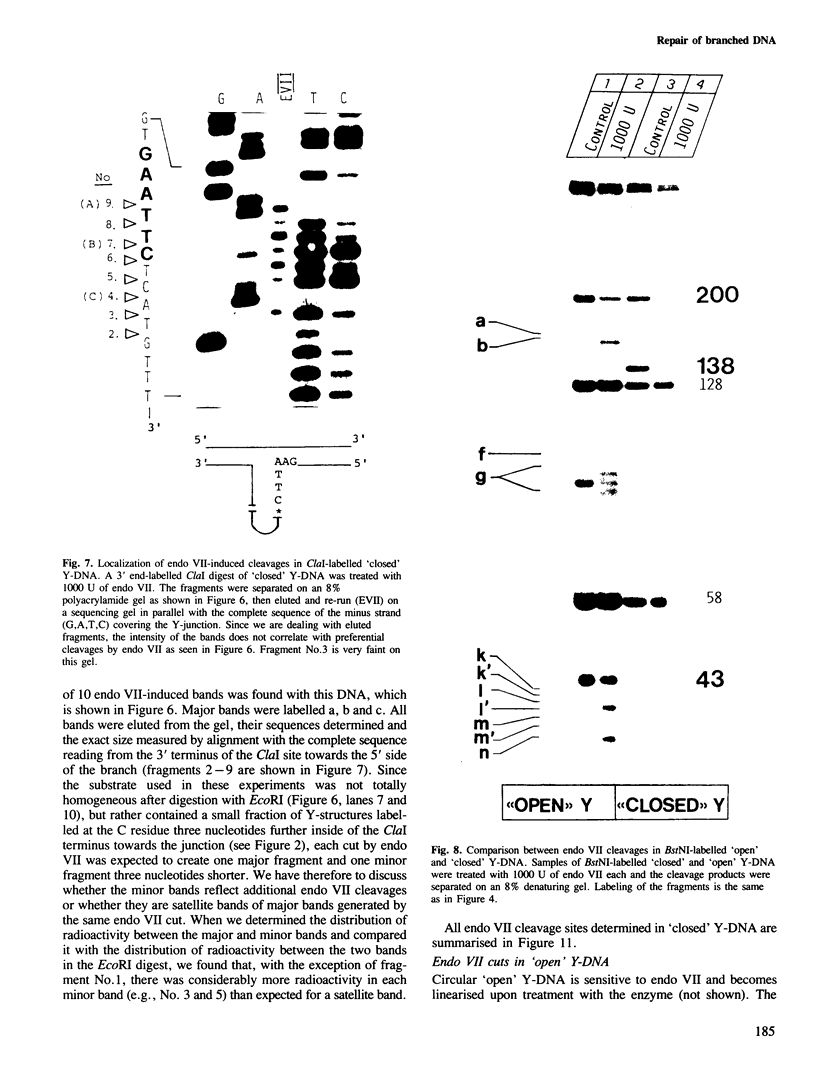
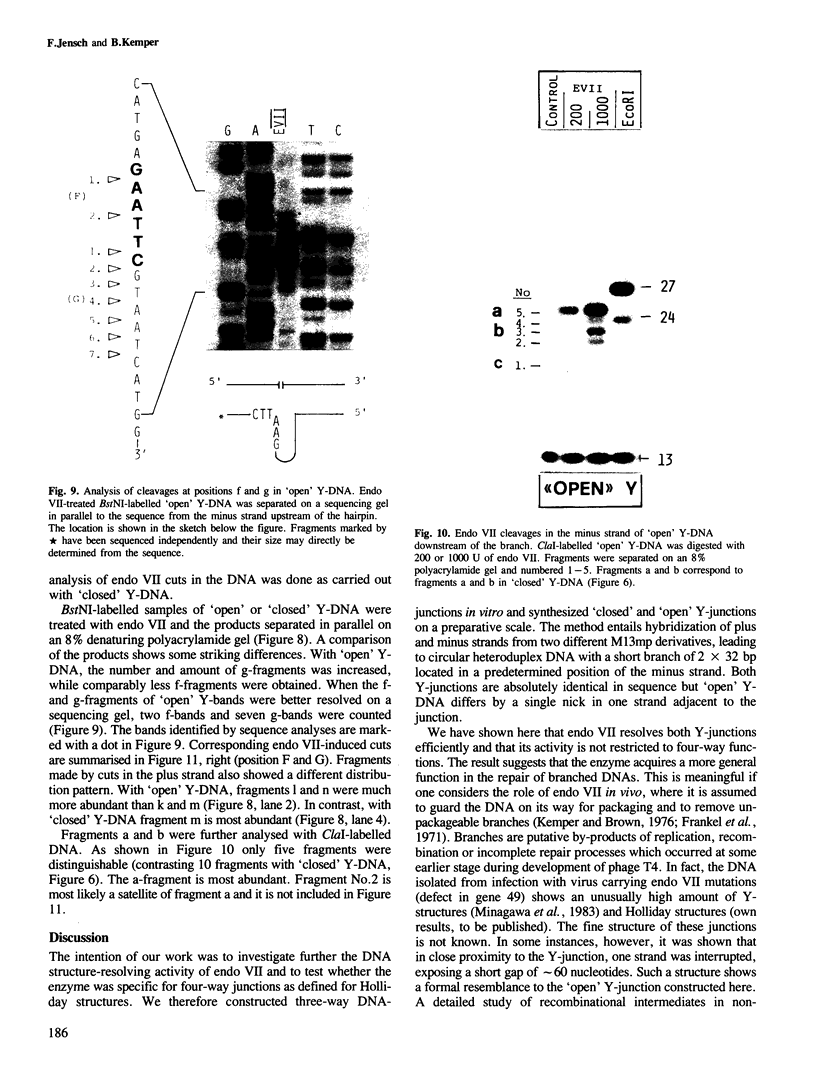
Abstract
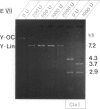
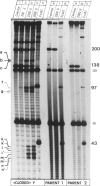
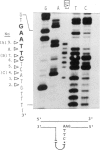
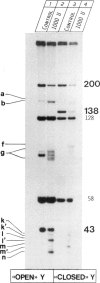
Endonuclease VII (gp 49 of phage T4) resolves four-way junctions in branched DNAs. We have extended our investigations of the specificity of endo VII and tested its activity with three-way junctions (Y-structures) constructed in vitro. Both 'closed' and 'open' Y-structures were made, absolutely identical in sequence but differing from each other by a single nick in one of the three arms. Pure Y-structures were obtained on a preparative scale by annealing plus and minus strands from two M13mp strains. One strain has an inverted repeat of 2 X 31 nucleotides cloned into the single EcoRI site while in the other strain this repeat is absent. The structures were used in reactions with endo VII, which recognizes the branch point of both structures and introduces a characteristic number of nicks, 3' to the junction in each arm of the structure. Strong and weak sites could be distinguished and the cleavage pattern differed significantly between the two structures. The observed resolution of Y-junctions by endo VII in vitro is compatible with a model for the resolution of recombinant Y-branches in DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broker T. R. An electron microscopic analysis of pathways for bacteriophage T4 DNA recombination. J Mol Biol. 1973 Nov 25;81(1):1–16. doi: 10.1016/0022-2836(73)90243-x. [DOI] [PubMed] [Google Scholar]
- Broker T. R., Doermann A. H. Molecular and genetic recombination of bacteriophage T4. Annu Rev Genet. 1975;9:213–244. doi: 10.1146/annurev.ge.09.120175.001241. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courage-Tebbe U., Kemper B. Construction of gapped circular DNA from phage M13 by in vitro hybridization. Biochim Biophys Acta. 1982 Apr 26;697(1):1–5. doi: 10.1016/0167-4781(82)90037-9. [DOI] [PubMed] [Google Scholar]
- Dressler D., Potter H. Molecular mechanisms in genetic recombination. Annu Rev Biochem. 1982;51:727–761. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- Frankel F. R., Batcheler M. L., Clark C. K. The role of gene 49 in DNA replication and head morphogenesis in bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):439–463. doi: 10.1016/0022-2836(71)90147-1. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Gronenborn B., Messing J. Methylation of single-stranded DNA in vitro introduces new restriction endonuclease cleavage sites. Nature. 1978 Mar 23;272(5651):375–377. doi: 10.1038/272375a0. [DOI] [PubMed] [Google Scholar]
- Heidecker G., Messing J., Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980 Jun;10(1):69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Kemper B., Brown D. T. Function of gene 49 of bacteriophage T4. II. Analysis of intracellular development and the structure of very fast-sedimenting DNA. J Virol. 1976 Jun;18(3):1000–1015. doi: 10.1128/jvi.18.3.1000-1015.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Garabett M. Studies on T4-head maturation. 1. Purification and characterization of gene-49-controlled endonuclease. Eur J Biochem. 1981 Mar 16;115(1):123–131. [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- Lee D., Sadowski P. D. Genetic recombination of bacteriophage T7 in vivo studied by use of a simple physical assay. J Virol. 1981 Dec;40(3):839–847. doi: 10.1128/jvi.40.3.839-847.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lundquist R. C., Olivera B. M. Transient generation of displaced single-stranded DNA during nick translation. Cell. 1982 Nov;31(1):53–60. doi: 10.1016/0092-8674(82)90404-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa T., Murakami A., Ryo Y., Yamagishi H. Structural features of very fast sedimenting DNA formed by gene 49 defective T4. Virology. 1983 Apr 15;126(1):183–193. doi: 10.1016/0042-6822(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Minagawa T., Ryo Y. Genetic control of formation of very fast sedimenting DNA of bacteriophage T4. Mol Gen Genet. 1979 Feb 16;170(1):113–115. doi: 10.1007/BF00268587. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Mosig G., Shaw M., Garcia G. M. On the role of DNA replication, endonuclease VII, and rII proteins in processing of recombinational intermediates in phage T4. Cold Spring Harb Symp Quant Biol. 1984;49:371–382. doi: 10.1101/sqb.1984.049.01.044. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Lau L. F., Bahl C. P., Narang S. A., Wu R. Synthetic adaptors for cloning DNA. Methods Enzymol. 1979;68:98–109. doi: 10.1016/0076-6879(79)68009-6. [DOI] [PubMed] [Google Scholar]
- Rüther U., Koenen M., Otto K., Müller-Hill B. pUR222, a vector for cloning and rapid chemical sequencing of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4087–4098. doi: 10.1093/nar/9.16.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier P. H., Bothwell A. L., Mueller-Hill B., Baltimore D. Multiple differences between the nucleic acid sequences of the IgG2aa and IgG2ab alleles of the mouse. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4495–4499. doi: 10.1073/pnas.78.7.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]