Summary
Background
Sudden Infant Death Syndrome (SIDS) is a leading cause of postneonatal mortality. The absence of specific biomarkers of SIDS diagnosis and risk leaves a significant gap in understanding SIDS pathophysiology. Metabolomics offers an avenue to better understand SIDS biology and identifying potential biomarkers.
Methods
Using Metabolon Inc., global discovery panel, we analysed 828 metabolites from post-mortem serum samples of infants from the Chicago Infant Mortality Study (CIMS) and the NIH NeuroBioBank (NBB). In total, 300 infants (195 SIDS; 105 non-SIDS) across multiple race/ethnicities (70% Black, 13% White, and 16% Hispanic) were included. Metabolite associations with SIDS were performed using Welch’s t-tests, linear and logistic regression, and network-cluster analyses.
Findings
We identified thirty-five significant metabolite predictors of SIDS after adjustment for age, sex, race and ethnicity, and post-mortem interval, including ornithine (OR 21.98; p-value 6.44e-7), 5-hydroxylysine (OR 19.48; p-value 6.78e-7), 1-stearoyl-2-linoleoyl-GPC (18:0/18:2) (OR 16.80; p-value 3.4e-7), ribitol (OR 8.19; p-value 4.2e-8), and arabitol/xylitol. Using Weighted Gene Co-expression Network Analysis (WGCNA), ten metabolite clusters were identified. Four exhibited significant associations with SIDS. The two most correlated clusters were enriched for metabolites in the tyrosine metabolism pathway and lipid (sphingomyelins) pathways.
Interpretation
We identified metabolite biomarkers within key biological pathways and processes (e.g., nitrogen metabolism, lipid and fatty acid metabolism, stress response, nerve cell communication, hormone regulation, oxidative stress) with potential implications in SIDS pathology. Further research is needed to validate these biomarkers in additional SIDS cohorts.
Funding
The Chicago Infant Mortality Study was funded by Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Institute on Deafness and Other Communication Disorders under contract number NO1-HD-3-3188, the Centers for Disease Control and Prevention and the Association of Teachers of Preventive Medicine under cooperative agreement number U50/CCU300860-06, and the Playmates in Heaven Foundation. The current analyses were funded by Eunice Kennedy Shriver National Institute of Child Health and Human Development under 5R01HD101518-04.
Keywords: Metabolomics, Biomarkers, Sudden infant death syndrome
Research in context.
Evidence before this study
Sudden Infant Death Syndrome (SIDS) is a significant cause of infant mortality, with its pathophysiology poorly understood despite the existence of the Triple Risk Model. Previous research has suggested metabolic disruptions in SIDS cases, but specific biomarkers remained unidentified. We searched PubMed for studies on metabolomics and SIDS, finding that while several studies proposed potential metabolic pathways associated with SIDS, conclusive biomarkers for prediction and diagnosis were lacking.
Added value of this study
This research significantly contributes to our understanding of SIDS by employing a comprehensive metabolomic analysis, leveraging advanced liquid chromatography-mass spectrometry to analyse post-mortem serum samples from infants diagnosed with SIDS and other causes of sudden death (non-SIDS). This study identifies potential metabolite biomarkers and clarifies their roles within critical biological pathways, such as lipid and fatty acid metabolism, stress response, and oxidative stress, directly associated with SIDS. This study applies a comprehensive metabolic framework to SIDS, offering new avenues for diagnosis and prevention.
Implications of all the available evidence
The findings from this study, combined with existing research, underscore the potential of metabolomics as a powerful tool in unravelling the complex aetiology of SIDS. By identifying specific metabolites and pathways, this research provides foundational knowledge to develop predictive biomarkers and preventive strategies. This paves the way for increased use of metabolomic screening in the clinical investigation of SIDS, highlighting the need for further validation studies and the potential for clinical application.
Introduction
Sudden Infant Death Syndrome (SIDS), the unexplained demise of an apparently healthy infant during sleep, remains a leading cause of postneonatal infant mortality worldwide.1 SIDS is characterized as a sudden, unexpected infant death (SUID) that remains unexplained after the autopsy, investigation of the death scene, and review of the medical history. Despite extensive research, SIDS continues to be an enigmatic condition, largely due to its multifactorial nature and the absence of clear, identifiable precursors.2 The Triple Risk Model,3 proposing an intersection of a vulnerable infant, a critical developmental period, and external stressors, provides a theoretical framework for understanding SIDS.4 However, this model lacks specific biomarkers to assist with SIDS diagnosis and risk, leaving a significant gap in understanding SIDS pathophysiology.5
The advent of metabolomics, with its capacity to systematically characterize small-molecule metabolites in biological systems, presents a promising avenue for advancing our understanding of SIDS.6 Global, unbiased metabolomics association studies have emerged as powerful tools in elucidating complex disease aetiologies, offering insights into underlying metabolic disruptions and potential diagnostic markers.7 In SIDS research, metabolomics can potentially identify unique metabolic signatures that differentiate infants at risk from healthy controls, providing a window into the physiological state preceding the tragic event.8 There is prior work using metabolomic data from brain9,10 and colon11 tissues attempting to discovery metabolic predictors of SIDS diagnosis. These studies suffer from small sample sizes of SIDS cases (n = 16) and controls (n = 7 to 13). They used machine learning approaches, like LASSO regression, to discovery metabolic predictors, but have not validated their findings in independent cohorts.
This study aims to leverage the sensitivity and comprehensiveness of metabolomics to explore metabolomic profiles in SIDS cases compared to non-SIDS controls in a much larger data set using samples from the Chicago Infant Mortality Study (CIMS) and the NIH NeuroBioBank (NBB). By conducting a metabolomics association study, we intend to uncover metabolic pathways and biomarkers that are distinct in SIDS, thereby contributing to the discovery of potential diagnostic biomarkers for SIDS.
Methods
Study design
This study is a retrospective, metabolomic-wide association study of infants who died with a SIDS diagnosis versus non-SIDS diagnosis. Clinical data and serum samples from two independent cohorts were included to assess the metabolite-by-metabolite associations with SIDS diagnosis.
Cohorts
Chicago Infant mortality study (CIMS)
The Chicago Infant Mortality Study (CIMS) was conducted between November 1993 and April 1996, and aimed to understand risk factors for SIDS, particularly among a high-risk, urban African American population. CIMS infants were all residents of Chicago and their autopsies were handled by the Office of the Medical Examiner of Cook County. Infants included were from birth to 1 year old for those who died from SIDS and 1 month to 1 year for infants who died from other causes. Infants for study inclusion were identified via a thorough case investigation encompassing a complete autopsy, examination of the death scene, and review of the clinical history. Tissue samples and other relevant data were collected through standardized protocols for autopsy, medical record review, and a detailed death scene investigation, which included an extensive questionnaire about the infant's and family's medical history, sleep environment, and circumstances leading to the infant's death; prior works provide more details of the methodology.12, 13, 14, 15
To achieve consensus on SIDS diagnosis, the cause of death for each infant was determined by the responsible medical examiner with input from the others. Additionally, one in four cases were reviewed by a blinded external review team, which included a forensic pathologist and paediatric pathologist with expertise in sudden infant death. Any difference in diagnosis between the medical examiner and the external review team was further discussed by a multidisciplinary committee to establish SIDS diagnosis by consensus.
For biospecimen collection, study technicians were present during the autopsies to handle the specimens; including blood, vitreous humour, urine swabs, liver, and heart. Specimens were frozen and sent in batches with dry ice to the University of Maryland Brain and Tissue Bank for Developmental Disorders within 48 h of time of death. All tissues, including serum, were stored at −80 °C, and there was no report of power failure or other compromise to the tissues.
NIH NeuroBioBank
The NIH NeuroBioBank (NBB) was established in September 2013 as a national resource for investigators utilizing human post-mortem brain tissue and related biospecimens for research to understand conditions of the nervous system (http://www.neurobiobank.nih.gov). The NBB receives donations from individuals who register before death and from next-of-kin who authorize a post-mortem donation. Brain and other tissues are eligible for post-mortem donation based on an assessment from a trained Brain and Tissue Repositories (BTR) staff member. All BTR sites had their policies and procedures reviewed and approved by their respective Institutional Review Boards. All donated biospecimens are collected within 24 h of death and prepared to have the widest use for research. Donated/extracted serum samples were flash frozen at −150 °C and stored at −80 Celsius. Serum samples were stored in glass tubes.
The cause of death determination for the NBB samples and the definition of SIDS is unknown because these cases were sent to the NBB from various medical examiners who used different definitions and criteria for death determination. To achieve consistency between NBB and CIMS, we requested serum from NBB infants who had a diagnosis of SIDS under age 1 and other causes of death ages 1 month to 1 year to match our study age criteria. Dr. Hauck and study coordinators reviewed all NBB infant deaths, including assigned cause of death and available notes, and adjudicated each cause of death using the SIDS definition: the sudden and unexpected death of an infant less than twelve months of age that remained unexplained after a review of the clinical history, a complete autopsy, and a death scene investigation.16 For infants with a diagnosis of possible SIDS or SUID, those infants were assigned SIDS as the cause of death, while infants with a diagnosis as possible or probable asphyxia were assigned asphyxia as the cause of death.
Due to small numbers, causes of death other than SIDS are summarized by combining CIMS and NBB and grouping into ten broader categories (genetic disorders, cardiac disorders, dehydration/malnutrition, accident/injury, infection, respiratory (including hypoxia), asphyxia/suffocation, prematurity/developmental, neurological (including seizures), and undetermined). Nearly one-quarter of the non-SIDS causes of death were due to infection (including pneumonia, sepsis, and other infections), while ∼19% were due to asphyxia/suffocation, ∼14% due to accident/injury, ∼12% undetermined, and ∼10% due to cardiac disorders (including congenital heart disease, hypertrophy, and myocarditis). Respiratory conditions accounted for ∼7% of causes of death, with the remaining causes (genetic, dehydration/malnutrition, prematurity/developmental, and neurological) accounting for less than 5% of deaths, respectively.
Ethics
The Chicago Infant Mortality Study (CIMS) was approved by a steering committee consisting of 14 representatives from participating institutions and other experts on infant mortality and urban minority health, with additional approval by the Chicago Department of Public Health and Office of the Cook County Medical Examiner. Protocols conducted through the Medical Examiner's Office (autopsy, death scene investigation, and medical record review) were routine parts of the investigation of sudden infant death and did not require informed consent. Informed consent was obtained from deceased infant family participants for follow-up interviews.12 The CIMS protocol was approved by the institutional review boards of the Loyola University Medical Center and University of Virginia.
The NIH-funded NeuroBioBank (NBB) was established as a national resource for investigators utilizing human post-mortem brain tissue and related biospecimens for their research to understand conditions of the nervous system. All NBB samples were procured, stored, and distributed according to applicable state and federal guidelines and regulations involving consent, protection of human subjects and donor anonymity.
Metabolites
Sample storage, preparation and data extraction
Post-mortem serum samples from CIMS and NBB were stored at −80 °C, and 200 uL aliquots were shipped to Metabolon, Inc. on dry ice. Global, untargeted metabolite profiles were generated via liquid chromatography-mass spectrometry using the DiscoveryHD4 platform at Metabolon, Inc., for 1237 metabolites, as previously described.17 In-house quality control protocols were utilized by Metabolon to account for instrument and sample variability.
Compound identification and curation
Raw data were extracted, peak-identified, and QC processed using Metabolon’s hardware and software. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Metabolon maintains a library based on authenticated standards that contain the retention time/index (RI), the mass-to-charge ratio (m/z), and chromatographic data (including MS/MS spectral data) on all molecules present in the library. Furthermore, biochemical identifications are based on three criteria: retention index within a narrow RI window of the proposed identification, accurate mass match to the library ± 10 ppm, and the MS/MS forward and reverse scores between the experimental data and authentic standards.
Curation procedures were also performed internally by Metabolon using proprietary visualization and interpretation software. This included checks of consistency of peak identification in the samples and compound library matches. Peaks were quantified using area-under-the-curve. Batch normalized metabolite data were used for subsequent analyses. For each metabolite, the raw values in the experimental samples were divided by the median of those samples in each instrument batch, giving each batch and metabolite a median of one. The Batch-normalized data reflect median-scaled raw data.
Statistical analysis
Pre-analysis QC
We examined the missing data pattern of the metabolites prior to imputation between SIDS cases and non-SIDS controls. Across serum samples, we found no significant relationship between the amount of missing metabolite values and the pathways in which they belong. Additionally, there was no difference (p = 0.2) in the proportion of missing metabolites within samples in SIDS cases median (IQR) 0.21 (0.19, 0.24) and non-SIDS controls 0.22 (0.19, 0.25). Because there is no relationship of missing due to metabolic pathway or SIDS diagnosis, we concluded that the left censoring of metabolite values due to the limits of detection are non-biasing. A common approach to address left-censoring is to impute missing values with a value less than the minimal observed value for that specific metabolite. We utilized half of the minimum observed value. We excluded any metabolite with greater than twenty percent missing values from all analyses. We chose this threshold to minimize the impact of data missingness on metabolite-wide associations in our primary and downstream analyses.
We conducted additional quality control to investigate batch effects and cohort differences between CIMS and NBB samples. We pruned highly correlated metabolites (r > 0.5) and then calculated the principal components of the remaining metabolites. We investigated evidence of clustering between cohort, age quartiles, sex, and quality control sample duplicates.
Primary metabolite-wide association analysis
Differences in metabolite levels between SIDS cases and non-SIDS controls
To identify metabolites with differences in normalized values between SIDS cases and non-SIDS controls, we used Welch’s t-test of unequal variances to estimate the degree of difference and its significance. We then conducted a linear regression adjusting for age in days, sex, race and ethnicity, post mortem interval (PMI) in hours, and (CIMS or NBB) cohort. To investigate heterogeneity between cohorts, we performed stratified analyses by cohort for both the Welch’s t-test and linear regression approaches. Significant associations were defined at the Bonferroni adjusted p-value (0.05/828 = 6e-5). We also performed stratified analyses using the CIMS only and NBB only data for both Welch’s t-tests and linear regression.
Metabolite predictors of SIDS cases
In order to find potential metabolite predictors of SIDS, we utilized logistic regression adjusting for important SIDS confounding variables. We estimated the odds of SIDS diagnosis by each metabolite independently adjusting for age in days, sex, race, ethnicity, PMI in hours, and cohort. Significant associations were defined at the Bonferroni adjusted p-value (0.05/828 = 6e-5).
Metabolite co-expression network analysis
To address metabolite correlation, we adapted the Weighted Gene Co-expression Network Analysis (WGCNA) R package18,19 to cluster metabolites together based on their correlations with each other. WGCNA uses an unbiased approach to cluster data based on similarity of expression patterns. We visualized several power thresholds for proper network algorithm optimization, which resulted in a power value of six. We set the minimum module size to 20 and selected signed Topological Overlap Matrix and Pearson correlation parameters. After clustering metabolites into modules (i.e., groups) and generating their representative eigen vectors, we investigated their Module Trait Relationships among SIDS diagnosis, sex, race and ethnicity, age in days, birth weight in grams, bed sharing, PMI in hours, and gestational age in weeks, separately.
Metabolite SIDS subgroup sensitivity analysis
In addition to SIDS case–control analyses, we explored differences in metabolite levels in SIDS cases with and without known SIDS risk factors: bed sharing, maternal smoking, and maternal drug or smoking, from the CIMS cohort using Welch’s t-tests. Analyses were performed for metabolites associated with SIDS diagnosis from the logistic regression analysis with a p-value less than 6e-5. The purpose was to determine if metabolites levels differed in SIDS cases by exposure level.
Role of funders
The Funders of this study had no role in study design, data collection, interpretation, or writing of this report.
Results
Demographics
A total of 300 unique individuals were included in downstream analyses composed of 266 infants from CIMS and 34 infants from NBB; see Table 1. We observed a mean difference in age of −37.4 days (95% CI −54.6 to −20.2; p < 0.001) between SIDS cases and non-SIDS controls (Median IQR 81 [54–125] days and 112 [74–181] days, respectively; p < 0.001). As expected, there were more males than females in both SIDS cases, and non-SIDS controls, and bed sharing had a greater prevalence in SIDS cases (Table 1). Race and ethnicity, PMI, maternal smoking, soft surface, and found on a pillow were balanced between SIDS cases and non-SIDS controls. PMI, specifically, had a median (IQR) of 25 h (Q1 20, Q3 28).
Table 1.
Demographics by cohort.
| Characteristic | CIMS |
NBB |
Combined |
||||
|---|---|---|---|---|---|---|---|
| SIDS Case N = 170 | Non-SIDS Control N = 96 | Overall N = 266 | SIDS Case N = 25 | Non-SIDS Control N = 9 | Overall N = 34 | Overall N = 300 | |
| Cohort n (%) | 170 (64%) | 96 (36%) | 266 | 25 (74%) | 9 (26%) | 34 | 300 |
| Age in days, Median (Q1,Q3) | 78 (50, 118) | 110 (73, 181) | 86 (58, 142) | 106 (88, 142) | 121 (99, 201) | 114 (88, 142) | 90 (61, 142) |
| Sex, n (%) | |||||||
| Male | 100 (59%) | 53 (55%) | 153 (58%) | 15 (63%) | 6 (67%) | 21 (64%) | 174 (58%) |
| Female | 70 (41%) | 43 (45%) | 113 (42%) | 9 (38%) | 3 (33%) | 12 (36%) | 125 (42%) |
| Unknown | 1 | – | 1 | 1 | |||
| Race/Ethnicity, n (%) | |||||||
| Black | 125 (74%) | 71 (74%) | 196 (74%) | 10 (40%) | 4 (44%) | 14 (41%) | 210 (70%) |
| Hispanic | 29 (17%) | 19 (20%) | 48 (18%) | – | – | – | 48 (16%) |
| White | 15 (8.8%) | 6 (6.3%) | 21 (7.9%) | 12 (48%) | 5 (56%) | 17 (50%) | 38 (13%) |
| Other | 1 (0.6%) | – | 1 (0.4%) | 3 (12%) | – | 3 (8.8%) | 4 (1.3%) |
| PMI, Median (Q1,Q3) | 25 (21, 28) | 24 (19, 28) | 25 (20, 28) | 25 (21, 29) | 21 (15, 27) | 25 (18, 29) | 25 (20, 28) |
| Unknown | 13 | 10 | 23 | 23 | |||
| Maternal Smoking, n (%) | 60 (35%) | 27 (28%) | 87 (33%) | – | – | – | 87 (33%) |
| Unknown | – | 1 | 1 | 25 | 9 | 34 | 35 |
| Bed Sharing, n (%) | 79 (48%) | 25 (32%) | 104 (43%) | – | – | – | 104 (43%) |
| Unknown | 4 | 18 | 22 | 25 | 9 | 34 | 56 |
| Soft Surface, n (%) | 78 (47%) | 33 (49%) | 111 (48%) | – | – | – | 111 (48%) |
| Unknown | 5 | 28 | 33 | 25 | 9 | 34 | 67 |
| Found on a pillow, n (%) | 46 (28%) | 22 (30%) | 68 (28%) | – | – | – | 68 (28%) |
| Unknown | 4 | 23 | 27 | 25 | 9 | 34 | 61 |
Metabolite-wide associations with SIDS diagnosis
We excluded metabolites with greater than 20% missing values, resulting in 828 metabolites subject to imputation and included in subsequent data analyses.
Primary results
Differential metabolite levels by SIDS diagnosis
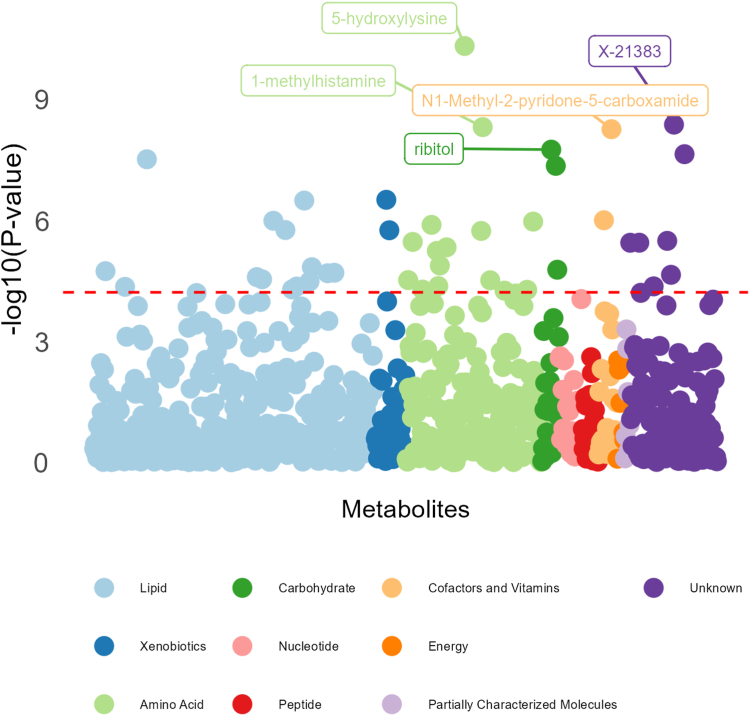
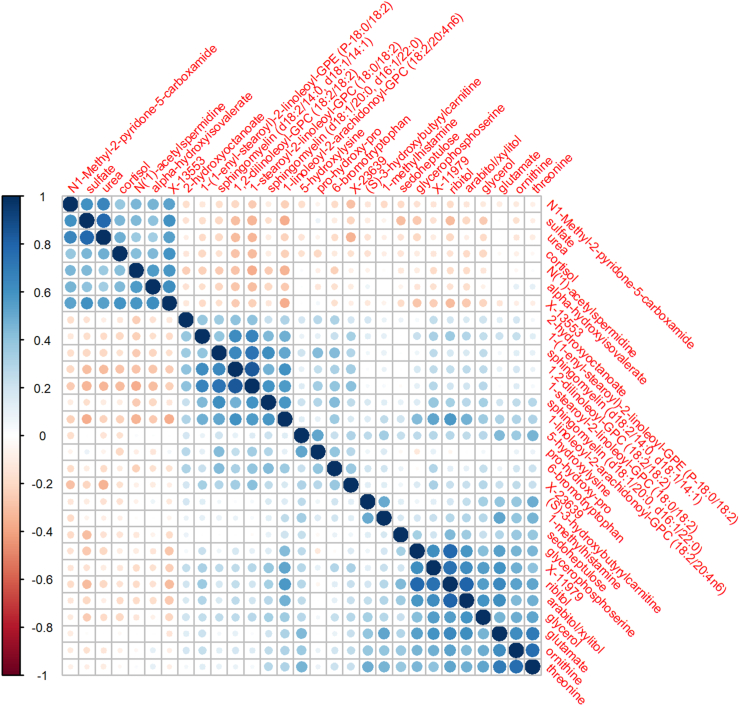
Forty-three of the 828 metabolites had significant differences between SIDS cases and non-SIDS controls after Bonferroni adjustment (0.05/828 = 6e-5), based on Welch’s t-test; Fig. 1. Supplemental Table S1 lists these significant metabolites, their statistics, and biological annotations. Twenty-nine of which were also significant after adjustment for age in days, sex, race and ethnicity, PMI, and cohort via linear regression analysis (Supplemental Table S2). Fig. 2 shows the metabolite–metabolite correlation and clustering of these twenty-nine metabolites.
Fig. 1.
CIMS and NBB combined Metabolite-wide associations between SIDS cases and non-SIDS controls from the Welch’s t-test. Legend: The red line represents the Bonferroni Correction p-value threshold for significance (0.05/828 = 6.04e-5) on the −log10 scale, which should be interpreted as any dot above the red line has a significant association with SIDS diagnosis.
Fig. 2.
Metabolite-by-Metabolite correlations of significantly associated candidate biomarkers. Legend: The figure is a correlation matrix of the twenty-nine significant metabolites from the Welch’s t-test that were also significant in linear regression analyses adjusting for confounders. The size of the circles and shade of colour represent the strength of the correlation and the gradient from red to blue shows the direction of the association from −1 to +1 as seen in the legend on the left side of the correlation matrix.
Metabolites as a biomarker of SIDS diagnosis
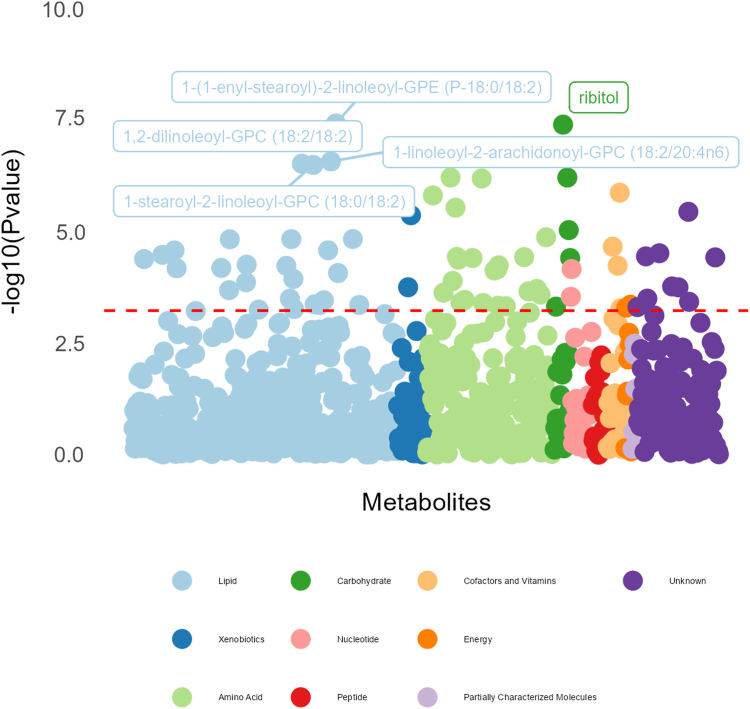
Subsequent logistic regression analyses, testing for metabolite predictors of SIDS diagnosis, identified thirty-five significantly associated metabolites after Bonferroni correction; see Supplemental Table S3 and Fig. 3. Of the thirty-five significantly associated metabolites, the five most significant associations were observed for ornithine (OR 21.98; p-value 6.4e-7), 5-hydroxylysine (OR 19.48; p-value 6.8e-7), 1-stearoyl-2-linoleoyl-GPC (18:0/18:2) (OR 16.80; p-value 3.4e-7), ribitol (OR 8.19; p-value 4.2e-8), and arabitol/xylitol (OR 7.70; p-value 9.6e-6). Twenty-six metabolites displayed consistent significant associations with metabolites identified from the combined Welch’s t-test analysis.
Fig. 3.
Metabolite-wide associations as predictors of SIDS diagnosis via logistic regression models. Legend: The red line represents the Bonferroni Correction p-value threshold for significance on the −log10 scale, which should be interpreted as any dot above the red line has a significant association with SIDS diagnosis. The logistic regression adjusted for age in days, sex, race and ethnicity, post-mortem interval (time from death to autopsy), maternal smoking exposure, and cohort.
Metabolite co-expression network analysis
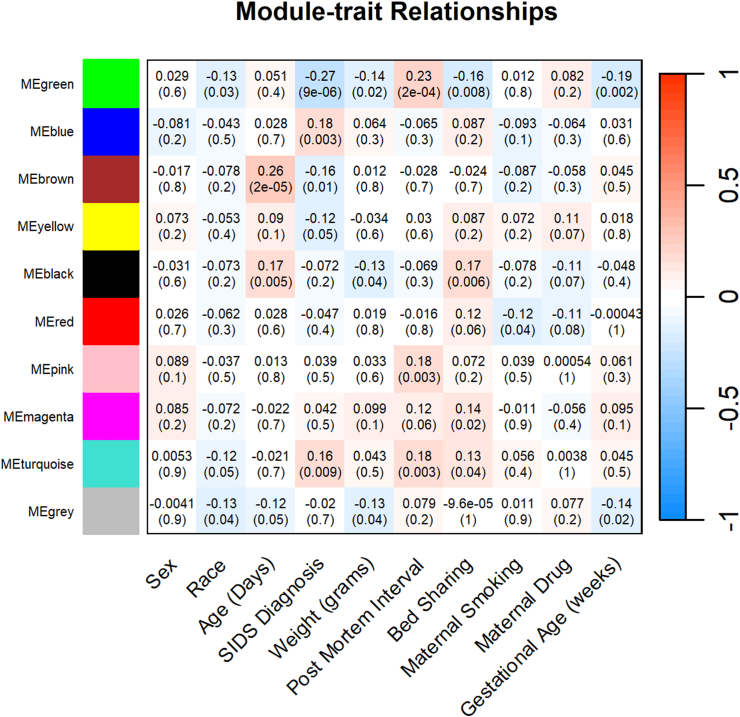
WGCNA cluster analyses resulted in ten different clustered groups (modules); Fig. 4. Each cluster is determined based on pairwise correlation of all metabolite values, with similarly expressed patterns grouped into modules. Correlation coefficients (r) between modules and phenotype data can be calculated to identify modules highly correlated with specific phenotypes. Fig. 4 shows the correlation coefficients and corresponding p-values for the 10 clusters/modules and SIDS diagnosis and other relevant SIDS phenotypes. Four clusters had significant correlations with SIDS diagnosis: MEgreen (51 metabolites, r = −0.27, p = 9e-6), MEblue (89 metabolites, r = 0.18, p = 0.003), MEbrown (88 metabolites, r = −0.16, p = 0.01), and MEturquoise (98 metabolites, r = 0.16, p = 0.009) (Supplemental Table S4). For those module correlations, positive r values indicate that SIDS diagnosis likelihood increases, while negative r values indicate that the likelihood of SIDS diagnosis decreases. MEgreen consisted of fifty-one metabolites where 43.1% were in the amino acid super pathway and 25.5% had an unknown associated super pathway. Furthermore, 15.75% of metabolites in the MEgreen cluster were related to Tyrosine metabolism. In relation to other SIDS risk factors, the MEgreen cluster had the same direction of association with race, birth weight in grams, bed sharing, and gestational age. In contrast, this cluster had an association with post-mortem interval in the opposite direction. Eighty-nine metabolites comprised the MEblue cluster, which 84.3% were lipids. Of the lipid metabolites, 37.3% were Sphingomyelins. Unlike the MEgreen cluster, the MEblue cluster appears to be uniquely associated with SIDS and not with other known risk factors. Similar to the MEblue cluster, the MEbrown cluster had a large proportion (55.7%, n = 49) of metabolites in the lipid super pathway, with the majority of those metabolites (n = 36) within the fatty acid metabolism/Acyl Carnitine sub pathway. The MEbrown metabolites were negatively associated with SIDS, but positively associated with age in days, while the MEturquiose cluster (n = 98 metabolites) had a positive association with post-mortem interval and bed sharing as well as SIDS, and had a large proportion of metabolites within the amino acid super pathway (52%).
Fig. 4.
Correlations of SIDS diagnosis and risk factors among Metabolite Clusters. Legend: This module-trait heatmap represents the correlations of the module eigengenes (ME) with various SIDS risk factors (sex, race and ethnicity, age in days, weight at birth in grams, post mortem interval (in hours), bed sharing, maternal smoking exposure, maternal drug exposure, and gestational age in weeks. Each module (cluster group) is depicted as a colour name (green, blue, brown, etc.) on the Y axis, with corresponding correlation coefficient and p-value for each trait (X axis) per cell. Colour intensities indicate significance (darker shade (intensity) indicating a more significant p-value; stronger correlation) with red indicating a positive correlation, white indicating no correlation, and blue indicating a negative correlation.
Metabolite SIDS subgroup sensitivity analysis
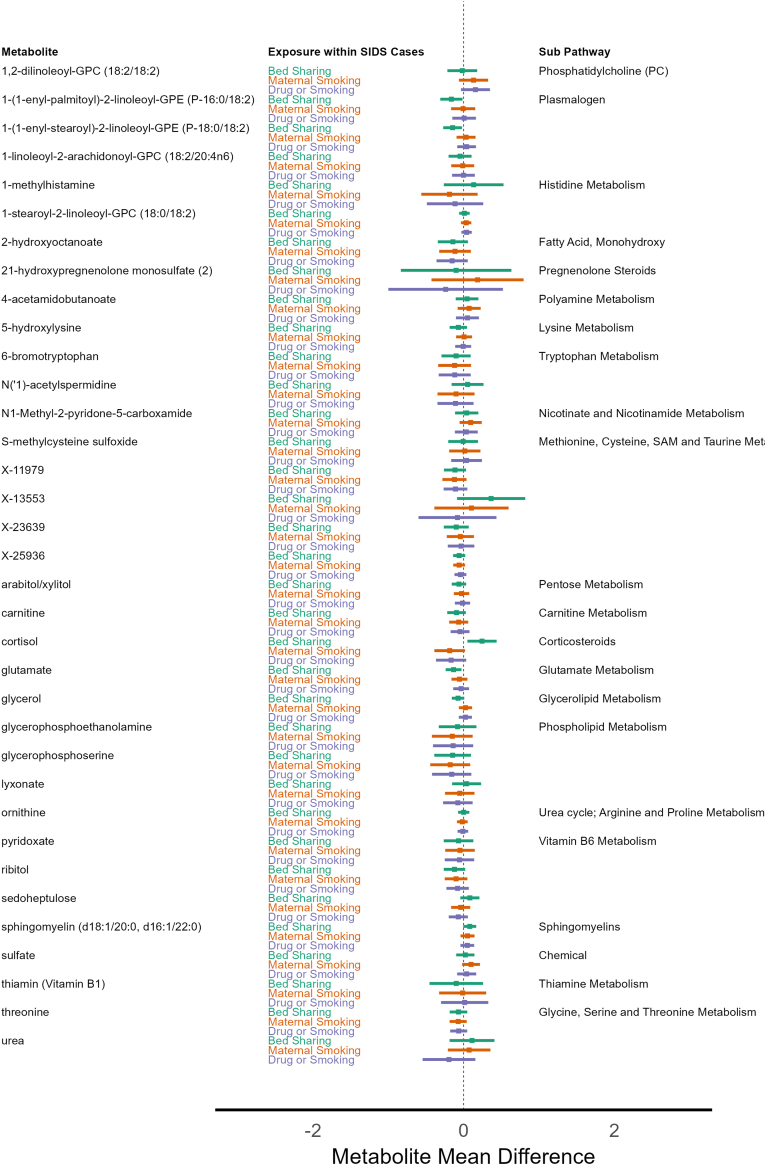
For exploration, we investigated if metabolites differed within SIDS cases from the CIMS cohort. We used the significant metabolites identified from the logistic regression models, then compared differences between those with and without known SIDS environmental risk factors: 1) bed sharing, 2) maternal smoking exposure, and 3) any maternal drug or smoking exposure. For bed sharing, four metabolites had mean differences with 95% confidence intervals that did not encompass zero. They were cortisol [0.24 95% CI 0.05, 0.44], glutamate [−0.13, 95% CI −0.24, −0.03], 1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (P-16:0/18:2) [−0.16, 95% CI −0.31, −0.02], and 1-(1-enyl-stearoyl)-2-linoleoyl-GPE (P-18:0/18:2) [−0.15 95% CI −0.27, −0.02]. Maternal smoking exposure was associated with one unknown metabolite (X-25790) [0.23 95% CI 0.01, 0.46], while no metabolites were associated with any maternal drug or smoking exposure. Fig. 5 shows the thirty-five metabolome-wide significant metabolites from the case versus control logistic regression analysis and their mean differences among each SIDS risk factor exposure, while Supplemental Table S5 shows the estimates for all metabolites.
Fig. 5.
Metabolite differences by SIDS risk factor exposures among SIDS Cases from the CIMS Cohort. Legend: Estimated metabolite mean group differences are depicted for each of the thirty-five significantly associated metabolites from the logistic regression analysis (left column) for three known SIDS environmental exposure risk factors: Bed Sharing (Yes/No, Green), Maternal Smoking Exposure (Yes/No, Orange), and Any Maternal Drug or Smoking Exposure (Yes/No, Purple) within CIMS SIDS cases only. Points greater than zero indicate mean metabolite values that are higher in SIDS cases, and mean metabolite values less than zero indicate lower levels in SIDS cases for the corresponding risk factor.
Discussion
SIDS remains a leading cause of postneonatal infant mortality worldwide.1 There is a significant need to have diagnostic biomarkers for SIDS to assist with accurate diagnosis as well as to provide insights into myriad causal candidates from the Triple Risk Model.3 Thus, we performed a large-scale metabolomics analysis of 828 metabolites to investigate unique metabolite signatures of a SIDS diagnosis and known SIDS risk factors. The study populations reflect the age and sex distributions of prior SIDS literature.20, 21, 22 However, the individuals include mostly infants identified as Black (70%).
Twenty-nine metabolites had significant differential levels in serum between SIDS cases and non-SIDS controls even after adjustment for confounding (Supplemental Table S2). We also observed thirty-five metabolites as significant predictors of SIDS diagnosis, after adjustment for confounders (Supplemental Table S3). The top five most predictive metabolites of SIDS diagnosis that overlap Supplemental Tables S2 and S3 are: 1) ornithine, 2) 5-hydroxylysine, 3) 1-stearoyl-2-linoleoyl-GPC (18:0/18:2), 4) ribitol, and 5) arabitol/xylitol.
Related to the urea cycle, ornithine (OR 21.98; 95% CI 6.51–74.23) is strongly associated with SIDS diagnosis. Ornithine plays a critical role in the disposal of ammonia. Ammonia (nitrogen) metabolism has been previously linked to SIDS risk,23,24 with implications of a hepatic metabolic profile in SIDS.25 Ornithine activates urea synthesis via the Ornithine Transcarbamylase (OTC) enzyme, which turns ammonia into urea within the kidneys prior to excretion.26 Interestingly, the OTC gene is located on the X-chromosome (Xp21.1) and its deficiency results in high levels of ornithine and ammonia,26 which is consistent with the direction of association for ornithine in this study. Although no consensus has been reached regarding the correlations or causality between OTC polymorphisms and SIDS, multiple case studies report the cooccurrence of OTC mutations and SIDS diagnoses.27, 28, 29 Furthermore, high levels of ornithine also occur when there is excessive lysine. The metabolite 5-hydroxylysine (OR 19.49; 95% CI 6.04–62.93), an amino acid derivative of lysine,30 is moderately correlated with ornithine (Fig. 2) and has a similar SIDS predictive strength as ornithine. However, it is not directly involved in urea metabolism, but collagen formation. Levels of 5-hydroxylysine can increase with the degradation of collagen post-mortem.31 Reassuringly, the PMI time did not differ between SIDS cases and non-SIDS controls with a mean difference of −0.27 min (95% CI −2.93 to 2.38). On the other hand, this does not rule out the potential confounding of the time death occurred to the discovery of the infant.
The third candidate metabolite biomarker is 1-stearoyl-2-linoleoyl-GPC (18:0/18:2) (OR 16.8; 95% CI 5.69–49.69). This metabolite contains linoleic acid (“2-linoleoyl”); an essential fatty acid obtained only through diet. Additionally, this metabolite is a phosphatidylcholine, which is integral for cerebral cortex lipid structure and neuronal membrane stability during development.32 This metabolite, also known as phosphatidylcholine diacyl C36:2, is a potential biomarker of foetal congenital heart defects during the first trimester.33
The last of the top five candidate metabolite biomarkers, ribitol and arabitol/xylitol, are positively correlated with each other (Fig. 2) and are both part of the same pentose interconversion and phosphate pathways (KEGG pathway C00379). In this study, we observed the same direction of SIDS prediction with each unit increase in ribitol levels having an increase of the odds ratio by 8.19 times (95% CI 3.86–17.37; p-value 4.2e-8). Ribitol is a normal by-product in urine.34 However, elevated levels of ribitol and D-arabitol (derivative of xylitol) in serum or urine can be indicative of transaldolase deficiency, which affects the pentose phosphate pathway leading to liver cirrhosis in children.35 The pentose phosphate pathway is an important metabolic pathway that functions as an alternate pathway to glycolysis36,37 and plays a critical role in minimizing oxidative stress through the production of NADPH. It is widely acknowledged that impaired energy metabolism is implicated in SIDS,38 and recent animal studies suggest that ribitol therapy reduces skeletal muscle pathology, cardiac fibrosis, and improved respiratory function in a muscular dystrophy model.39
In addition to individual metabolite associations, WGCNA analyses identified several metabolite clusters associated with SIDS and related phenotypes, with the two most correlated clusters enriched for metabolites in the tyrosine metabolism (MEgreen) and lipid (sphingomyelins) pathways (MEblue). For the MEgreen cluster associated with SIDS diagnosis (r = −0.27, p = 9e-6), eight of the 51 metabolites (15.75%) in this cluster were related to tyrosine metabolism (homovanillate, vanillylmandelate, N-acetyltyrosine, phenol sulphate, homovanillate sulphate, 3-methoxytyramine sulphate, vanillactate, and dopamine 3-O-sulfate). Tyrosine, a nonessential amino acid, is a precursor for hormones and neurotransmitters such as catecholamines (e.g., dopamine, norepinephrine, and epinephrine) that are important for stress response,40 nerve cell communication,41 and hormone regulation. Tyrosine has also been shown to reduce physiological arousal,42 while defects in arousal have long been suspected to contribute to SIDS.43,44 Furthermore, polymorphisms in genes within the tyrosine metabolism pathway, in particular the tyrosine hydroxylase gene, have shown previous associations with SIDS.45,46 Our WGCNA findings further support a role for tyrosine metabolism in SIDS.
The MEblue cluster was solely associated with SIDS diagnosis and no other known SIDS risk factors and was enriched for lipid metabolites. Sphingomyelins represent a specific type of sphingolipid that has many important functions and roles that are relevant to SIDS. Sphingolipids are critical for brain development and sphingomyelin, specifically, is a major component of the myelin sheath that surrounds nerve axons. While causality has not been established, the role of myelination in SIDS has long been postulated.47 In addition to brain development and function, sphingomyelins are important for lung function, as a component of surfactant. Surfactant itself has ongoing interest as a biomarker of SIDS.48, 49, 50, 51, 52 However, our sphingomyelins of interest play a large role in cell signalling via lipid rafts,53 which initiate neural signalling and are highly relevant to the development and function of neural circuits. Even though phosphatidylcholines have distinct functions from sphingomyelins, they were highly correlated with sphingomyelins and clustered together by WGCNA. Similar to sphingomyelins, phosphatidylcholines have links to surfactant abnormalities in SIDS,48,49,53 as well as a known component of cerebral cortex neuronal membrane phospholipids.32 Although not found in the MEblue cluster, 1-oleoyl-2-linoleoyl-GPE (18:1/18:2) was associated with SIDS (OR 2.105 95% CI 1.39–3.19), which is a lipid part of the phosphatidylethanolamine sub pathway. In an animal study,54 a choline-deficient diet was associated with decreased linoleic acids in liver phosphatidylethanolamine, which plays a role in pulmonary surfactants. The authors showed that inositol supplements altered the composition of the surfactant phospholipids and reduced the need for oxygen therapy.
Another metabolite in MEblue, succinate has high biological relevance, because it is a component of the tricarboxylic acid (TCA, or citric acid) cycle. The TCA cycle has previously been implicated in SIDS pathology over twenty years ago.55 A more recent study demonstrated succinate as a potential biomarker of SIDS versus death from other causes in an age- and sex-matched cohort using brain tissue from the NBB.8 In our current study, we found succinate to have a moderate to large odds ratio of 6.73 (95% CI 2.20–16.90; p-value 5.20e-4) for SIDS diagnosis, but it did not reach our Bonferroni corrected threshold of 6e-5. One reason for the lack of replication of succinate as a biomarker of SIDS may be the difference in tissue types. The metabolomics data from the current study were generated from serum instead of brain tissue. This is an important distinction since succinate levels are affected by asphyxiation.56 It is possible that succinate levels in the brain tissue are more sensitive due to asphyxiation than those measured from serum.
Similar to prior work, we observed many amino acids and short chain fatty acids having associations with SIDS57, 58, 59 even after adjusting for confounders. The WGCNA analysis revealed an enrichment for two clusters (MEgreen and MEturquoise) that show a high prevalence of amino acids. Interestingly, these clusters have opposing correlation coefficients with similar effect sizes (−0.23 versus 0.18); Fig. 3. These clusters also correlate with the PMI and the bed sharing risk factor for SIDS. In addition, the MEbrown cluster was enriched for metabolites in the fatty acid metabolism/Acyl Carnitine sub pathway. Fatty acid oxidation disorders have long been suspected as primary causes of SIDS.60,61
The CIMS cohort measured several important confounders and SIDS risk factors. One is maternal prenatal smoking exposure, which has great interest62 as a SIDS risk factor. Our second strongest metabolite-SIDS association was for N1-Methyl-2-pyridone-5-carboxamide (2PY). 2PY is involved in niacin metabolism and is a known uremic toxin.63,64 Smoking exposure is a determinant of this metabolite.64 This is a unique finding as no prior work on 2PY in SIDS exists. We further investigated differences in metabolite levels, in infants who died from SIDS in the CIMS cohort, for known SIDS environmental risk factors: 1) bed sharing, 2) maternal smoking exposure, and 3) any maternal drug or smoking exposure (Fig. 5). Notably, cortisol, a human stress hormone, showed differences in CIMS infants who died of SIDS who were bed sharing versus those who were exposed to smoking or drugs. It has been hypothesized that some SIDS deaths are due to uncontrolled inflammatory reactions and cortisol levels may be an indicator for stress and/or a lack of inflammatory control in SIDS.65,66
Our study has limitations. This metabolomics study was conducted using post-mortem biospecimens. Recent studies demonstrated that post-mortem material is robust and beneficial in metabolomic studies, especially when material was collected within 48 h of the time of death.67,68 We identified candidate metabolite biomarkers; we do not have an immediate causal interpretation. However, we applied a conservative Bonferroni p-value correction to narrow the pool of potential metabolite biomarkers for SIDS. Furthermore, we applied a correlation-weighted clustering algorithm to understand general metabolomic profiles associated with SIDS and incorporated each metabolite’s biological pathway and sub-pathway information to gain insight into metabolic processes associated with a SIDS diagnosis.
Another limitation is that this study combines two cohorts with notable differences in race and ethnicity composition, sample sizes, and detailed phenotype and environmental exposure data. For example, the NBB database did not contain detailed information regarding SIDS risk factors such as maternal smoking and bed sharing. This limited our ability to adjust for these confounders in the combined CIMS and NBB analyses. Additionally, our sensitivity analyses for maternal smoking and drug exposure and bed sharing only pertain to the CIMS cohort due to the lack of clinical data available for the NBB biospecimens. Furthermore, the CIMS cohort consisted of 74% Black infants as compared to only 41% Black infants in NBB. To account for potential bias due to differences in race/ethnicity distribution in CIMS and NBB, we performed Welch’s t-tests for combined (all CIMS and NBB), CIMS only, NBB only, and combined race/ethnicity, and stratified (Black only, White only, Hispanic only) as shown in Supplemental Table S1. Although NBB only and race/ethnicity stratified analyses were underpowered (no significant associations for NBB and Hispanic only analyses), CIMs only analysis identified 32 statistically significant metabolites, while 31 and 2 metabolites were significant in the Black only and White only stratified analyses, respectively. Ribitol (p-value 4.99e-05) and 2-hydroxybutyrate/2-hydroxyisobutyrate (p-value 4.27e-05) were the two top associations in White only analyses. Thus, our results may be more specific to the CIMS cohort or driven by infants identified as Black. Recent work by Galván-Tejada et al. demonstrated univariate associations with SIDS, specifically acetic, butyric, hexanoic, and valeric acids having the best ability to predict SIDS diagnosis.59 In contrast, none of our metabolites that are derivatives of these acids were significant predictors of SIDS. However, these derivatives are present in both the green and turquoise clusters. This discrepancy may be due to the difference between univariate associations with SIDS and those after adjustment for important confounders like post-mortem interval. The current interpretation of our findings remains limited as most previous literature focusing on SIDS and metabolites are few and older than a 1995 publication date. While there are recent efforts9, 10, 11,69 of metabolomic investigations, they suffer from very small sample sizes. We believe this underscores the need for larger metabolomics studies focused on SIDS to investigate the metabolomic contributions to important SIDS subgroups and risk factors.
In conclusion, we performed an unbiased discovery of metabolites finding new candidate biomarkers for SIDS. Moreover, several metabolomics profiles specific to SIDS arose having consistency with the Triple Risk model, e.g., pulmonary surfactant composition. Some pointed to a potential biological mechanism of central nervous circuitry via sphingomyelins, which are critical for insulating neurons and transferring their impulses from one to another. However, direct biological and causal interpretation remains limited and beyond the scope of this study. Thus, it is imperative for future work to replicate these findings and add other “omics” to tease out causality.
Contributors
CMA: formal analysis, methodology, visualization, writing - original draft, writing – review & editing, accessed and verified the underlying data; KLK: conceptualisation, funding acquisition, methodology, supervision, writing – original draft, writing – review & editing, accessed and verified the underlying data; CAN: data curation, writing – review & editing, accessed and verified the underlying data; JCM: funding acquisition, supervision, writing – review & editing; FRH: conceptualisation, data curation, funding acquisition, writing – review & editing, accessed and verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
The de-identified participant data used to generate the results in this article will be made available through the database of Genotypes and Phenotypes (dbGaP; Study: Chicago Infant Mortality Study; phs accession phs003790.v1.p1) and shared with researchers on reasonable request to the corresponding or senior authors.
Declaration of interests
All authors declare no conflicts of interest relevant to this work.
Acknowledgements
We wish to acknowledge all families who participated in the Chicago Infant Mortality Study and the NIH NeuroBioBank which made this study possible in hopes of benefiting at risk infants and to prevent other families from experiencing a SIDS loss. Research funding provided by Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Institute on Deafness and Other Communication Disorders under contract number NO1-HD-3-3188 and 5R01HD101518-04, the Centers for Disease Control and Prevention and the Association of Teachers of Preventive Medicine under cooperative agreement number U50/CCU300860-06, and the Playmates in Heaven Foundation.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105484.
Appendix ASupplementary data
References
- 1.Moraga P., GBD 2016 Causes of Death Collaborators Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell E.A., Tuohy P.G., Brunt J.M., et al. Risk factors for sudden infant death syndrome following the prevention campaign in New Zealand: a prospective study. Pediatrics. 1997;100:835–840. doi: 10.1542/peds.100.5.835. [DOI] [PubMed] [Google Scholar]
- 3.Filiano J.J., Kinney H.C. A perspective on neuropathologic findings in victims of the sudden infant death syndrome: the triple-risk model. Biol Neonate. 1994;65:194–197. doi: 10.1159/000244052. [DOI] [PubMed] [Google Scholar]
- 4.Spinelli J., Collins-Praino L., Van Den Heuvel C., Byard R.W. Evolution and significance of the triple risk model in sudden infant death syndrome. J Paediatr Child Health. 2017;53:112–115. doi: 10.1111/jpc.13429. [DOI] [PubMed] [Google Scholar]
- 5.Baruteau A.-E., Tester D.J., Kapplinger J.D., Ackerman M.J., Behr E.R. Sudden infant death syndrome and inherited cardiac conditions. Nat Rev Cardiol. 2017;14:715–726. doi: 10.1038/nrcardio.2017.129. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 6.Young S.P., Wallace G.R. Metabolomic analysis of human disease and its application to the eye. J Ocul Biol Dis Infor. 2009;2:235–242. doi: 10.1007/s12177-009-9038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap I.K.S., Brown I.J., Chan Q., et al. Metabolome-wide association study identifies multiple biomarkers that discriminate north and south Chinese populations at differing risks of cardiovascular disease: INTERMAP study. J Proteome Res. 2010;9:6647–6654. doi: 10.1021/pr100798r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R., Zhang T., Ali A.M., Al Washih M., Pickard B., Watson D.G. Metabolomic profiling of post-mortem brain reveals changes in amino acid and glucose metabolism in mental illness compared with controls. Comput Struct Biotechnol J. 2016;14:106–116. doi: 10.1016/j.csbj.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham S.F., Chevallier O.P., Kumar P., Türko Gcaron Lu O., Bahado-Singh R.O. Metabolomic profiling of brain from infants who died from Sudden Infant Death Syndrome reveals novel predictive biomarkers. J Perinatol Off J Calif Perinat Assoc. 2017;37:91–97. doi: 10.1038/jp.2016.139. [DOI] [PubMed] [Google Scholar]
- 10.Graham S.F., Turkoglu O., Kumar P., et al. Targeted metabolic profiling of post-mortem brain from infants who died from sudden infant death syndrome. J Proteome Res. 2017;16:2587–2596. doi: 10.1021/acs.jproteome.7b00157. [DOI] [PubMed] [Google Scholar]
- 11.Terry J., Dyer R.A. Aberrant colon metabolome and the sudden infant death syndrome. Pediatr Res. 2024;95(3):634–640. doi: 10.1038/s41390-023-02847-0. [DOI] [PubMed] [Google Scholar]
- 12.Hauck F.R., Moore C.M., Herman S.M., et al. The contribution of prone sleeping position to the racial disparity in sudden infant death syndrome: the Chicago infant mortality study. Pediatrics. 2002;110:772–780. doi: 10.1542/peds.110.4.772. [DOI] [PubMed] [Google Scholar]
- 13.Hauck F.R., Tanabe K.O., Blackstone S.R. Risk of postneonatal infant mortality associated with prior founded allegations of Child abuse. Child Maltreat. 2022;27:185–193. doi: 10.1177/10775595211069923. SAGE Publications Inc. [DOI] [PubMed] [Google Scholar]
- 14.Hauck F.R., Herman S.M., Donovan M., et al. Sleep environment and the risk of sudden infant death syndrome in an urban population: the Chicago infant mortality study. Pediatrics. 2003;111:1207–1214. [PubMed] [Google Scholar]
- 15.Mychaleckyj J.C., Normeshie C., Keene K.L., Hauck F.R. Organ weights and length anthropometry measures at autopsy for sudden infant death syndrome cases and other infant deaths in the Chicago infant mortality study. Am J Hum Biol. 2024;36 doi: 10.1002/ajhb.24126. Epub. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein R.D., Kinney H.C., Willinger M. Sudden unexpected death in fetal life through early childhood. Pediatrics. 2016;137 doi: 10.1542/peds.2015-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis Armstrong N.M., Spragley K.J., Chen W.M., et al. Multi-omic analysis of stroke recurrence in african Americans from the vitamin intervention for stroke prevention (VISP) clinical trial. PLoS One. 2021;16 doi: 10.1371/journal.pone.0247257. https://pubmed.ncbi.nlm.nih.gov/33661917/ PloS One [online serial]. Accessed at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langfelder P., Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- 20.Moon R.Y., Horne R.S.C., Hauck F.R. Sudden infant death syndrome. Lancet Lond Engl. 2007;370:1578–1587. doi: 10.1016/S0140-6736(07)61662-6. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira AM. de F., Andrade PR de, Pinheiro E.M., Avelar A.F.M., Costa P., Belela-Anacleto A.S.C. Risk and protective factors for sudden infant death syndrome. Rev Bras Enferm. 2020;73 doi: 10.1590/0034-7167-2019-0458. Associação Brasileira de Enfermagem. [DOI] [PubMed] [Google Scholar]
- 22.Jhun I., Mata D.A., Nordio F., Lee M., Schwartz J., Zanobetti A. Ambient temperature and sudden infant death syndrome in the United States. Epidemiol Camb Mass. 2017;28:728–734. doi: 10.1097/EDE.0000000000000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiklund L., George M., Nord C.E., Ronquist G., Saldeen T. Sudden infant death syndrome and nitrogen metabolism: further development of a hypothesis. Eur J Clin Invest. 1998;28:958–965. doi: 10.1046/j.1365-2362.1998.00385.x. [DOI] [PubMed] [Google Scholar]
- 24.Tyler J.W. Cot-death: the ammonia factor. Med Hypotheses. 1985;16:61–64. doi: 10.1016/0306-9877(85)90039-8. [DOI] [PubMed] [Google Scholar]
- 25.Vawter G.F., McGraw C.A., Hug G., Kozakewich H.P.W., McNaulty J., Mandell F. An hepatic metabolic profile in sudden infant death (SIDS) Forensic Sci Int. 1986;30:93–98. doi: 10.1016/0379-0738(86)90003-4. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto S., Häberle J., Kido J., Mitsubuchi H., Endo F., Nakamura K. Urea cycle disorders—update. J Hum Genet. 2019;64:833–847. doi: 10.1038/s10038-019-0614-4. [DOI] [PubMed] [Google Scholar]
- 27.Hartung B., Temme O., Neuen-Jacob E., Ritz-Timme S., Hinderhofer K., Daldrup T. Ornithine transcarbamylase deficiency of a male newborn with fatal outcome. Int J Legal Med. 2016;130:783–785. doi: 10.1007/s00414-015-1311-2. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein A.S., Hoogenraad N.J., Johnson J.D., et al. Metabolic and genetic studies of a family with ornithine transcarbamylase deficiency. Pediatr Res. 1974;8:5–12. doi: 10.1203/00006450-197401000-00002. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 29.Popowska E., Ciara E., Rokicki D., Pronicka E. Three novel and one recurrent ornithine carbamoyltransferase gene mutations in Polish patients. J Inherit Metab Dis. 1999;22:92–93. doi: 10.1023/a:1005476021549. [DOI] [PubMed] [Google Scholar]
- 30.NCI Thesaurus [online] https://ncithesaurus.nci.nih.gov/ncitbrowser/ConceptReport.jsp?dictionary=NCI_Thesaurus&ns=ncit&code=C118878 Accessed at:
- 31.Euli D., Colombo L., Bruno A., Mussini E. Assay for 5-hydroxylysine and L-lysine in human and rat urine and in bone by gas chromatography. J Chromatogr B Biomed Sci App. 1999;724:373–379. doi: 10.1016/s0378-4347(98)00539-8. [DOI] [PubMed] [Google Scholar]
- 32.Farquharson J., Jamieson E.C., Abbasi K.A., Patrick W.J., Logan R.W., Cockburn F. Effect of diet on the fatty acid composition of the major phospholipids of infant cerebral cortex. Arch Dis Child. 1995;72:198–203. doi: 10.1136/adc.72.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahado-Singh R.O., Ertl R., Mandal R., et al. Metabolomic prediction of fetal congenital heart defect in the first trimester. Am J Obstet Gynecol. 2014;211:240.e1–240.e14. doi: 10.1016/j.ajog.2014.03.056. [DOI] [PubMed] [Google Scholar]
- 34.Haga H., Nakajima T. Determination of polyol profiles in human urine by capillary gas chromatography. Biomed Chromatogr BMC. 1989;3:68–71. doi: 10.1002/bmc.1130030206. [DOI] [PubMed] [Google Scholar]
- 35.Verhoeven N.M., Huck J.H., Roos B., et al. Transaldolase deficiency: liver cirrhosis associated with a new inborn error in the pentose phosphate pathway. Am J Hum Genet. 2001;68:1086–1092. doi: 10.1086/320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.TeSlaa T., Ralser M., Fan J., Rabinowitz J.D. The pentose phosphate pathway in health and disease. Nat Metab. 2023;5:1275–1289. doi: 10.1038/s42255-023-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonham J.R., Downing M. Metabolic deficiencies and SIDS. J Clin Pathol. 1992;45:33–38. [PubMed] [Google Scholar]
- 38.Reid G.M. Sudden infant death syndrome (SIDS): oxygen utilization and energy production. Med Hypotheses. 1993;40:364–366. doi: 10.1016/0306-9877(93)90219-g. [DOI] [PubMed] [Google Scholar]
- 39.Cataldi M.P., Lu P., Blaeser A., Lu Q.L. Ribitol restores functionally glycosylated α-dystroglycan and improves muscle function in dystrophic FKRP-mutant mice. Nat Commun. 2018;9:3448. doi: 10.1038/s41467-018-05990-z. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eiden L.E. Neuropeptide–Catecholamine interactions in stress. Adv Pharmacol San Diego Calif. 2013;68:399–404. doi: 10.1016/B978-0-12-411512-5.00018-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brumovsky P., Shi T.S., Landry M., Villar M.J., Hökfelt T. Neuropeptide tyrosine and pain. Trends Pharmacol Sci. 2007;28:93–102. doi: 10.1016/j.tips.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Mathar D., Erfanian Abdoust M., Marrenbach T., Tuzsus D., Peters J. The catecholamine precursor Tyrosine reduces autonomic arousal and decreases decision thresholds in reinforcement learning and temporal discounting. PLoS Comput Biol. 2022;18 doi: 10.1371/journal.pcbi.1010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillipson E.A., Sullivan C.E. Arousal: the forgotten response to respiratory stimuli. Am Rev Respir Dis. 1978;118:807–809. doi: 10.1164/arrd.1978.118.5.807. American Thoracic Society - AJRCCM. [DOI] [PubMed] [Google Scholar]
- 44.Kahn A., Groswasser J., Franco P., et al. Sudden infant deaths: stress, arousal and SIDS. Early Hum Dev. 2003;75(Suppl):S147–S166. doi: 10.1016/j.earlhumdev.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 45.Klintschar M., Reichenpfader B., Saternus K.-S. A functional polymorphism in the tyrosine hydroxylase gene indicates a role of noradrenalinergic signaling in sudden infant death syndrome. J Pediatr. 2008;153:190–193. doi: 10.1016/j.jpeds.2008.02.032. [DOI] [PubMed] [Google Scholar]
- 46.Courts C., Madea B. Significant association of TH01 allele 9.3 and SIDS. J Forensic Sci. 2011;56:415–417. doi: 10.1111/j.1556-4029.2010.01670.x. [DOI] [PubMed] [Google Scholar]
- 47.Kinney H.C., Ann Brody B., Finkelstein D.M., Vawter G.F., Mandell F., Gillies F.H. Delayed central nervous system myelination in the sudden infant death syndrome. J Neuropathol Exp Neurol. 1991;50:29–48. doi: 10.1097/00005072-199101000-00003. [DOI] [PubMed] [Google Scholar]
- 48.James D., Berry P.J., Fleming P., Hathaway M. Surfactant abnormality and the sudden infant death syndrome--a primary or secondary phenomenon? Arch Dis Child. 1990;65:774–778. doi: 10.1136/adc.65.7.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hills B.A., Masters I.B., Vance J.C., Hills Y.C. Abnormalities in surfactant in sudden infant death syndrome as a postmortem marker and possible test of risk. J Paediatr Child Health. 1997;33:61–66. doi: 10.1111/j.1440-1754.1997.tb00993.x. [DOI] [PubMed] [Google Scholar]
- 50.Stray-Pedersen A., Vege A., Stray-Pedersen A., Holmskov U., Rognum T.O. Post-neonatal drop in alveolar SP-A expression: biological significance for increased vulnerability to SIDS? Pediatr Pulmonol. 2008;43:160–168. doi: 10.1002/ppul.20750. [DOI] [PubMed] [Google Scholar]
- 51.Goetzman E.S., Alcorn J.F., Bharathi S.S., et al. Long-chain acyl-CoA dehydrogenase deficiency as a cause of pulmonary surfactant dysfunction. J Biol Chem. 2014;289:10668–10679. doi: 10.1074/jbc.M113.540260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reid G.M. Sudden infant death syndrome and lipoproteins. Med Hypotheses. 1989;29:77–80. doi: 10.1016/0306-9877(89)90065-0. [DOI] [PubMed] [Google Scholar]
- 53.Watson A.D. Thematic review series: systems Biology Approaches to Metabolic and Cardiovascular Disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. Elsevier. [DOI] [PubMed] [Google Scholar]
- 54.Tinoco J., Shannon A., Miljanich P., Babcock R., Lyman R.L. Liver LIPIDS of choline-deficient rats. Biochem J. 1965;94:751–754. doi: 10.1042/bj0940751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson K.M., Christensen E., Jakobs C., et al. The clinical phenotype of succinic semialdehyde dehydrogenase deficiency (4-hydroxybutyric aciduria): case reports of 23 new patients. Pediatrics. 1997;99:567–574. doi: 10.1542/peds.99.4.567. [DOI] [PubMed] [Google Scholar]
- 56.Solberg R., Enot D., Deigner H.-P., et al. Metabolomic analyses of plasma reveals new insights into asphyxia and resuscitation in pigs. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Atkinson Bourke. Sudden infant death syndrome: possible link with impaired gut ureolysis and metabolic alkalosis. Eur J Clin Invest. 1998;28:966–968. doi: 10.1046/j.1365-2362.1998.00384.x. [DOI] [PubMed] [Google Scholar]
- 58.Machaalani R., Waters K.A. Neurochemical abnormalities in the brainstem of the sudden infant death syndrome (SIDS) Paediatr Respir Rev. 2014;15:293–300. doi: 10.1016/j.prrv.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Galván-Tejada C.E., Villagrana-Bañuelos K.E., Zanella-Calzada L.A., et al. Univariate analysis of short-chain fatty acids related to sudden infant death syndrome. Diagnostics. 2020;10:896. doi: 10.3390/diagnostics10110896. Multidisciplinary Digital Publishing Institute. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cederbaum S.D. SIDS and disorders of fatty acid oxidation: where do we go from here? J Pediatr. 1998;132:913–914. doi: 10.1016/s0022-3476(98)70381-6. [DOI] [PubMed] [Google Scholar]
- 61.Lundemose J.B., Kølvraa S., Gregersen N., Christensen E., Gregersen M. Fatty acid oxidation disorders as primary cause of sudden and unexpected death in infants and young children: an investigation performed on cultured fibroblasts from 79 children who died aged between 0-4 years. Mol Pathol. 1997;50:212. doi: 10.1136/mp.50.4.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duncan J.R., Garland M., Myers M.M., et al. Prenatal nicotine-exposure alters fetal autonomic activity and medullary neurotransmitter receptors: implications for sudden infant death syndrome. J Appl Physiol. 2009;107:1579–1590. doi: 10.1152/japplphysiol.91629.2008. American Physiological Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deen C.P.J., van der Veen A., Gomes-Neto A.W., et al. Urinary excretion of N1-methyl-2-pyridone-5-carboxamide and N1-methylnicotinamide in renal transplant recipients and donors. J Clin Med. 2020;9:437. doi: 10.3390/jcm9020437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lenglet A., Liabeuf S., Bodeau S., et al. N-methyl-2-pyridone-5-carboxamide (2PY)—major metabolite of nicotinamide: an update on an old uremic toxin. Toxins. 2016;8:339. doi: 10.3390/toxins8110339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gordon A.E., Madani O.A., Weir D.M., Busuttil A., Blackwell C. Cortisol levels and control of in£ammatory responses to toxic shock syndrome toxin-1 (TSST-1): the prevalence of night-time deaths in sudden infant death syndrome (SIDS) FEMS Immunol Med Microbiol. 1999;25(1-2):199–206. doi: 10.1111/j.1574-695X.1999.tb01344.x. [DOI] [PubMed] [Google Scholar]
- 66.Naeye R.L., Fisher R., Rubin H.R., Demers L.M. Selected hormone levels in victims of the sudden infant death syndrome. Pediatrics. 1980;65:1134–1136. [PubMed] [Google Scholar]
- 67.Elmsjö A., Vikingsson S., Söderberg C., Kugelberg F.C., Green H. Post-mortem metabolomics: a novel approach in clinical biomarker discovery and a potential tool in death investigations. Chem Res Toxicol. 2021;34:1496–1502. doi: 10.1021/acs.chemrestox.0c00448. American Chemical Society. [DOI] [PubMed] [Google Scholar]
- 68.Steuer A.E., Wartmann Y., Schellenberg R., et al. Postmortem metabolomics: influence of time since death on the level of endogenous compounds in human femoral blood. Necessary to be considered in metabolome study planning? Metabolomics. 2024;20:51. doi: 10.1007/s11306-024-02117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Villagrana-Bañuelos K.E., Galván-Tejada C.E., Galván-Tejada J.I., et al. Machine learning model based on lipidomic profile information to predict sudden infant death syndrome. Healthcare. 2022;10:1303. doi: 10.3390/healthcare10071303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.