Abstract
Breast cancer (BC) is a malignant tumor, that damages the physical health of female patients. It is crucial to develop new treatment strategies for BC, as this disease significantly affects the quality of life of women in both developing and developed countries, despite the existence of effective treatment options to reduce mortality. Recently, several researchers have been studying circular RNAs (circRNAs) in BC due to their stability and sponge function. In the present study, a new circRNA, hsa_circ_0001508, was discovered using Gene Chip's prediction. This gene is important because it is novel in BC. However, due to financial constraints, the study was designed as a pilot study for future research. The present study included several common biomedical experimental techniques, such as reverse transcription-quantitative PCR, flow cytometry, cell counting, cell invasion, wound healing assay and western blotting. It also involves the use of DNA microarray (gene chip) and prediction of microRNA and mRNA interactions using bioinformatics tools such as TargetScan. The findings revealed that hsa_circ_0001508 exhibits carcinogenic properties that facilitate the progression of BC. Furthermore, potential binding sites were identified as crucial areas for future investigation.
Keywords: breast cancer, hsa_circ_0001508, microRNA-505-3p, high mobility group box 1, vestigial-like family member 3
Introduction
Breast cancer (BC) has ranked as the second-prevalent malignant tumor worldwide and poses a substantial threat to women's health (1). Ahmad (1) reported that 25% of cancer cases in 2020 were new BC cases. Various treatment methods were applied in the field of therapy for patients with BC with different stages (from 0 to IV), including surgery, chemotherapy and targeted therapy. The following therapy techniques have been proven to effectively reduce both the occurrence and mortality rates among patients at all BC stages, ranging from stage 0 to stage IV (2). Nevertheless, 42% of patients with BC are found to encounter difficulties in achieving full recovery since they may experience treatment-related side effects such as fatigue, hair loss and increased risk of infection, among others. These side effects can significantly impair the quality of life for patients (2). Hence, it is imperative to enhance the efficacy of current treatment strategies for BC. To attain this goal, numerous researchers highlighted the potential of circular RNAs (circRNAs) in BC diagnosis, treatment and prognosis (3).
CircRNAs circular structure (a closed-loop structure devoid of a 5'-cap or a 3'-polyadenylated tail) is more stable compared with the linear structure of microRNAs (miRNAs or miRs). Additionally, circRNAs modulate gene expression and cancer progression through four major mechanisms, including miRNA sponging, transcriptional regulation, protein scaffolding and translation (3-6). Moreover, numerous researchers have recently focused on the sponge function of circRNAs. For example, circRNAs act as competing endogenous RNAs, sequestering miRNAs to prevent degradation of targeted mRNAs by bonding with them. Exon-derived circRNAs (for example, ciRS-7 and circ-ITCH) could act as miRNA sponge for RNA binding, resulting in negative regulation of miRNA and downstream target genes (7,8).
In the BC field, researchers have found pathways, such as the circRNA-miRNA-mRNA, that are associated with tumorigenesis, progression and metastasis. For example, hsa_circ_0003645-miR-139-3p/high mobility group box 1 (HMGB1) has been shown to promote the proliferation of BC cells (9). Moreover, the Gene Expression Omnibus Database (GEO; https://www.ncbi.nlm.nih.gov/geo/ is an authority database supported by the National Center for Biotechnology Information (NCBI), and it can be used to characterize gene function (10,11). Via GEO analysis, a new circRNA was found, namely hsa_circ_0001508, showing high expression in BC tissue. To the best of our knowledge, there is no research focusing on this new gene across PubMed and Embase databases. Consequently, hsa_circ_0001508 has become a focus of the present study.
Additionally, TargetScan (https://www.targetscan.org/) is an online available bioinformatics tool useful to predict pathways by finding possible binding sites, and its application only has a low false positive rate of 30% (12). The present study identified two possible pathways through the application of TargetScan, including hsa_circ_0001508/miR-505-3p/HMGB1 as well as hsa_circ_0001508/miR-505-3p/vestigial-like family member 3 (VGLL3), in which there are some possible binding sites between these genes.
Moreover, previous research indicates that miR-505-3p (tumor suppressor in BC), HMGB1 (oncogene) and VGLL3 (oncogene) are associated with BC (13,14). miR-505-3p was found to have binding sites for HMGB1 mRNA [for example, 3'-untranslated regions (3'-UTR) in HMGB1 mRNA position 417-424]; miR-505-3p can also negatively regulate the expression of HMGB1, leading to hepatocellular carcinoma cell decreasing (15). However, to the best of our knowledge, there are no previous studies focused on the binding sites between miR-505-3p and VGLL3.
In summary, in the BC field, the circRNA-miRNA-targeted gene axis has already been studied for a long time, but several new genes still have not been investigated. Based on the results of previous predictions via TargetScan and GEO analysis, a pilot study was conducted, including the biological function of hsa_circ_0001508 in BC and its possible mechanism in hsa_circ_0001508/miR-505-3p/HMGB1 as well as hsa_circ_0001508/miR-505-3p/VGLL3.
Materials and methods
Gene Chip data collection and analyses
CircRNAs data of BC were obtained from the NCBI database (accession no. GSE182471; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182471). The raw data obtained were subjected to two types of analysis: Functional enrichment analysis and biological information analysis using various software tools including CRAFT v1.0 (https://github.com/annadalmolin/CRAFT), miRbase (https://www.mirbase.org), the Circular RNA interactome database (https://circinteractome.nia.nih.gov), circMIR (https://www.bio-inf.cn/circmir/) and TargetScan (https://www.targetscan.org/).
Cell lines
BC cell lines, including HCC1806, HCC1937 and CAL-51, and MCF10A cells (non-cancerous cell line), were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. Cells were cultured in a medium containing RPMI-1640 supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin and 1% streptomycin for Cell Culture (Beyotime Institute of Biotechnology) in an incubator at 37˚C with 5% CO2.
The HCC1937 cell line was chosen for the next experimental steps since hsa_circ_0001508 in HCC1937 cells presented the highest expression compared with the rest of the cell lines (HCC1806, CAL-51 and MCF10A cells). Furthermore, the HCC1937 cell line is BRCA1-mutated, which sets it apart from the HCC1806 and CAL-51 cell lines. Additionally, the combined expression of miR-505-3p and BRCA1 could be utilized to differentiate between different types of BC, indicating its potential clinical diagnostic value (16). As a result, the present study focused on the HCC1937 cell line due to its potential clinical implications.
Based on the results of, the HCC1937 cell line was chosen as a major cell line for the next experimental steps as the expression of hsa_circ_0001508 in HCC1937 cells exhibited the highest expression compared with the other cell lines (HCC1806, CAL-51 and MCF10A cells). Furthermore, the HCC1937 cell line is BRCA1-mutated, which distinguishes it from the HCC1806 and CAL-51 cell lines. Additionally, the combined expression of miR-505-3p and BRCA1 could be utilized to differentiate between different types of BC, indicating its potential clinical diagnostic value (16). As a result, the present study focused on the HCC1937 cell line due to its potential clinical implications. Consequently, the present study focused on the HCC1937 cell line due to its potential clinical implications.
Transfection
Aiming to knockdown hsa_circ_0001508, HCC1937 cells were transfected with small interfering (si)RNAs for circ_0001508 (si-circ_0001508; 5'-ACATAAATG-ATAAATCTGTATTATTTGG-3'; 10 nM) and si-negative control (siNC; 5'-TTCTCC-GAACGTGT-CACGT-3'; 10 nM). All vectors were purchased from Sangon Biotech Co., Ltd., and all transfections were performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's protocol. Subsequently, the cells were incubated at 37˚C with 5% CO2 for 12 h, the culture medium was replaced with fresh medium containing 10% FBS and the subsequent experimental process was conducted within 24 h.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using the TriQuick reagent (Thermo Fisher Scientific, Inc.) and reverse-transcribed into cDNA using the Quantscript RT Kit (Tiangen Biotech Co., Ltd.), according to the manufacturer's protocol. The reaction condition for the reverse transcription was 37˚C for 60 min. Subsequently, qPCR was performed using RealMaster Mix (SYBR Green; Tiangen Biotech Co., Ltd.) on an ABI 7500 system (Thermo Fisher Scientific, Inc.) with the following thermocycling conditions: Initial denaturation at 95˚C for 10 min followed by 40 cycles at 94˚C for 15 sec and 60˚C for 60 sec. The expression of genes was calculated by using the 2-ΔΔCq method (17). Both U6 and GAPDH were used as references for the expression of hsa_circ_0001058 and miRNA, respectively. The sequences of the primers used for qPCR were as follows: Circ_0001508 forward, 5'-AATGGAAGGCACCCTGTGATT-3' and reverse, 5'-CAGCTTTTGTGCTTCCAACCT-3'; miR-505-3p forward, 5-CGTCAACACTTGCTGGTTTCCTA-3' and reverse, 5'-ACGAATTTGCGTGTCATCC-3'; HMGB1 forward, 5'-TCAAAGGAGAACATCCTGGCCTGT-3' and reverse, 5'-CTGCTTGTCATCTGCAGCAGTGTT-3'; VGLL3 forward, 5'-CCAACTACAGTCACCTCTGCTAC-3' and reverse, 5'-ACCACGGTGATTCCTTACTCTTG-3'; GAPDH forward, 5'-GGAGTCCACTGGCGTCTTCA-3' and reverse, 5'-GTCATGAGTCCTTCCA CGATACC-3'; and U6 forward, 5'-CTCGCTTCGGCAGCACA-3'and reverse, 5'-AACGCTTCACGAATTTGCGT-3'.
Cell counting MTT assay
HCC1937 cells were seeded into 96-well plates (1.0x103 cells/well) and incubated at 37˚C with 5% CO2 for 12, 24, 48, 72 and 96 h. A total of 20 µl of MTT reagent (Procell Life Science & Technology Co., Ltd.) was added into each well and subsequently incubated for 4 h at 37˚C. The absorbance of each cell was measured at a wavelength of 490 nm by using a microplate reader (Thermo Fisher Scientific, Inc.).
Cell invasion assay
The invasion of HCC1937 cells was observed by using a 24-well Transwell chamber. A total of 10 µl Matrigel (BD Biosciences) was placed in the lower compartment of a Transwell chamber and incubated at 37˚C for 2 h. HCC1937 cells (1x104 cells/well) were cultured for 12 h at 37˚C in the upper compartment. Subsequently, 100 µl FBS was added to the lower compartment culture medium. After 24 h, the cell invasion in the lower surface of the compartment was observed with a light microscope (magnification, x100; Olympus Corporation) after staining with crystal violet staining solution (cat. no. C0121; Beyotime Institute of Biotechnology). A total of five visual fields (upper, lower, central, left, and right) were randomly selected for counting. The pore diameter of Transwell membrane is 0.8 µm.
Wound healing assay
The transfected HCC1937 cells were cultured in a 6-well plate with serum-free culture medium. After scratching, the cells were incubated for 24 h at 37˚C. Images were captured at the same site at the start of the experiment and after 24 h.
Flow cytometry assay
Cells (3x105) were prepared following the instructions provided in the reagent kit (BD Biosciences). Subsequently, the cells were resuspended after mixed 100 µl of binding buffer. A total of 4-5 µl of FITC-Annexin V and 5 µl of PI working solution was used to stain the cells, which were stored on ice with dark conditions for 15 min. Cell apoptosis analysis was performed using a flow cytometer (DxFLEX Flow Cytometer; CytExpert™ Software; Beckman Coulter, Inc.).
Western blot assay
The protein extraction buffer (cell lysis buffer for Western and IP, Biosharp; PMSF, Beijing Solarbio Science & Technology Co., Ltd.) was used. Total protein was collected and transferred onto a PVDF membrane, which was subsequently blocked for 1-2 h. The membrane was then incubated overnight at 4˚C with the primary antibodies (HMGB1 from rabbit and VGLL3 from mouse) followed by incubation at room temperature with the secondary antibody for 2 h. The bicinchoninic acid method was used to determine protein concentration. A total of 20 µg of protein were loaded per lane and subjected to SDS-PAGE (percentage of gel, 20%). Blocking was performed using non-fat dried milk in blocking reagent (5% TBST with 05% Tween) at 37˚C. The visualisation reagent used was SuperKine™ West Femto Maximum Sensitivity Substrate Abbkine Scientific Co., Ltd.) and Gel imaging system, Tanon 4600SF (Tanon Science and Technology Co., Ltd.) was used for densitometric analysis. Antibodies used were as follows: HMGb1 (cat. no. 3935s; Cell Signaling Technology, Inc.), secondary antibody anti-rabbit IgG (HRP-linked; cat. no. 7074; Cell signaling Technology, Inc.), VGLL3 (cat. no. ab68262; Abcam), secondary antibody goat Anti-Mouse IgG H&L (HRP-linked; cat. no. ab205719; Abcam) and β-actin [(13E5) rabbit mAb (HRP Conjugate); cat. no. 5125; Cell Signaling Technology, Inc.]. The dilution of primary and secondary antibodies was 1:1,000 and 1:2,000, respectively.
Statistical analysis
All data were collected and analyzed with SPSS software (version 26.0; IBM Corp.) and GraphPad Prism software (version 5.0; Dotmatics). Data are presented as the mean ± standard deviation. Differences in the two groups were assessed using one-way ANOVA followed by LSD (≤3 groups) or Bonferroni (>3 groups) as post hoc tests. P<0.05 was considered to indicate a statistically significant difference.
Results
Gene Chip analysis
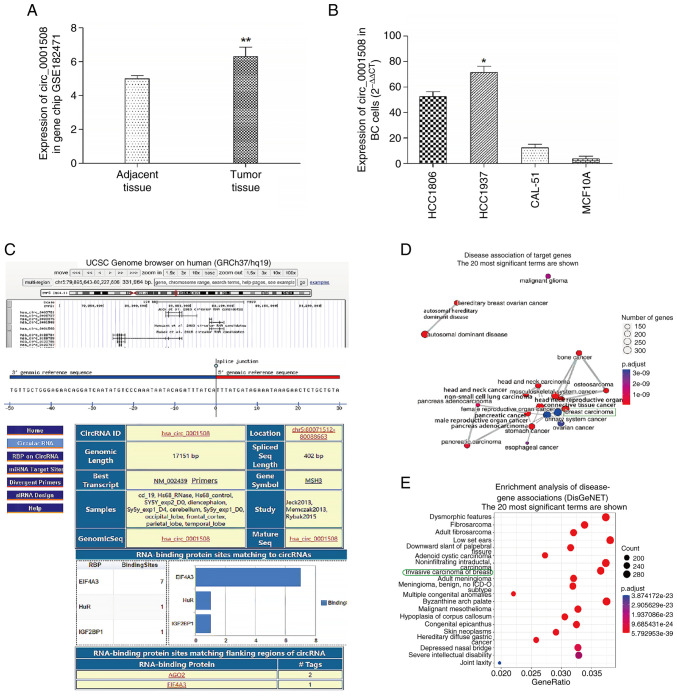
BC-related data were retrieved from the NCBI website and gene expression data from the GSE182471 dataset was extracted. The expression levels of hsa_circ_0001508 were significantly higher in BC tissues compared with adjacent BC normal tissues with fold changes of 7 and 4.5, respectively (Fig. 1A).
Figure 1.
Previous prediction of hsa_circ_0001508 in BC. (A) Expression of hsa_circ_0001508 in BC tissue from gene chip (GSE182471). (B) Expression of hsa_circ_0001508 in BC cell lines (HCC1806, HCC1937 and CAL-51) and MCF10A cell line. (C) Information about hsa_circ_0001508 from the circinteractome website. (D and E) Function of hsa_circ_0001508 through cluster analysis using CRAFT v1.0. *P<0.05 and **P<0.01 vs. adjacent tissue, MCF10A. BC, breast cancer; circRNA, circular RNA.
hsa_circ_0001508 basic information
The genomic location of hsa_circ_0001508 on chromosome 5 (chr5.80071513-80088663) and its length of 17,151 base pairs is demonstrated in Fig. 1C. These results were sourced from the Circular RNA interactome database (https://circinteractome.nia.nih.gov/api/v2/circsearch?circular_rna_query=hsa_circ_0001508+&gene_symbol_query=&submit=circRNA+Search). Additionally, the functional analysis of hsa_circ_0001508 performed by using cluster analysis software (CRAFT v1.0; https://github.com/annadalmolin/CRAFT) revealed a higher relevance of hsa_circ_0001508 to BC compared with other malignant tumors (Fig. 1D and E).
Identification of hsa_circ_0001508 expression in each cell line
The results of the RT-qPCR assay showed various expression levels for hsa_circ_0001508 in different cell lines, including the BC cell lines (HCC1806, HCC1937 and CAL-51) and the non-cancerous cell line MCF10A. The HCC1937 cells showed the highest expression of hsa_circ_0001508 among all the cell lines used in the present study (Fig. 1B). For example, the expression level of hsa_circ_0001508 in HCC1937 cells was 25-fold higher than that observed in MCF10A cells. Consequently, HCC1937 cells were selected for subsequent experiments.
Binding interaction between hsa_circ_0001508 and miR-505-3p
The potential sequestering of binding sites of miR-505-3p within target genes were predicted using the circMIR software (https://www.bio-inf.cn/circmir/). It was found that there are two binding sites between hsa_circ_0001508 and miR-505-3p, including 3'-UTR of miR-505 positions 223 and 224 (Table I).
Table I.
hsa_circ_0001508-miR-505-3p interactions were predicted using the circMIR.
| Circular RNA ID | miRNA ID | Number of binding sites predicted | Specific binding sites predicted |
|---|---|---|---|
| hsa_circ_0001508 | miR-505-3p | 2 | 223,224 |
miR or miRNA, microRNA.
hsa_circ_0001508 knockdown inhibits the proliferation and motility of HCC1937 cells
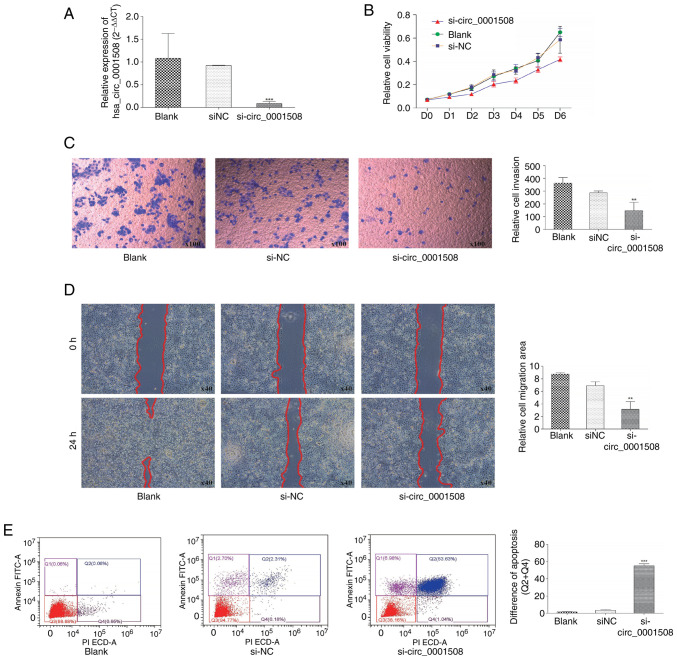
Knockdown experiments were conducted in the present study using si-circ_0001508. Subsequently, to explore the biological function of hsa_circ_0001508 in BC cells, the expression of hsa_circ_0001508 was detected using RT-qPCR in three experimental groups: si-circ_0001508; si-NC and blank. A significant decrease was observed in the expression of hsa_circ_0001508 in the si-circ_0001508 group compared with both the blank and si-NC groups (Fig. 2A).
Figure 2.
Silencing of circ_0001508 significantly inhibits the proliferation and other biological functions of BC HCC1937 cells. (A) Reverse transcription-quantitative PCR analysis of hsa_circ_0001508 expression levels in different experimental groups: Si-circ_0001508, si-NC, and blank. (B) MTT analysis of HCC1937 viability after transfection. (C) HCC1937 cell invasion through Matrigel assay. (D) Wound-healing assay for HCC1937 cell migration analysis. (E) Flow cytometry assay of HCC1937 cell apoptosis. **P<0.01 and ***P<0.001 vs. si-NC and blank groups. si-circ_0001508, small interfering RNA of circ_0001508; si-NC, small-interfering RNA negative control.
MTT analysis results revealed a significant decrease in the proliferation of HCC1937 cells when hsa_circ_0001508 was knocked down (si-circ_0001508 group), compared with the si-NC and Blank group (Fig. 2B). Moreover, Transwell and wound-healing assays indicated the involvement of hsa_circ_0001508 in HCC1937 cell migration. For instance, the si-circ_0001508 group exhibited fewer positively stained cells (Fig. 2C), suggesting the invasion of the cells in the si-circ_0001508 group was inhibited. Additionally, would-healing assays which were conducted 24 h after transfection indicated a significant delay in wound closure in the hsa_circ_0001508 knockdown group compared with the si-NC group (Fig. 2D).
The result of flow cytometry (Fig. 2E) revealed that the percentage of apoptotic cells, including early and late apoptotic, in the si-circ_0001508 group was significantly higher than in the si-NC group and blank group. Additionally, when the hsa_circ_0001508 was silenced, the percentage of early and late apoptotic cells was found to be 53.63 and 1.04%, respectively. By contrast, the si-NC group showed an early apoptotic rate of 2.31% and a late apoptotic rate of 0.18%. These findings suggested that hsa_circ_0001508 may play an oncogenic role in the progression of BC.
The aforementioned findings suggested an association between hsa_circ_0001508 expression levels and the proliferation and migration of BC cells. Notably, hsa_circ_0001508 may play an oncogenic effect on HCC1937 BC cells, meaning that its overexpression may promote BC tumorigenesis.
hsa_circ_0001508 affects the expression of target genes HMGB1 and VGLL3 by regulating miR-505-3p
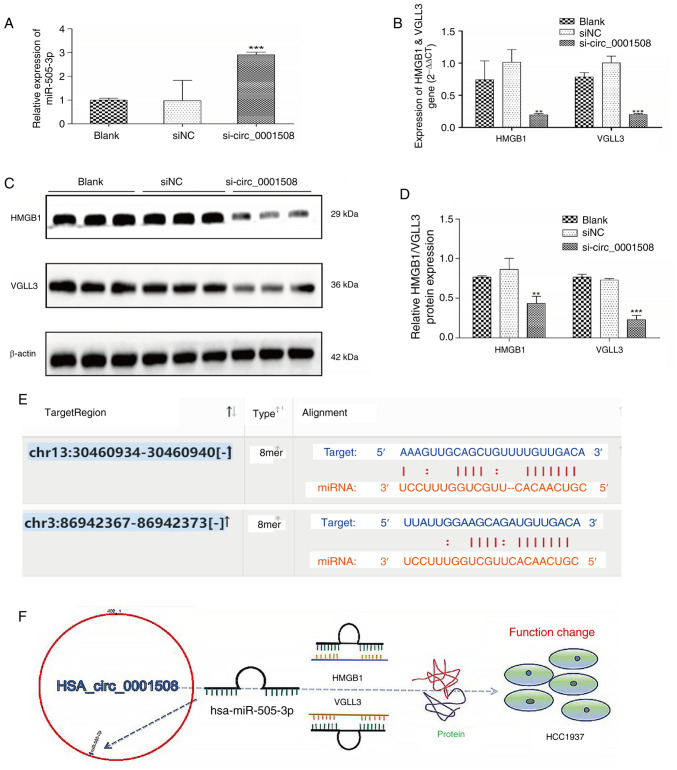
Bioinformatics analysis results were conducted with the TargetScan tool, which indicated the putative binding sites of miR-505-3p for the HMGB1 and VGLL3 genes (Fig. 3E). Additionally, the expression of miR-505-3p was significantly increased in the si-circ_0001508 group compared with the si-NC and blank groups (Fig. 3A). Similarly, the gene expression analysis performed by RT-qPCR showed that the expression levels of HMGB1 and VGLL3 were significantly lower in the si-circ_0001508 group compared with the si-NC and blank groups (Fig. 3B). The western blotting demonstrated a significant decrease in the protein expression levels of HMGB1 and VGLL3 in the si-circ_0001508 group (Fig. 3C and D).
Figure 3.
Expression of miR-505-3p and protein HMGB1 and VGLL3 in HCC1937 cells after hsa_circ_0001508 knockdown. (A) Expression of miR-505-3p was the highest in si-circ_0001508 group vs. si-NC and blank groups as identified using RT-qPCR. (B) Expression of HMGB1 and VGLL3 was lower in si-circ_0001508 group vs. siNC and blank groups as demonstrated using RT-qPCR. (C and D) Protein expression of HMGB1 and VGLL3 was suppressed in the si-circ_0001508 group vs. si-NC and blank groups as determined using the western blot assay. (E) Binding sites between miR-505-3p and HMGB1 and VGLL3 predicted using the TargetSCAN website. (F) Schematic diagram of hsa_circ_0001508 function in HCC1937 cells. **P<0.01 and ***P<0.001 vs. si-NC and blank groups. miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; si-circ_0001508, small interfering RNA of circ_0001508; si-NC, small-interfering RNA negative control.
The possible mechanism of action of hsa_circ_0001508 in HCC1937 cells is shown in Fig. 3F; it is suggested that hsa_circ_0001508 may act as carcinogenic based on biological function and related molecular regulation.
Discussion
The present results suggested that hsa_circ_0001508 serves as an oncogene in promoting BC progression. The findings of the Transwell and wound-healing assays, MTT analysis, as well as flow cytometry, revealed different expression levels of target proteins in cells within three different experiment groups: si-hsa_circ_0001508, si-NC and blank. These results demonstrate the cancer-promoting function of hsa_circ_0001508 in BC. Furthermore, there are no previous studies regarding the biological roles of hsa_circ_0001508 in major biomedical databases (PubMed). The present study provides empirical evidence to support the cancer-promoting function of hsa_circ_0001508 in BC. Therefore, the oncogene function of hsa_circ_0001508 could be a valuable focus for future research in the BC field, including diagnosis, treatment and prognosis.
hsa_circ_0001508 may play a vital role in regulating HCC1937 cells through the hsa_circ_0001508/miR-505-3p/HMGB1 axis. When hsa_circ_0001508 is silenced, the expression of the targeted gene HMGB1 decreases. While other researchers have reported a binding position [for example, 417-424(15)] between miR-505-3p and HMGB1, there is no research focusing on possible binding sites in HCC1973 cell lines. Additionally, the TargetScan results (Fig. 3E) predict one possible binding site, such as 3'UTR of HMGB1 mRNA position 934-940. In conclusion, it would be valuable to conduct a dual-luciferase assay to identify the binding site.
Furthermore, the expression of VGLL3 was significantly lower when hsa_circ_0001508 was silenced. Additionally, TargetScan (Fig. 3E) also indicated a potential binding site between miR-505-3p and VGLL3, specifically within the 3'UTR of HMGB1 mRNA at position 367-373. As there is no existing research on miR-505-3p/VGLL3 in BC, these findings could be valuable for future research. In addition to miR-505-3p and its targeted possible binding sites, the circMIR software (Table I) reported that there are two binding sites located at position 223 and 224 in the 3'UTR of miR-505, although there is no research supporting the competitive endogenous RNA crosstalk between hsa_circ_0001508 and miR-505-3p, resulting in negative regulation of miR-505 (inhibition) via sponge function to upstream target genes (HMGB1 and VGLL3) in the development of BC.
Nevertheless, the present study does provide some empirical evidence (for example, the expression of miR-505, HMGB1 and VGLL3 changed after hsa_circ_001508 silencing) to support the research value of hsa_circ_0001508 and its related axis (for example, hsa_circ_0001508/miR-505-3p/HMGB1 and hsa_circ_0001508/miR-505-3p/VGLL3).
The study provides empirical evidence using biomedical experimental tools and computer tools (for example, target prediction, and miRNA and mRNA interaction analysis) to support the biological function of hsa_circ_0001508 (BC activator) and two possible circRNAs/miRNAs/mRNA axes. These findings are valuable for researchers looking to expand their knowledge of the BC field. However, due to financial limitations, there is a lack of validation experiments in the present pilot study to investigate these signaling pathways, such as dual-luciferase assay. However, hsa_circ_0001508/miR-505-3p/HMGB1 and hsa_circ_0001508/miR-505-3p/VGLL3 have research value for further research.
BC has become a focal point in the biomedical field, with several researchers focusing on circRNAs, especially circRNAs/miRNAs/mRNA axis. The present study discovered a new circRNA (hsa_circ_0001508) using data from Gene Chip analysis and its novelty makes it valuable for research in the BC field. Certain potential binding sites were found between hsa_circ_0001508/miR-505-3p/HMGB1 and hsa_circ_0001508/miR-505-3p/VGLL3, providing empirical evidence to support that hsa_circ_0001508 could influence the expression of miR-505-3p, HMGB1 and VGLL3. Based on these findings, hsa_circ_0001508/miR-505-3p/HMGB1 and hsa_circ_0001508/miR-505-3p/VGLL3 are considered valuable pathways for future research to contribute more knowledge to the BC field.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be found in the Gene Expression Omnibus under accession number GSE182471 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182471. The data generated in the present study may be found in the Circular RNA Interactome database under accession number hsa_circ_0001508 or at the following URL: https://circinteractome.nia.nih.gov/api/v2/circsearch?circular_rna_query=hsa_circ_0001508+&gene_symbol_query=&submit=circRNA+Search.
Authors' contributions
ZS played a leadership position, being responsible for conducting the experimental process. FG was in another leadership position (principal investigator), developing research protocols and overseeing the data analysis. WD conducted bioinformatics analysis. JQ and LC performed statistical analysis and visual presentations of data. All authors read and approved the final version of the manuscript. ZS and FG confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ahmad A. Breast cancer: Current perspectives on the disease status. In: Breast Cancer Metastasis and Drug Resistance. Springer International Publishing AG, pp51-64, 2019. [Google Scholar]
- 2.Caswell-Jin JL, Sun LP, Munoz D, Lu Y, Li Y, Huang H, Hampton JM, Song J, Jayasekera J, Schechter C, et al. Analysis of breast cancer mortality in the US-1975 to 2019. JAMA. 2024;331:233–241. doi: 10.1001/jama.2023.25881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, Song C, Zhang J, Zhu L, Tang H. Circular RNAs in breast cancer diagnosis, treatment and prognosis. Oncol Res. 2023;32:241–249. doi: 10.32604/or.2023.046582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, Zhong Y, Wang X, Shen J, An W. Advances in circular RNA and its applications. Int J Med Sci. 2022;19:975–985. doi: 10.7150/ijms.71840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(30) doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G, Morrone A, Certo F, Broggi G, Caltabiano R, et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers (Basel) 2019;11(194) doi: 10.3390/cancers11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(11215) doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q, Zhang W. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(19) doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang M, Bai X, Zeng X, Liu J, Liu F, Zhang Z. circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta. 2021;523:120–130. doi: 10.1016/j.cca.2021.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Clough E, Barrett T. doi: 10.1007/978-1-4939-3578-9_5. The gene expression omnibus database. In: Methods in molecular biology. Springer, New York, NY, pp93-110, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Zhang Y, Gao Y, Cui Y, Liu H, Li M, Tian Y. Downregulation of HNF1 homeobox B is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;32:979–988. doi: 10.3892/or.2014.3297. [DOI] [PubMed] [Google Scholar]
- 12.Rasheed Z. Bioinformatics approach: A powerful tool for microRNA research. Int J Health Sci (Qassim) 2017;11:1–3. [PMC free article] [PubMed] [Google Scholar]
- 13.Dhumale SS, Waghela BN, Pathak C. Quercetin protects necrotic insult and promotes apoptosis by attenuating the expression of RAGE and its ligand HMGB1 in human breast adenocarcinoma cells. IUBMB Life. 2015;67:361–373. doi: 10.1002/iub.1379. [DOI] [PubMed] [Google Scholar]
- 14.Takakura Y, Hori N, Terada N, Machida M, Yamaguchi N, Takano H, Yamaguchi N. VGLL3 activates inflammatory responses by inducing interleukin-1α secretion. FASEB J. 2021;35(e21996) doi: 10.1096/fj.202100679RR. [DOI] [PubMed] [Google Scholar]
- 15.Yan J, Ying S, Cai X. MicroRNA-mediated regulation of HMGB1 in human hepatocellular carcinoma. Biomed Res Int. 2018;2018(2754941) doi: 10.1155/2018/2754941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanic M, Yanowski K, Gómez-López G, Socorro Rodriguez-Pinilla M, Marquez-Rodas I, Osorio A, Pisano DG, Martinez-Delgado B, Benítez J. MicroRNA expression signatures for the prediction of BRCA1/2 mutation-associated hereditary breast cancer in paraffin-embedded formalin-fixed breast tumors. Int J Cancer. 2015;136:593–602. doi: 10.1002/ijc.29021. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated in the present study may be found in the Gene Expression Omnibus under accession number GSE182471 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182471. The data generated in the present study may be found in the Circular RNA Interactome database under accession number hsa_circ_0001508 or at the following URL: https://circinteractome.nia.nih.gov/api/v2/circsearch?circular_rna_query=hsa_circ_0001508+&gene_symbol_query=&submit=circRNA+Search.