Abstract
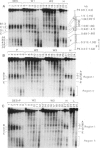
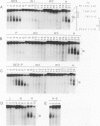
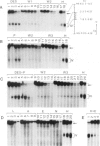
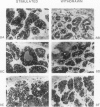
Four DNase I-hypersensitive regions (I-IV) occur in the 5'-flanking region of the ovalbumin gene in hen oviducts. One is centered close to the cap site (position +1) of transcription and the others at -0.8, -3.3 and -6.0 kb. The correlation of hypersensitivity with transcription was determined for each region in oviducts of chicks, where expression of the ovalbumin gene can be controlled by administration and withdrawal of steroid hormones. DNase I-hypersensitive regions were mapped by the indirect end-labeling technique and ovalbumin mRNA levels were determined by the dot blot assay. Diethylstilbestrol (DES) induces the appearance of hypersensitive regions I-IV whereas progesterone induces regions I, II and IV, but not region III. Upon withdrawal, regions II and III, and most of regions I and IV, disappear. A weak zone of hypersensitivity in region I near the cap site persists during withdrawal and a new zone of hypersensitivity appears between regions I and II. There is strong correlation between hypersensitivity at regions I-IV and gene transcription; ovalbumin mRNA levels are high in stimulated chicks, where hypersensitive regions are present, and drop to very low levels in withdrawn chicks, where the hypersensitivity is absent. We suggest that proteins, perhaps hormone receptors acting together with tissue-specific factors, induce DNAse I-hypersensitive regions I-IV of the ovalbumin gene.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Becker P., Renkawitz R., Schütz G. Tissue-specific DNaseI hypersensitive sites in the 5'-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J. 1984 Sep;3(9):2015–2020. doi: 10.1002/j.1460-2075.1984.tb02084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breindl M., Harbers K., Jaenisch R. Retrovirus-induced lethal mutation in collagen I gene of mice is associated with an altered chromatin structure. Cell. 1984 Aug;38(1):9–16. doi: 10.1016/0092-8674(84)90521-x. [DOI] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Herbomel P., Jouanneau J., Saragosti S., Katinka M., Bourachot B., de Crombrugghe B., Yaniv M. Structure and function of the promoter-enhancer region of polyoma and SV40. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):935–944. doi: 10.1101/sqb.1983.047.01.107. [DOI] [PubMed] [Google Scholar]
- Chambon P., Dierich A., Gaub M. P., Jakowlev S., Jongstra J., Krust A., LePennec J. P., Oudet P., Reudelhuber T. Promoter elements of genes coding for proteins and modulation of transcription by estrogens and progesterone. Recent Prog Horm Res. 1984;40:1–42. doi: 10.1016/b978-0-12-571140-1.50005-0. [DOI] [PubMed] [Google Scholar]
- Chandler V. L., Maler B. A., Yamamoto K. R. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983 Jun;33(2):489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- Chung S. Y., Folsom V., Wooley J. DNase I-hypersensitive sites in the chromatin of immunoglobulin kappa light chain genes. Proc Natl Acad Sci U S A. 1983 May;80(9):2427–2431. doi: 10.1073/pnas.80.9.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R. S., Meselson M. Inducible transcription and puffing in Drosophila melanogaster transformed with hsp70-phage lambda hybrid heat shock genes. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5509–5513. doi: 10.1073/pnas.81.17.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. G., Schrader W. T., O'Malley B. W. DNA sequence preference of the progesterone receptor. Proc Natl Acad Sci U S A. 1983 Jan;80(1):16–20. doi: 10.1073/pnas.80.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costlow N. A., Simon J. A., Lis J. T. A hypersensitive site in hsp70 chromatin requires adjacent not internal DNA sequence. Nature. 1985 Jan 10;313(5998):147–149. doi: 10.1038/313147a0. [DOI] [PubMed] [Google Scholar]
- Costlow N., Lis J. T. High-resolution mapping of DNase I-hypersensitive sites of Drosophila heat shock genes in Drosophila melanogaster and Saccharomyces cerevisiae. Mol Cell Biol. 1984 Sep;4(9):1853–1863. doi: 10.1128/mcb.4.9.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. C., Gope R., Knoll B. J., Riser M. E., O'Malley B. W. A similar 5'-flanking region is required for estrogen and progesterone induction of ovalbumin gene expression. J Biol Chem. 1984 Aug 25;259(16):9967–9970. [PubMed] [Google Scholar]
- Dean D. C., Knoll B. J., Riser M. E., O'Malley B. W. A 5'-flanking sequence essential for progesterone regulation of an ovalbumin fusion gene. Nature. 1983 Oct 6;305(5934):551–554. doi: 10.1038/305551a0. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Dudler R., Travers A. A. Upstream elements necessary for optimal function of the hsp 70 promoter in transformed flies. Cell. 1984 Sep;38(2):391–398. doi: 10.1016/0092-8674(84)90494-x. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. Anatomy of hypersensitive sites. Nature. 1984 May 17;309(5965):213–214. doi: 10.1038/309213a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig R., Muraskowsky R., Mandel J. L. The ovalbumin gene family. The 5' end region of the X and Y genes. J Mol Biol. 1982 Mar 25;156(1):1–19. doi: 10.1016/0022-2836(82)90455-7. [DOI] [PubMed] [Google Scholar]
- Heilig R., Perrin F., Gannon F., Mandel J. L., Chambon P. The ovalbumin gene family: structure of the X gene and evolution of duplicated split genes. Cell. 1980 Jul;20(3):625–637. doi: 10.1016/0092-8674(80)90309-8. [DOI] [PubMed] [Google Scholar]
- Hynes N. E., Groner B., Sippel A. E., Jeep S., Wurtz T., Nguyen-Huu M. C., Giesecke K., Schütz G. Control of cellular content of chicken egg white protein specific RNA during estrogen administration and withdrawal. Biochemistry. 1979 Feb 20;18(4):616–624. doi: 10.1021/bi00571a011. [DOI] [PubMed] [Google Scholar]
- Hynes N., van Ooyen A. J., Kennedy N., Herrlich P., Ponta H., Groner B. Subfragments of the large terminal repeat cause glucocorticoid-responsive expression of mouse mammary tumor virus and of an adjacent gene. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3637–3641. doi: 10.1073/pnas.80.12.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Jost J. P., Seldran M., Geiser M. Preferential binding of estrogen-receptor complex to a region containing the estrogen-dependent hypomethylation site preceding the chicken vitellogenin II gene. Proc Natl Acad Sci U S A. 1984 Jan;81(2):429–433. doi: 10.1073/pnas.81.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. S., Bellard M., Dretzen G., Bellard F., Chambon P. A close association between sites of DNase I hypersensitivity and sites of enhanced cleavage by micrococcal nuclease in the 5'-flanking region of the actively transcribed ovalbumin gene. EMBO J. 1984 May;3(5):1137–1144. doi: 10.1002/j.1460-2075.1984.tb01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll B. J., Zarucki-Schulz T., Dean D. C., O'Malley B. W. Definition of the ovalbumin gene promoter by transfer of an ovalglobin fusion gene into cultured cells. Nucleic Acids Res. 1983 Oct 11;11(19):6733–6754. doi: 10.1093/nar/11.19.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok K., Snippe L., Ab G., Gruber M. Nuclease-hypersensitive sites in chromatin of the estrogen-inducible apoVLDL II gene of chicken. Nucleic Acids Res. 1985 Jul 25;13(14):5189–5202. doi: 10.1093/nar/13.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMeur M., Glanville N., Mandel J. L., Gerlinger P., Palmiter R., Chambon P. The ovalbumin gene family: hormonal control of X and Y gene transcription and mRNA accumulation. Cell. 1981 Feb;23(2):561–571. doi: 10.1016/0092-8674(81)90152-5. [DOI] [PubMed] [Google Scholar]
- Majors J., Varmus H. E. A small region of the mouse mammary tumor virus long terminal repeat confers glucocorticoid hormone regulation on a linked heterologous gene. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5866–5870. doi: 10.1073/pnas.80.19.5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- McKnight G. S., Palmiter R. D. Transcriptional regulation of the ovalbumin and conalbumin genes by steroid hormones in chick oviduct. J Biol Chem. 1979 Sep 25;254(18):9050–9058. [PubMed] [Google Scholar]
- McKnight G. S., Pennequin P., Schimke R. T. Induction of ovalbumin mRNA sequences by estrogen and progesterone in chick oviduct as measured by hybridization to complementary DNA. J Biol Chem. 1975 Oct 25;250(20):8105–8110. [PubMed] [Google Scholar]
- McKnight G. S. The induction of ovalbumin and conalbumin mRNA by estrogen and progesterone in chick oviduct explant cultures. Cell. 1978 Jun;14(2):403–413. doi: 10.1016/0092-8674(78)90125-3. [DOI] [PubMed] [Google Scholar]
- Mirault M. E., Southgate R., Delwart E. Regulation of heat-shock genes: a DNA sequence upstream of Drosophila hsp70 genes is essential for their induction in monkey cells. EMBO J. 1982;1(10):1279–1285. doi: 10.1002/j.1460-2075.1982.tb00025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill E. R., LePennec J. P., Chambon P. Chicken oviduct progesterone receptor: location of specific regions of high-affinity binding in cloned DNA fragments of hormone-responsive genes. Cell. 1982 Mar;28(3):621–632. doi: 10.1016/0092-8674(82)90217-3. [DOI] [PubMed] [Google Scholar]
- Oka T., Schimke R. T. Interaction of estrogen and progesterone in chick oviduct development. II. Effects of estrogen and progesterone on tubular gland cell function. J Cell Biol. 1969 Oct;43(1):123–137. doi: 10.1083/jcb.43.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Carey N. H. Rapid inactivation of ovalbumin messenger ribonucleic acid after acute withdrawal of estrogen. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2357–2361. doi: 10.1073/pnas.71.6.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Christensen A. K., Schimke R. T. Organization of polysomes from pre-existing ribosomes in chick oviduct by a secondary administration of either estradiol or progesterone. J Biol Chem. 1970 Feb 25;245(4):833–845. [PubMed] [Google Scholar]
- Palmiter R. D., Moore P. B., Mulvihill E. R. A significant lag in the induction of ovalbumin messenger RNA by steroid hormones: a receptor translocation hypothesis. Cell. 1976 Aug;8(4):557–572. doi: 10.1016/0092-8674(76)90224-5. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell. 1975 Mar;4(3):189–189. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Payvar F., DeFranco D., Firestone G. L., Edgar B., Wrange O., Okret S., Gustafsson J. A., Yamamoto K. R. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell. 1983 Dec;35(2 Pt 1):381–392. doi: 10.1016/0092-8674(83)90171-x. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982 Sep;30(2):517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Picard D., Schaffner W. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 1984 Jan 5;307(5946):80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- Pospelov V. A., Klobeck H. G., Zachau H. G. Correlation between DNase I hypersensitive sites and putative regulatory sequences in human immunoglobulin genes of the kappa light chain type. Nucleic Acids Res. 1984 Sep 25;12(18):7007–7021. doi: 10.1093/nar/12.18.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Renkawitz R., Schütz G., von der Ahe D., Beato M. Sequences in the promoter region of the chicken lysozyme gene required for steroid regulation and receptor binding. Cell. 1984 Jun;37(2):503–510. doi: 10.1016/0092-8674(84)90380-5. [DOI] [PubMed] [Google Scholar]
- Royal A., Garapin A., Cami B., Perrin F., Mandel J. L., LeMeur M., Brégégègre F., Gannon F., LePennec J. P., Chambon P. The ovalbumin gene region: common features in the organisation of three genes expressed in chicken oviduct under hormonal control. Nature. 1979 May 10;279(5709):125–132. doi: 10.1038/279125a0. [DOI] [PubMed] [Google Scholar]
- Shepherd J. H., Mulvihill E. R., Thomas P. S., Palmiter R. D. Commitment of chick oviduct tubular gland cells to produce ovalbumin mRNA during hormonal withdrawal and restimulation. J Cell Biol. 1980 Oct;87(1):142–151. doi: 10.1083/jcb.87.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Spelsberg T. C., Littlefield B. A., Seelke R., Dani G. M., Toyoda H., Boyd-Leinen P., Thrall C., Kon O. L. Role of specific chromosomal proteins and DNA sequences in the nuclear binding sites for steroid receptors. Recent Prog Horm Res. 1983;39:463–517. doi: 10.1016/b978-0-12-571139-5.50016-1. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Chao M. V., Axel R. The structure of the thymidine kinase gene promoter: nuclease hypersensitivity correlates with expression. Cell. 1982 Dec;31(2 Pt 1):347–353. doi: 10.1016/0092-8674(82)90128-3. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T., Fan H. Mapping of DNase I-hypersensitive sites in the 5' and 3' long terminal repeats of integrated moloney murine leukemia virus proviral DNA. Mol Cell Biol. 1985 Apr;5(4):601–609. doi: 10.1128/mcb.5.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Woo S. L., Beattie W. G., Catterall J. F., Dugaiczyk A., Staden R., Brownlee G. G., O'Malley B. W. Complete nucleotide sequence of the chicken chromosomal ovalbumin gene and its biological significance. Biochemistry. 1981 Oct 27;20(22):6437–6446. doi: 10.1021/bi00525a024. [DOI] [PubMed] [Google Scholar]
- Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984 Sep 6;311(5981):81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]