Abstract
Metastasis is a significant contributor to cancer-related mortality and a critical issue in cancer. Monitoring the changes in circulating tumor cells (CTCs) with metastatic potential is a valuable prognostic and predictive biomarker. CTCs are a rare population in the peripheral blood of patients with cancer. The enrichment process is extremely important for the isolation of clinically significant CTC subpopulations, which can then be used for further analysis. The present study postulates that the buffer serves as an essential field for immunomagnetic separation, thereby enhancing the efficacy of CTC enrichment in peripheral blood. This, in turn, facilitates CTC detection. Here, we describe the design of buffers for developing a novel immunomagnetic-negative separation method for CTC enrichment. During the design process, the buffer properties of the floating and cell coatings had a synergistic effect on the efficiency of cell enrichment in blood samples. The efficacy of the method was evaluated using peripheral blood samples from patients with non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). The developed method enriched clinically relevant CTC subpopulations that expressed the epithelial-mesenchymal transition (EMT)-related molecule vimentin and/or the cancer immune checkpoint marker programmed death ligand 1 (PD-L1). Furthermore, it was applicable as a part of the enrichment process in a TelomeScan® (OBP-401)-based CTC detection assay with high sensitivity and specificity. From the perspective of methodological approaches, the design of buffer properties can be useful for developing a highly versatile enrichment method for handling CTC heterogeneity.
Keywords: Buffer property, CTC enrichment, Liquid biopsy, Epithelial-mesenchymal transition, Immunomagnetic separation and lung cancer
Graphical Abstract
Highlights
-
•
A novel method for the enrichment of circulating tumor cell based on immunomagnetic-negative separation.
-
•
Design of physical properties of a buffer that serves as an essential filed for immunomagnetic separation.
-
•
Utilization of both a solvent for cell coating and a density gradient medium as buffer components.
-
•
Revealing a synergistic effect of the floating and cell coating of the buffer on the efficiency of cell enrichment.
-
•
Applicability of buffer design for the development of the versatile methods that are not affected by CTC heterogeneity.
1. Introduction
Metastasis is a major cause of death in cancer patients and remains a challenge for cancer treatment and detection [1]. Circulating tumor cells (CTCs) are live cancer cells released from a primary lesion into blood vessels and possess significant potential for metastasis to distant organs [2]. Higher CTC counts correlate with unfavorable outcomes such as therapeutic failure and cancer recurrence in various cancers, including breast [3], prostate [3], colorectal [3], lung [3], [4], pancreatic [5], gastric [6] and brain metastases [7]. Recent technological innovations such as next-generation sequencing (NGS) can accurately analyze gene expression profiles, leading to the discovery of valuable information for assessing clinical status [8]. Therefore, CTC alterations have great potential as prognostic and predictive biomarkers.
The enrichment of CTCs in blood samples is extremely important for obtaining clinically valuable information. CTCs are rare cells that circulate among a large number of normal cells, including immune cells, red blood cells, and platelets [9]. CTC detection tests such as the US Food and Drug Administration (FDA) CellSearch System (Veridex, LLC, Raritan, NJ) involve two processes: 1) enrichment and 2) characterization of CTC subpopulations [4]. Technologies with a high potential for cell enrichment are favorable for application as part of the enrichment process in CTC detection tests. Extensive studies have aimed to improve the efficiency of cell enrichment in the analysis of clinically important CTC subpopulations associated with cancer recurrence and poor prognosis [10], [11], [12]. Epithelial-mesenchymal transition (EMT) is the process by which epithelial cells acquire mesenchymal properties and promote CTC generation by acquiring cell mobility [13]. EMT-transformed CTCs are characterized by the loss of epithelial cell markers, such as epithelial cell adhesion molecule (EpCAM), and gain of mesenchymal markers, such as vimentin and N-cadherin [13]. An association between the expression and related signaling of programmed death ligand 1 (PD-L1) and EMT has been reported [14]. In addition to tumor-related antigens, cellular characteristics such as size [15], deformability [16], and density [17] are fundamental to the development of CTC enrichment methods. CTC enrichment methods can be broadly categorized into positive and negative selection methods. Positive selection depends on tumor-related antigens and captures CTCs directly from blood samples using an antibody that binds to a tumor-related antigen. The CellSearch system uses a positive-selection-based strategy to target EpCAM-positive CTC [4]. Positive selection directly captures CTC and was initially considered as the ideal approach for CTC enrichment. However, positive selection cannot deal with changes in tumor-related antigens. For example, EpCAM-based positive selection cannot capture EMT-transformed CTCs owing to the loss of EpCAM expression [13]. In contrast, negative selection is independent of tumor-related antigens and leaves CTCs by removing normal cells from the blood samples. The antibodies expressed on immune cells such as CD45 and CD66b are used in combination with centrifugation and microfluidic technology to remove normal cells [18], [19]. Negative selection can collect CTC subpopulations independent of changes in tumor-related antigens and has recently become a standardized method for CTC enrichment [12], [18], [19]. CTCs comprise diverse subpopulations and there is currently no ideal CTC marker available. An enrichment method was developed based on cellular properties. CTC heterogeneity leads to the formation of diverse subpopulations with varying molecular and physical properties. This implies that a single method is inadequate for enriching all CTC subpopulations. In other words, each CTC enrichment method has its cell enrichment properties and researchers must select a method according to each clinical or research objective. Consequently, the development of novel CTC enrichment methodologies remains a significant challenge for addressing the heterogeneity of CTCs.
In this study, we described a novel CTC enrichment method based on the design of a buffer for immunomagnetic-negative separation. Immunomagnetic-negative separation removes normal cells from blood samples of cancer patients diluted in a buffer, which results in the floating of CTCs for enrichment. The buffer serves as a place for CTC after both antigen–antibody reactions targeting surface antigens expressed on normal cells and magnetic separation. From the perspective of the underlying separation principle, the buffer used for immunomagnetic separation may affect cell enrichment efficiency. In the present study, we postulated that the physical properties of the buffer are crucial for efficient cell enrichment during immunomagnetic-negative separation. During the design process, the buffer properties of the floating and cell coatings had a synergistic effect on the enrichment of spiked cancer cells in blood samples. Furthermore, the present study demonstrated that the developed method was applicable for the enrichment of EMT-related CTC subpopulations in patients diagnosed with non-small cell lung cancer (NSCLC) or small cell lung cancer (SCLC).
2. Materials and methods
2.1. Ethical approval of the study protocol and patient characteristics
This study was conducted at Juntendo University Hospital (Tokyo, Japan), and has been carried out in accordance with the Declaration of Helsinki. A total of 42 patients with lung cancer at Juntendo University Hospital provided peripheral venous blood samples between August 2022 and June 2023. The histological types of the patients who participated in the present study are summarized in Table 1. The study protocol was approved by the Institutional Review Board of the Juntendo University Hospital (approval ID: E21–0222). Written informed consent was obtained from all the patients. Additionally, blood samples (6 mL) were obtained from patients with noncancerous lung diseases, including nontuberculous mycobacteria (NTM), interstitial pneumonia (IP), and from healthy volunteers. The pathological and clinical stage was determined in accordance with the current tumor, node, and metastasis (TNM) classification.
Table 1.
Characteristics of patients with lung cancer and a summary of CTC positivity as enrichment capability of the developed method.
| (More than 1 CTC in 6 mL of peripheral blood defined as positive) | |||||
|---|---|---|---|---|---|
| CTC phenotype | CTC | PD-L1 (+)EMT (-) CTC | PD-L1 (-)EMT (+) CTC | PD-L1 (+)EMT (+)CTC | Total PD-L1 (+) CTC* |
| All patients (n = 42) | 30 (71.4%) | 14 (33.3%) | 11 (26.2%) | 19 (45.2%) | 25 (59.5%) |
| NSCLC | |||||
| All patients (n = 35) | 26 (74.3%) | 12 (34.3%) | 8 (22.9%) | 16 (45.7%) | 22 (62.9%) |
| Histological type | |||||
| Adenocarcinoma (n = 18) | 12 (66.7%) | 3 (16.7%) | 6 (33.3%) | 9 (50.0%) | 10 (55.6%) |
| Squamous cell carcinoma (n = 9) | 9 (100%) | 5 (55.6%) | 2 (22.2%) | 6 (66.7%) | 8 (88.9%) |
| Others (n = 8) | 5 (62.5%) | 4 (50.0%) | 0 (0%) | 1 (12.5%) | 4 (50.0%) |
| SCLC | |||||
| All patients (n = 6) | 3 (50.0%) | 1 (16.7%) | 2 (33.3%) | 2 (33.3%) | 2 (33.3%) |
| Pleomorphic carcinoma | |||||
| All patients (n = 1) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) |
| Stage | |||||
| 0 (n = 1) | 1 (100%) | 0 (0%) | 1 (100%) | 1 (100%) | 1 (100%) |
| I (n = 11) | 9 (81.8%) | 6 (54.5%) | 2 (18.2%) | 5 (45.5%) | 8 (72.7%) |
| II (n = 3) | 2 (66.7%) | 1 (33.3%) | 1 (33.3%) | 2 (66.7%) | 2 (66.7%) |
| III (n = 10) | 8 (80.0%) | 4 (40.0%) | 3 (30.0%) | 4 (40.0%) | 6 (60.0%) |
| IV (n = 17) | 10 (58.8%) | 3 (17.6%) | 4 (23.5%) | 7 (41.2%) | 8 (47.1%) |
| Postoperative recurrence in NSCLC | |||||
| All patients (n = 4) | 3 (75.0%) | 1 (25.0%) | 1 (25.0%) | 2 (50.0%) | 2 (50.0%) |
The CTC positivity was calculated based on the detection of CTCs and the EMT-related CTC subpopulations (≥1 CTC), *calculated using the sum of PD-L1(+)EMT(-) CTC and PD-L1(+)EMT(+) CTCs.
2.2. Cell culture
Human lung cancer cell line A549 was cultured in Dulbecco’s modified Eagle’s medium (DMEM) (FUJIFILM Wako Pure Chemical, Osaka, Japan) supplemented with 10% heat-inactivated fetal calf serum (FCS, Cosmo Bio, Tokyo, Japan), 100 U/mL penicillin, and 100 μg/mL streptomycin (FUJIFILM Wako Pure Chemical, Osaka, Japan). Cells were cultured at 37 °C in a humidified incubator with 5% CO2 for subsequent analyses.
2.3. Spike-in experiments for evaluating the effect of buffer density
Cultured A549 cells were labeled with the fluorescent dye PKH26, which is used for general cell membrane labeling, using Red Fluorescent Cell Linker Kit (Sigma-Aldrich, St. Louis, MO, USA), and then fixed with 4% paraformaldehyde (4% PFA, FUJIFILM Wako Pure Chemical, Osaka, Japan). After washing with 2% FCS D-PBS(-), the fluorescently labeled A549 cells were used for the spike-in experiments. The EasySep™ Direct Human CTC Enrichment Cocktail (STEMCELL Technologies, Vancouver, Canada) was utilized as an antibody cocktail to remove hematopoietic cells and platelets. The buffer was designed using the following density gradient media: LymphoprepTM (d=1.077 medium, Abbott Diagnostics Technologies, IL, USA) with a density of 1.077 g/mL, Ficoll-Paque PREMIUM 1.084 (d=1.084 medium, Sigma-Aldrich) with a density of 1.084 g/mL, and a lymphocyte separation solution (d=1.119 medium, NACALAI TESQUE, Kyoto, Japan) with a density of 1.119 g/mL. On-chip T-buffer (On-chip Biotechnologies, Tokyo, Japan) was used as a solvent to dilute the density gradient medium. Details of the spike-in experiments are summarized in Supplementary Fig. S2. The separation buffer was prepared by combining each density gradient medium with 2% FCS T-buffer and 50 mM ethylenediaminetetraacetic acid (EDTA)/D-PBS(-) at a ratio of 2:1:0.5, respectively (density gradient medium/2%FCS T-buffer/EDTA). Fluorescently labeled A549 cells were spiked into either 6 mL of separation buffer or 6 mL of a blood sample diluted with 6 mL of separation buffer. The resuspension buffer was prepared by combining the density gradient medium with 2% FCS T-buffer at a ratio of 2:1 (density gradient medium/2% FCS T-buffer). All the procedures were performed at room temperature. The fluorescently labeled A549 cells were spiked into a tube containing 6 mL of the separation buffer, which was further supplemented with 100 µL of human FcR Blocking Reagent (Miltenyi Biotec, Bergisch Gladbach, Germany) and incubated for 10 min. Upon spiking the labeled A549 cells into 6 mL of the blood sample diluted with 6 mL of the separation buffer, an additional 300 µL of the EasySep™ Direct Human CTC Enrichment Cocktail (STEMCELL Technologies) was additionally added to the reaction tube and further incubated for 5 min. Subsequently, 300 µL of EasySep™ Direct RapidSpheres™ 50300 magnetic beads (STEMCELL Technologies) were added to the reaction tube and incubated for 10 min. The tube was then placed into “The Big Easy" EasySep™ Magnet stand (STEMCELL Technologies) for the initial magnetic separation. After additional incubation for 10 min, the supernatant was collected in a 50 mL tube containing 25 mL of 2% FCS/D-PBS(-). The pellet was resuspended in 6 mL of resuspension buffer, and the tube was placed on a magnetic stand for a second magnetic separation. After incubation for 10 min, the supernatant was collected in the same 50 mL tube utilized during the initial magnetic separation. The third magnetic separation step was conducted in a manner similar to that previously described. The magnetic separation process was repeated thrice. The tube containing all the supernatants was centrifuged at 200g for 10 min. Subsequently, the pellet was resuspended in 1 mL of 2% FCS T-buffer following centrifugation at 500g for 5 min. Following the removal of the supernatant, the pellet was resuspended in 200 µL of 2% FCS T-buffer. Enriched-labeled A549 cells were seeded in a 96-well glass-bottomed plate. The images were captured using an all-in-one fluorescence Microscope BZ-X800 (KEYENCE, Osaka, Japan). A BZX TRITC dichroic filter (OP-87764, KEYENCE) was used to examine the PKH26 fluorophore in the enriched A549 cells. The recovery rate of labeled A549 cells was calculated using the following equation: number of enriched A549 cells/number of spiked A549 cells × 100 (%).
2.4. Spike-in experiments for evaluating the effect of both d= 1.119 medium and 2%FCS T-buffer as buffer components
The details of the spike-in experiments are summarized in Supplementary Fig. S3. To examine the effect of 2% FCS T-buffer, a separation buffer was prepared using 2% FCS T-buffer and 50 mM EDTA/D-PBS(-) at a ratio of 1: 0.5. A 2% FCS T-buffer was used as the resuspension buffer. To examine the effect of the additional use of d= 1.119 medium, a separation buffer was prepared using d= 1.119 medium, 2% FCS T-buffer, and 50 mM EDTA/D-PBS(-) at a ratio of 2:1:0.5, respectively. The resuspension buffer was prepared using d= 1.119 medium and 2% FCS T-buffer at a ratio of 2:1. The remaining procedure was conducted in a manner analogous to that described above, except for the collection of the supernatants after each magnetic separation. After each magnetic separation step, the supernatant was collected and used to calculate the recovery rate of the spiked cells.
2.5. Density and viscosity measurements
Density measurements were performed at 22–24 °C using a standard specific gravity meter (TOA KEIKI MFG, Tokyo, Japan). The density was corrected using temperature compensation. To measure the effect on buffer viscosity changes, polyoxyethylene (20) sorbitan monolaurate (Tween 20; FUJIFILM Wako Pure Chemical) was diluted with either 2% FCS T-buffer or 2% FCS D-PBS (-) in a 1:1 ratio. Viscosity was measured at room temperature using a Visco Tester VT-06 (RION, Tokyo, Japan).
2.6. CTC enrichment from patients with lung cancer using the developed method and analysis of its subtype
The same procedure depicted in Supplementary Fig. S3B was used for CTC enrichment, which represents the final iteration of the developed method. The details of all procedures for the CTC enrichment of blood samples, green fluorescent protein (GFP) labeling of cells by TelomeScan, and immunostaining are summarized in the Supplementary Information. CTC- and EMT-related subpopulations were identified based on the threshold levels of each fluorophore and cell shape obtained from image analysis. DAPI-, GFP-positive-, and CD45-negative cells were defined as CTCs. PD-L1(+) Vimentin(-) DAPI(+) GFP(+) CD45(-) cells, PD-L1(-) Vimentin(+) DAPI(+) GFP(+) CD45(-) cells, and PD-L1(+)Vimentin(+) DAPI(+) GFP(+) CD45(-)cells were defined as PD-L1(+) EMT(-) CTC, PD-L1(-) EMT(+) CTC, and PD-L1(+) EMT(+) CTC, respectively. The total number of PD-L1(+) CTC was expressed as the sum of PD-L1(+) EMT(-) CTC and PD-L1(+) EMT(+) CTC. Image analysis was conducted using Python image-processing libraries such as OpenCV and Fiji software [20].
2.7. Statistics
The Mann–Whitney U test was used for comparison between two groups whose sample distributions were asymmetrical. To conduct multiple comparisons, the Mann–Whitney U test was applied, followed by the Holm–Sidak method, which was conducted in two groups with asymmetrical sample distributions. The Kruskal–Wallis test followed by Dunn’s multiple comparison test was used to compare groups with asymmetrical sample distributions. Statistical significance was set at p < 0.05. All statistical analyses were performed using GraphPad Prism9 for Windows (GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. Density of the buffer affects cell enrichment efficiency
In a medium with a range of densities, the cells either sink or float to a location where their density is equal to that of the surrounding medium [21]. Blood samples have a higher viscosity than water [22]. Cell death releases cellular DNA and debris, which can cause cell aggregation. Therefore, the buffer plays a role in reducing the viscosity of the blood sample and in the loss of enriched cells by non-specific absorption. The molecular components of the buffer can determine its physical properties, such as pH, density, and viscosity [23], [24].
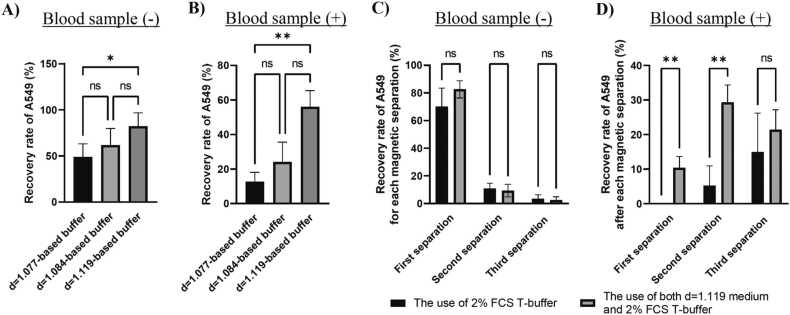
It was postulated that buffer density might influence the ratio of cells remaining in the buffer following magnetic separation. In addition, the physical properties of the cell coating were considered in relation to the cell density. Cationic polymers can prevent cellular aggregation by affecting the electrostatic interactions between cells [25]. The on-chip T-buffer (T-buffer) serves as a solvent to prevent cell adhesion and thus cell loss [26]. Subsequently, we used a density gradient medium and T-buffer as the buffer components to devise a buffer for immunomagnetic-negative separation. The buffer was designed based on spike-in experiments. A summary of the components of the spike-in experiments is provided in Supplementary Fig. S1. The human lung cancer cell line A549 was fluorescently labeled and used to calculate the recovery rate. The antibody cocktails and magnetic beads in EasySep™ Direct Human CTC Enrichment kit were used to remove hematopoietic cells and platelets from the blood sample. First, the effect of buffer density on the recovery rate was evaluated. Details of the spike-in experiments are summarized in Supplementary Fig. S2. The separation buffer (density gradient medium/2% FCS T-buffer/EDTA) contained EDTA to reduce the effects of calcium ions, which can cause aggregation in blood samples. A resuspension buffer (density gradient medium/2% FCS T-buffer) was used to suspend the pellet to facilitate repeated magnetic separation. Three density gradient media (d=1.077, d=1.084, and d=1.119) were chosen for buffer preparation. Figs. 1A and 1B show the total recovery rates of the spiked cells after repeating the magnetic separation process. The use of d= 1.119 medium as buffer components demonstrated a higher recovery rate of the spiked cells than that of d= 1.077 medium, resulting in a 1.7-fold higher recovery rate of the spike cells (Fig. 1A). When the cells were spiked into the blood sample, the effect of density on the recovery rate exhibited a notable disparity between the use of d= 1.119 and d= 1.077 media, resulting in a 4.4-fold higher recovery rate of spiked cells (Fig. 1B). Among the components of the buffer, the d= 1.119 medium was more suitable than the other density gradient media.
Fig. 1.
Use of the most-dense gradient medium as buffer components showed highest cell enrichment efficiency by a synergistic effect with the T-buffer. Recovery rate of A549 cells spiked in either A) buffer only or B) the blood sample diluted with the buffer. A, B) Data are expressed as the mean ± SD (N = 6). *p < 0.05, * *p < 0.01, ns (non-significantly different), compared by Kruskal–Wallis test followed by Dunn‘s multiple comparison test. The recovery rate of the spiked A549 cells in either C) only the 2% FCS T-buffer or D) the buffer containing d= 1.119 medium and 2% FCS T-buffer at either each magnetic separation. Spike-in experiments were performed in either buffer or a blood sample diluted with buffer. C, D) Data are expressed as the mean ± SD (N = 6), * *p < 0.01, ns (non-significantly different), compared by Mann–Whitney U test followed by the Holm–Sidak method for comparison of the recovery rate in each magnetic separation.
3.2. Use of both the density gradient medium and T-buffer as buffer components shows a synergistic effect on cell enrichment efficiency from blood samples
Using d= 1.119 medium as a buffer component enhanced the cell enrichment efficiency. However, it remains unclear whether the buffer density or the properties of the cell coating exert a more pronounced influence on cell enrichment efficiency. Subsequently, the spike-in experiments were conducted using either 2% FCS T-buffer or a buffer containing d= 1.119 medium and 2% FCS T-buffer. The details of the spike-in experiments are summarized in Supplementary Fig. S3. The recovery rate of the spiked cells was calculated at each step (Figs. 1C and 1D) or overall (Supplementary Fig. S4A and S4B) after three magnetic separations. When the cells were spiked into only the buffer without the blood sample, there was no significant effect of d= 1.119 medium in 2% FCS T-buffer on the recovery rate of the spiked cells at each step (Fig. 1C) or overall (Supplementary Fig. S4A). Conversely, the effect of the d= 1.119 medium was observed when the cells were spiked into the blood sample (Fig. 1D and Supplementary Fig. S4B). The use of both d= 1.119 medium and 2% FCS T-buffer helped recover 10% of the total spiked cells from the blood sample after the first magnetic separation, whereas the use of the 2% FCS T-buffer/EDTA failed to enrich the spiked cells after the first separation (Fig. 1D). Repetition of the magnetic separation procedure resulted in an enhanced recovery rate of the spiked cells from the blood sample (Supplementary Fig. S4B). The addition of a density gradient medium altered the physical properties of the buffer, thereby facilitating the diffusion of spiked cells trapped in the buffer from the antibody-magnetic bead complexes.
These results demonstrate that the use of both d= 1.119 medium and T-buffer as buffer components had a synergistic effect on cell enrichment efficiency. The T-buffer can coat the surface of cells and prevent cell adhesion [26], which may lead to a decrease in the viscosity of blood samples. The effect of the T-buffer on the buffer viscosity was also evaluated indirectly in an experiment using a highly viscous liquid instead of a blood sample (Supplementary Table S1). Whole blood has both hydrophilic and hydrophobic properties owing to its cellular and molecular composition [27]. Polyoxyethylene (20) sorbitan monolaurate (Tween 20) is a surfactant with a hydrophilic ethylene glycol head group and a hydrophobic alkyl tail. Tween 20 is a highly viscous liquid compound with a viscosity of 4.4 dPa⸳s (Supplementary Table S1). For viscosity measurements, Tween 20 was diluted with either 2% FCS T-buffer or 2% FCS D-PBS (-). Addition of 2% FCS T-buffer reduced the viscosity of Tween 20 from 4.4 dPa⸳s to 1.3 dPa⸳s (Supplementary Table S1). The T-buffer reduces the viscosity of the highly viscous liquid. The effect of the T-buffer on viscosity was similar to that caused by the addition of 2% FCS D-PBS (-) (Supplementary Table S1). These results indicate the major role of the T-buffer in the designed buffer to coat the cells and prevent cell loss by reducing nonspecific absorption (Supplementary Fig. S4B and Supplementary Table S1).
The spiked-in experiments demonstrated the importance of the presence of a d= 1.119 medium for cell enrichment efficiency (Fig. 1). To evaluate the molecular mechanisms underlying the synergistic effects on cell enrichment, the densities of the designed buffers were measured. The densities of the designed buffers are summarized in Supplementary Table S2. The density of 2% FCS T-buffer was 1.010 g/mL (Supplementary Table S2). The addition of EDTA solution had little effect on the density of 2% FCS T-buffer. In contrast, the addition of d= 1.119 medium in 2% FCS T- buffer resulted in a difference in density value of approximately 0.07. The densities of the separation (d=1.119 medium/2% FCS T-buffer/EDTA) and resuspension buffers (d=1.119 medium/2% FCS T-buffer) were 1.075 and 1.085 g/mL, respectively (Supplementary Table S2). These results indicate that the addition of d= 1.119 medium not only changed the density of the designed buffer but also maintained the cell-coating property of the T-buffer.
3.3. The developed method enriches CTC and the EMT-related subpopulations from peripheral blood of patients with lung cancer
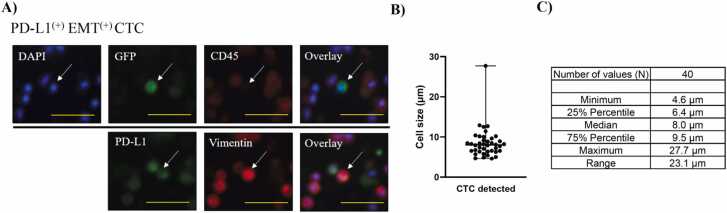
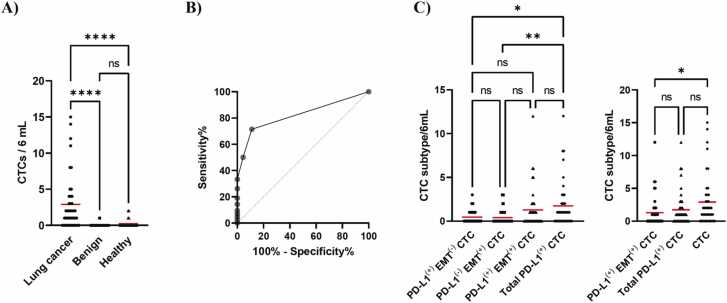
The efficacy of the developed method for enriching CTCs was evaluated using peripheral blood samples obtained from patients with lung cancer. The same procedure depicted in Supplementary Fig. S3B was used for CTC enrichment, which represents the final iteration of the developed method. Fluorescence labeling and immunostaining are frequently used for CTC characterization [28]. A high telomerase activity is a hallmark of cancer cells [29]. In the present study, we used GFP labeling of CTCs with TelomeScan OBP-401 [30], [31], and immunostaining to define CTCs as GFP(+) DAPI(+) CD45(-) cells. Thus, TelomeScan has the potential to detect highly positive CTCs in patients [31]. The developed method successfully captured CTCs from patients with lung cancer, and CTC positivity is summarized in Table 1. The positivity of CTC (≥1 CTC) was calculated based on the number of the CTC including PD-L1(+) EMT(-) CTC, PD-L1(-)EMT(+) CTC, PD-L1(+) EMT(+) CTC, and total PD-L1(+) CTC. Peripheral blood samples (6 mL) were obtained from 35 patients diagnosed with NSCLC and six patients diagnosed with SCLC. CTC enrichment was performed in a patient diagnosed with pleomorphic carcinoma, a rare histological subtype of lung cancer, using a newly developed method. The presence of CTC was identified in 74.3% of patients with NSCLC and 50% of patients with SCLC, respectively (Table 1). The developed method enriched CTC, including EMT-related subpopulations that exist in major histological types of patients with NSCLC and SCLC (Table 1). The developed method was also employed to enrich CTC from the blood samples of patients diagnosed with pleomorphic carcinoma, as well as from the blood samples of 75% of the patients who underwent postoperative recurrence (Table 1). The mean number of CTCs (≥1 CTC) with standard deviation (SD) is presented as follows: CTC (4.1 ± 4.1 cells/6 mL), PD-L1(+) EMT(-) CTC (1.4 ± 0.6 cells/6 mL), PD-L1(-) EMT(+) CTC (1.5 ± 0.8 cells/6 mL), PD-L1(+) EMT(+) CTC (2.8 ± 2.9 cells/6 mL) and total PD-L1(+) CTC (2.9 ± 2.9 cells/6 mL). Fig. 2A shows the fluorescent images of PD-L1(+) EMT(+) CTC. The size distribution of the enriched CTCs was obtained from image analysis and is shown in Fig. 2B. The median cellular size was 8.0 µm with a range of 23.1 µm (Fig. 2C). These results indicated that the developed method was capable of capturing CTC and clinically significant EMT-related subpopulations.
Fig. 2.
The developed enrichment method captures CTCs including the EMT-related subpopulations. A) Fluorescent images of PD-L1(+) EMT(+) CTC defined as PD-L1(+)Vimentin(+)DAPI(+)GFP(+)CD45(-) were summarized, bar size: 25 µm. B) Size distribution (N = 40) of the enriched CTCs was plotted in a graph. C) Mean, min, max, and percentile vales of the size was summarized in a table.
3.4. The developed method has applicability as part of the enrichment process in a TelomeScan-based CTC detection assay
A CTC detection test comprises two distinct steps 1) enrichment and 2) characterization of CTCs. To assess the applicability of the developed method as a component of the enrichment process in a TelomeScan-based CTC detection assay, we also applied the method to analyze blood samples from non-cancerous individuals to calculate the specificity and ROC curve, which reflects its diagnostic accuracy. Fig. 3A presents a summary of the number of CTC in 42 patients with lung cancer and the number of false-positive cells detected in 45 non-cancerous individuals, including 20 patients with benign disease and 25 healthy donors. In the present study, we used image analysis to distinguish between CTCs (data not shown). Although a few number of false positive cells were detected in non-cancerous individuals, the use of the developed method as part of the CTC detection test yielded 88.9% of specificity (Fig. 3A). The ROC curve was generated using a cutoff of one CTC (Fig. 3B). The area under the curve (AUC) value was 0.8209 (95% CI: 0.7280–0.9138, p < 0.0001). These results indicate that the developed method is applicable to a portion of the enrichment step in CTC detection tests. Additionally, the number of CTCs was plotted for each stage from I to IV (Supplementary Fig. S5A). There was no significant difference in the number of CTC between each stage and stage intervals of 0-I, II-III, and IV (Supplementary Fig. S5B), as partially reported by Togo et al. [31]. Finally, we summarized the number of CTCs detected in the CTC subpopulations of 42 patients with lung cancer (Fig. 3C). The developed method captured CTCs with high efficiency, including those belonging to EMT-related subpopulations. Based on these findings, we devised a novel CTC enrichment method that leveraged the properties of the buffer.
Fig. 3.
Potential of the developed enrichment method as part of an enrichment process in a TelomeScan-based CTC detection assay. A) Number of CTC detected in 6 mL of the peripheral blood sample from patients with and without cancer was plotted and red bars expressed the mean of data, * ** *p < 0.0001, ns (non-significantly different), compared by Kruskal–Wallis test followed by Dunn's multiple comparison test. B) ROC curve was calculated with CTC cutoff value of 1. C) Number of CTC re-plotted at each CTC subpopulation is shown. Each red bar expresses the mean of the data, *p < 0.05, * *p < 0.01, ns (non-significantly different), compared by Kruskal–Wallis test followed by Dunn's multiple comparison test.
4. Discussion
Density gradient media has been used to enrich cells, including immune [32] and cancer cells [17]. In a medium with a range of densities, cells either sink or float to a location where their density is equal to that of the surrounding medium [21]. The density of a density gradient medium is affected by the physical properties of the molecules in the medium [24]. Initially, density gradient media such as albumin [33] and silicon-polyvinylpyrrolidone (PVP) medium [34] were used to collect malignant cells from patients with cancer. Subsequently, polysucrose-based density gradient media, such as Ficoll [35], and a density gradient centrifugation-based system, Oncoquick [36], have been used to enrich cancer cells. Polysucrose is a hydrophilic sucrose polymer that is used to prepare various density gradient media by diluting polysucrose solution [37].
The density gradient method is quite simple; however, the rarity and heterogeneity of CTC have necessitated the enrichment method to use combined technologies rather than simple technology to prevent the loss of diverse CTC subpopulations. More recently, immunomagnetic separation has become the standardized method for cell enrichment owing to its flexibility of development [38]. This separation method is based on assays consisting of antigen-antibody reactions and magnetic separation performed in the buffer used to dilute the blood sample. Most previous studies focused on the type of antibody that binds to the antigen in either CTCs or normal cells for immunomagnetic separation [39], [40], [41]. Previous studies have utilized typical buffers such as phosphate buffered saline (PBS) and a lysis buffer containing NH4Cl and KHCO3 to dilute blood samples and perform immunomagnetic separation [42], [43]. Based on these studies, we postulated that the physical properties of the buffer are critical for improving the efficiency of cell enrichment during immunomagnetic separation. In accordance with the fundamental principles of the separation process, the buffer must possess the properties required to facilitate cell floatation during magnetic separation.
In the present study, we used a Ficoll-based density gradient medium and solvent to coat the cells and prevent loss of CTC. In addition to the density gradient medium, we anticipated that the cell-coating property would be useful for diffusing CTCs into the buffer during magnetic separation. On-chip T-buffer (T-buffer) coats cells, thereby preventing nonspecific absorption [26]. The present study used spike-in experiments to evaluate the effect of buffer components on cell enrichment efficiency (Supplementary Fig. S1). The antibody cocktails were utilized in conjunction with the EasySep™ Direct Human CTC Enrichment Cocktail to facilitate removal of normal cells. The antibody-magnetic bead complex bound to CD2, CD14, CD16, CD19, CD45, CD61, CD66b, and glycophorin A surface markers, resulting in the removal of hematopoietic cells and platelets from blood samples. The use of a density gradient medium with a density of 1.119 g/mL (d=1.119 medium) as a buffer component resulted in a higher recovery rate of spiked cells than that observed with a d= 1.077 medium (Fig. 1A). The significance of d= 1.119 medium as a buffer component was clearly demonstrated when performing spike-in experiments on the blood sample (Fig. 1B).
In principle, spiked A549 cancer cells could be retained in the supernatant during immunomagnetic separation. However, only 2% FCS T-buffer as the buffer component was not sufficient for the high recovery rate of the spiked cells. Upon spiking the cells into the blood sample, the effect of repeated suspension of the pellet after each magnetic separation was more pronounced in the buffer containing d= 1.119 medium and 2% FCS T-buffer than in the 2% FCS T-buffer alone (Fig. 1D and Supplementary Fig. S4B). The repeated steps of suspension and magnetic separation of the pellet resulted in an increased total recovery rate of spiked A549 cells captured in the antibody-magnetic bead complexes. The addition of a density gradient medium to the T-buffer changed the physical properties of the buffer and allowed the spiked cells to float to the supernatant.
The specific gravity of cells is an important factor when discussing cell enrichment methods based on density gradient media. The specific gravities of red blood cells, leukocytes, and cancer cells were 1.092, 1.065, and 1.056, respectively. Therefore, a medium with a density of 1.075 g/mL was chosen to centrifuge denser cells such as blood cells and polymorphonuclear neutrophil leukocytes at a higher rate than cancer cells [34]. The density gradient method, using a typical density gradient Lymphoprep medium at a density of 1.077 g/mL, has been demonstrated to enrich the CTCs in NSCLC with a reported CTC positivity of 13% [10]. Heterogeneity in CTCs may result in the formation of subpopulations with densities > 1.056 g/mL. Cell density can be altered by several factors, including the cell cycle, metabolic status, differentiation, and pathological conditions [44]. Consequently, the use of a density gradient medium alone may not be sufficient to effectively float CTCs in a buffer [10]. The designed buffer exhibited a density comparable to that of a previous study (1.075 g/mL and 1.085 g/mL, respectively), yet demonstrated superior capacity for CTC enrichment (71.4%) in NSCLC patients (Table 1) than the previous study [10]. The addition of the d= 1.119 medium to 2% FCS T-buffer resulted in a difference in the density value of approximately 0.07, while maintaining the property of cell coating in the designed buffer (Supplementary Table S2). The T-buffer serves as a solvent to prevent cell adhesion and thus cell loss [26]. The addition of T-buffer reduced the viscosity of the blood samples, which was supported by the results of the Tween 20 assay (Supplementary Table S1). Cationic polymers can prevent cellular aggregation by affecting the electrostatic interactions between cells [25]. The T-buffer in the designed buffer might exert its potential through an action similar to that of cationic polymers [25]. Compared to the use of only the density gradient medium [10], the supplementary property of cell coating with T-buffer resulted in a reduction in the nonspecific absorption and viscosity of the blood sample, thereby facilitating the diffusion of cells into the buffer.
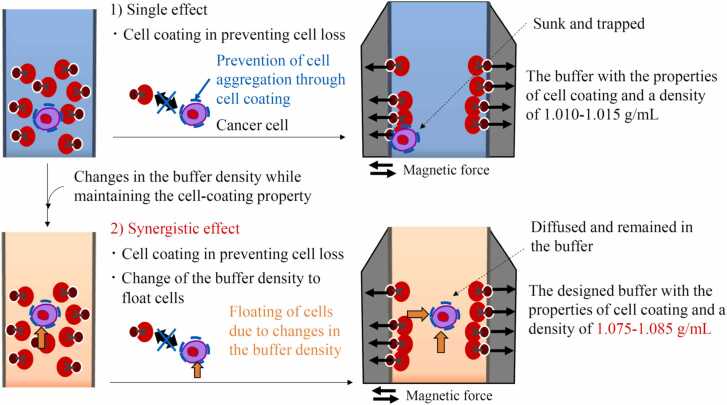
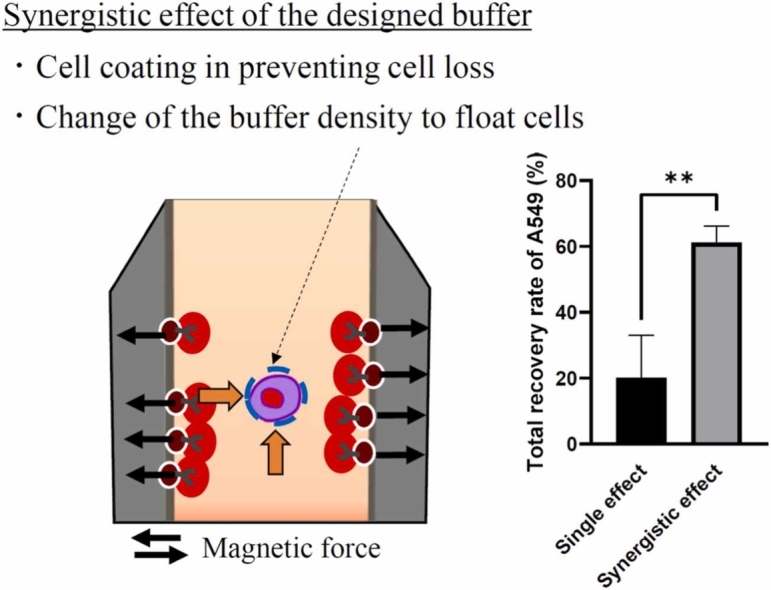
From the results of the spike-in experiments and the CTC positivity in the blood sample from patients with lung cancer, a summary of the potential model underlying the synergistic effect of the designed buffer on CTC enrichment is shown in Fig. 4. The T-buffer plays a role in coating cells and preventing cell loss. In addition, the density gradient medium changed the buffer density suitable for CTC enrichment while maintaining the cell-coating properties of the T-buffer. The utilization of both density gradient medium (d=1.119) and T-buffer resulted in the generation of a novel buffer property, thereby enhancing the efficiency of cell enrichment through the diffusion of cells during the magnetic separation process. The developed method captured the CTC populations with a median size of 8.0 µm and a range of 23.1 µm (Fig. 2C). The heterogeneous nature of the CTCs results in a size range of 7 µm to 30 µm, as observed in previous studies [12], [45], [46]. Additionally, smaller CTCs, with a diameter of less than 5 µm, have been previously documented [38]. The developed method was applicable to the enrichment of CTCs of varying sizes, including those of small and large magnitudes (Fig. 2C). Previous studies have utilized typical buffers, such as phosphate buffered saline (PBS) and a lysis buffer containing NH4Cl and KHCO3, to dilute blood samples and perform immunomagnetic separation [42], [43]. No information was available regarding the buffer components used in the CellSearch system. In contrast, this study was initiated with a buffer design based on the characteristics of cell coating and floating. This study is the first to propose a model based on the synergistic effect of cell coating and floating in immunomagnetic-negative separation for CTC enrichment.
Fig. 4.
A possible mechanism underlying the synergistic effect of the designed buffer on CTC enrichment.
Given the heterogeneity of CTCs, researchers must select the appropriate enrichment method according to the specific research or clinical objective in question. The developed method in the present study was employed as a component of enrichment process in a TelomeScan-based CTC detection assay, resulting in a positivity rate of 71.4% and a sensitivity rate of 88.9% (Fig. 3A). Additionally, this method exhibited a high AUC (Fig. 3B). The Food and Drug Administration-approved CellSearch system employs EpCAM-dependent immunomagnetic-positive separation to enrich epithelial CTC subpopulations [4]. This enrichment method has the capacity to capture EpCAM-positive CTCs with a positivity rate of 83% and a high PD-L1 status in the NSCLC patients [47]. In contrast, our method was suitable for enriching the EMT-related mesenchymal CTC subpopulations (Table 1) that were not enriched using the EpCAM-based method. Recent studies have reported a correlation between PD-L1 expression and EMT during osimertinib treatment [14], [48], and the significance of PD-L1 positive CTC monitoring during treatment with immune checkpoint inhibitors, including nivolumab, pembrolizumab, and atezolizumab [47], [49]. Immune checkpoint inhibitors have become the standard of care for all cancers [50]. These recent studies highlight the importance of analyzing the EMT process and PD-L1 expression in a clinical context. Consequently, an enrichment method capable of capturing both PD-L1-single positive and PD-L1/vimentin-double-positive EMT-transformed CTC subpopulations is of significant value for further analysis with the potential to uncover new insights into metastasis and poor response to treatment. The immunomagnetic-negative separation method is not dependent on the presence of tumor-related antigens on CTCs. Buffer properties are critical factors determining the efficiency of cell enrichment during immunomagnetic-negative separation.
The present study clarifies the important factors related to CTC enrichment based on immunomagnetic-negative separation for the further development of a highly versatile CTC enrichment method. The buffer component must reduce CTC loss and maintain CTC in the supernatant. The properties of a buffer are determined by the chemical and physical properties of the molecules dissolved in the buffer [23]. The hydrophilic/hydrophobic balance of polymers influences their interactions with tumor cells [51]. Selecting a molecule with high cell-coating properties can improve the efficiency of cell enrichment.
The type of antibody used in the immunomagnetic-negative separation may support the development of a method for capturing other EMT-related CTCs. In the present study, an antibody-magnetic bead complex bound to the surface markers CD2, CD14, CD16, CD19, CD45, CD61, CD66b, and glycophorin A was utilized as a component of the developed method. EMT-related CTCs circulate as heteroclusters surrounded by immune cells [9]. Vascular cell adhesion molecule 1 (VCAM1), which mediates adhesion between CTCs and neutrophils, is associated with cancer metastasis and EMT [52], [53]. Neutrophils express CD45 and CD66b on their surfaces. The use of antibodies bound to CD45 and CD66b may remove EMT-related heterotypic CTC clusters via immunomagnetic-negative separation. Changing the antibody cocktail may be an option for treating other EMT-related CTCs, including heteroclusters. From a methodological perspective, a buffer design for immunomagnetic-negative separation could be useful for developing versatile methods that are not affected by CTC heterogeneity.
5. Conclusion
A novel CTC enrichment method was developed using a design approach based on the buffering properties of negative immunomagnetic separation. The developed method was highly effective for enriching CTCs and EMT-related mesenchymal subpopulations from the blood samples of patients with lung cancer, including those with NSCLC and SCLC. Buffer property design is a crucial methodological approach that can be leveraged to develop a highly versatile enrichment immunomagnetic separation method capable of addressing CTC heterogeneity.
Funding
This study was supported by Oncolys BioPharma, Inc. (Tokyo, Japan), and a Grant-in-Aid for Scientific Research B (research project number: 21H02928).
Author statement
This manuscript has not been published or presented elsewhere in part or in entirety and is not under consideration by another journal. We have read and understood your journal’s policies, and we believe that neither the manuscript nor the study violates any of these.
CRediT authorship contribution statement
Shoko Saiwaki: Validation, Methodology, Investigation, Formal analysis, Data curation. Kazuhisa Takahashi: Supervision, Funding acquisition, Conceptualization. Shinsaku Togo: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization. Yasuo Urata: Supervision, Resources. Yasinjan Hashim: Writing – original draft, Methodology, Investigation, Data curation. Kanae Abe: Resources. Kazuaki Hoshi: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Yoko Tabe: Supervision, Conceptualization. Rihyang Heo: Visualization, Data curation. Chieko Miyoshi: Validation, Data curation. Yusuke Ochi: Investigation, Formal analysis. Shun Nakazawa: Investigation, Formal analysis, Data curation. Issei Sumiyoshi: Visualization, Methodology, Data curation. Hiroaki Motomura: Validation, Methodology, Investigation, Data curation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was conducted in the Laboratory of Cell Biology, Biomedical Research Core Facilities, Juntendo University Graduate School of Medicine (Tokyo, Japan). We would like to thank Editage (www.editage.jp) for English language editing.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.11.033.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Dillekas H., Rogers M.S., Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8(12):5574–5576. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dasgupta A., Lim A.R., Ghajar C.M. Circulating and disseminated tumor cells: harbingers or initiators of metastasis? Mol Oncol. 2017;11(1):40–61. doi: 10.1002/1878-0261.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krebs M.G., Hou J.M., Ward T.H., Blackhall F.H., Dive C. Circulating tumour cells: their utility in cancer management and predicting outcomes. Ther Adv Med Oncol. 2010;2(6):351–365. doi: 10.1177/1758834010378414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truini A., Alama A., Bello M.G.Dal, Coco S., Vanni I., Rijavec E., et al. Clinical applications of circulating tumor cells in lung cancer patients by cellsearch system. Front Oncol. 2014;4:242. doi: 10.3389/fonc.2014.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ankeny J.S., Court C.M., Hou S., Li Q., Song M., Wu D., et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer. 2016;114(12):1367–1375. doi: 10.1038/bjc.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huong P.T., Gurshaney S., Binh N.T., Pham A.G., Nguyen H.H., Nguyen X.T., et al. Emerging role of circulating tumor cells in gastric cancer. Cancers. 2020;12(3) doi: 10.3390/cancers12030695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klotz R., Yu M. Insights into brain metastasis: recent advances in circulating tumor cell research. Cancer Rep-Us. 2022;5(4) doi: 10.1002/cnr2.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radfar P., Aboulkheyr Es H., Salomon R., Kulasinghe A., Ramalingam N., Sarafraz-Yazdi E., et al. Single-cell analysis of circulating tumour cells: enabling technologies and clinical applications. Trends Biotechnol. 2022;40(9):1041–1060. doi: 10.1016/j.tibtech.2022.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Lin D., Shen L., Luo M., Zhang K., Li J., Yang Q., et al. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther. 2021;6(1):404. doi: 10.1038/s41392-021-00817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papadaki M.A., Sotiriou A.I., Vasilopoulou C., Filika M., Aggouraki D., Tsoulfas P.G., et al. Optimization of the enrichment of circulating tumor cells for downstream phenotypic analysis in patients with non-small cell lung cancer treated with anti-PD-1 immunotherapy. Cancers. 2020;12(6) doi: 10.3390/cancers12061556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rushton A.J., Nteliopoulos G., Shaw J.A., Coombes R.C. A review of circulating tumour cell enrichment technologies. Cancers. 2021;13(5) doi: 10.3390/cancers13050970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harouaka R.A., Nisic M., Zheng S.Y. Circulating tumor cell enrichment based on physical properties. J Lab Autom. 2013;18(6):455–468. doi: 10.1177/2211068213494391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francart M.E., Lambert J., Vanwynsberghe A.M., Thompson E.W., Bourcy M., Polette M., et al. Epithelial-mesenchymal plasticity and circulating tumor cells: travel companions to metastases. Dev Dyn. 2018;247(3):432–450. doi: 10.1002/dvdy.24506. [DOI] [PubMed] [Google Scholar]
- 14.Jeong H., Koh J., Kim S., Song S.G., Lee S.H., Jeon Y., et al. Epithelial-mesenchymal transition induced by tumor cell-intrinsic PD-L1 signaling predicts a poor response to immune checkpoint inhibitors in PD-L1-high lung cancer. Br J Cancer. 2024 doi: 10.1038/s41416-024-02698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasantharajan S.S., Barnett E., Gray E.S., Rodger E.J., Eccles M.R., Pattison S., et al. Size-based method for enrichment of circulating tumor cells from blood of colorectal cancer patients. Methods Mol Biol. 2023;2588:231–248. doi: 10.1007/978-1-0716-2780-8_15. [DOI] [PubMed] [Google Scholar]
- 16.Shaw Bagnall J., Byun S., Begum S., Miyamoto D.T., Hecht V.C., Maheswaran S., et al. Deformability of tumor cells versus blood cells. Sci Rep. 2015;5:18542. doi: 10.1038/srep18542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gertler R., Rosenberg R., Fuehrer K., Dahm M., Nekarda H., Siewert J.R. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 2003;162:149–155. doi: 10.1007/978-3-642-59349-9_13. [DOI] [PubMed] [Google Scholar]
- 18.Lustberg M., Jatana K.R., Zborowski M., Chalmers J.J. Emerging technologies for CTC detection based on depletion of normal cells. Recent Results Cancer Res. 2012;195:97–110. doi: 10.1007/978-3-642-28160-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karabacak N.M., Spuhler P.S., Fachin F., Lim E.J., Pai V., Ozkumur E., et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9(3):694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norouzi N., Bhakta H.C., Grover W.H. Sorting cells by their density. Plos One. 2017;12(7) doi: 10.1371/journal.pone.0180520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nader E., Skinner S., Romana M., Fort R., Lemonne N., Guillot N., et al. Blood rheology: key parameters, impact on blood flow, role in sickle cell disease and effects of exercise. Front Physiol. 2019;10:1329. doi: 10.3389/fphys.2019.01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sa B., Mukherjee S., Roy S.K. Effect of polymer concentration and solution pH on viscosity affecting integrity of a polysaccharide coat of compression coated tablets. Int J Biol Macromol. 2019;125:922–930. doi: 10.1016/j.ijbiomac.2018.12.101. [DOI] [PubMed] [Google Scholar]
- 24.Mastronardi M.L., Hennrich F., Henderson E.J., Maier-Flaig F., Blum C., Reichenbach J., et al. Preparation of monodisperse silicon nanocrystals using density gradient ultracentrifugation. J Am Chem Soc. 2011;133(31):11928–11931. doi: 10.1021/ja204865t. [DOI] [PubMed] [Google Scholar]
- 25.Ribeiro R.D.C., Pa D., Jamieson D., Rankin K.S., Benning M., Dalgarno K.W., et al. Temporary single-cell coating for bioprocessing with a cationic polymer. ACS Appl Mater Inter. 2017;9(15):12967–12974. doi: 10.1021/acsami.6b16434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Rutte J., Dimatteo R., Zhu S., Archang M.M., Di Carlo D. Sorting single-cell microcarriers using commercial flow cytometers. SLAS Technol. 2022;27(2):150–159. doi: 10.1016/j.slast.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yayapour N., Nygren H. Interactions between whole blood and hydrophilic or hydrophobic glass surfaces: kinetics of cell adhesion. Colloid Surf B. 1999;15(2):127–138. [Google Scholar]
- 28.Keomanee-Dizon K., Shishido S.N., Kuhn P. Circulating tumor cells: high-throughput imaging of CTCs and bioinformatic analysis. Recent Results Cancer Res. 2020;215:89–104. doi: 10.1007/978-3-030-26439-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 30.Kojima T., Hashimoto Y., Watanabe Y., Kagawa S., Uno F., Kuroda S., et al. A simple biological imaging system for detecting viable human circulating tumor cells. J Clin Invest. 2009;119(10):3172–3181. doi: 10.1172/JCI38609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Togo S., Katagiri N., Namba Y., Tulafu M., Nagahama K., Kadoya K., et al. Sensitive detection of viable circulating tumor cells using a novel conditionally telomerase-selective replicating adenovirus in non-small cell lung cancer patients. Oncotarget. 2017;8(21):34884–34895. doi: 10.18632/oncotarget.16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui C., Schoenfelt K.Q., Becker K.M., Becker L. Isolation of polymorphonuclear neutrophils and monocytes from a single sample of human peripheral blood. STAR Protoc. 2021;2(4) doi: 10.1016/j.xpro.2021.100845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fawcett D.W., Vallee B.L., Soule M.H. A method for concentration and segregation of malignant cells from bloody, pleural, and peritoneal fluids. Science. 1950;111(2872):34–36. doi: 10.1126/science.111.2872.34. (illust) [DOI] [PubMed] [Google Scholar]
- 34.Seal S.H. Silicone flotation: a simple quantitative method for the isolation of free-floating cancer cells from the blood. Cancer. 1959;12(3):590–595. doi: 10.1002/1097-0142(195905/06)12:3<590::aid-cncr2820120318>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 35.Ntouroupi T.G., Ashraf S.Q., McGregor S.B., Turney B.W., Seppo A., Kim Y., et al. Detection of circulating tumour cells in peripheral blood with an automated scanning fluorescence microscope. Br J Cancer. 2008;99(5):789–795. doi: 10.1038/sj.bjc.6604545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg R., Gertler R., Friederichs J., Fuehrer K., Dahm M., Phelps R., et al. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49(4):150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 37.Okeda T., Ono J., Takaki R., Todo S. Simple method for the collection of pancreatic islets by the use of Ficoll-Conray gradient. Endocrinol Jpn. 1979;26(4):495–499. doi: 10.1507/endocrj1954.26.495. [DOI] [PubMed] [Google Scholar]
- 38.Frenea-Robin M., Marchalot J. Basic principles and recent advances in magnetic cell separation. Magnetochemistry. 2022;8(1) [Google Scholar]
- 39.Gao S., Li X., Hu Z., Wang Z., Hao X. Dual targeting negative enrichment strategy for highly sensitive and purity detection of CTCs. Front Chem. 2024;12:1400988. doi: 10.3389/fchem.2024.1400988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Li Y., Shao W., Li Z., Zhao R., Ye Z. Strategies for enrichment of circulating tumor cells. Transl Cancer Res. 2020;9(3):2012–2025. doi: 10.21037/tcr.2020.01.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun C., Hsieh Y.P., Ma S., Geng S., Cao Z., Li L., et al. Immunomagnetic separation of tumor initiating cells by screening two surface markers. Sci Rep. 2017;7:40632. doi: 10.1038/srep40632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou H.W., Warkiani M.E., Khoo B.L., Li Z.R., Soo R.A., Tan D.S., et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z., Fusi A., Klopocki E., Schmittel A., Tinhofer I., Nonnenmacher A., et al. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med. 2011;9:70. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neurohr G.E., Amon A. Relevance and regulation of cell density. Trends Cell Biol. 2020;30(3):213–225. doi: 10.1016/j.tcb.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendelaar P.A.J., Kraan J., Van M., Zeune L.L., Terstappen L., Oomen-de Hoop E., et al. Defining the dimensions of circulating tumor cells in a large series of breast, prostate, colon, and bladder cancer patients. Mol Oncol. 2021;15(1):116–125. doi: 10.1002/1878-0261.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J., Kulasinghe A., Bogseth A., O'Byrne K., Punyadeera C., Papautsky I. Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel. Micro Nanoeng. 2019;5 doi: 10.1038/s41378-019-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolazzo C., Raimondi C., Mancini M., Caponnetto S., Gradilone A., Gandini O., et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep. 2016;6:31726. doi: 10.1038/srep31726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ntzifa A., Strati A., Kallergi G., Kotsakis A., Georgoulias V., Lianidou E. Gene expression in circulating tumor cells reveals a dynamic role of EMT and during osimertinib treatment in NSCLC patients. Sci Rep-Uk. 2021;11(1) doi: 10.1038/s41598-021-82068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dall'Olio F.G., Gelsomino F., Conci N., Marcolin L., De Giglio A., Grilli G., et al. PD-L1 expression in circulating tumor cells as a promising prognostic biomarker in advanced non-small-cell lung cancer treated with immune checkpoint inhibitors. Clin Lung Cancer. 2021;22(5):423–431. doi: 10.1016/j.cllc.2021.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Tan S., Day D., Nicholls S.J., Segelov E. Immune checkpoint inhibitor therapy in oncology: current uses and future directions: JACC: cardiooncology state-of-the-art review. JACC CardioOncol. 2022;4(5):579–597. doi: 10.1016/j.jaccao.2022.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramamurthi P., Zhao Z.C., Burke E., Steinmetz N.F., Müllner M. Tuning the hydrophilic-hydrophobic balance of molecular polymer bottlebrushes enhances their tumor homing properties. Adv Health Mater. 2022;11(12) doi: 10.1002/adhm.202200163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuster E., Taftaf R., Reduzzi C., Albert M.K., Romero-Calvo I., Liu H.P. Better together: circulating tumor cell clustering in metastatic cancer. Trends Cancer. 2021;7(11):1020–1032. doi: 10.1016/j.trecan.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D., Bi J., Liang Q., Wang S., Zhang L., Han F., et al. VCAM1 promotes tumor cell invasion and metastasis by inducing EMT and transendothelial migration in colorectal cancer. Front Oncol. 2020;10:1066. doi: 10.3389/fonc.2020.01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material