Summary
Background
We aimed to evaluate the incremental predictive value of metabolomic biomarkers for assessing the 10-year risk of type 2 diabetes when added to the clinical Cambridge Diabetes Risk Score (CDRS).
Methods
We utilized 86,232 UK Biobank (UKB) participants (recruited between 13 March 2006 and 1 October 2010) for model derivation and internal validation. Additionally, we included 4383 participants from the German ESTHER cohort (recruited between 1 July 2000 and 30 June 2002 for external validation). Participants were followed up for 10 years to assess the incidence of type 2 diabetes. A total of 249 NMR-derived metabolites were quantified using nuclear magnetic resonance (NMR) spectroscopy. Metabolites were selected with LASSO regression and model performance was evaluated with Harrell's C-index.
Findings
11 metabolomic biomarkers, including glycolysis related metabolites, ketone bodies, amino acids, and lipids, were selected. In internal validation within the UKB, adding these metabolites significantly increased the C-index (95% confidence interval (95% CI)) of the clinical CDRS from 0.815 (0.800, 0.829) to 0.834 (0.820, 0.847) and the continuous net reclassification index (NRI) with 95% CI was 39.8% (34.6%, 45.0%). External validation in the ESTHER cohort showed a comparable statistically significant C-index increase from 0.770 (0.750, 0.791) to 0.798 (0.779, 0.817) and a continuous NRI of 33.8% (26.4%, 41.2%). A concise model with 4 instead of 11 metabolites yielded similar results.
Interpretation
Adding 11 metabolites to the clinical CDRS led to a novel type 2 diabetes prediction model, we called UK Biobank Diabetes Risk Score (UKB-DRS), substantially outperformed the clinical CDRS. The concise version with 4 metabolites performed comparably. As only very few clinical information and a blood sample are needed for the UKB-DRS, and as high-throughput NMR metabolomics are becoming increasingly available at low costs, these models have considerable potential for routine clinical application in diabetes risk assessment.
Funding
The ESTHER study was funded by grants from the Baden-Württemberg state Ministry of Science, Research and Arts (Stuttgart, Germany), the Federal Ministry of Education and Research (Berlin, Germany), the Federal Ministry of Family Affairs, Senior Citizens, Women and Youth (Berlin, Germany), and the Saarland State Ministry of Health, Social Affairs, Women and the Family (Saarbrücken, Germany). The UK Biobank project was established through collaboration between various entities including the Wellcome Trust, the Medical Research Council, Department of Health, Scottish Government, and the Northwest Regional Development Agency. Additional funding was provided by the Welsh Assembly Government, British Heart Foundation, Cancer Research UK, and Diabetes UK, with support from the National Health Service (NHS). The German Diabetes Center is funded by the German Federal Ministry of Health (Berlin, Germany) and the Ministry of Culture and Science of the state North Rhine-Westphalia (Düsseldorf, Germany) and receives additional funding from the German Federal Ministry of Education and Research (BMBF) through the German Center for Diabetes Research (DZD e.V.).
Keywords: Type 2 diabetes, Metabolomics, Risk score, Prediction model, Metabolite
Research in context.
Evidence before this study
Existing diabetes risk prediction models primarily relied on traditional risk factor. The potential of a broader range of metabolomic biomarkers for enhancing risk prediction had not been fully explored in large-scale settings. We searched PubMed up until July 2024 for studies using the terms “type 2 diabetes”, “risk prediction”, “metabolomics”, “metabolite”, and “risk score”. Most studies focused on small, specific populations or included a limited number of metabolites. Furthermore, very few studies had performed external validation of their findings, a critical step for confirming the generalizability of risk prediction models.
Added value of this study
This study leverages a comprehensive set of 249 metabolites analyzed through high-throughput nuclear magnetic resonance (NMR) spectroscopy, significantly expanding the range of biomarkers considered in diabetes risk prediction. Our development and external validation of the UK Biobank Diabetes Risk Score (UKB-DRS) across two large cohorts, including over 90,000 participants, distinguishes this work from prior efforts. The UKB-DRS integrates both traditional risk factors and a novel panel of 11 metabolites, providing a more nuanced understanding of metabolic disturbances leading up to type 2 diabetes. For translation purposes into clinical routine, we also developed a concise model with 4 metabolites, which had a comparable predictive performance. This is the largest study of its kind to date and the first to perform such extensive external validation. Additionally, we examined the incremental value of each metabolite to the model's predictive capability across both cohorts. Remarkably consistent results were observed, which underscores the reproducibility and robustness of the metabolomic enhancements across different populations, further enhancing the clinical applicability of our findings.
Implications of all the available evidence
The findings from this study suggest that integrating metabolomic data into diabetes risk prediction models significantly enhances their predictive accuracy. This could lead to earlier and more precise identification of individuals at high risk for type 2 diabetes, enabling targeted preventive interventions. Given the increasing accessibility and affordability of NMR-based metabolomic profiling, the practical implementation of such advanced prediction models in clinical settings is becoming feasible. This advancement has the potential to transform preventive healthcare by allowing for more personalized risk assessments and tailored intervention strategies.
Introduction
The global prevalence of type 2 diabetes is on a significant upward trend, associated with increased mortality, diminished quality of life, and substantial economic burden.1, 2, 3 Early identification of individuals at elevated risk is essential, given the effectiveness of preventative measures in mitigating or delaying disease onset.4 Although current models for predicting type 2 diabetes risk effectively differentiate between individuals with low and high future risks, their clinical applicability is limited by a lack of specificity and an incomplete representation of the complex risk factors.5, 6, 7
Recent advances in metabolomics, particularly through the use of nuclear magnetic resonance (NMR) spectroscopy, offer promising insights into the early detection of type 2 diabetes.8,9 The comprehensive metabolomic profiling enabled by NMR spectroscopy, including its ability to measure a wide array of metabolites in a single assay, provides a more nuanced view of the metabolic disturbances preceding type 2 diabetes.10,11 Furthermore, the high-throughput nature of NMR spectroscopy, coupled with its low operational costs and minimal batch-to-batch variation, makes it an ideal tool for large-scale epidemiological studies.12
Despite the potential of metabolomics to enhance risk prediction models, previous studies have often been limited to investigating a small number or single subclasses of metabolomic biomarkers, leaving the value of metabolomic analysis in predicting the risk of type 2 diabetes uncertain.13, 14, 15, 16 This study is the first aiming to derive and externally validate a NMR metabolomics data based risk score for type 2 diabetes in two large, population-based cohort studies.
Methods
Study population
The UK Biobank (UKB) is a large prospective cohort study with 502,493 participants, aged 37–73 years, recruited from 13 March 2006 to 1 October 2010 across 22 assessment sites in England, Scotland, and Wales.17
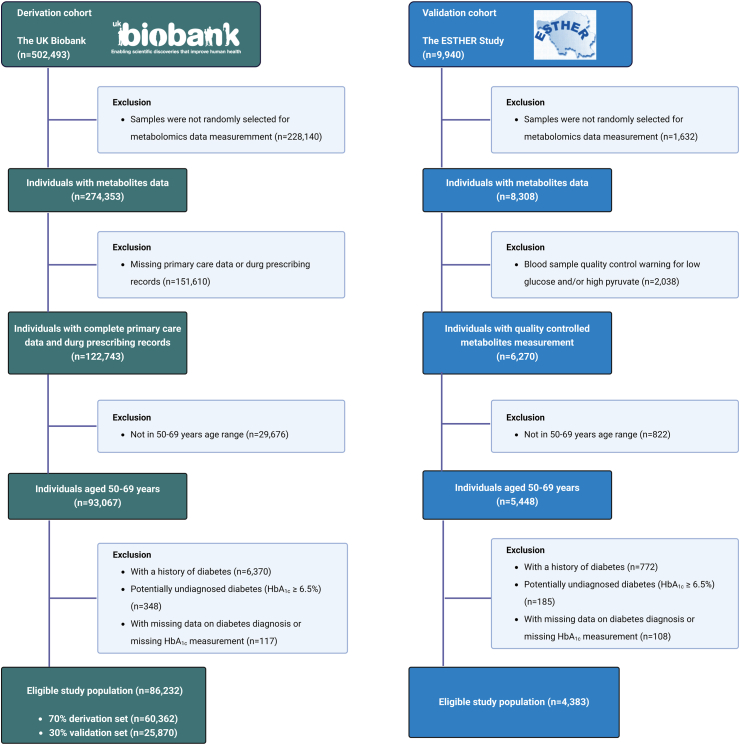
The ESTHER study is an ongoing population-based cohort study conducted in Saarland, a federal state in South-West Germany, with 9940 participants, aged 50–75 years. The recruitment occurred during standard health checkups by general practitioners (GPs) from 1 July 2000 to 30 June 2002. Follow-ups were conducted every two to three years thereafter.18 The inclusion and exclusion criteria for the analyzed study population are shown in Fig. 1 and Supplemental Text S1.
Fig. 1.
Flow-charts for participant inclusion and exclusion.
Metabolomic profiling
Nightingale Health's high-throughput NMR metabolomics platform was used to measure 250 metabolites from randomly selected baseline plasma samples of UKB participants, alongside all baseline serum samples from the ESTHER cohort with sufficient blood sample quality.19 Glycerol was excluded because it could not be measured in most of the participants of both cohorts, leaving N = 249 metabolites for the analyses. The nomenclature and completeness of these metabolites are shown in Supplemental Table S1.
Variables of the clinical CDRS and type 2 incidence ascertainment
The CDRS is a predictive tool used to assess the risk of individuals for future type 2 diabetes development. This scoring system includes age, sex, body mass index (BMI), family history of diabetes, smoking status, prescription of anti-hypertensive medication and steroids.20 If blood samples can be taken, the clinical CDRS is recommended to use, which additionally includes the HbA1c.21 The assessment methods of the variables of the clinical CDRS and of type 2 incidence in both cohorts are shown in Supplemental Text S2 and Supplemental Table S2.
Statistical analyses
General remarks
All analyses were performed using R software version 4.3.0 (R Foundation for Statistical Computing, Vienna, Austria), and statistical significance for two-sided tests was set at a P-value of <0.05. Missing values were imputed using the Random Forest estimation method in the r package missForest (version 1.5).22 This method estimates missing values based on observed data patterns in the original data set without creating multiple datasets. The variables of the clinical CDRS and all 249 metabolites were used in the imputation model. The completeness of each variable within the UKB and ESTHER cohorts is detailed in Supplemental Table S1. The highest proportion of missing values was 6% (family history of diabetes in the UKB).
Metabolite selection and model derivation
To stabilize variance and reduce the influence of outliers, concentrations of all metabolites were log-transformed. Although normality is not strictly required for predictors in models like Cox proportional hazards or Least Absolute Shrinkage and Selection Operator (LASSO), log transformation helps mitigate the skewness commonly seen in metabolomics data. After log transformation, the metabolite values were scaled to standard deviation units within each cohort, facilitating comparisons across metabolites with different distributions. The UKB cohort was divided into a training set (70%) and a test set (30%). The ESTHER study served as the external validation set. To perform variable selection, we employed the LASSO method using Cox proportional hazards models with the r package ‘glmnet’ (version 4.1-7). The variables of the clinical CDRS and all metabolite concentrations were included as independent variables, and incident type 2 diabetes was the dependent variable. Within the training set, we conducted a bootstrap procedure with 1000 resamples to enhance the robustness and generalizability of the variable selection process. For each bootstrap sample, we performed ten-fold cross-validation to identify the optimal value of the regularization parameter λ that minimizes the cross-validation error. The LASSO Cox proportional hazards model was fitted using the optimal λ in each bootstrap sample, which shrinks some coefficients towards zero and others exactly to zero.23 We recorded, which metabolites were selected because they had non-zero coefficients in the final Cox proportional hazards model in each bootstrap sample. After completing all 1000 bootstrap samples, we calculated the selection frequency for each metabolite in these 1000 bootstrap samples as the percentage of times it was selected. Metabolites were subsequently ranked based on their selection frequency, ranging from 0% to 100%. Metabolites selected by LASSO in at least 95% of the 1000 bootstrap samples were selected, a threshold shown previously to enhance model generalizability and minimize model overfitting.24 The selected metabolites were subsequently incorporated into the clinical CDRS to construct a new prediction model.
Validation of the model's predictive performance
The 30% UKB subsample was utilized as a test set, and the ESTHER study served as an external validation set to evaluate the predictive performance of the derived models. The model's predictive performance was assessed with discrimination, risk stratification and model calibration statistics. Discrimination was assessed using Harrell's C-index and the confidence intervals were estimated with bootstrapping with 500 resamples (r package ‘survival’ (version 3.5-5)). Additionally, we assessed the receiver operating characteristic curve (ROC). To determine whether the addition of all selected metabolites improved model discrimination compared to the clinical CDRS, the statistical significance of improvements in the C-index was determined using the ‘compareC’ package (version 1.3.2) in R, which applies the approach of Kang L et al.25 Based on the incremental predictive value of each selected metabolite, we additionally developed a concise version of the UKB-DRS using the clinical CDRS variables and only those metabolites with a substantial C-index increase >0.005 in the external validation cohort. The continuous net reclassification index (NRI) and the integrated discrimination index (IDI) were assessed to evaluate risk reclassification.26 The continuous NRI can range from −2 to 2, where 0 indicates no net reclassification improvement, positive values indicate improved reclassification, and negative values indicate worse reclassification. The detailed calculation method for the continuous NRI can be found in Supplemental Text S3. In addition, the C-index change, NRI and IDI of each of the selected metabolite was assessed separately by adding them individually to the clinical CDRS model. All of these analyses were performed for both the internal validation set and the external validation set.
To test, whether adding metabolites to the clinical CDRS is especially useful (or not useful) for patients with specific characteristics, we also computed the model discrimination metrics for subgroup analyses by age (</≥65 years), sex, obesity (BMI </≥ 30 kg/m2), and clinical CDRS risk (</≥ median).
Calibration of the predicted probabilities was performed using the Platt Scaling method, comparing the observed incidence rate of type 2 diabetes events in deciles of absolute predicted risk to their corresponding predicted event rates.27
Associations of selected metabolites with incident type 2 diabetes
To derive the hazard ratios (HRs) and 95% confidence intervals (CIs) of selected metabolites for 10-year type 2 diabetes risk, metabolites were individually added to Cox proportional hazards regression models in the test set and the external validation set. These models were adjusted for the variables of the CDRS, using the r-package survival (version 3.5-5). HRs and 95% CIs were estimated per one standard deviation (SD) increase with the metabolite concentrations in a log-transformed and standardized format. Log transformation was applied to remove the skewness of the metabolite concentration distributions. Standardization to a mean of zero and a SD of one was subsequently applied to facilitate comparability across metabolites with different units and scales.
Ethics
The UKB received ethical approval from the North-West Multicentre Research Ethics Committee (REC reference: 11/NW/03820). The ESTHER study was approved by the Ethics Committee of the Medical Faculty of the University of Heidelberg (Application number: S-58/2000). Both UKB and ESTHER study are conducted in accordance with the 1964 Helsinki declaration and its later amendments. All study participants of UKB and ESTHER study gave written informed consent.
Role of the funding source
The funders had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Baseline characteristics
Table 1 summarizes the characteristics of the clinical CDRS variables among 86,232 participants in the UKB cohort and 4383 participants in the ESTHER study. The average age and sex distribution were similar across the two cohorts, with participants in the UKB having an average age of 59.9 years (SD 4.4) and comprising 44.3% males. In the ESTHER cohort, the mean age was 60.2 years (SD 5.5), with 42.7% of the participants being male. Furthermore, the BMI and HbA1c levels were comparable. Compared to the UKB, a higher proportion of participants in the ESTHER study were current smokers (17.5% in ESTHER vs. 9.3% in the UKB), more had a family history of diabetes (36.9% vs. 16.8%), and more were prescribed anti-hypertensive medication (37.8% in ESTHER vs. 13.3% in the UKB). Steroid prescriptions were equally rare in both cohorts (0.8% in ESTHER vs. 1.3% in UKB).
Table 1.
Baseline characteristics of selected participants from the UK Biobank and ESTHER study.
| Baseline characteristics | Derivation cohort |
Validation cohort |
|---|---|---|
| UK Biobank | ESTHER | |
| Number of participants | 86,232 | 4383 |
| Male sex, n (%) | 39,442 (44.3) | 1871 (42.7) |
| Age (years), mean (SD) | 59.9 (5.4) | 60.2 (5.5) |
| HbA1c (%), mean (SD) | 3.1 (0.4) | 3.2 (0.5) |
| BMI (kg/m2), mean (SD) | 27.4 (4.5) | 27.4 (4.3) |
| BMI category, n (%) | ||
| <25 kg/m2 | 27,626 (32.0) | 1302 (29.7) |
| 25–27.49 kg/m2 | 21,529 (25.0) | 1193 (27.2) |
| 27.5–29.99 kg/m2 | 16,666 (19.3) | 958 (21.7) |
| ≥30 kg/m2 | 20,411 (23.7) | 984 (22.4) |
| Cigarette smoking, n (%) | ||
| Never | 46,150 (53.5) | 2233 (50.9) |
| Ex-smoker | 32,075 (37.2) | 1385 (31.6) |
| Current smoker | 8007 (9.3) | 765 (17.5) |
| Family history of diabetes, n (%) | ||
| None | 71,783 (83.2) | 2768 (63.2) |
| Parent or sibling | 12,982 (15.1) | 1384 (31.6) |
| Parent and sibling | 1467 (1.7) | 231 (5.3) |
| Prescribed medication, n (%) | ||
| Anti-hypertensive | 11,502 (13.3) | 1658 (37.8) |
| Steroid | 1104 (1.3) | 33 (0.8) |
Abbreviations: BMI, body mass index; HbA1c, glycated hemoglobin; SD, standard deviation.
Associations of selected metabolites with incident type 2 diabetes
Over a follow-up of up to 10 years, 3537 of 86,232 study participants of the UKB developed diabetes (incidence rate (IR) per 10,000 person-years (PY), 55.0) and 495 of 4383 participants of the ESTHER study (IR per 10,000 PY, 145.2).
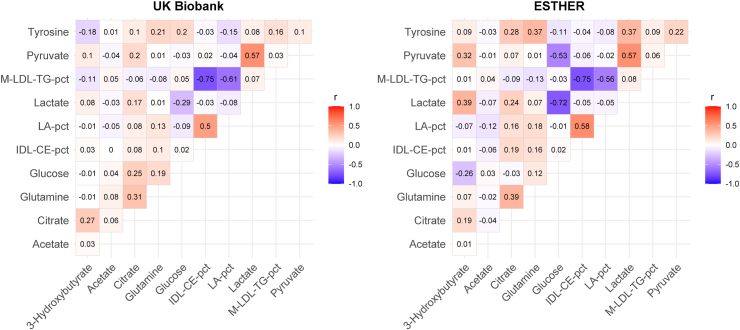
Through LASSO analyses and bootstrapping, 11 metabolites were selected to enhance the predictive power of the clinical CDRS for the risk of type 2 diabetes in the training set. These metabolites include four glycolysis related metabolites (citrate, glucose, lactate, and pyruvate), two ketone bodies (3-hydroxybutyrate and acetate), two amino acids (glutamine and tyrosine), two lipoprotein related metabolites (IDL-CE-pct (cholesteryl esters to total lipids in IDL percentage) and M-LDL-TG-pct (triglycerides to total lipids in medium LDL percentage)), and a fatty acid-related metabolite LA-pct (linoleic acid to total fatty acids percentage). Fig. 2 displays the Pearson correlation matrix of the selected 11 metabolites in their log-transformed and standardized format, which transformed their distributions close to normal distributions. Three biomarkers of glycolysis, pyruvate, glucose, and lactate, were highly correlated with each other (all one-on-one Pearson's r > 0.5 in ESTHER and for pyruvate and lactate in UKB). Furthermore, the three lipoprotein related biomarkers M-LDL-TG-pct, IDL-CE-pct, and LA-pct were highly correlated in both cohorts (all Pearson's r > 0.5). Except for these two biomarker clusters, the correlations were low to moderate.
Fig. 2.
Correlation matrix of Pearson correlation coefficients for the 11 selected metabolites. Abbreviations: IDL-CE-pct, cholesteryl esters to total lipids in IDL percentage; LA-pct, linoleic acid to total fatty acids percentage; M-LDL-TG-pct, triglycerides to total lipids in medium LDL percentage.
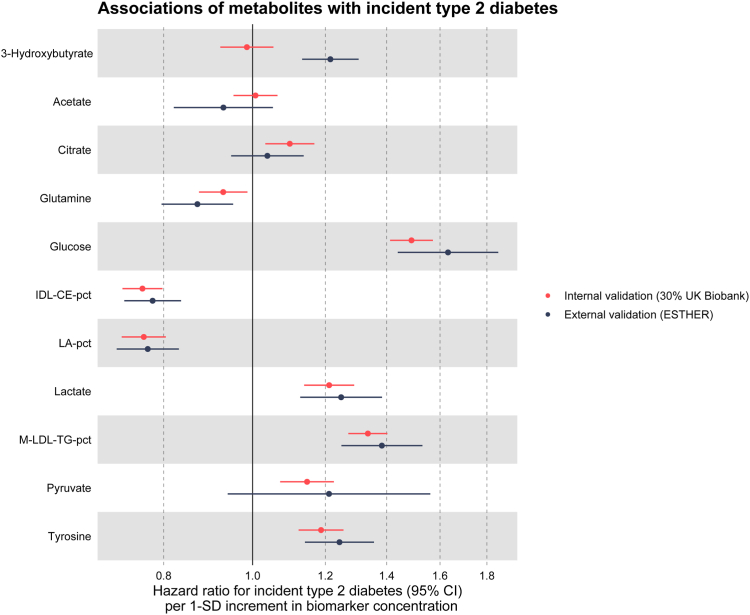
The 11 selected metabolites alone (without any covariates like age and sex) already predicted incident diabetes cases in the next 10 years well, with a C-index (95% CI) of 0.733 (0.715, 0.750) in the internal validation set and a C-index of 0.735 (0.713, 0.757) in the external validation set. Fig. 3 presents the associations between the 11 selected metabolites and incident type 2 diabetes within the test set of the UKB and the ESTHER cohort, adjusted for the variables of the clinical CDRS graphically, while the HRs with 95% CIs per SD increments and P-values can be found in Supplemental Table S3. All biomarkers except the ketone bodies 3-hydroxybutyrate and acetate were statistically significantly associated with type 2 diabetes in the UKB. Except for 3-hydroxybutyrate, for which confidence intervals did not overlap, the results were remarkably similar in the ESTHER study. Results were also comparable for citrate and pyruvate although these biomarkers lacked statistical significance in the ESHTER study.
Fig. 3.
Associations between selected metabolites and incident type 2 diabetes in the test set (30% of UK Biobank, N = 25,870) and external validation set (ESTHER, N = 5904). Notes: Hazard ratios are expressed per 1 standard deviation of the respective metabolite concentration and are adjusted for the variables of the clinical CDRS. The exact HRs with 95% CIs and P-values per SD increments and the SDs of the metabolites are shown in Supplemental Table S3. Abbreviations: CI, confidence interval; IDL-CE-pct, cholesteryl esters to total lipids in IDL percentage; LA-pct, linoleic acid to total fatty acids percentage; M-LDL-TG-pct, triglycerides to total lipids in medium LDL percentage; SD, standard deviation.
Improvements in type 2 diabetes risk prediction by metabolomic biomarkers
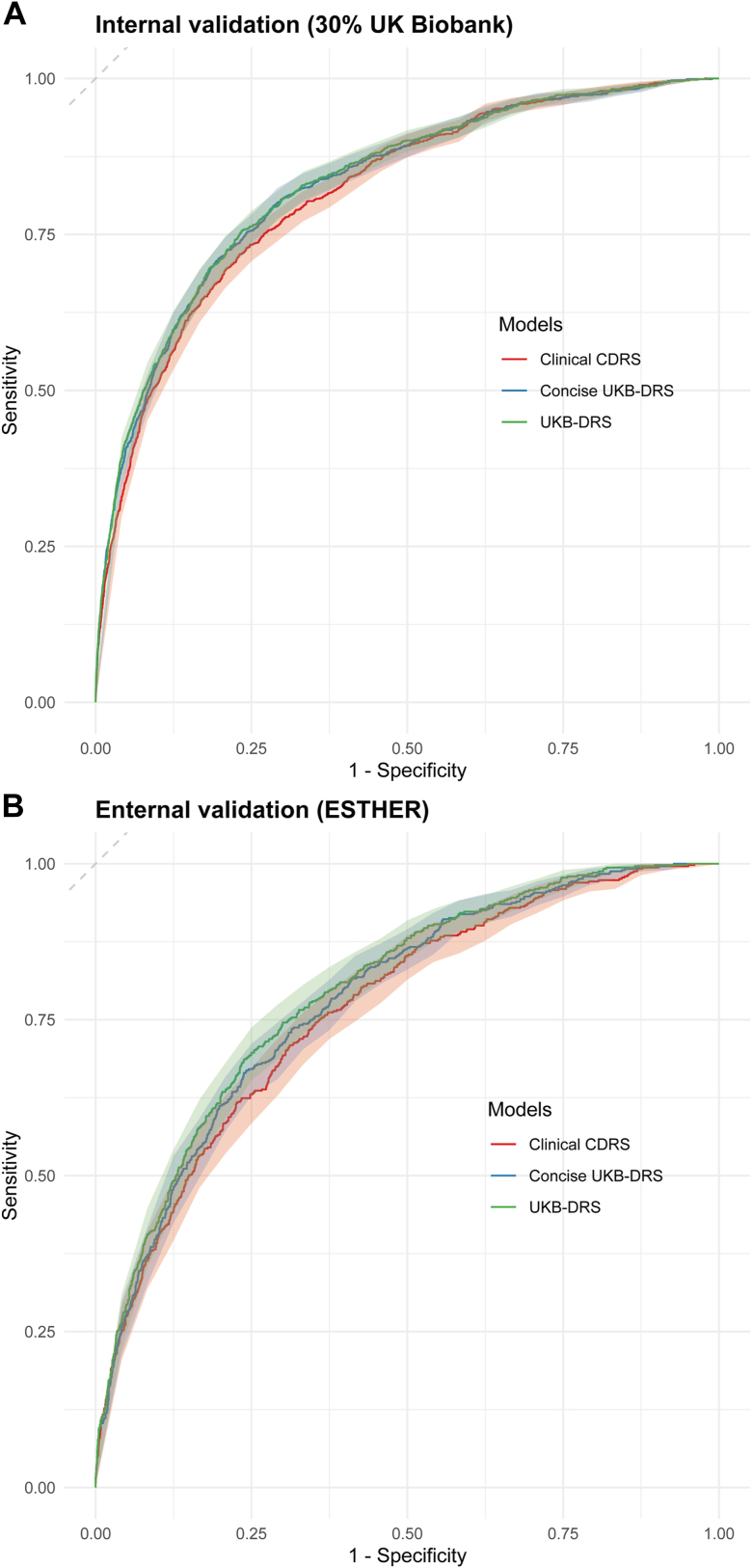
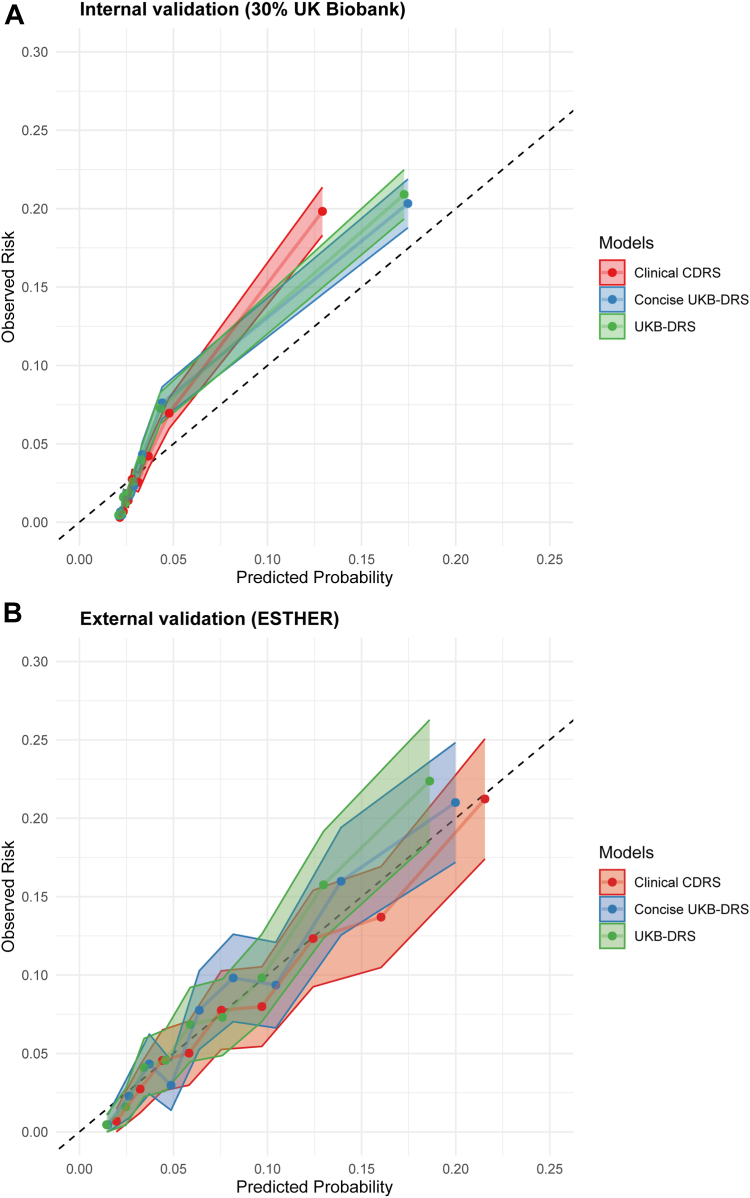
In the training set, the C-index for the clinical CDRS [0.830 (0.821, 0.839)] was much higher than for the CDRS [0.740 (0.731, 0.749)]. Thus, we compared the predictive performance of the novel model of the clinical CDRS extended by 11 metabolites to the clinical CDRS, only. We call the novel model “UK Biobank Diabetes Risk Score (UKB-DRS)” in the following. Coefficients for all variables in the UKB-DRS are listed in Supplemental Table S4. In the training set, test set and external validation set, the UKB-DRS consistently had a statistically significantly (P < 0.001) higher C-index (95% CI) than the clinical CDRS, with comparable C-index increases of 0.017, 0.019, and 0.028 units, respectively (Table 2). The receiver operating curves (ROC) are shown in Fig. 4. Moreover, an improved risk stratification ability was observed, with a continuous NRI (95% CI) of 39.8% (34.6%, 45.0%) and an IDI (95% CI) of 0.0216 (0.0170, 0.0262) in internal validation and a continuous NRI of 33.8% (26.4%, 41.2%), and an IDI of 0.0016 (0.0012, 0.0019) in external validation. Supplemental Text S3 provides the calculation how we derived at a continuous NRI (95% CI) of 39.8% (34.6%, 45.0%). The model calibration of both the clinical CDRS and the UKB-DRS was good in the internal and the external validation set and the confidence interval bands overlapped (Fig. 5).
Table 2.
Comparison of the predictive performance of the clinical Cambridge Diabetes Risk Score and the novel developed UK Biobank Diabetes Risk Score for 10-year type 2 diabetes risk prediction.
| Metrics | |
|---|---|
| Derivation set (70% of UK Biobank) | |
| Sample size/incident type 2 diabetes | N = 60,362/2477 |
| C-index (Clinical CDRSa) | 0.830 (0.821, 0.839) |
| C-index (Concise UKB-DRSb) | 0.846 (0.837, 0.856) |
| C-index (UKB-DRSc) | 0.847 (0.838, 0.855) |
| ΔC-index (UKB-DRSc vs. Clinical CDRSa); P-value | +0.017; P < 0.0001 |
| Internal validation set (30% of UK Biobank) | |
| Sample size/incident type 2 diabetes | N = 25,870/1059 |
| C-index (Clinical CDRSa) | 0.815 (0.800, 0.829) |
| C-index (Concise UKB-DRSb) | 0.830 (0.815, 0.844) |
| C-index (UKB-DRSc) | 0.834 (0.820, 0.847) |
| ΔC-index (UKB-DRSc vs. Clinical CDRSa); P-value | +0.019; P < 0.0001 |
| Continuous NRI (%) (UKB-DRSc vs. Clinical CDRSa); P-value | 39.8 (34.6, 45.0); P < 0.0001 |
| IDI (UKB-DRSc vs. Clinical CDRSa); P-value | 0.0216 (0.0170, 0.0262); P < 0.0001 |
| External validation set (ESTHER) | |
| Sample size/incident type 2 diabetes | N = 4383/495 |
| C-index (Clinical CDRSa) | 0.770 (0.750, 0.791) |
| C-index (Concise UKB-DRSb) | 0.786 (0.766, 0.805) |
| C-index (UKB-DRSc) | 0.798 (0.779, 0.817) |
| ΔC-index (UKB-DRSc vs. Clinical CDRSa); P-value | +0.028; P < 0.0001 |
| Continuous NRI (%) (UKB-DRSc vs. Clinical CDRSa); P-value | 33.8 (26.4, 41.2); P < 0.0001 |
| IDI (UKB-DRSc vs. Clinical CDRSa); P-value | 0.0016 (0.0012, 0.0019); P < 0.0001 |
Abbreviations: CDRS, Cambridge Diabetes Risk Score; IDI, integrated discrimination improvement; NRI, net reclassification improvement, UKB-DRS, UK Biobank Diabetes Risk Score.
The clinical CDRS consists of the following diabetes risk factors: (age, sex, BMI, smoking status, family history of diabetes, prescription of anti-hypertensive medication and steroids, and HbA1c).
The concise UKB-DRS is based on the traditional diabetes risk factors of the clinical CDRS (see above) and the following 4 metabolites: glucose, IDL-CE-pct (cholesteryl esters to total lipids in intermediate density lipoproteins percentage), LA-pct (linoleic acid to total fatty acids percentage), and M-LDL-TG-pct (triglycerides to total lipids in medium low-density lipoproteins percentage).
The UKB-DRS is based on the traditional diabetes risk factors of the clinical CDRS (see above) and the following 11 metabolites: glucose, citrate, lactate, pyruvate, 3-hydroxybutyrate, acetate, glutamine, tyrosine, IDL-CE-pct, LA-pct, and M-LDL-TG-pct (see footnote above for Abbreviations).
Fig. 4.
ROC curves with 95% confidence interval bands for the UKB-DRS, concise UKB-DRS, and clinical CDRS in internal validation (30% UK Biobank, N = 25,870) and external validation (ESTHER, N = 4383). Abbreviations: ROC, receiver operating characteristic; CDRS, Cambridge Diabetes Risk Score; UKB-DRS, UK Biobank Diabetes Risk Score.
Fig. 5.
Calibration plots for the UKB-DRS, Concise UKB-DRS, and Clinical CDRS in internal validation (30% UK Biobank, N = 25,870) and external validation (ESTHER, N = 4383). Notes: Confidence intervals are represented by shaded regions for each model. Abbreviations: CDRS, Cambridge Diabetes Risk Score; UKB-DRS, UK Biobank Diabetes Risk Score.
To check, whether glucose is the main source of the model improvement, we compared the predictive performance metrics of the UKB-DRS with the clinical CDRS augmented with glucose as the reference model (Supplemental Table S5). The UKB-DRS had a higher C-index (95% CI) [0.834 (0.820, 0.847) vs. 0.820 (0.805, 0.834) in internal and 0.798 (0.779, 0.817) vs. 0.773 (0.755, 0.791) in external validation set] and a statistically significant NRI (95% CI) [26.3% (21.2%, 31.4%) in internal and 13.2% (7.1%, 19.2%) in external validation set] compared to the clinical CDRS extended by glucose.
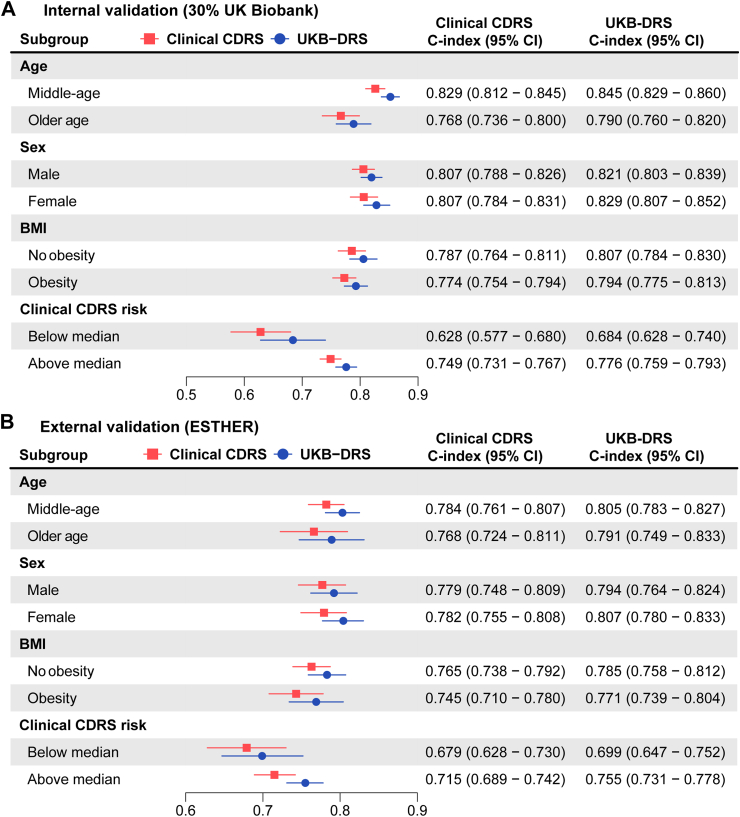
To further explore whether the addition of the 11 metabolites was particularly useful in specific subgroups, we conducted subgroup analyses by age, sex, obesity, and clinical CDRS risk. As shown in Fig. 6, the UKB-DRS consistently outperformed the clinical CDRS across all subgroups in both internal and external validation. There was no subgroup with a particularly low or high C-index change.
Fig. 6.
Subgroup analyses comparing the C-index of the UKB-DRS and the clinical CDRS in internal validation (30% UK Biobank, N = 25,870) and external validation (ESTHER, N = 4383). Notes: Middle-aged is defined as <65 years and older age as ≥65 years. Obesity is defined as a BMI ≥ 30 kg/m2. Abbreviations: CDRS, Cambridge Diabetes Risk Score; UKB-DRS, UK Biobank Diabetes Risk Score; CI, confidence interval.
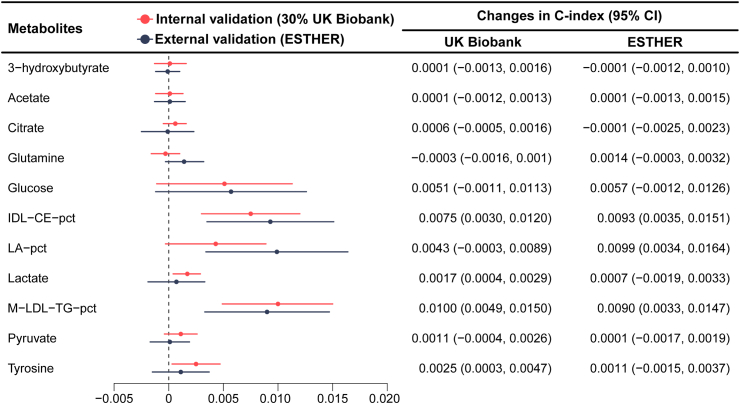
Fig. 7 illustrate the incremental improvement in the C-index and Supplemental Table S6 the NRI and IDI for each of the 11 selected metabolites after their incorporation into the clinical CDRS in internal and external validation. Especially glucose, IDL-CE-pct, LA-pct, and M-LDL-TG-pct enhanced the discriminative ability of the model in both cohorts with C-index changes >0.005 in the external validation cohort. The other seven metabolites showed low increases in C-statistic at best. Thus, we additionally developed a simplified model, termed the concise UKB-DRS, which incorporated only these four key metabolites (glucose, IDL-CE-pct, LA-pct, and M-LDL-TG-pct) alongside the traditional CDRS variables. With a C-index (95% CI) of 0.830 (0.815, 0.844) in internal validation and 0.786 (0.766, 0.805) in external validation, the concise UKB-DRS had a comparable discriminative performance compared to the full UKB-DRS, including 11 metabolites. Fig. 4 shows that, compared to the ROC curves of the full UKB-DRS, the ROC curves of the concise UKB-DRS were almost identical (in internal validation) or just a little lower (in external validation). The NRI (95% CI) and IDI (95% CI) of the concise UKB-DRS in internal validation (36.7 (31.4, 41.9) and 0.0205 (0.0162, 0.0248), respectively) and external validation (24.4 (16.7, 32.2) and 0.0011 (0.0008, 0.0015), respectively) were also just a little weaker than for the full UKB-DRS. Moreover, the calibration curves of the full and concise UKB-DRS were also comparable (Fig. 5).
Fig. 7.
The incremental discrimination of each metabolite for the model after the selected metabolites were added to the clinical Cambridge Diabetes Risk Score in the test set (30% of UK Biobank, N = 25,870) and external validation set (ESTHER, N = 4383). Abbreviations: CI, confidence interval; IDL-CE-pct, cholesteryl esters to total lipids in IDL percentage; LA-pct, linoleic acid to total fatty acids percentage; M-LDL-TG-pct, triglycerides to total lipids in medium LDL percentage.
Discussion
To our knowledge, this study represents the largest-scale investigation to date on the predictive utility of metabolomic biomarkers for type 2 diabetes risk, leveraging data from two large European cohorts comprising over 90,000 middle-aged and older adults of whom more than 4000 developed type 2 diabetes during 10 years of follow-up. We developed and validated the UKB-DRS that integrates an established risk prediction model (clinical CDRS) with 11 metabolites for enhanced predictive performance of 10-year type 2 diabetes risk. In both internal and external validation, the UKB-DRS demonstrated significant improvements in the C-index compared to the clinical CDRS, with increases of 0.019 and 0.028, respectively. Additionally, we examined the incremental value of each metabolite to the model's predictive capability across both cohorts, finding similar results in internal and external validation for all 11 metabolites. These findings confirm the robustness and generalizability of the new algorithm in enhancing the predictive accuracy for the risk of type 2 diabetes.
The association between circulating metabolomic biomarkers and the risk of type 2 diabetes has been extensively documented. Ahola-Olli et al. investigated four Finnish cohorts and identified that approximately half of the 229 NMR-based metabolomic biomarkers examined were significantly associated with incident type 2 diabetes.28 Similarly, Seah et al. observed that multiple metabolites were linked to an increased risk of type 2 diabetes in cohorts with Asian ethnicity.29 Moreover, to enhance the accuracy of existing prediction models for type 2 diabetes risk, numerous studies have investigated the incremental value of metabolomic biomarkers.30 Many traditional prediction models already achieve high predictive accuracy, making significant improvements upon these models challenging.5,6 Previous studies often focused on the impact of specific types of metabolomic biomarkers, with several studies finding that incorporating pre-selected metabolomic biomarkers into traditional risk prediction models did not enhance risk identification capabilities.31,32 However, other studies have shown that screening among a large number of metabolomic biomarkers tends to improve model discrimination.15,33 The European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam study, which measured 163 metabolites for 800 incident type 2 diabetes cases (with an average follow-up of 7 years) and 2282 controls, showed that adding 14 metabolomic biomarkers selected from 163 metabolite candidates to a traditional risk model significantly improved the model's discriminative ability (C-index increased from 0.901 to 0.912).15 The EPIC-Potsdam study is the second largest study after the UK Biobank using untargeted metabolomics data. Prior to our study, a study from the UKB, which did not have the full number of metabolomics data available had the largest sample size (N = 65,684 participants analyzed, including 1719 type 2 diabetes cases).33 The authors conducted internal validation after incorporating metabolomic data into the Framingham Diabetes Risk Score, significantly enhancing the model's C-index (95% CI) from 0.802 (0.791, 0.812) to 0.830 (0.822, 0.841) (ΔC-index = 0.028). However, a lack of external validation remains a major barrier to translating metabolomic based models into clinical applications, which we overcame by our joint analysis of the UKB and ESTHER cohort. Furthermore, we included more than twice the number of incident type 2 diabetes case in our UKB analysis (N = 3536) compared to the previous UKB study by including more study participants with metabolomics data (N = 86,232) and by the additional use of primary care records for the outcome assessment.
The ensemble of metabolites identified in our study captures a broad spectrum of biochemical pathways integral to the metabolic dysregulation that precedes type 2 diabetes. The identified metabolites stem from the tricarboxylic acid cycle (citrate, glucose, lactate, and pyruvate), are ketone bodies (3-hydroxybutyrate and acetate), amino acids (glutamine and tyrosine), lipoproteins (IDL-CE-pct and M-LDL-TG-pct), and a fatty acid (LA-pct). This metabolic signature characterizes an impaired glucose and lipid metabolism that is detectable in blood samples up to 10 years prior diabetes diagnosis and might also give new insights into the mechanisms of type 2 diabetes pathogenesis.
Citrate is a key metabolite in the tricarboxylic acid cycle and not only facilitates energy production but also plays a vital role in fatty acid synthesis, thereby linking glycolytic processes with lipid metabolism.9 Concurrently, glucose as a direct participant in glycolysis, provides critical insights into the disrupted glucose homeostasis that is characteristic of diabetes.34 The presence of lactate and pyruvate, products of anaerobic and aerobic glycolysis respectively, signals a shift towards inefficient energy utilization, a common observation in insulin-resistant states. These shifts are believed to reflect the underlying mitochondrial adaptations that compromise cellular energy management and exacerbate hyperglycemic conditions.35
Ketone bodies, including 3-hydroxybutyrate and acetate, accumulate during increased fatty acid oxidation—a compensatory response to impaired glucose utilization in diabetes. Elevated levels of these ketone bodies indicate the body's attempt to maintain energy balance under conditions of insulin resistance.36
Furthermore, the amino acids glutamine and tyrosine are noted for their dual roles in metabolic and neurotransmitter pathways. Glutamine is involved in gluconeogenesis and plays a crucial role in maintaining glucose homeostasis. Tyrosine is associated with catecholamine synthesis and may influence stress responses that exacerbate beta-cell dysfunction and insulin resistance.34
Lipoprotein-related metabolites, including IDL-CE-pct and M-LDL-TG-pct, reflect changes in lipoprotein metabolism that contribute to a dyslipidemia pattern commonly associated with insulin resistance.37 Additionally, the percentage of linoleic acid, a crucial component in inflammatory pathways, has been associated with variations in dietary intake and metabolic status, influencing cellular functions pivotal to insulin sensitivity.38
Overall, the identified metabolic profile comprising glycolysis-related metabolites, ketone bodies, amino acids, lipoprotein-related metabolites, and fatty acids not only contributes evidence to the biological pathways from early metabolic disturbances to type 2 diabetes development but also adds predictive power to traditional risk factor models, which will allow to detect those better who are at risk to develop type 2 diabetes and profit most from early preventive interventions.
This study stands out from previous research primarily due to its substantial sample size and the confirmation of our model's robustness through external validation. We employed a well-established, clinically approved targeted NMR metabolomics platform with absolute quantification of 250 biomarkers. This feature does not only facilitate comparative analyses across different populations but also enhances the potential for clinical translation.
However, our study is not without limitations. Although we only included UKB participants with available primary care records to ascertain the incidence of type 2 diabetes, the IR remained significantly lower than in the ESTHER study (55.0 vs. 145.2 per 10,000 person-years, respectively). This difference is likely due to an underreporting in the UKB and a more complete ascertainment in the ESTHER study, in which the GPs of the study participants were asked to provide their medical records and these were screened for diabetes diagnoses and glucose-lowering drugs. In addition, there are differences between the two cohorts regarding the blood samples. The UKB used plasma and ESTHER serum samples to determine the metabolites. Furthermore, only a small proportion of blood samples in the UK Biobank cohort were collected under fasting conditions, defined as no consumption of food or drink for at least 8 h (3.3%), whereas the majority of participants in the ESTHER study were not fasting (90.6%). However, a comparison of the levels of the 11 selected metabolites by fasting status in the UKB and ESTHER study shows that the differences in metabolite concentrations between fasting and non-fasting individuals are small in both cohorts (see Supplemental Table S7). This is in agreement with a previous study, which has shown that the duration of fasting has little impact on the variability of these metabolites.39 Thus, the novel UKB-DRS is robust towards fasting status and use of either serum or plasma samples. However, it is unknown how the model will perform in different populations. As its derivation and validation are based on British and German populations aged 50–69 years, extending its application to ethnically diverse populations or other age groups requires further validation.
Although the absolute improvement in the C-index for the UKB-DRS over the clinical CDRS alone was modest, this improvement is clinically meaningful for several reasons. First, even small gains in the discriminative power of risk prediction models can have substantial population-level implications when applied to large populations. In the context of type 2 diabetes, earlier and more accurate identification of individuals at risk can enable timely intervention strategies, potentially preventing thousands of cases annually.40 Second, in a clinical setting, the use of metabolomic biomarkers as an adjunct to traditional risk factors may help to reclassify individuals whose risk of developing diabetes was previously underestimated or overestimated. In this context, the moderately increased continuous NRI of >30% of the UKB-DRS, is more clinically relevant than the modest C-index increase. These reclassifications could refine decision-making processes related to lifestyle interventions or pharmacotherapy.34 Lastly, the metabolomic biomarkers selected in this study were derived from NMR spectroscopy, a high-throughput and cost-effective platform that, compared to liquid chromatography with tandem mass spectrometry (LC-MS/MS), is more suitable to be used in large-scale clinical routine settings.41 In this context, the concise UKB-DRS with just 4 added metabolites is likely the most cost-effective approach.
In conclusion, this study provides large-scale evidence from the UKB that a specific metabolomic profile indicative of alterations in glucose and lipid metabolism has additional value for the prediction of type 2 diabetes compared to a traditional risk factor-based model including the HbA1c. The derived novel UKB-DRS, which combines these traditional risk factors with 11 metabolomic biomarkers was robust in external validation using an independent, population-based German cohort. Furthermore, a concise UKB-DRS was developed as well, using only four key metabolites, which maintained considerable high predictive accuracy, making it an attractive, clinically feasible alternative. These findings have important future translational implications. With increasing clinical accessibility to high-throughput, targeted metabolomics analyses with the NMR technology, the use of these biomarkers in clinical risk prediction models is feasible. This can be useful for risk communication aiming at lifestyle changes and prioritizing cost-intensive preventive interventions (e.g., semaglutide injections for weight loss) to those at high risk for future type 2 diabetes development.
Contributors
H.B. designed and led the ESTHER cohort. R.X. and B.S. generated the idea for the study and formulated the analytical plan. R.X. performed the data analyses and drafted the manuscript. B.S. revised it. All authors contributed valuable intellectual content to the discussion. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. R.X. and B.S. had full access to UK Biobank and ESTHER data used for this study and are the guarantors of the manuscript and accepts full responsibility for the work and/or the conduct of the study.
Data sharing statement
Data from the UK Biobank are available to bona fide researchers upon application through the UK Biobank Access Management System (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). The ESTHER study data cannot be made publicly available due to data protection regulations but may be available upon reasonable request to the corresponding author for research purposes in line with the informed consent forms signed by the study participants and if the data protection rules of the country of the applicant are equally strict or stricter than those of the European Union (to be decided by the data security officer of the German Cancer Research Center).
Declaration of interests
No potential conflicts of interest were disclosed.
Acknowledgements
We would like to thank all participants of the ESTHER and UK Biobank cohort as well as the GPs of the ESTHER study and the staff of the UK Biobank assessment centers for their contribution to the studies this research is based on. Part of this research was conducted using the UK Biobank Resource under application 101633. The ESTHER study was funded by grants from the Baden-Württemberg state Ministry of Science, Research and Arts (Stuttgart, Germany), the Federal Ministry of Education and Research (Berlin, Germany), the Federal Ministry of Family Affairs, Senior Citizens, Women and Youth (Berlin, Germany), and the Saarland State Ministry of Health, Social Affairs, Women and the Family (Saarbrücken, Germany). The UK Biobank project was established through collaboration between various entities including the Wellcome Trust, the Medical Research Council, Department of Health, Scottish Government, and the Northwest Regional Development Agency. Additional funding was provided by the Welsh Assembly Government, British Heart Foundation, Cancer Research UK, and Diabetes UK, with support from the National Health Service (NHS). The German Diabetes Center is funded by the German Federal Ministry of Health (Berlin, Germany) and the Ministry of Culture and Science of the state North Rhine-Westphalia (Düsseldorf, Germany) and receives additional funding from the German Federal Ministry of Education and Research (BMBF) through the German Center for Diabetes Research (DZD e.V.). The first author (R.X.) was supported by the China Scholarship Council (No. 202208430012).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2024.102971.
Appendix A. Supplementary data
References
- 1.Ong K.L., Stafford L.K., McLaughlin S.A., et al. Global, regional, and national burden of diabetes from 1990 to 2021, with projections of prevalence to 2050: a systematic analysis for the Global Burden of Disease Study 2021. Lancet. 2023;402(10397):203–234. doi: 10.1016/S0140-6736(23)01301-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad E., Lim S., Lamptey R., Webb D.R., Davies M.J. Type 2 diabetes. Lancet. 2022;400(10365):1803–1820. doi: 10.1016/S0140-6736(22)01655-5. [DOI] [PubMed] [Google Scholar]
- 3.Tomic D., Shaw J.E., Magliano D.J. The burden and risks of emerging complications of diabetes mellitus. Nat Rev Endocrinol. 2022;18(9):525–539. doi: 10.1038/s41574-022-00690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herman W.H., Ye W., Griffin S.J., et al. Early detection and treatment of type 2 diabetes reduce cardiovascular morbidity and mortality: a simulation of the results of the Anglo-Danish-Dutch study of intensive treatment in people with screen-detected diabetes in primary care (ADDITION-Europe) Diabetes Care. 2015;38(8):1449–1455. doi: 10.2337/dc14-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbasi A., Peelen L.M., Corpeleijn E., et al. Prediction models for risk of developing type 2 diabetes: systematic literature search and independent external validation study. BMJ. 2012;345 doi: 10.1136/bmj.e5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hippisley-Cox J., Coupland C. Development and validation of QDiabetes-2018 risk prediction algorithm to estimate future risk of type 2 diabetes: cohort study. BMJ. 2017;359 doi: 10.1136/bmj.j5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buijsse B., Simmons R.K., Griffin S.J., Schulze M.B. Risk assessment tools for identifying individuals at risk of developing type 2 diabetes. Epidemiol Rev. 2011;33(1):46–62. doi: 10.1093/epirev/mxq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buergel T., Steinfeldt J., Ruyoga G., et al. Metabolomic profiles predict individual multidisease outcomes. Nat Med. 2022;28(11):2309–2320. doi: 10.1038/s41591-022-01980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Q., Ma R.C.W. Metabolomics in diabetes and diabetic complications: insights from epidemiological studies. Cells. 2021;10(11) doi: 10.3390/cells10112832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z.Z., Gerszten R.E. Metabolomics and proteomics in type 2 diabetes. Circ Res. 2020;126(11):1613–1627. doi: 10.1161/circresaha.120.315898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guasch-Ferré M., Hruby A., Toledo E., et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 2016;39(5):833–846. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart D.S., Cheng L.L., Copié V., et al. NMR and metabolomics-a roadmap for the future. Metabolites. 2022;12(8) doi: 10.3390/metabo12080678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu G., Zheng Y., Wang H., et al. Plasma metabolomics identified novel metabolites associated with risk of type 2 diabetes in two prospective cohorts of Chinese adults. Int J Epidemiol. 2016;45(5):1507–1516. doi: 10.1093/ije/dyw221. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J., Zhu Y., Hyun N., et al. Novel metabolic markers for the risk of diabetes development in American Indians. Diabetes Care. 2015;38(2):220–227. doi: 10.2337/dc14-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floegel A., Stefan N., Yu Z., et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang Z., Dong X., Jia J., Liu Z., Huang T., Qi L. Sleep patterns, plasma metabolome, and risk of incident type 2 diabetes mellitus. J Clin Endocrinol Metab. 2023;108(10):e1034–e1043. doi: 10.1210/clinem/dgad218. [DOI] [PubMed] [Google Scholar]
- 17.Sudlow C., Gallacher J., Allen N., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xuan Y., Gào X., Anusruti A., et al. Association of serum markers of oxidative stress with incident major cardiovascular events, cancer incidence, and all-cause mortality in type 2 diabetes patients: pooled results from two cohort studies. Diabetes Care. 2019;42(8):1436–1445. doi: 10.2337/dc19-0292. [DOI] [PubMed] [Google Scholar]
- 19.Soininen P., Kangas A.J., Würtz P., Suna T., Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8(1):192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
- 20.Rahman M., Simmons R.K., Harding A.H., Wareham N.J., Griffin S.J. A simple risk score identifies individuals at high risk of developing type 2 diabetes: a prospective cohort study. Fam Pract. 2008;25(3):191–196. doi: 10.1093/fampra/cmn024. [DOI] [PubMed] [Google Scholar]
- 21.Carrasco-Zanini J., Pietzner M., Wheeler E., Kerrison N.D., Langenberg C., Wareham N.J. Multi-omic prediction of incident type 2 diabetes. Diabetologia. 2024;67(1):102–112. doi: 10.1007/s00125-023-06027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stekhoven D.J., Bühlmann P. MissForest--non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28(1):112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 23.Bach F.R. 2008. Bolasso: model consistent lasso estimation through the bootstrap; pp. 33–40. [Google Scholar]
- 24.Huang Z., Klaric L., Krasauskaite J., et al. Combining serum metabolomic profiles with traditional risk factors improves 10-year cardiovascular risk prediction in people with type 2 diabetes. Eur J Prev Cardiol. 2023;30(12):1255–1262. doi: 10.1093/eurjpc/zwad160. [DOI] [PubMed] [Google Scholar]
- 25.Kang L., Chen W., Petrick N.A., Gallas B.D. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med. 2015;34(4):685–703. doi: 10.1002/sim.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pencina M.J., D'Agostino R.B., Sr., Steyerberg E.W. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30(1):11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng S., Rhee E.P., Larson M.G., et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–2231. doi: 10.1161/circulationaha.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahola-Olli A.V., Mustelin L., Kalimeri M., et al. Circulating metabolites and the risk of type 2 diabetes: a prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia. 2019;62:2298–2309. doi: 10.1007/s00125-019-05001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seah J.Y.H., Hong Y., Cichońska A., et al. Circulating metabolic biomarkers are consistently associated with type 2 diabetes risk in Asian and European populations. J Clin Endocrinol Metabol. 2022;107(7):e2751–e2761. doi: 10.1210/clinem/dgac212. [DOI] [PubMed] [Google Scholar]
- 30.Satheesh G., Ramachandran S., Jaleel A. Metabolomics-based prospective studies and prediction of type 2 diabetes mellitus risks. Metab Syndr Relat Disord. 2020;18(1):1–9. doi: 10.1089/met.2019.0047. [DOI] [PubMed] [Google Scholar]
- 31.Tillin T., Hughes A.D., Wang Q., et al. Diabetes risk and amino acid profiles: cross-sectional and prospective analyses of ethnicity, amino acids and diabetes in a South Asian and European cohort from the SABRE (Southall And Brent REvisited) Study. Diabetologia. 2015;58:968–979. doi: 10.1007/s00125-015-3517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fall T., Salihovic S., Brandmaier S., et al. Non-targeted metabolomics combined with genetic analyses identifies bile acid synthesis and phospholipid metabolism as being associated with incident type 2 diabetes. Diabetologia. 2016;59(10):2114–2124. doi: 10.1007/s00125-016-4041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bragg F., Trichia E., Aguilar-Ramirez D., Bešević J., Lewington S., Emberson J. Predictive value of circulating NMR metabolic biomarkers for type 2 diabetes risk in the UK Biobank study. BMC Med. 2022;20(1):159. doi: 10.1186/s12916-022-02354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz-Martínez M., González-González M., Martagón A.J., Hlavinka V., Willson R.C., Rito-Palomares M. Recent developments in biomarkers for diagnosis and screening of type 2 diabetes mellitus. Curr Diab Rep. 2022;22(3):95–115. doi: 10.1007/s11892-022-01453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konrad T., Vicini P., Kusterer K., et al. alpha-Lipoic acid treatment decreases serum lactate and pyruvate concentrations and improves glucose effectiveness in lean and obese patients with type 2 diabetes. Diabetes Care. 1999;22(2):280–287. doi: 10.2337/diacare.22.2.280. [DOI] [PubMed] [Google Scholar]
- 36.Laffel L. Ketone bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab Res Rev. 1999;15(6):412–426. doi: 10.1002/(sici)1520-7560(199911/12)15:6<412::aid-dmrr72>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 37.Mackey R.H., Mora S., Bertoni A.G., et al. Lipoprotein particles and incident type 2 diabetes in the multi-ethnic study of atherosclerosis. Diabetes Care. 2015;38(4):628–636. doi: 10.2337/dc14-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zong G., Liu G., Willett W.C., et al. Associations between linoleic acid intake and incident type 2 diabetes among US men and women. Diabetes Care. 2019;42(8):1406–1413. doi: 10.2337/dc19-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li-Gao R., Hughes D.A., le Cessie S., et al. Assessment of reproducibility and biological variability of fasting and postprandial plasma metabolite concentrations using 1H NMR spectroscopy. PLoS One. 2019;14(6) doi: 10.1371/journal.pone.0218549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillies C.L., Lambert P.C., Abrams K.R., et al. Different strategies for screening and prevention of type 2 diabetes in adults: cost effectiveness analysis. BMJ. 2008;336(7654):1180–1185. doi: 10.1136/bmj.39545.585289.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soininen P., Kangas A.J., Würtz P., et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134(9):1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.