Abstract
Polyethylene Terephthalate (PET) is a petroleum-based plastic polymer that, by design, can last decades, if not hundreds of years, when released into the environment through plastic waste leakage. In the pursuit of sustainable solutions to plastic waste recycling and repurposing, the enzymatic depolymerization of PET has emerged as a promising green alternative. However, the metabolic potential of the resulting PET breakdown molecules, such as the two-carbon (C2) molecule ethylene glycol (EG), remains largely untapped. Here, we review and discuss the current state of research regarding existing natural and synthetic microbial pathways that enable the assimilation of EG as a carbon and energy source for Escherichia coli. Leveraging the metabolic versatility of E. coli, we explore the viability of this widely used industrial strain in harnessing EG as feedstock for the synthesis of target value-added compounds via metabolic and protein engineering strategies. Consequently, we assess the potential of EG as a versatile alternative to conventional carbon sources like glucose, facilitating the closure of the loop between the highly available PET waste and the production of valuable biochemicals. This review explores the interplay between PET biodegradation and EG metabolism, as well as the key challenges and opportunities, while offering perspectives and suggestions for propelling advancements in microbial EG assimilation for circular economy applications.
Keywords: Escherichia coli, Ethylene glycol, Metabolic engineering, Synthetic biology, Protein engineering, PET biodegradation
Highlights
-
•
Review of PET depolymerization and its green alternative, enzymatic breakdown.
-
•
Explores E. coli's potential to metabolize ethylene glycol for bioproduction of high value compounds.
-
•
Assesses microbial pathways and protein engineering for EG assimilation.
-
•
Discusses EG as a sustainable carbon source, enhancing circular economy efforts.
1. Introduction
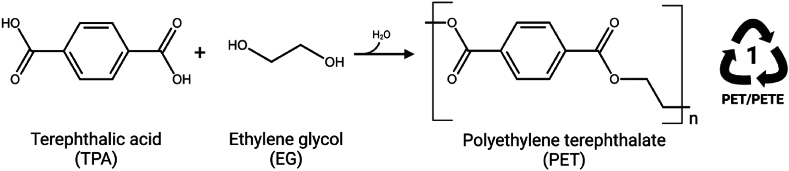
Polyethylene Terephthalate (PET) is a petroleum-based plastic polymer that is designed to endure for decades, if not centuries, which is highly disadvantageous when released into the environment via plastic waste leakage. PET is produced from a condensation reaction between ethylene glycol (EG) and terephthalic acid (TPA) (Fig. 1), two chemicals commonly derived from petroleum feedstock (Benyathiar et al., 2022). In 2021, it was estimated that global production reached 390.7 Mt of plastic, with 90.2% still originating from fossil-based feedstock (Plastics Europe, 2022). Although PET accounted for 6.2% of the annual global plastic production, this polymer is primarily and extensively used in single-use packaging, such as beverage bottles and food containers, as well as in textile manufacturing (polyester fibres), which are highly prone to being downcycled or mismanaged (Grant et al., 2022; Plastics Europe, 2022). In Europe, despite PET being one of the most recyclable plastics, the majority of PET waste is not currently managed within a circular model (Grant et al., 2022).
Fig. 1.
Chemical reaction between terephthalic acid (TPA) and ethylene glycol (EG) to form polyethylene terephthalate (PET). The PET polymer has the Resin Identification Code 1 and can be identified with acronym PET or PETE.
Consequently, plastic waste management and recycling are widely discussed topics today due to environmental concerns. The most commonly used recycling method is mechanical recycling, which entails grounding and melting the plastic waste into plastic pellets by extrusion. This process can cause undesirable changes in polymer properties in every cycle, limiting its reusability (Achilias and Karayannidis, 2004; Benyathiar et al., 2022). Alternatively, PET can be chemically recycled, partially broken down into small oligomers and other chemical substances, or completely into its monomer units, which can be repolymerized into a new oligomer. However, chemical recycling processes such as methanolysis and glycolysis are energy-intensive, requiring high pressures and temperatures, as well as downstream separation and purification steps. Additionally, the reaction catalysts employed are often difficult to remove from the final reaction, cannot be reused, and pose environmental threats (Achilias and Karayannidis, 2004; Benyathiar et al., 2022). Moreover, recycled PET faces challenges in competing with virgin PET in terms of availability, price, and quality, resulting in only a small fraction of the PET produced worldwide being recycled (Grant et al., 2022). In 2019, 91.25% of plastic was still either buried in landfills, incinerated for energy generation, or mismanaged (Benyathiar et al., 2022; OECD, 2023). The Great Pacific Garbage Patch serves as just one striking example of plastic waste being lost into the environment (National Geographic, 2024). Plastic waste pollution has thus been recognized as one of the most significant global environmental challenges of our lifetime.
In the quest for more sustainable alternatives to PET recycling and repurposing, researchers have focused on the biodegradation of PET using enzymes and engineered microorganisms. This pursuit aims to develop a biological recycling process, marking a significant area of research in recent years. Many examples include the application of enzymes in vitro that are capable of degrading the PET polymer along with derived engineered enzyme variants with catalytically improved PET breakdown capabilities (Khairul Anuar et al., 2022; Sui et al., 2023). Some of the most prominent cases include FAST-PETase, a mutant PETase from Ideonella sakaiensis engineered using a machine learning algorithm, which can completely degrade a number of different untreated postconsumer-PET products within one week, and even as quickly as 24 h (Lu et al., 2022). Another example is a variant of the leaf-branch compost cutinase (LCC), which degrades 90% of pretreated post-consumer PET in just 10 h (Tournier et al., 2020). Notably, some cases are already taking their first steps in industrial application (Carbios, 2021). Microorganisms have also been engineered to biodegrade PET in vivo, with some examples including E. coli (Benavides Fernández et al., 2022) but also some other species such as the non-conventional yeast Yarrowia lipolytica (Kosiorowska et al., 2022). Recent research has demonstrated how TPA resulting from in vitro enzymatic digestion can be repolymerized to create a new PET film with similar properties to virgin PET (Tournier et al., 2020). However, production of new PET from biologically obtained monomers has only recently started to be studied and is still not well developed. This process faces several associated hurdles, particularly regarding purity requirements. For instance, purifying EG from enzymatically obtained PET hydrolysate can be difficult and costly due to its high solubility in water and elevated boiling temperature, thus requiring complex and energy-intensive separation processes and distillation steps (Tournier et al., 2020; Wong et al., 2023). In alternative, if PET waste can be cost-effectively biodegraded, the resultant hydrolysed PET monomers could be directly used by microorganisms to fuel their metabolism and produce useful molecules and target compounds. Particularly, the two-carbon (C2) molecule EG holds great promise for fuelling microbial metabolism as a next-generation feedstock (NGF) for bioprocesses, even being considered a viable candidate for replacing commonly used microbial feedstocks such as glucose (Pandit et al., 2017). Thus, PET could potentially be recycled into new added-value compounds in a circular economy manner through microbial assimilation of EG. However, for efficient assimilation and/or upcycling of PET-derived EG, it is crucial to understand the EG assimilating pathways, as well as comprehend how bacteria can be engineered for improved EG assimilation and/or conversion.
This review aims to survey the known natural and synthetic pathways for microbial EG assimilation and recent advancements in metabolic engineering (ME) strategies. Specifically, our focus lies on the exploitation of synthetic pathways for consuming EG as a carbon source to support microbial growth and produce compounds of interest, focusing on the industrially well-established bacterium Escherichia coli. We further identify and discuss possible metabolic bottlenecks as well as other associated challenges with an EG assimilation bioprocess using PET as feedstock.
2. Production of EG
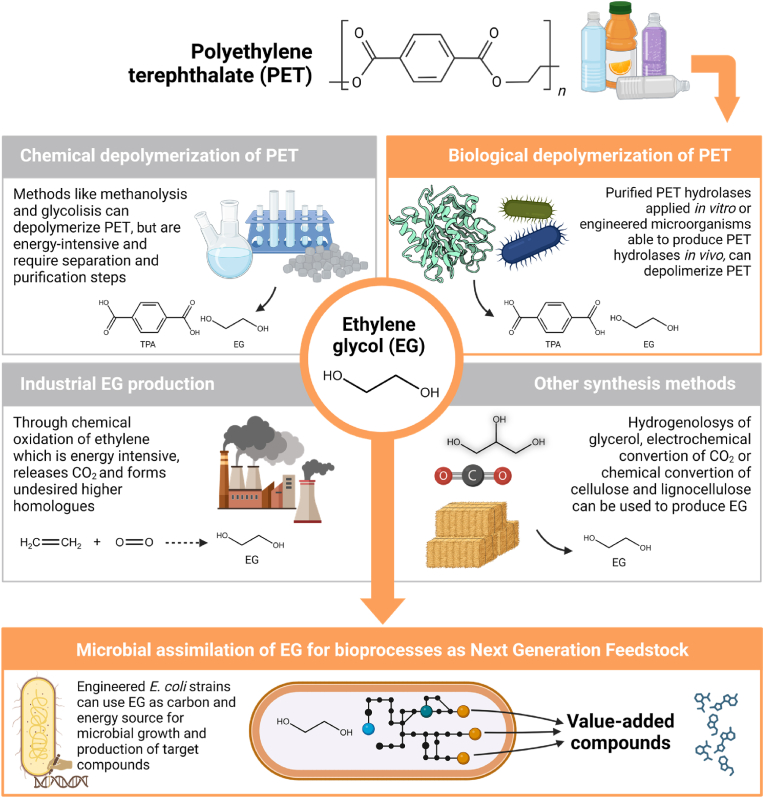
The potential use of EG as a starting compound for cellular metabolism and/or production of target compounds using ME strategies has only recently begun to be explored. EG is a commodity chemical that can be produced in a few different ways including chemical depolymerization and biological depolymerization of PET, industrial EG production, and other synthesis methods (Fig. 2). It is primarily produced by the chemical oxidation of ethylene, a process that requires high-energy consumption, resulting in a significant amount of CO2 emission and formation of undesired higher homologues (Kandasamy et al., 2019; Rebsdat and Mayer, 2000). Furthermore, the EG precursor ethylene is primarily obtained by the petrochemical industry through steam cracking of hydrocarbons from natural gas and crude oil (Rebsdat and Mayer, 2000; Yue et al., 2012). Besides being used for the production of PET plastics and polyester fibres, EG is primarily employed in antifreeze formulations, such as those used in automotive and industrial cooling systems (Rebsdat and Mayer, 2000; Yue et al., 2012). In 2023, the industry market of EG was estimated to reach 60.06 million tonnes per annum, with an expected average annual growth rate of more than 5%, between 2023 and 2028 (GlobalData, 2024). Due to its different uses and applications, a significant amount of EG enters the environment, for example through industrial manufacturing effluents or runoff water from airports, where it is used for aircraft de-icing (ATSDR, 2022; Staples et al., 2001). Hence, it is crucial to establish new methods for EG manufacturing that substitute toxic petrochemical materials and decrease environmental impact. This has prompted the scientific community to explore renewable alternatives for EG production that valorise waste feedstocks. Some examples include the hydrogenolysis of glycerol, a waste from the biodiesel industry (Kandasamy et al., 2019), the electrochemical conversion of CO2 (Chen et al., 2021; Li et al., 2020; Tamura et al., 2015), synthesis of EG using syngas (Satapathy et al., 2018), and chemical conversion of cellulosic and lignocellulosic biomass (Pang et al., 2011; Wong et al., 2023). The latter technique has been the focus of extensive research in recent years, but has not yet been commercialised mainly due to the need of complex post-reaction separation processes and high purifying cost of EG (Wong et al., 2023).
Fig. 2.
Overview of the methods used for producing ethylene glycol (EG) as feedstock and its integration into Escherichia coli metabolism to produce value-added compounds. The different routes for EG production, including chemical PET depolymerization, biological PET depolymerization by engineered microorganisms or enzymes, industrial production, and other alternative synthesis methods are shown. Created with BioRender.com.
Alternatively, EG can be biologically obtained from the enzymatic depolymerization of PET plastic, a highly abundant waste material. The biodegradation of PET and assimilation of the resulting EG could even be envisioned as a simultaneous process carried out by a single microbial cell factory. Microbial cell factories (MCFs) leverage the use of microorganisms to metabolically convert renewable raw sources into target products in an efficient manner via fermentation. These processes operate under mild conditions in terms of temperature and pressure, bypassing the use of toxic solvents and catalysts typically associated with the conventional chemical processes (Cho et al., 2022). The efficient development of MCFs often requires the application of rationally-designed ME strategies to rewire the native metabolism of the cell for the desired industrial application. This plays special importance when utilizing different carbon sources as substrates to produce non-native metabolites (Cho et al., 2022). In this respect, the model organism E. coli is one of the most widely used hosts for designing MCFs due to its extensively studied metabolism and well-established molecular tools for engineering (Lee et al., 2019). Concerning current biotechnological processes, EG has been pinpointed as a viable candidate to replace commonly used sugars such as glucose and sucrose. Sugar feedstocks used in fermentation are still commonly derived from hydrolysis of starch retrieved from crops, such as wheat and corn, which currently pose concerns such as competition with food sources in the face of a growing world population, or the intensive use of water and energy in crop processing (Tubb, 2023; Zhang et al., 2022). Lignocellulose, a non-edible residue from crop processing, has been widely studied as an alternative feedstock (second generation), however, due to its recalcitrant nature, extensive pre-treatment to release the sugars is usually required, which requires high energy consumption and often leads to the release of toxic by-products. This leads to low yields and consequently increases cost and competitiveness compared to starch processing (Ning et al., 2021; Salim et al., 2019). The use of NGFs such as C1 (methane, CO2, CO, methanol and formate) and C2 (acetate and ethanol) feedstocks has also been a key area of interest across a range of industries. However, though abundant and low-cost, the key challenge of using these molecules as feedstocks lies in the absence of native assimilation pathways in most biotechnologically relevant microorganisms, although recent advances are paving the way towards creation of C1 and C2-utilizing microorganisms (Zhang et al., 2022).
The future bioeconomy will depend on various bioprocesses that utilize carbon feedstocks. These processes will play a crucial role in the bioeconomy, generating a wide range of products, including alternative proteins, chemicals, and therapeutic pharmaceuticals. In this sense, discovering alternative feedstocks that are inexpensive, sustainable, and noncompeting with the food industry remains an important challenge yet to be overcome. In this regard, the PET-derived EG emerges as a potential source of inexpensive renewable C2 feedstock that could directly fuel bacterial metabolism. PET could be recycled and repurposed via EG for the bioproduction of high-added value compounds, aligning with the principles of a circular economy. We consider EG to be an important and relevant NGF. In the following sections, we will review the known natural EG assimilation pathways employed by microorganisms, as well as recently developed ME and synthetic biology strategies that propose new pathways for EG assimilation for biomass and value-added products in E. coli.
3. Natural EG assimilation
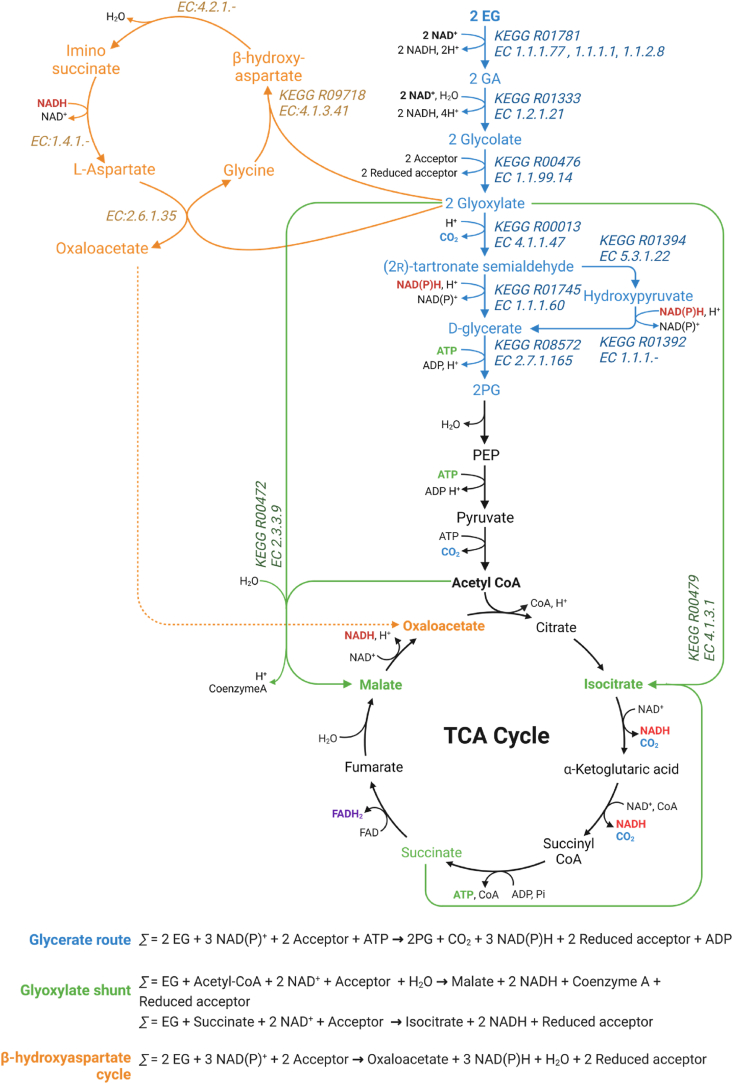
EG is considered readily biodegradable in the environment (Staples et al., 2001), and to date, several microbial species have been identified as capable of using EG as carbon and energy source, under both aerobic and anaerobic conditions. Examples include, but are not limited to, Flavobacterium sp., Clostridium glycolicum and Pseudomonas putida (Child and Willetts, 1978; Fincher and Payne, 1962; Gonzalez et al., 1972; Hartmanis and Stadtman, 1986; Kataoka et al., 2001; Mückschel et al., 2012; Toraya et al., 1979). Under aerobic conditions, EG is converted to glyoxylate in three successive steps (Boronat et al., 1983; Mückschel et al., 2012). Then, glyoxylate can be used to generate biomass through the glycerate route (Boronat et al., 1983; Li et al., 2019), or the β-hydroxyaspartate (BHA) cycle (Schada von Borzyskowski et al., 2019). Glyoxylate can also combine with acetyl-CoA to form malate or with succinate to form isocitrate, through the glyoxylate shunt (Li et al., 2019; Mückschel et al., 2012). These natural EG assimilating pathways will be discussed in this Section, and a schematic representation of each pathway is provided in Fig. 3.
Fig. 3.
Overview of the natural microbial ethylene glycol (EG) assimilation pathways. The metabolic EG assimilation pathways, namely the oxidative route (blue), the glyoxylate shunt (green), and the β-hydroxyaspartate (BHA) cycle (orange), and their integration with central metabolism (black) is shown. Each step of these three pathways is annotated with its corresponding Enzyme Commission (EC) number and Kyoto Encyclopaedia of Genes and Genomes (KEGG) reaction number, when available, along with the stoichiometric equations for each pathway. The alternative conversion of tartronate semialdehyde into glycerate via hydroxypyruvate is not considered for the stoichiometric equation of the glycerate route. Abbreviations: EG, ethylene glycol; GA, glycolaldehyde; 2 PG, 2-phospho-d-glycerate; PEP, phosphoenolpyruate; TCA, tricarboxylic acid cycle. Created with BioRender.com.
3.1. The EG oxidative pathway
The EG oxidative pathway (pathway depicted in blue in Fig. 3) exists in different bacteria, including E. coli, Pseudomonas and Flavobacterium species, where EG is aerobically oxidized into molecules that can enter central metabolism using nicotinamide cofactors, or other types of cofactors depending on the organism (such as pyrroloquinoline quinone – PQQ – cofactors in the first EG oxidation step in P. putida), and ATP (Boronat et al., 1983; Mückschel et al., 2012; Willetts, 1981). In this route, EG is first converted to glycolaldehyde (GA) by a propanediol/EG oxidoreductase or other alcohol dehydrogenases (refer to Section 5 for a discussion about enzymes involved in this first step of EG oxidation). Then, GA is converted into glyoxylate via the intermediate glycolate by the chained action of the enzymes GA dehydrogenase and glycolate dehydrogenase (Fig. 3) (Caballero et al., 1983; Lord, 1972). These enzymes employ cofactors such as NAD+ and proton acceptor quinones that direct electrons into the electron transport chain (Boronat et al., 1983; Mückschel et al., 2012). Both glycolate and glyoxylate molecules have economic interest, being routinely used in industrial applications. Particularly, glycolate is a C2 α-hydroxy acid that, due to its versatile properties of both alcohol and carboxylic acid, has many different applications in cosmetics and dermatology (Babilas et al., 2012). Moreover, this compound is used in the synthesis of the biodegradable packaging material polyglycolic acid and in biomedical applications (Fredenberg et al., 2011). While glyoxylate has fewer direct industrial applications, it still serves as a building block in the synthesis of many compounds used in perfumery, flavour, pharmaceuticals and agrochemicals (Jin et al., 2003), with one significant example being the production of vanillin (Kalikar et al., 1986; Mao et al., 2020). Both compounds are currently synthetized from petrochemical resources with the biotechnological production of glycolate receiving significant interest in recent years (Cabulong et al., 2021; Jin et al., 2003; Salusjärvi et al., 2019; Schad et al., 2023). Hence, the microbial conversion of EG to glycolate and glyoxylate may offer a sustainable alternative to produce these two industrially relevant compounds. In particular, the engineered production of glycolate from EG in E. coli has been reported (Pandit et al., 2021; Yan et al., 2024) and will be discussed in Section 4.2. Glyoxylate can then enter the central carbon metabolism by three different routes (Fig. 3).
3.1.1. The glycerate pathway
On the first route, the glycerate pathway (pathway depicted in blue in Fig. 3), glyoxylate can be used as the sole carbon and energy source. Two molecules of glyoxylate are first converted to (2r)-tartronate semialdehyde and CO2 by a glyoxylate carboligase, which is then converted to d-glycerate by the action of tartronate semialdehyde reductase (Chang et al., 1993; Gotto and Kornberg, 1961; Gupta and Vennesland, 1964; Njau et al., 2000). Additionally, tartronate semialdehyde can also be transformed into d-glycerate indirectly via hydroxypyruvate by the enzymes hydroxypyruvate isomerase and glyoxylate/hydroxypyruvate reductase (Ashiuchi and Misono, 1999; Nuñez et al., 2001). Finally, d-glycerate 2-kinase converts d-glycerate to 2-phopho-d-glycerate (2 PG), further entering in lower glycolysis (Fig. 3) (Zelcbuch et al., 2015). It should be noted that there are glycerate 2-kinases that convert d-glycerate into 3-phopho-d-glycerate (3 PG), however the one from E. coli produces 2 PG instead of 3 PG as initially claimed (Bartsch et al., 2008; Zelcbuch et al., 2015). In E. coli, the glycerate pathway is seemingly essential for growth on glyoxylate, since a deletion of the native glyoxylate carboligase gene (gcl) prevents growth on this substrate (Aslan et al., 2020; Ornston and Ornston, 1969). In terms of carbon molecules, the pathway from EG (C2) to the central carbon metabolite 2 PG (C3) yields 0.75 mol/mol (2 PG/EG) at the expense of one ATP molecule, releasing one molecule of CO2 and two of NADH.
3.1.2. The glyoxylate shunt
The glyoxylate shunt (pathway depicted in green in Fig. 3) acts as an alternative pathway to metabolize EG. In this pathway, glyoxylate and acetyl-CoA are converted into (s)-malate, releasing one molecule of coenzyme A, by the action of malate synthase (Dixon et al., 1960; Maloy and Nunn, 1982; Mückschel et al., 2012). Malate can enter directly into the tricarboxylic acid cycle (TCA) cycle (Boronat et al., 1983). Additionally, glyoxylate and succinate can also be fused to form isocitrate by isocitrate lyase (Fig. 3) (Ashworth and Kornberg, 1964; Maloy and Nunn, 1982; Mückschel et al., 2012).
Interestingly, studies have reported differential expression of the enzymes constituting the glyoxylate shunt in E. coli strains. During growth on glucose, E. coli strain BL21 exhibited constitutive expression of proteins isocitrate lyase (aceA) and malate synthase (aceB), while the inverse happens with E. coli JM106 due to expression of the isocitrate lyase repressor (iclR) (Phue and Shiloach, 2004). This implies that there is a higher flux through the glyoxylate shunt that, together with the also attested lower expression of isocitrate dehydrogenase in E. coli BL21, is pointed as the reason for lower acetate accumulation in glucose-based fermentations (Liu et al., 2017; Phue and Shiloach, 2004). This different expression level of the glyoxylate shunt might also impact the cell growth on EG, as will be further considered is Section 6. The glyoxylate shunt might play a complementary role to the glycerate pathway since mutant strains unable to produce malate synthase G were able to grow on glyoxylate but showed a slower growth rate than the wild-type (WT) cells (Ornston and Ornston, 1969). Also, E. coli mutants without isocitrate lyase activity were able to grow on glycolate (Ashworth and Kornberg, 1964). The oxidative route can be considered essential for growth on EG since it feeds into glycolysis and subsequently serves as a source of acetyl-CoA, which in its turn is essential for the glyoxylate shunt (Tiso et al., 2022). The glyoxylate shunt enables utilization of glyoxylate as carbon source but requiring acetyl-CoA or succinate. However, the assimilated carbon atoms would be totally lost as CO2 when acetyl-CoA and succinate are recovered through oxidative decarboxylation reactions. Consequently, these two pathways are unable to donate carbon atoms, but they can supply reducing equivalents and energy (Panda et al., 2023).
3.1.3. The β-hydroxyaspartate cycle
An alternative pathway with potential for EG metabolization is the recently characterized BHA cycle (pathway depicted in orange in Fig. 3) (Kornberg and Morris, 1963; Schada von Borzyskowski et al., 2019). This cycle was first discovered in Paracoccus denitrificans (formally Micrococcus denitrificans) (Kornberg and Morris, 1963) and it was recently fully characterized in Paracoccus denitrificans DSM413 (Schada von Borzyskowski et al., 2019). This cycle enables the direct production of oxaloacetate, a TCA cycle intermediate, from glyoxylate through four enzymatic steps. First, an l-aspartate-glyoxylate aminotransferase transaminates one molecule of glyoxylate into glycine using aspartate as the preferred amino group donor, generating oxaloacetate in the process. Then, a BHA aldolase catalyses the condensation of another glyoxylate molecule with glycine into BHA, which is then converted to iminosuccinate by BHA dehydratase. Finally, iminosuccinate is reduced to l-aspartate by iminosuccinate reductase, thus regenerating the amino donor for the first step of the cycle. Additionally, iminosuccinate can spontaneously decay into free ammonia and oxaloacetate in solution (Fig. 3) (Schada von Borzyskowski et al., 2019). Overall, the BHA cycle converts two molecules of glyoxylate (C2) into one molecule of oxaloacetate (C4) with no carbon loss, consuming one reducing equivalent, and leading to the regeneration of the catalytic amino donor l-aspartate (Schada von Borzyskowski et al., 2019). The analysis of marine metagenomes from the Tara Oceans expedition online database demonstrated this cycle is ubiquitous in marine Proteobacteria and that this route is globally distributed. On average, it was 20-fold more abundant than the glycerate route in the tested dataset (Schada von Borzyskowski et al., 2019).
Although P. denitrificans is the only organism where this pathway has been fully characterized, Schada von Borzyskowski and colleagues have implemented the BHA cycle into P. putida KT2440 with the goal of assessing whether the implementation of this cycle would permit better growth than the natural EG oxidative pathway (Schada von Borzyskowski et al., 2023). In their work, a P. putida strain unable to assimilate EG through their native glycerate pathway – due to a deletion of the glyoxylate carboligase gene (gcl) – and expressing the BHA cycle enzymes was subjected to adaptive laboratory evolution (ALE). The evolved strain was able to grow 35% faster on 20 mM EG with a 20% higher biomass yield when compared to another P. putida strain evolved to grow on EG with the glycerate pathway (Li et al., 2019; Schada von Borzyskowski et al., 2023). Interestingly, before application in P. putida, the authors implemented part of this pathway in E. coli SIJ488 (Schada von Borzyskowski et al., 2023). The expression of BhcC (BHA aldolase), BhcB (BHA dehydratase) and BhcD (iminosuccinate reductase), a linear metabolic module that transforms glyoxylate and glycine into aspartate, was able to support biomass formation in an E. coli Δgcl after ALE. The deletion of the gcl gene means that this strain is not able to grow on glycolate and glycine, unless the BHA proteins BhcCBD are used. After prolonged incubation in M9 medium with glycolate and glycine, spontaneous mutants capable of growing on both carbon sources were obtained. These mutants add acquired point mutations in genes encoding for enzymes of the TCA cycle, suggesting the need for slight adaptations in the native metabolic network to support growth using the BHA cycle (Schada von Borzyskowski et al., 2023). Thus, it would be interesting to, in the future, connect this strain to EG as feedstock.
3.2. Dehydratase route
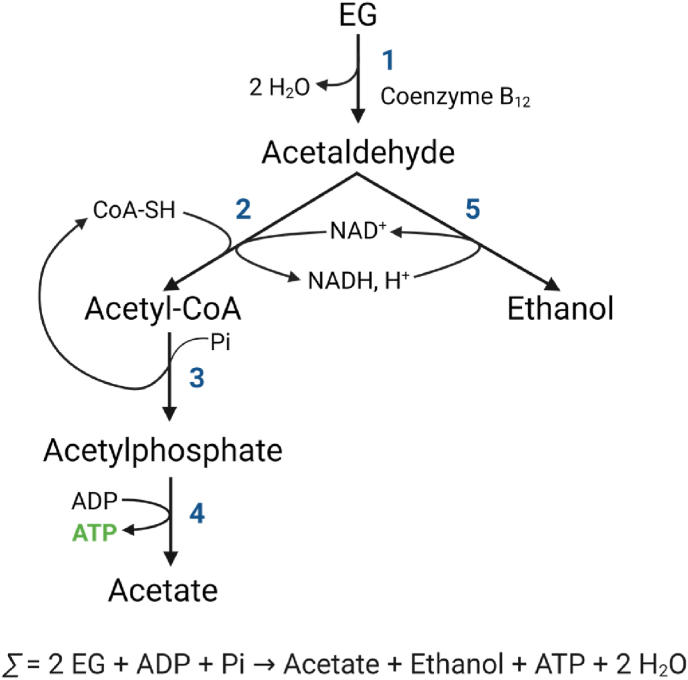
The dehydratase route (Fig. 4) is typically found in some Clostridium, Salmonella and Klebsiella species, and a few other anaerobic or facultative anaerobic organisms due to the oxygen sensitivity of the diol-dehydratase involved in this pathway (Crowley et al., 2010; Hartmanis and Stadtman, 1986; Toraya et al., 1979; Trifunović et al., 2016; Wiegant and De Bont, 1980). In this alternative route for EG assimilation, EG is dehydrated to acetaldehyde by a diol-dehydratase (Wiegant and De Bont, 1980). Acetaldehyde molecules are then used to form the alcohol ethanol and acetyl-CoA. The latter is further converted to acetic acid in two steps that are coupled to ATP formation by substrate-level phosphorylation. More specifically, a CoA and NAD+ dependent aldehyde dehydrogenase catalyses the oxidation of acetaldehyde into acetyl-CoA, which is then converted to acetylphosphate by phosphotransacetylase with regeneration of CoA. Finally, acetylphospate is hydrolysed to acetate by acetate kinase with generation of one molecule of ATP (Fig. 4) (Hartmanis and Stadtman, 1986; Toraya et al., 1979).
Fig. 4.
Metabolic pathway of ethylene glycol (EG) fermentation through the dehydratase route. The metabolization of EG through this pathway is dependent of a coenzyme B12-dependent diol-dehydratase (1), an aldehyde dehydrogenase (2), a phosphotransacetylase (3), an acetate kinase (4) and an alcohol dehydrogenase (5) (Hartmanis and Stadtman, 1986; Toraya et al., 1979). Image adapted from Toraya et al. (1979). Created with BioRender.com.
In the earlier studies, diol-dehydratase (EC 4.2.1.28) is reported to be extremely oxygen-sensitive (Hartmanis and Stadtman, 1986; Toraya et al., 1979) and strictly dependent on coenzyme B12 (Toraya et al., 1979; Wiegant and De Bont, 1980). It is also reported to be strongly associated with the cell membrane, and to require monovalent or divalent ions for its activity (Hartmanis and Stadtman, 1986; Toraya et al., 1979). Furthermore, diol-dehydratase has also been identified in Salmonella enterica (Crowley et al., 2010) and Acetobacterium woodie (Trifunović et al., 2016) inside a microcompartment (Crowley et al., 2010). This compartment encapsulates the first two steps of the dehydratase route preventing cytosolic exposure of the toxic aldehyde resulting from the first diol-dehydratase reaction, which can lead to chromosomal mutations and cytotoxicity, together with enzymes required to maintain the active form of the B12 cofactor at the expense of ATP (Crowley et al., 2010). Although the dehydratase route enables the anaerobic two-steps conversion of EG into acetyl-CoA, and E. coli is capable of anaerobic growth, the challenges involved in diol-dehydratase activity would pose difficulties when expressing this pathway in industrially relevant E.coli strains. For example, E. coli, although possessing a few enzymes of the de novo biosynthetic coenzyme B12 pathway, is only capable of synthetizing coenzyme B12 if the intermediate adenosylcobinamide is supplied (Lawrence and Roth, 1995). Alternatively, the remaining enzymes lacking in the coenzyme B12 production pathway would need to be added to E. coli as well as other relevant proteins, meaning that up to 28 genes would need to be heterologously expressed in E. coli to confer ability to synthesize coenzyme B12 (Fang et al., 2018).
4. Metabolic engineering and synthetic assimilation of EG in E. coli
EG, though an unconventional feedstock, holds promise as a sustainable and renewable precursor for producing different chemical compounds of industrial interest. Recently, Pandit et al. have explored the idea of designing orthogonal network structures, which operate with minimal interaction between chemical-producing pathways and biomass-producing pathways for more efficient chemical production. According to this study, EG shows a high orthogonality score to produce target chemicals. The orthogonality score is a quantitative measure inversely proportional to the interconnectedness between pathways responsible for producing a target chemical and those involved in biomass production. It must be added that this approach is contrary to the notion of growth-coupled strain design, where cell growth is linked to product formation by removing reactions that produce biomass without producing the target chemical. In this way, biomass production and chemical production become coupled. Here, the authors argue that growth-coupled approaches may invariably lead to suboptimal production of biomass and target compounds due to the complex nature of metabolism and that orthogonal pathways may be an advantageous alternative (refer to Pandit et al., 2017 for a more detailed discussion about this topic). EG is highly promising when used as substrate for orthogonal production of a variety of chemicals since it minimizes interactions with biomass-producing pathways (Pandit et al., 2017). It is even suggested that EG should be used as an alternative substrate to sugars, such as glucose, that have significant overlap between biomass and chemical production pathways (Pandit et al., 2017).
Although E. coli, a commonly used host for ME, possesses the genetic inventory to express the EG oxidative pathway, it cannot naturally use EG as carbon source (Panda et al., 2021). However, either by genetically modifying its metabolism or by performing ALE experiments, it is possible to enable E. coli's growth on EG. Up until recently, only one study had reported EG utilization by E. coli (Boronat et al., 1983). In that study, a strain able to use EG was selected from evolved propylene glycol (or l-1,2-propanediol) utilizing mutants. In the EG-assimilating strain, the authors detected an increment of enzymatic activities of l-1,2-propanediol oxidoreductase (FucO), GA dehydrogenase (AldA) and glycolate dehydrogenase (GlcDEF). Enzyme promiscuity is an inherent and relevant element that lead to enzymatic activity with substrates others than the natural ones, including both native and non-native compounds (Khersonsky and Tawfik, 2010). In this case, enzymes that act on l-1,2-propanediol (hereafter referred to as propanediol) and other alcohols, have shown ability to accept EG as substrate as well (Child and Willetts, 1978; Hartmanis and Stadtman, 1986; Mückschel et al., 2012). In recent years, more articles reporting EG consumption by E. coli have been published. For example, a recent study demonstrated that overexpression of fucO and aldA in E. coli K-12 MG1655 strain conferred the ability to grow on EG, when 4 g/L glycerol was also added, as well as on propanediol (Szappanos et al., 2016). Other studies leverage the overexpression of FucO and AldA as a basis for metabolic engineering in EG metabolization, leading to the production of target compounds. These ME strategies will be further discussed in Section 4.2.
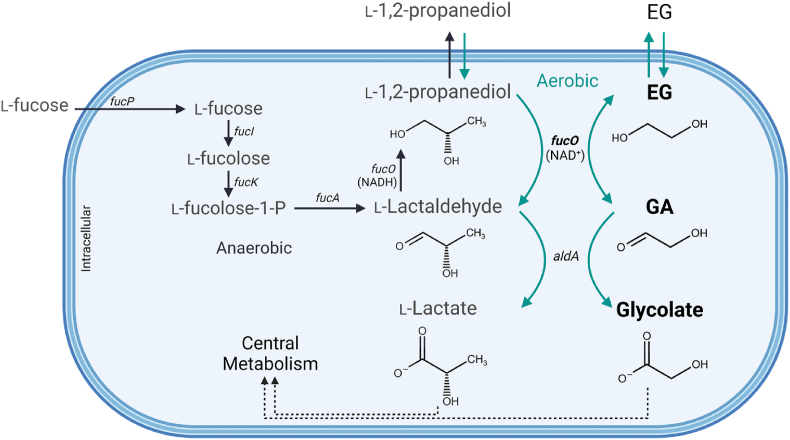
4.1. The FucO enzyme
FucO (EC 1.1.1.77) is encoded by the gene fucO, a member of the fuc regulon that controls l-fucose consumption in E. coli (Fig. 5). Under fermentation, FucO is responsible for generating NAD+ by reducing l-lactaldehyde, a product from l-fucose fermentation, to propanediol. Thus, functioning as a lactaldehyde oxidoreductase under fermentation on l-fucose. FucO also catalyses the inverse reaction converting propanediol into l-lactaldehyde yielding NADH (Fig. 5) (Boronat and Aguilar, 1979; Cocks et al., 1974). Induction of the fuc regulon, and consequently of fucO, requires the presence of transcriptional activator FucR and its effector l-fuculose-1-phosphate (Bartkus and Mortlock, 1986; Chen et al., 1987). Thus, propanediol or EG cannot solely be used aerobically as carbon source since these compounds cannot induce the expression of the fuc regulon. To allow growth from propanediol or EG, the expression of FucO will need to be constitutive or induced. FucO can convert EG into GA, which is then irreversibly converted to glycolate by the action of AldA (Baldomà and Aguilar, 1987). Finally, the native GlcDEF, an oxidoreductase complex present in the cytoplasmic membrane, transforms glycolate into glyoxylate and reduces an electron-transfer quinone (Lord, 1972; Pellicer et al., 1996; Sallal and Nimer, 1989).
Fig. 5.
Metabolic pathway of thel-fucose system and its intersection with ethylene glycol (EG) metabolism. The metabolization of l-fucose is dependent of l-fucose permease (fucP), l-fucose isomerase (fucI), l-fuculose kinase (fucP), l-fuculose-1-phosphate aldolase (fucA), and NADH dependent l-1,2-propanediol oxidoreductase (fucO). Under anaerobiosis, FucO reduces l-lactaldehyde to propanediol which is excreted via a facilitator (Lu et al., 1998). Under aerobic conditions, l-lactaldehyde is instead oxidized to l-lactate by a NAD+-dependent oxidoreductase (aldA). l-lactate is further oxidized to pyruvate and enters central metabolism. Due to enzyme promiscuity, FucO can oxidize both propanediol and EG into their aldehydes, l-lactaldehyde and glycolaldehyde, respectively. Thus, when EG is available, it is oxidized to GA by the NAD+-dependent dehydrogenase function of FucO, and GA is converted to glycolate by AldA. Glycolate can then enter central metabolism via the oxidation pathway and/or the glyoxylate shunt. Abbreviations: EG, ethylene glycol; GA, glycolaldehyde; l-fucolose-1-P, l-fuculose-1-phosphate. Created with BioRender.com.
FucO, which is Fe2+-dependent, can be irreversibly inactivated by metal-catalysed oxidation (MCO) (Cabiscol et al., 1992, 1994). Although this enzyme oxidizes EG into GA aerobically, oxygen and NADH can result in an intrinsically catalysed Fenton reaction that damages the protein, which leads to diminished efficiency of this enzyme (Lu et al., 1998). Thus far, two variants of FucO have been reported in the literature as being more oxygen-stable. One variant contains the mutations I7L and L8V at the N-terminal, identified in mutant strains that acquired the ability to grow in propanediol, demonstrating increased resistance to MCO without significant effect on catalysis (Lu et al., 1998). In a second report, another FucO variant containing a single L8M mutation is reported, also identified in evolved propanediol strains, that might alleviated MCO toxicity (Lee and Palsson, 2010). Interestingly, it was also discovered that these and other evolved E. coli strains obtained from continued selection with propanediol, were able to express fucO constitutively as a result of an IS5 insertion in the promoter region between the two fuc regulons, fucAO and fucPIKUR, simultaneously disabling ability to grow on fucose (Chen et al., 1989; Lee and Palsson, 2010; Lu et al., 1998). In the report by Lee and Palsson, it was verified that the propanediol assimilating phenotype of the evolved strain was a result of the combination of the mutation fucOL8M together with the IS5 insertion that led to the constitutive expression of FucOL8M. Interestingly, when the mutation fucOL8M and IS5 insertion were introduced in the genome of the non-evolved WT strain, together with two other identified mutations in ilvG and ylbE in the evolved strain, the propanediol assimilating phenotype could be reconstructed (Lee and Palsson, 2010). A recent in vivo comparison between FucOI7L/L8V and FucOL8M mutants report that the first leads to a markedly higher EG assimilation rate and growth rate/yield, although still maintaining some oxygen sensitivity (Pandit et al., 2021). In particular, the mutant FucOI7L/L8V has been used in a few recent EG metabolization strategies in E. coli, as will be described in the following sections. A more comprehensive discussion about the enzyme FucO is provided is Section 5.
Naturally, some of the studies focusing on engineering EG assimilation in E. coli have targeted the overexpression of FucO and AldA, making use of the existing natural EG assimilation routes. Alternatively, other studies have focused on the development of whole new synthetic routes for EG assimilation. These EG assimilation studies in E. coli will be reviewed and discussed in the following Sections 4.2, 4.3.
4.2. Overexpressing the FucO and AldA enzymes
A number of studies have successfully modified E. coli to use EG as a carbon source, using the overexpression of the native enzymes FucO and AldA as basis of the ME strategies applied (Panda et al., 2021, 2023; Pandit et al., 2021). An overview of these reports is provided in Table 1. In a recent study, orthogonality principles were used to demonstrated that EG has higher orthogonality scores then glucose, xylose, and formate when used to simulate production of four chemicals with industrial importance: succinate, ethanol, glycolate, and 2,3-butanediol (Pandit et al., 2021). Since the route converting EG into glycolate had the highest orthogonality, the authors engineered E. coli for glycolate production using EG. Flux balance analysis (FBA) results suggested that micro-aerobic conditions are required for accumulation of glycolate. The conversion of glycolate to glyoxylate is catalysed by the glycolate dehydrogese GlcDEF, reducing a proton acceptor quinone (Fig. 3). Then, the resulting quinol utilises oxygen as final electron acceptor. As such, under anaerobic conditions, the regeneration of quinone is blocked and, consequently, the activity of GlcDEF is inhibited, leading to accumulation of glycolate. Thus, oxygen levels can serve as a control mechanism to switch metabolism from cell growth to glycolate accumulation. This controllable switch for product synthesis is desired in an orthogonal branched pathway and beneficial in a two-step industrial fermentation process. (Pandit et al., 2017, 2021). Bioreactor studies with EG as main carbon source and with an E. coli strain overexpressing FucOI7L/L8V and AldA demonstrated that glycolate accumulation could be induced through a reduction in the aeration (Table 1). More specifically, two strategies were ultimately tested by these authors in fed-batch reactors with minimal media supplemented with yeast extract at 2 g/L. The first one consisting of a biomass production phase with increased aeration (600 mL/min) followed by a drop in aeration (100 mL/min) during the glycolate production phase. The second strategy used a lower continuous aeration rate (50 mL/min) with a faster agitation speed during initial growth, and a slower speed during the glycolate production stage to reduce the oxygen transfer rate. The rotor speed was decreased until the respiratory quotient (RQ) reached ∼ 0.4 since FBA results demonstrated a correlation between the RQ and glycolate production. Particularly, a RQ value between 0.15 and 0.4 was optimal for higher glycolate productivity yield on the substrate (Pandit et al., 2021). The second strategy led to the highest glycolate titres (10.4 g/L vs 6.8 g/L) and yield from EG during production phase (0.8 g/g vs 0.75 g/g) (Table 1). Finally, while oxygen limitation might benefit glycolate production and the reaction catalysed by the oxygen-sensitive FucO, it might also limit regeneration of NAD+ if the oxygen level is too low, which would limit the flux through the EG assimilation pathway. So, a trade-off between the verified oxygen sensitivity of FucO and the oxygen dependent regeneration of NAD+ would need to be fine-tuned to maximize glycolate production (Pandit et al., 2021).
Table 1.
Characteristics, experimental setup of the best result and identified challenges of the EG assimilation strategies applied to Escherichia. Coli that are based on the natural oxidative Ethylene Glycol (EG) assimilation pathway. These pathways are based on the overexpression of the first two enzymes of the natural EG oxidative pathway, FucO and AldA, except for the study by Yan et al. where Gox0313 is used instead of FucO. Abbreviations: EG, ethylene glycol; CSM, Complete Supplement mixture; P_gyrA, promoter gyrA; FucO, propanediol oxidoreductase; FucOI7L/L8V, propanediol oxidoreductase mutant I7L/L8V; AldA, glycolaldehyde dehydrogenase.
| Reference | Objective | Strategy | Carbon source | Best result | Identified challenges |
|---|---|---|---|---|---|
| Pandit et al. (2021) | Production of glycolate from EG |
E. coli MG1655 strain expressing FucOI7L/L8V and AldA under trc promoter in high copy number plasmid pTRC99a Glycolate production performed in bioreactor with two stage fermentation: aerobic growth phase followed by an oxygen limiting glycolate production phase |
M9 minimal medium supplemented with 2 g/L yeast extract with addition of EG during fermentation as to maintain 5–10 g/L EG | 10.4 g/L glycolate after 140 h at 37 °C and pH 7.0, corresponding to a yield of 0.8 g/g EG during production phase | - Oxygen sensitivity during early exponential growth phase when using FucO that is only in part alleviated by using FuCOI7L/L8V - NAD+ generation might be limited if oxygen level is too low, which limits EG uptake and EG assimilation - Many regulatory changes that occur under oxygen-limiting condition are not accounted for in metabolic models |
| Panda et al. (2021) | Improve the growth of E. coli on EG |
E. coli MG1655 DE3 expressing FucO and AldA under constitutive P_gyrA in medium copy number plasmid pEG03 Growth assays performed in shake flasks without antibiotic supplementation |
Modified M9 minimal medium with 20 g/L of EG supplemented with 2 g/L CSM | Total consumption of 10 g EG after 72 h at 30 °C, reaching an OD600 of ∼6 after 24 h | - Supplementation with glycerol (0.1 g/L) of CSM was required for efficient EG utilization, furthermore, no family of amino acids (aspartate, aromatic, pyruvate and others) could be excluded - Almost no EG is consumed at 37 °C - High EG concentrations (100 g/L) inhibited growth and only a small amount of EG was consumed |
| Panda et al. (2023) | Production of L-tyrosine from EG (synthesis of L-phenylalanine and p-coumaric acid from EG at gram per litre titre is also reported) |
E. coli MG1655 DE3 ΔtyrR ΔpheA containing a plasmid expressing aroG and tyrA under lac promoter and a second medium copy number plasmid pEG03∗ overexpressing FucOI7L/L8V and AldA under constitutive P_gyrA L-tyrosine production performed in shake flasks without antibiotic supplementation |
Modified M9 minimal medium with 2 g/L NH4+ and 20 g/L EG supplemented with 1.6 g/L CSM after | 3 g/L L-tyrosine was produced from 17 g/L EG after 96 h at 30 °C | - Supplementation with CSM was required for efficient EG utilization |
| Yan et al. (2024) | Production of glycolate from EG |
E. coli MG1655 ΔfucO ΔyqhD expressing Gox0313 (instead of FucO) and AldA under trc promoter in high copy number plasmid pTrc99a Glycolate production performed in shake flasks |
M9 minimal medium with 10 g/L EG supplemented with 2 g/L yeast extract | 5.1 g/L glycolate from 6.8 g/L EG at a yield of 0.75 g/g EG after 120 h at 37 °C, having a maximum productivity of 0.158 g/L/h during the initial 24 h | - High concentration of glycolate inhibit growth and EG uptake |
In another study by the same research group, an EG-consuming E. coli MG1655 DE3 strain was obtained by systematically optimizing the fucO and aldA gene expression together with the medium composition (Panda et al., 2021). It was verified that a low concentration of glycerol (0.1 g/L) was required for more efficient EG utilization as carbon source. Without glycerol, an initial strain overexpressing fucO and aldA under an l-arabinose inducible promoter consumed only 6% of the EG in the media (5 g/L EG) within 72 h, while adding 0.1 g/L glycerol doubled cell density and increased the percentage of EG consumed to 17%. Glycerol, in addition to being a carbon source, may stimulate the expression of transporters and enzymes related to polyol degradation which might aid in EG import, as both glycerol and EG are polyols (Panda et al., 2021). Furthermore, glycerol has a higher redox potential then EG, providing more reducing equivalents and energy which benefits cellular processes and might create a more favourable intercellular environment for EG assimilation. During process optimization, the authors saw that using a strong constitutive promoter (PgyrA) proved to be significantly better than inducible promoters in driving expression of FucO and AldA, resulting in the further tripling of cell density and 80% EG consumption. Switching between different origins of replication and different antibiotic resistance markers had no substantial effect on EG utilization. Additionally, antibiotic was not needed to maintain the plasmid expressing fucO and AldA due to the selective pressure exerted by the need of expressing these enzymes to use EG as carbon source. Additionally, the authors had initially tested another host strain (E. coli strain BW25113), which proved to be slightly outperformed by E. coli strain MG1655 DE3 in terms of cell growth and EG consumption, thus ultimately only strain MG1655 DE3 is used as best strain. While cells were able to use EG at 30 °C, almost no EG was consumed at 37 °C. Further medium optimization showed that glycerol supplementation could be substituted with a Complete Supplement mixture (CSM, a mixture of amino acids and nucleobases; 1 g/L) with an even higher cell density and almost complete consumption of EG in the media as a result. Interestingly, fucO and aldA had higher expression levels (200% and 70% higher, respectively) with CSM supplementation, compared to glycerol supplementation. Finally, the EG using strain under all verified optimal conditions, in medium containing 20 g/L EG and 2 g/L CSM, could consume all EG in 72 h (Table 1). However, when fed 100 g/L EG, the strain consumed only a small amount, suggesting that high EG concentrations inhibit growth (Panda et al., 2021). Although CSM supplementation adds to industrial fermentation costs, these are encouraging results. Future research lines could include engineering a strain able to use EG efficiently without amino acid supplementation and testing improved FucO mutants such as FucOI7L/L8V.
In a more recent work by Panda and colleagues, an E. coli strain was engineered to produce l-tyrosine from EG (Panda et al., 2023). In this study, the mutant FucOI7L/L8V was tested instead of the WT FucO (Lu et al., 1998; Pandit et al., 2021). Using FucOI7L/L8V resulted in slightly faster growth on EG. Subsequently, FucOI7L/L8V and AldA under control of PgyrA, were introduced in an E. coli MG1655 DE3 strain previously engineered for l-tyrosine overproduction from glucose (Ma et al., 2020). Specifically, this strain contains the deletion of two genes: tyrR (to alleviate the transcriptional repression of l-tyrosine production) and pheA (to prevent formation of the competition aromatic amino acid phenylalanine), and a plasmid overexpressing feedback-resistant aroG and tyrA (Ma et al., 2020). In this work, the authors added a lower concentration of isopropyl β-d-1-thiogalactopyranoside (IPTG; 0.01 mM) upon inoculation avoiding the monitorization of cell growth to induce gene expression, which increased biomass growth and l-tyrosine titre and productivity (Panda et al., 2023). Also, when supplementing 20 g/L EG, the cells stopped growing after 48 h, point at which 1.5 g/L l-tyrosine was achieved and around 10 g/L EG was left. By considering that the nitrogen and/or phosphorous sources might become depleted after 48 h, the authors doubled the initial ammonium concentration to 2 g/L, observing an improvement in growth and l-tyrosine production (Panda et al., 2023). Overall, the developed strain yielded a titre of 3 g/L l-tyrosine after 96 h, when supplementing the medium with 20 g/L EG and 1.6 g/L CSM (Table 1). These conditions led to the maximum titres outperforming the value (1.2 g/L) achieved when growing the cells solely on glucose (Panda et al., 2023).
Interestingly, in this work transcriptome analysis was used to investigate the reasons for the increased l-tyrosine synthesis using EG as carbon source compared to glucose (Panda et al., 2023). It was revealed that fucO and aldA were upregulated by 2-fold in the presence of EG and that most of the transcripts came from the plasmid, which may suggest that PgyrA is either activated by EG or repressed by glucose. Also, when growing on EG, the glycolate dehydrogenase encoding genes (glcDEF) and the genes in the EG oxidative pathway from glyoxylate up until 2 PG – glyoxylate carboligase (gcl), tartronate semialdehyde reductase (glxR) and glycerate 2-kinase (glxK/garK) – were upregulated by > 100-fold. Malate synthase (glcB or aceB) and isocitrate lyase (aceA) were upregulated by > 20-fold, suggesting that the glyoxylate shunt also operate during growth on EG (Panda et al., 2023). During growth on glucose, through glycolysis, enolase (eno) converts 2 PG into phosphoenolpyruvate (PEP), a precursor for both l-tyrosine and acetyl-CoA synthesis. PEP is converted into pyruvate, which is primarily transformed into acetyl-CoA by pyruvate dehydrogenase (PDH) complex (aceEF/lpd). However, when growing on EG, an upregulation of two alternative routes to convert pyruvate into acetyl-CoA were identified (Panda et al., 2023). The first route involves the conversion of pyruvate to acetate by PoxB, which was upregulated by 17-fold, followed by its transformation into acetyl-CoA by acetyl-CoA synthetase (ACS), which was upregulated by 44-fold. In the second route, upregulated by 2-fold, pyruvate is converted to acetyl-CoA and formate by pyruvate formate-lyase (PflB). Since the transcription of eno and aceEF/lpd was halved, the expression of enzymes might be inactivated during growth on EG. This would divert pyruvate from glycolysis and facilitate the accumulation of PEP, resulting in a higher l-tyrosine titre (Panda et al., 2023). Furthermore, in this study, E. coli was also engineered to synthesize l-phenylalanine, achieving 1.5 g/L phenylalanine from 10 g/L EG (Panda et al., 2023). EG-derived production of p-coumaric (pCA) was also engineered by additionally expressing tyrosine ammonia lyase (Tal), which transforms l-tyrosine into pCA (∼1 g/L pCA from 10 g/L EG) (Table 1) (Panda et al., 2023). Finally, it was demonstrated that the grade of EG used in these experiments could be replaced by EG derived from acid-hydrolysed PET bottles, with similar titres of l-tyrosine, supporting the concept that plastic waste can be transformed into value-added chemicals (Panda et al., 2023).
Lastly, a recent report successfully constructed an E. coli MG1655 strain able to produce glycolate from EG (Yan et al., 2024). Different enzyme candidates were tested for the initial EG oxidation step into GA, concluding that expressing the NAD+-dependent alcohol dehydrogenase Gox0313 from Gluconobacter oxydan also capable of oxidizing EG, in place of FucO, could lead to a complete assimilation of 10 g/L EG as major carbon source in 96 h (further discussion about the enzyme Gox0313 and results obtained regarding the comparison of different enzyme candidates for EG oxidation reported by Yan et al. is provided in Section 5). Thus, a new strain was engineered by heterologous expression of Gox0313, by overexpressing aldA, and by knocking out the endogenous GA reductase genes fucO and yqhD to prevent the reconversion of GA into EG. This strain achieved a glycolate titre of 5.1 g/L and a yield of 0.75 g/g EG in shake flaks with M9 medium with 10 g/L EG as major carbon source (Table 1) (Yan et al., 2024). Although the maximum productivity reached 0.158 g/L/h during the initial 24 h, as growth continued, the reported strain could not fully use 10 g/L EG and eventually stopped consuming it, which was confirmed to be caused by growth inhibition when high concentrations of glycolate are achieved. In fact, growth could be inhibited by exogenous glycolate in a linear manner (Yan et al., 2024). The high glycolate titre of the engineered strain where related to high levels of gox0313 transcription, which was promoted by the deletion of fucO and yqhD and the overexpression of aldA. The transcriptional level of aldA was also high and, while it was boosted by deletion of fucO and yqhD, it was hindered by knocking out the endogenous glcDEF. Genes glcDEF were also overexpressed and their transcription was promoted by the fucO and yqhD deletions and the aldA overexpression (Yan et al., 2024). This engineered strain had a higher content of various amino acids such as l-tryptophan and l-tyrosine, and curiously produced a new compound rosmarinic acid (Yan et al., 2024) which might hold interest for further production of derived value-added compounds.
4.3. Glycolaldehyde as a metabolic hub
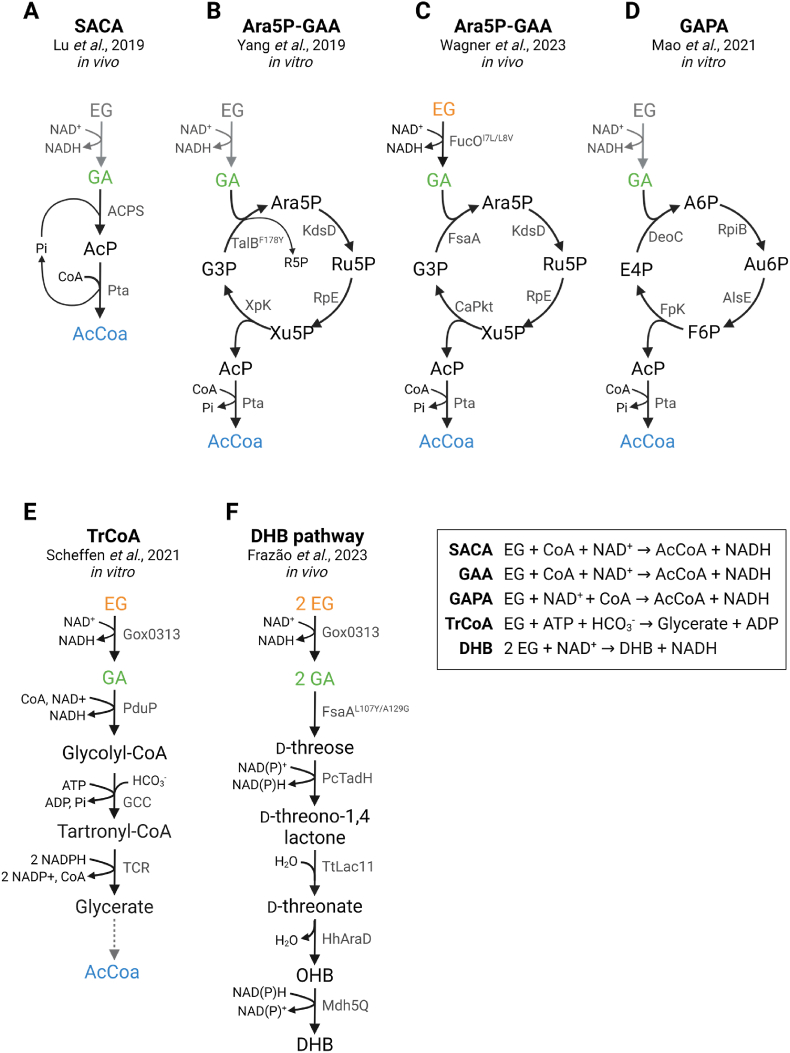
In E. coli, the microbial assimilation of EG proceeds via the intermediate GA through the natural oxidative pathway discussed previously. GA can be considered a crucial molecular intermediate and metabolic hub for EG metabolism, from which carbon can be used to fuel metabolism. A few recent studies have developed new synthetic pathways that use GA as intermediate or precursor to produce, for example, acetyl-CoA in vitro (Mao et al., 2021; Yang et al., 2019) or in vivo (Lu et al., 2019; Wagner et al., 2023) using E. coli, as well as other compounds of interest such as glycerate (Scheffen et al., 2021) or 2,4-dihydroxybutyric acid (DHB) (Frazão et al., 2023). It is worth noting that some of these studies focus on the use of GA as an intermediate primarily for the assimilation of one-carbon (C1) molecules due to the recent development of a novel engineered enzyme termed GA synthase (Gals), which converts two C1 formaldehyde (FA) molecules into GA (Lu et al., 2019; Yang et al., 2019). Since different C1 compounds, such as methanol and methane, are all converted to FA before being further assimilated by natural pathways that inherently have poor carbon yields, improving FA assimilation through novel pathways can lead to more efficient assimilation of C1 compounds (Yang et al., 2019). Thus, the discovery of the Gals enzyme opened new possibilities for C1 feedstock assimilation trough GA (Lu et al., 2019; Mao et al., 2021; Yang et al., 2019). Nevertheless, the reaction that converts EG into GA, if coupled to these novel synthetic GA-assimilating pathways, can lead to novel ways of assimilating PET by microorganisms such as E. coli, and for that reason they will be reviewed and discussed in this section. The different pathways discussed throughout this Section are outlined in Fig. 6. Also, the pathways applied in vivo are described in more detail in Table 2.
Fig. 6.
Synthetic ethylene glycol (EG) assimilation pathways that use glycolaldehyde (GA) as key intermediate. The pathways (A) Synthetic Acetyl-CoA (SACA) pathway, (B) Ara5P-dependent GA assimilation (GAA) pathway, (C) Ara5P-GAA pathway applied in vivo, (D) GA-allose 6-phosphate assimilation (GAPA) pathway, (E) The tartronyl-CoA (TrCoA) pathway and (F) 2,4-dihydroxybutyric acid (DHB) production pathway are shown, along with the stoichiometric equations for each pathway where the EG to GA reaction is hypothetically considered. When the EG to GA pathway is depicted in grey means that this reaction was not included in the referred report. List of abbreviations in alphabetical order: A6P, allose 6-phosphate; AcCoa, acetly-CoA; AcP, acetyl-phosphate; ACPS, acetyl-phosphate synthase from Actinobacteria; AlsE, d-allulose-6-phosphate 3-epimerase from Escherichia coli; Ara5P, d-arabinose 5-phosphate; Au6P, d-allulose 6-phosphate; CaPkt, phosphoketolase from Clostridium acetobutylicum; DeoC, 2-deoxy-d-ribose-5-phosphate aldolase from E. coli; DHB, 2,4-dihydroxybutyric acid; E4P, erythrose 4-phosphate; EG, ethylene glycol; F6P, d-fructose 6-phosphate; FA, formaldehyde; FpK, F6P phosphoketolase from Bifidobacterium adolescentis; FsaA, d-fructose 6-phosphate aldolase from E. coli; FucO, EG oxidoreductase from E. coli; G3P, glyceraldehyde-3-phophate; GA, glycolaldehyde; Gals, GA synthase from Pseudomonas putida; GCC, engineered glycolyl-CoA carboxale from Methylorubrum extorquens; Gox0313, alcohol dehydrogenase from Gluconobacter oxidant; HCO3−, hydrogen carbonate; HhAraD, l-fuconate dehydratase from Herbaspirillum huttiense; KdsD, Ara5P isomerase from E. coli; Mdh5Q, malate dehydrogenase mutant from E. coli; OHB, 2-oxo-4-hydroxybutyrate; PcTadH, d-threo-aldose 1-dehydrogenase from Paraburkholderia caryophylli; PduP, aldehyde dehydrogenase of Rhodopseudomonas palustris; Pta, phosphate acetyltransferase from E. coli; R5P, ribose 5-phosphate; RpE, d-ribulose 5-phosphate epimerase from E. coli; RpiB, allose-6-phosphate isomerase/R5P isomerase B from E. coli; Ru5P, ribulose 5-phosphate; TalBF178Y, transaldolase B mutant F178Y from E. coli; TCR, tartronyl-CoA reductase from Chloroflexus aurantiacu; TtLac11, gluconolactonase from Thermogutta terrifontis; XpK, xylulose 5-phosphate phosphoketolase from Pseudomonas stutzeri; Xu5P, d-xylulose 5-phosphate. Created with BioRender.com.
Table 2.
Characteristics, experimental setup of the best result and identified challenges of the synthetic glycolaldehyde (GA) or ethylene glycol (EG) assimilating pathways reported in the literature and that were tested in in vivo. Abbreviations: FA, formaldehyde; GA, glycolaldehyde; MVA, melavonate; DHB, 2,4-dihydroxybutyric acid; Gals, GA synthase; ACPS, acetyl-phosphate synthase; Pta, phosphate acetyltransferase; FucOI7L/L8V, propanediol oxidoreductase mutant I7L/L8V; FsaA, D-fructose 6-phosphate (F6P) aldolase; CaPkt, phosphoketolase from Clostridium acetobutylicum; KdsD, D-arabinose 5-phosphate (Ara5P) isomerase; Rpe, D-ribulose 5-phosphate epimerase; Gox0313, alcohol dehydrogenase from Gluconobacter oxydan; FsaAL107Y/A129G, F6P aldolase mutant L107Y/A129G; Mdh5Q, malate dehydrogenase; HhAraD, L-fuconate dehydratase from Herbaspirillum huttiense; Re.kdgT, D-threonate importer system.
| Pathway | Objective | Type of pathway | Strategy | Carbon source | Best result | Identified challenges | Reference |
|---|---|---|---|---|---|---|---|
| SACA pathway | Production of acetyl-CoA from FA (through GA) | Linear with 2 steps from GA to acetyl-CoA |
E. coli BL21 DE3 expressing Gals, ACPS and Pta in plasmid pET-28a Growth assays performed in shake flasks |
LB medium with 0.4 g/L GA | Strain grew better with GA supplementation and reached an OD600 of ∼7 (vs ∼ 5.5 without GA) after 20 h at 37 °C | - ACPS might have low affinity for GA (Km = 51 mM) | Lu et al. (2019) |
| Ara5P-GAA pathway | Production of acetyl-CoA from EG | Cyclic with 6 steps from EG to acetyl-CoA |
E. coli MG1655 ΔyqhD ΔaldA overexpressing FucOI7L/L8V, FsaA and CaPkt in plasmid pZS23 and KdsD, Rpe and Pta in plasmid pZS13 (for GA metabolization); strain contains plasmid pMEV-7 harbouring the MVA pathway genes Acetyl-CoA production in shake flasks |
M9 minimal medium with 500 mM EG, 250 mM glycerol, 5 g/L yeast extract and 100 mM CaCO3 | 0.82% of MVA from EG after complete glycerol consumption at 46 h and 30 °C, with a maximum of 1.24% MVA at 22 h | - No growth detected at elevated GA concentrations - The main by-products were EG and D-threose - Extremely low acetyl-CoA yield |
Wagner et al. (2023) |
| DHB production pathway | Production of DHB from EG | Linear with 6 steps from EG to DHB |
E. coli MG1655 ΔyqhD ΔaldA ΔlldD expressing Gox0313, FsaAL107Y/A129G, PcTadH, TtLac11 in plasmid pACT3, expressing Mdh5Q, HhAraD in plasmid pEXT22, and Re.kdgT in plasmid pEXT21; all genes are under the control of the tac promoter DHB production in concentrated IPTG-induced pre-cultures |
M9 minimal medium with 320 mM (∼20 g/L) EG (no co-substrate) LB medium with 320 mM EG and supplemented with L-cysteine and FeCl3 |
2.14 mM of DHB from EG after 48 h at 37 °C in the M9 medium 6.75 mM (1 g/L) of DHB from EG after 48 h at 37 °C in the LB medium |
- High GA concentrations led to increased accumulation of D-threose and almost no DHB production - Substrate affinities of FsaAL107Y/A129G and PcTadH may be low - The main by-products were EG and D-threose - Low production rates and titres are obtained from EG or GA |
Frazão et al. (2023) |
4.3.1. Acetyl-CoA yielding pathways
The natural oxidative EG-assimilation route yields only 0.5 mol acetyl-CoA per mole of EG, at the expense of 1 molecule of ATP and 2 molecules of NAD+. Besides being a critical intermediate metabolite for the cell, acetyl-CoA can serve as a precursor for many industrially relevant products, such as biofuels (Guo et al., 2015) and bioplastics like polyhydroxyalkanoates (PHA) (Kudo et al., 2023). Hence, this prompts the search for more effective aerobic routes for converting EG (via GA) into acetyl-CoA using pathways that are carbon conserving, independent of energy or reducing equivalents, and have higher carbon yield. A few synthetic pathways have been developed to produce acetyl-Coa (Lu et al., 2019; Mao et al., 2021; Wagner et al., 2023; Yang et al., 2019) that have the potential to be applied in connected to a bioprocess of EG metabolization. These pathways will be discussed throughout this Section and a schematic representation of each pathway can be found in Fig. 6A–D.
4.3.1.1. Synthetic Acetyl-CoA (SACA) pathway
The Synthetic Acetyl-CoA (SACA) pathway (Fig. 6A) converts GA into acetyl-CoA without carbon loss and is independent of energy and reducing equivalents (Lu et al., 2019). In this linear pathway, two molecules of FA are first condensed into GA by Gals from P. putida. Then, GA is converted into acetyl-phosphate (AcP) in a single step by a novel reaction catalysed by a phosphoketolase (Pkt) from Actinobacteria, termed acetyl-phosphate synthase (ACPS), using inorganic phosphate. E. coli's phosphate acetyltransferase (Pta) then converts AcP into acetyl-CoA (Fig. 6A). A calculated total Gibbs energy change suggested that this pathway is highly thermodynamically favourable (−96.7 kJ/mol). Although a yield of over 80% AcP from GA was achieved in vitro, only 33% acetyl-CoA yield was reached starting from FA, possibly due to inhibition of ACPS by FA, and to instability of acetyl-CoA and AcP in solution (Lu et al., 2019). Pathway expression in E. coli BL21 DE3 confirmed that the SACA pathway is functional in vivo and leads to production of acetyl-CoA and derived metabolites such as oxaloacetate from FA or GA (Lu et al., 2019). To mitigate the cellular toxicity arising from FA supplementation, the methanol dehydrogenase gene from Bacillus stearothermophilus was added to the engineered strain. This way, methanol is instead supplemented which is converted into FA by the methanol dehydrogenase, helping maintain a constant but low FA concentration. Still, the SACA pathway only contributed to around 3% biomass from methanol. One of the reasons for this low yield might be the low substrate affinities of the enzymes Gals (Km = 165 mM) and ACPS (Km = 51 mM) to methanol and GA, respectively. Nonetheless, and since the goal of this review is to contemplate EG metabolization, if we consider GA as starting compound, that can be derived from EG, this synthetic pathway does enable the carbon-conserved and ATP-independent conversion of EG into acetyl-CoA through only three steps (Fig. 6A). Interestingly, by supplementing rich medium with more than 0.4 g/L GA resulted in a remarkably higher OD600 for strains expressing the SACA pathway than for those without GA or the SACA pathway (Table 2) (Lu et al., 2019). In this context, increasing the substrate affinity of ACPS through protein engineering would be one relevant way to improve the production of acetyl-CoA from GA, and eventually from EG.
4.3.1.2. Ara5P-dependent GA assimilation (GAA) pathway
Two other studies have computationally predicted two cyclic synthetic pathways for the assimilation of GA that circumvents CO2 loss and ATP/NAD(P)H consumption. One of these examples is the Ara5P-dependent GA assimilation (GAA) pathway (Fig. 6B) that converts FA into acetyl-CoA with a calculated standard Gibbs free energy change of −86.8 kJ/mol (Yang et al., 2019). In this pathway, FA is first condensed to GA by Gals. The pathway progresses through the aldol addition of GA into the acceptor molecule glyceraldehyde-3-phophate (G3P) by an engineered transaldolase B from E. coli (TalBF178Y), yielding d-arabinose 5-phosphate (Ara5P) (Fig. 6B). This process also generates the isomers ribulose 5-phosphate (Ru5P) and ribose 5-phosphate (R5P). Although R5P could be converted to Ru5P by R5P isomerase (Rpi) from E. coli, the small amount of R5P produced could be converted without Rpi due to the promiscuous activities of the other enzymes in the pathway. Then, after two isomerization reactions, Ara5P is converted into d-xylulose 5-phosphate (Xu5P) via d-ribulose 5-phosphate (Ru5P), by chaining the Ara5P isomerase (KdsD) and d-ribulose 5-phosphate epimerase (Rpe) enzymes from E. coli. Finally, the xylulose 5-phosphate Pkt (Xpk) from Pseudomonas stutzeri cleaves Xu5P into AcP and 3 GP, thus regenerating the precursor and closing the loop. AcP can be then converted to acetyl-CoA by Pta from E. coli (Fig. 6B). The viability of the full pathway was only demonstrated in vitro, reaching 88% product carbon yield. (Yang et al., 2019).
Recently, the Ara5P-GAA pathway has been implemented in vivo by Wagner et al., to convert EG to acetyl-CoA, using E. coli as host (Wagner et al., 2023). Some modifications to the original pathway designed by Yang et al. were made, such as the replacement of TalBF178Y with the F6P aldolase FsaA from E. coli that catalyses the cross-aldol addition of GA to G3P without formation of the by-product R5P (Fig. 6C). Also, a Pkt from Clostridium acetobutylicum (CaPkt) was used instead of Xpk. In vitro testing resulted in a low yield of 18.1% AcP from GA, possibly due to aldehyde-induced enzyme(s) inactivation in vitro (Wagner et al., 2023). Subsequently, a mutant E. coli strain – whose genes encoding YqhD, a major endogenous GA reductase, and AldA were deleted to direct GA assimilation through the synthetic pathway – was used as host for the expression of the Ara5P-GAA pathway together with the less oxygen-sensitive FucOI7L/L8V. Glycerol was used as co-substrate for growth and labelled 13C EG and GA were used to assess the pathway for the formation of the acetyl-CoA proxy melavonate (MVA). After enzymes expression optimization, 16.12% of MVA was originated from GA. However, the pathway performance was extremely poor when EG was used as major carbon source, with a maximum of 1.24% of MVA originated from EG (Table 2) (Wagner et al., 2023). Although able to function in vivo, the Ara5P-GAA pathway led to an extremely low acetyl-CoA yield, and a few possible bottlenecks hindering carbon flow were considered. The FucOI7L/L8V oxidation reaction is suggested to be thermodynamically unfavourable (ΔrG’° = 23.7 kJ/mol, Wagner et al., 2023), potentially constraining metabolic flux. Despite employing a high EG concentration to promote initial GA production and assessing adequate gene expression levels, the authors were not able to further improve MVA formation (Noor et al., 2014; Wagner et al., 2023). As such, to increase the metabolic flux towards acetyl-CoA, it is hypothesized that the GA affinity of the next enzyme in the pathway must be very high. However, the GA affinity of FsaA might not be sufficient to enable carbon flux (Wagner et al., 2023). In fact, Yang's study reports a high Km of 11.59 mM and very low kcat of 0.08 s−1 for FsaA when using GA as substrate (Yang et al., 2019). Also, FsaA catalyses the reversible dimerization of GA both as donor and acceptor, into threose, which was one of the by-products detected when GA was used as substrate (Wagner et al., 2023). Interestingly, the other major reaction that drained GA was EG formation, which might suggest the existence of unspecific GA reductases that convert GA to EG. As comparison, AldA from the natural EG oxidative pathway has a very low Km of 0.14–0.38 mM for GA, allowing flow of EG as carbon source (Baldomà and Aguilar, 1987; Rodríguez-Zavala et al., 2006). Overall, the second enzyme in this cyclic pathway needs to be carefully selected, specially targeting a high affinity for GA as donor. Secondly, a better understanding of the apparent limiting role of Pta is currently missing. Although this enzyme was thought to be naturally present in excess, the overexpression of the Pta enzyme led to an increase in acetyl-CoA production, potentially indicating an unforeseen bottleneck (Wagner et al., 2023).
4.3.1.3. GA-allose 6-phosphate assimilation (GAPA) pathway
The other cyclic pathway is the GA-allose 6-phosphate (A6P) assimilation (GAPA) pathway (Fig. 6D), developed by Mao and colleagues (Mao et al., 2021). This pathway is mechanistically similar to the GAA pathway and also relies on an aldolase that condenses the donor GA and an acceptor, which in this case is erythrose 4-phosphate (E4P). First, FA is converted into GA by Gals. Then, 2-deoxy-d-ribose-5-phosphate aldolase (DeoC) from E. coli catalyses the aldol reaction between GA and E4P into A6P. Next, A6P also goes through two isomerization reactions into d-fructose 6-phosphate (F6P) via d-allulose 6-phosphate (Au6P), by the chained activities of allose-6-phosphate isomerase/R5P isomerase B (RpiB) and d-allulose-6-phosphate 3-epimerase (AlsE) from E. coli. The acetyl-CoA precursor AcP and E4P are then produced from F6P, by the enzyme F6P Pkt (Fpk) from Bifidobacterium adolescentis (Mao et al., 2021). AcP is then converted to Acetyl-CoA by Pta (Fig. 6D). This novel pathway was tested in vitro with E4P and GA as reaction substrates and a high carbon yield of 94% was achieved (Mao et al., 2021). However, both the GAA and GAPA pathway are subject to a kinetic trap caused by the broad substrate activity of X/Fpk, with both reports revealing a low but non-neglectable breakdown of other substrates, such as Ru5P, R5P, E4P and GA, into AcP and G3P/E4P. Although it is reported that these unwanted activities were very low compared with those for Xu5P or F6P, they can still cause pathway imbalance. Thus, the yield of these pathways could benefit from future enzyme engineering to increase substrate specificity by X/Fpk, and/or from screening for alternative enzymes.
Overall, these cyclic pathways (Fig. 6B–D) are complex, and any kinetic or stoichiometric imbalance has the potential to trigger the collapse of GA assimilation. Also, since they rely on the recycling of a metabolite (G3P and E4P) that can be diverted into other metabolic reactions, it might be very difficult to implement these pathways in vivo with a significant production yield. Particularly, this was observed in the case of the Ara5P-GAA pathway, where the extremely low acetyl-CoA yields obtained in vivo from EG suggest the requirement for a co-substrate, such as glycerol, to replenish G3P and support cellular growth (Wagner et al., 2023). Overall, further research is essential to enhance the applicability of GA-assimilating circular pathways within industrial context. Conversely, the adoption of a linear GA assimilating pathway like the SACA pathway may offer greater efficacy due to its simplicity and its independence from an acceptor molecule, and further reports are highly expected.
4.3.2. The tartronyl-CoA (TrCoA) pathway
Although not specifically targeting acetyl-CoA production, the tartronyl-CoA (TrCoA) pathway theoretically converts GA to glycerate in four linear reactions (Trudeau et al., 2018). Since glycerate is a direct route to acetyl-CoA production in vivo, this pathway will also be considered in this section (Fig. 6E). This pathway was first developed and tested in vitro by Trudeau et al. as an effort to find alternative synthetic pathways to bypass the energy dissipation and CO2 release from photorespiration. Recently, this pathway was adapted and tested in vitro for EG conversion (Scheffen et al., 2021). EG is converted to GA by FucO, then GA is converted into glycolyl-CoA by aldehyde dehydrogenase of Rhodopseudomonas palustris (PduP). Glycolyl-CoA is converted into glycerate via tartonyl-CoA, by the chained action of a novel glycolyl-CoA carboxale (GCC) obtained after protein engineering of a propionyl-CoA carboxylase from Methylorubrum extorquens and of the bifunctional malonyl-CoA reductase from Chloroflexus aurantiacus (tartronyl-CoA reductase; TCR) (Fig. 6E). After in vitro implementation, only 77 μM (approximately 8.16 mg/L) of glycerate after 2 h were produced. By replacing FucO with Gox0313 (refer to Section 5 for further discussion about the enzyme Gox0313), adding a water forming NADH oxidase to maintain a high NAD+ concentration, and including an efficient ATP regeneration system, a glycerate yield of 485 μM (approximately 51.4 mg/L) from 100 mM EG (approximately 6.2 g/L) was reached. Although this reaction requires ATP and reducing equivalents, the reaction catalysed by GCC fixates carbon by using hydrogen carbonate (HCO3−) as substrate. Overall, the in vivo application of this pathway in E. coli is anticipated.
4.3.3. C4 carbon molecules yielding pathways
A recently proposed synthetic route established the production of the non-natural chemical 2,4-dihydroxybutyric acid (DHB) from GA and EG in E. coli (Frazão et al., 2023). DHB is an important industrial precursor for the chemical synthesis of the amino acid methionine analogue 2-hydroxy-4-(methylthio)butyrate (HTMB) (Walther et al., 2017), or to produce 1,3-propanediol (Frazão et al., 2019). In this pathway, GA is converted to DHB via five carbon conserving steps (Fig. 6F). First, two molecules of GA are fused by FsaAL107Y/A129G, a mutant FsaA with increased efficacy for self-aldol GA addition yielding the C4 compound d-threose (thus, a d-threose aldolase). The resulting four-carbon sugar is oxidized into d-threono-1,4-lactone by a d-threo-aldose 1-dehydrogenase from Paraburkholderia caryophylli with d-threose dehydrogenase activity (PcTadH), which is then converted to d-threonate in a reaction catalysed by a gluconolactonase from Thermogutta terrifontis with d-threono-1,4-lactonase activity (TtLac11). A l-fuconate dehydratase from Herbaspirillum huttiense (HhAraD) with activity on d-threonate converts this molecule to 2-oxo-4-hydroxybutyrate (OHB), which is later converted into DHB by an OHB reductase, which in the case was a malate dehydrogenase mutant from E. coli (Mdh5Q) (Fig. 6F) (Frazão et al., 2023). To connect EG to this pathway, Gox0303 was selected to perform the initial conversion of EG to GA. The computed standard Gibbs free energy indicates the thermodynamic viability of this synthetic sequence, either starting from GA (−116.2 kJ/mol) or EG (−92.5 kJ/mol) (Frazão et al., 2023).
To achieve DHB accumulation in vivo, an E. coli MG1655 ΔyqhD ΔaldA lacIq mutant strain that is unable to use the natural EG assimilating pathway, due to the knockout of AldA and YqhD, was used as host strain (Frazão et al., 2023; Wagner et al., 2023). Bioreactor cultivation with defined mineral medium containing glucose and supplemented with GA in a way that its concentration is adjusted to 10 mM in each feeding cycle, lead to an accumulation of 8.2 mM (1 g/L) DHB after 24 h. As for DHB production starting from EG, the highest yield of 6.75 mM of DHB was verified after 48 h when using concentrated pre-cultures (resting cells) in LB medium supplemented with 320 mM EG, l-cysteine and FeCl3 (Table 2). The two last compounds were supplemented to the media considering that HhAraD is a Fe−S dependent dehydratase and on the hypothesis that Fe−S cluster biogenesis may be limiting inside the cells (Frazão et al., 2023). Accumulation of pathway intermediates at relevant amounts, notably d-threonate/d-threano-1,4-lactone, was verified (Frazão et al., 2023). Although being the first study reporting production of a non-natural value-added compound from EG, low DHB yields were obtained. The efficiency of DHB production was believed to be due to several factors, including the unfavourable thermodynamic equilibrium of the initial conversion of EG into GA. Also, the substrate affinities of FsaAL107Y/A129G and PcTadH might be low and further characterization of these enzymes is needed. The accumulation of d-threonate when cells enter stationary phase might also suggest that the d-threonate dehydratase enzyme (HhAraD) may become inactivated. Unidentified side reactions might also be diverting carbon away from the DHB biosynthetic pathway (Frazão et al., 2023).
Another route to obtain C4 molecules is the conversion of GA into the C4 sugar erythrulose by the action of an engineered enzyme reported by Güner and colleagues (Güner et al., 2021). In their study, rational protein engineering led to the discovery of a mutant benzaldehyde lyase, or formolase (Fls), able to catalyse a C–C bond formation between two GA molecules into erythrulose. The mutant FlsH29I/Q113S yielded 24.6 g/L erythrulose in vitro from 25 g/L GA after 16 h, translating into an outstanding theoretical yield of 98%. Although FLSH29I/Q113S has a high Km (159.8 mM) and a low kcat (0.25 s−1) for GA, this enzyme is robust and did not undergo significant degradation or conformational change during the in vitro reaction. Further extension of this reaction, starting from EG, and converting erythrulose into another metabolite or value-added compound in vivo, is highly expected.
5. Initial EG oxidation: a metabolic bottleneck
The enzymatic conversion of EG to GA, the first step in EG biodegradation, represents a metabolic bottleneck in any of the previously mentioned EG assimilation pathways. Moreover, few alcohol dehydrogenases are known to catalyse this reaction (Table 3). In E. coli, the reaction catalysed by the endogenous FucO is reportedly thermodynamically unfavourable with a positive ΔrG’° of 23.7 kJ/mol (Wagner et al., 2023). This can be evidenced, for example, by the poor performance of the pathway Ara5P-GAA in E. coli (discussed in Section 4.3.1) when EG was used as substrate, but which significantly increased with GA as substrate (Wagner et al., 2023). The poor FucO performance means that, to enable carbon flux the subsequent enzyme in the pathway should have a high substrate affinity for GA. Indeed, the AldA enzyme has a Km of 0.14–0.38 mM (Baldomà and Aguilar, 1987; Rodríguez-Zavala et al., 2006) and performs the irreversible oxidation of GA to glycolate with a favourable ΔrG'° of −17.0 kcal/mol. FucO is a dimeric Fe2+-dependent alcohol dehydrogenase made of two identical subunits, each containing a N-terminal NAD(H)-binding domain and a C-terminal Fe2+-binding domain (Montella et al., 2005; Zavarise et al., 2023). The active centre is found near the bound nicotinamide moiety of NAD(H) and is inside a deep narrow cleft shaped between both domains. The subunits can exist in both open (in the absence of cofactor) and closed (in the presence of cofactor) states in which the relative position of the two domains is different. Recently, the reaction mechanism of FucO was examined regarding the NADH-dependent reduction of its physiological substrate (more specifically, the inverse reaction from l-lactaldehyde to propanediol, Fig. 5) and a detailed description can be found in the report by Zavarise et al. (2023). FucO is only active in the presence of Fe2+ and is inactivated by Zn2+, Cu2+ and Cd2+ (Montella et al., 2005; Sridhara et al., 1969). Furthermore, FucO is highly enantioselective, has preference for short 2–4 carbons substrates, and displays strict regiospecificity for oxidation of primary alcohols (Blikstad and Widersten, 2010).
Table 3.
Characteristics of the known alcohol dehydrogenases that perform the oxidative conversion of ethylene glycol (EG) into glycolaldehyde (GA). Abbreviations: FucO, propanediol oxidoreductase from E. coli; FucOI7L/L8V, propanediol oxidoreductase mutant I7L/L8V; Gox0313, alcohol dehydrogenase from Gluconobacter oxydan; PedE and PedH, alcohol dehydrogenases from P. putida; PQQ, pyrroloquinoline quinone; AgaA, alcohol dehydrogenase from Rhodococcus jostii RHA1; MFT, mycofactocin.
| Enzyme | Source | Cofactor | Ion | Km (EG) | Challenges |
|---|---|---|---|---|---|
| FucO | E. coli | NAD+ | Fe2⁺ | 7–51 mM | Thermodynamically unfavourable, oxygen sensitive, low affinity for EG, more affinity for the inverse reduction reaction, product release and nucleotide dissociation are suggested to be rate limiting |
| FucOI7L/L8V | E. coli (engineered) | NAD+ | Fe2⁺ | – | Still faces thermodynamic challenges, engineering effects of the double mutations are not well studied |
| Gox0313 | G. oxydans | NAD+ | Zn2⁺ | 2.4–964 mM | More affinity for the inverse reduction reaction, still leads to an unfavourable thermodynamic equilibrium in vivo compared to FucO, no crystallographic structures available |
| PedE | P. putida | PQQ | Ca2⁺ | – | E. coli does not naturally produce PQQ. Structural data and kinetic data with EG lacking. |
| PedH | P. putida | PQQ | Lanthanides | – | |
| EgaA | R. Jostii RHA1 | MFT | – | – | E. coli does not naturally produce MFT de novo. Structural data and kinetic data with EG lacking. |
To this day, a few reports have attempted to improve the FucO catalysed reduction of l-lactaldehyde. Recently, laboratory-directed evolution using the bulkier aldehyde substrate phenylacetaldehyde resulted in a FucO double mutant, N151G/L259V, with 9000-fold increase in the ratio kcat/Km for this compound. This double mutant also showed an 80-fold decrease in activity for l-lactaldehyde and lost the inverse oxidative activity for propanediol (Sridhar et al., 2023). By examining its crystal structure co-crystallized with a bulky substrate analog, it was possible to determine that the two amino acid substitutions provided more space in the region where the substrate binds (Sridhar et al., 2023). In another example, the pocket where the coenzyme's adenine side is positioned, distal to the site of catalysis, was engineered through site-saturation mutagenesis (Cahn et al., 2015). Two FucO mutants M185A and M185C were identified that had increased kcat for the cofactor NADH, consequently improving reductive detoxification of the toxic aldehyde furfural in vivo. The solved structure of FucOM185C revealed that substitution with a smaller cysteine led to a realignment of the cofactor in a way that might improve catalytic spatial organization (Cahn et al., 2015).
Regarding the oxidative reaction of FucO, reports are fewer and focus mostly on propanediol as the substrate. For propanediol, the reaction is thermodynamically unfavoured and has a catalytic efficiency lower than the reduction reaction (Blikstad and Widersten, 2010; Boronat and Aguilar, 1979; Sridhar et al., 2023). Blikstad and Widersten report a 40-fold higher kcat/Km for the reduction of propanal, an aldehyde for which FucO has comparable activity to propanediol, compared to the oxidation of 1-propanol, and a Km that is 3–4 fold lower for NADH than for NAD+ (Blikstad and Widersten, 2010). Oxidation by FucO is also pH dependent, displaying highest oxidation efficiency at pH values around 9.5–10 while reduction is more insensitive to changes in pH (Blikstad and Widersten, 2010; Boronat and Aguilar, 1979). Furthermore, product release and nucleotide dissociation are suggested to be rate limiting in both reaction directions (Blikstad and Widersten, 2010). The Km values at pH 10 for propanediol and EG are 5.4 mM and 51 mM, respectively, suggesting that the enzyme affinity for EG is considerably lower (Blikstad and Widersten, 2010).
To the best of our knowledge, reports analysing the reaction mechanism and kinetic parameters of the NAD+-dependent dehydrogenation of EG into GA are lacking, although the existence of the promiscuous EG oxidation of FucO is well known. As discussed in Section 4.2, a few recent studies targeting EG biodegradation employ the less oxygen-sensitive mutant FucOI7L/L8V, with one of these studies (Panda et al., 2021) reporting a slightly faster E. coli growth when using this double mutant instead of the WT version. Nevertheless, these mutations are positioned in the N-terminal amino acid sequence responsible for dimerization, away from the catalytic site, and the reason for their positive effect on catalysis and oxygen sensitivity are not studied in detail. Considering that FucO's catalytic site cavity consists mainly of uncharged residues (Blikstad and Widersten, 2010) and that it has been previously engineered to change selectivity for other types of substrates, such as larger aldehydes (Sridhar et al., 2023), it might hold high potential as target for substrate specificity manipulation through protein engineering and/or directed evolution towards increased EG catalysis. We also suggest that in silico studies of FucO with EG as substrate could guide a rational engineering strategy to improve EG preference and favour the oxidation reaction by, for example, increasing enzyme affinity for NAD+. Furthermore, studies are needed to understand why the binding of Zn2+ to the enzyme instead of Fe2+ makes it inactive. By examining the solved structure 5BR4 of FucOM185C complexed with Zn2+ and NAD+ (Cahn et al., 2015), it is possible to observe, for example, that the Zn2+ is positioned away from the recognized Fe2+ position, such that the distance between the Zn2+ and the C5 atom of the nicotinamide is 4 Å instead of 3 Å (Sridhar et al., 2023). Rational engineering strategies guided by examination of structural and functional differences between Fe2+ and Zn2+-dependent alcohol dehydrogenases and/or directed evolution approaches might help shift the ion preference and alleviate the oxygen sensitivity of FucO. An earlier study was able to produce a mutant FucO with a cysteine residue insertion in position 263 that, although catalytically inactive, had lost capacity to bind Fe2+ while retaining the ability to bind Zn2+ (Obradors et al., 1998).
Recently another NAD+-dependent EG dehydrogenase – the alcohol dehydrogenase from G. oxydans (Gox0313) – was characterized (Zhang et al., 2015). Gox0313 is a Zn2+-dependent homotetrameric alcohol dehydrogenase. This enzyme has a broad pH optimum in EG oxidation from 6.5 to 9.0, with a maximum of 8.5, and maintained >95% activity over 24 h at pH 7.0 (Zhang et al., 2015). With a Km and Vmax for EG oxidation of 2.4 mM and 4.82 U/mg, respectively, and for GA reduction of 1.20 mM and 10.26 U/mg, respectively, this enzyme displays higher affinity and catalytic rate for GA reduction. Gox0313 also displayed higher turnover and catalytic efficiency for GA compared to EG, which also agreed with the Km value of NADH (0.015 mM) being lower than that for NAD+ (0.058 mM). Nevertheless, when combined with NADH oxidase-2 (NOX-2) from Lactobacillus brevis ATCC367 for NAD + regeneration, and after process optimization, a 96.8% GA conversion yield from 500 mM EG was obtained in vitro (Zhang et al., 2015). Recently, FucOWT and FucOI7L/L8V were compared to Gox0313 regarding their ability to convert EG to GA (Frazão et al., 2023). When expressed with a high-copy plasmid in E. coli TW64, assays with cell extracts confirmed that the three enzymes were active on EG, with FucOI7L/L8V having the highest activity (0.203 μmol/min/mg) and Gox0313 having the lowest (0.076 μmol/min/mg) (Frazão et al., 2023). However, when tested in vivo in E. coli MG1655 ΔydhD, expression of Gox0313 resulted in the fastest cell growth and was therefore selected for inclusion in the synthetic DHB production pathway. Nonetheless, Gox0313 still led to an unfavourable thermodynamic equilibrium of the initial EG oxidation step restricting GA accumulation (Frazão et al., 2023). Additionally, as discussed in Section 4.3.2, by replacing FucO with Gox0313 in the TrCoA pathway (designed for glycerate production from EG), an improved glycerate yield could be achieved in vitro (Scheffen et al., 2021). Further understanding the structural (currently, there is no crystallographic structure available) and kinetic characteristics of this oxygen insensitive Zn2+-dependent Gox0313 could, in the future, provide valuable insights into optimizing EG assimilation in E. coli by using this enzyme. However, it must be further noted that the use of different in vitro reaction setups leads to varying reported Km values for Gox0313, where values of 2.4 mM (Zhang et al., 2015) and 964 mM (Scheffen et al., 2021) can be found. In the case of FucO, values between 7 mM (Sridhara et al., 1969) and 51 mM (Blikstad and Widersten, 2010) are also reported.
Two other enzymes that are known to oxidize EG into GA are the pyrroloquinoline quinone (PQQ) dependent PedE and PedH periplasmic alcohol dehydrogenases from P. putida, which use Ca2+ or lanthanides as cofactor, respectively (Mückschel et al., 2012; Wehrmann et al., 2017). These two enzymes are also the ones used by the PET metabolizing bacterium I. sakaiensis (Hachisuka et al., 2022). Unlike E. coli, P. putida can naturally use EG as carbon and energy source and either PedE and PedH can perform irreversible EG oxidation (Mückschel et al., 2012). Reports have shown that P. putida can use EG to form biomass (Li et al., 2019; Mückschel et al., 2012) or other compounds such as PHA (Franden et al., 2018) or β-ketoadipic acid (Werner et al., 2021). However, having PQQ as the prosthetic group limits the expression of these enzymes in hosts that produces PQQ, which is not the case for E. coli. Thus, PedE and PedH expression in E. coli would require co-expression with heterologous PQQ biosynthetic genes, such as the five genes of the pqq cluster of G. oxydans ATCC 9937 (Yang et al., 2010), which would considerably increase the complexity and burden of such an EG consumption ME strategy. Furthermore, as is the case of Gox0313, further knowledge about the structural and kinetic characteristics of PedE and PedH using EG as substrate is still lacking. In the recent report by Yan et al. discussed in Section 4.2, different alcohol dehydrogenases including PedE, along with PQQ supplementation, FucO, FucOI7L/L8V, Gox0313 were tested and compared as candidates for the first step of EG oxidation in the in vivo production of glycolate in E. coli MG1655 (Yan et al., 2024). When growing in M9 medium containing 10 g/L EG as major carbon source, strains expressing PedE, FucO and FucOI7L/L8V could only use a small amount of EG, while the strain containing Gox0313 completely consumed available EG for growth within 96 h (Yan et al., 2024).
Recent studies have highlighted the potential of other enzymes such as Gox0313, which, although less studied, shows promise. Future research should focus on detailed structural and kinetic analyses of these enzymes that, together with in silico modelling and rational design and/or directed evolution efforts, could help identify robust and stable variants with improved catalytic efficiency for EG. Also, the efficient and cost-effective application of these enzymes is significantly influenced by the availability of cofactors. Cofactors’ high cost has limited the use of many cofactor-dependent enzymes in industrial bioprocesses in the past (Bachosz et al., 2023). However, the use of enzymatic cofactor regeneration systems has gained renewed attention due to the growing interest in sustainable bioreactions requiring cofactors. In fact, several cofactor recycling systems have been tested in biocatalysis and applied using ME strategies to maximize production of target compounds (Bachosz et al., 2023; Sun et al., 2023). Cofactors such as NAD(P)+/NAD(P)H play crucial roles in microbial cells and are frequently required in enzymatic reactions, and their availability limits the full catalytic activity of enzymes. When cofactors are synthetized and consumed, their intracellular redox state and balance is disrupted, negatively affecting cellular growth and biosynthesis (Sun et al., 2023). During EG oxidation mediated by FucO or Gox0313, a stoichiometric amount of NAD+ is reduced into NADH, thus the use of an efficient and fine-tuned NAD+ regeneration system that maintains balanced cofactor concentrations is needed. The use of NOX-2 as a partner enzyme that regenerates the cofactor by converting O2 to H2O is just one example (Zhang et al., 2015) among many strategies that should be considered in this case. Additionally, mining for new alcohol dehydrogenases from diverse microbial sources could uncover novel enzymes with superior characteristics for EG oxidation. For example, a mycofactocin (MFT)-associated alcohol dehydrogenase from Rhodococcus jostii RHA1 (a species capable of assimilating EG via glycolate) named EgaA that oxidizes EG into GA has been recently identified (Shimizu et al., 2024). This finding points out that there may still be many EG oxidizing enzymes to discover. By overcoming these challenges, it will be possible to create more efficient and robust E. coli strains capable of converting EG derived from PET into valuable bioproducts.
6. Challenges and future perspectives
The microbial biodegradation of EG holds tremendous promise for addressing environmental plastic pollution and advancing the circular economy agenda. However, the complexity associated with EG bioconversion in E. coli still presents numerous challenges and requires a profound comprehension of metabolic pathways, regulatory mechanisms, strain engineering, and process optimization (Fig. 7).
Fig. 7.
Overview of the main identified challenges associated with the full bioprocess of PET biodegradation, monomers Ethylene Glycol (EG) and Terephthalic acid (TPA) assimilation and production of value-added compounds in Escherichia coli. Represented as valuable chemicals are the molecules already reported to be synthetized from either EG or TPA by metabolic engineering strategies in E. coli and referenced in this review. Abbreviations: PDC, 2-pyrone-4,6-dicarboxylic acid. Created with BioRender.com.
One of these challenges is the strain adaptation and inherent heterogeneity among E. coli strains in their ability to metabolize EG-derived substrates. The variability in growth kinetics, lag phases, and adaptation periods among different E. coli strains when exposed to EG or other derived compounds like glycolate has been demonstrated in several reports here reviewed. Particularly, a recent study by Höhmann et al. has underscored this heterogeneity, demonstrating that while various E. coli strains can utilize glycolate as a sole carbon and energy source, they exhibit significant differences in their ability and time to adapt to this compound (Höhmann et al., 2024). For example, after cultivation in the complex medium LB, E. coli W was able to grow on glycolate after a lag phase of about 4 h with a μmax of 0.20 h−1. In contrast, other strains such as E. coli BW25113, JM101 and BL21 DE3 showed significantly longer lag phases of 20, 40 and 100 h, respectively, with a μmax < 0.1 h−1 (Höhmann et al., 2024). As mentioned in Section 3.1, E. coli BL21 DE3 has demonstrated higher expression levels of glyoxylate shunt genes (Phue and Shiloach, 2004). This increased flux through the glyoxylate shunt might explain the extended lag-phase observed, as the strain may need a prolonged period to adjust its metabolism to efficiently catabolize glycolate and achieve anabolism (Höhmann et al., 2024). Supporting this assumption, while E. coli BW25113 and JM101 required about 10 generations of adaptive batch cultivations with glycolate to exhibit a short lag phase of 1–2 h, E. coli BL21 DE3 needed 30 generations and still did not achieved a stable growth phenotype (Höhmann et al., 2024).
Thus, the selection of E. coli strains for metabolic engineering tasks must be carefully considered, taking into account their metabolic diversity and the final aim. Furthermore, understanding the genetic basis of strain-specific adaptations and elucidating the regulatory mechanisms and possible bottlenecks governing EG metabolism are essential for developing robust chassis across different E. coli strains. Systems biology methodologies such as evolutionary algorithms and simulation methods using genome-scale metabolic models may also offer valuable insights into the complex genetic and metabolic networks underlying these strain-specific responses (Monk et al., 2013), aiding in the rational design of optimized microbial cell factories. Targeted genetic interventions can redirect the metabolism of E. coli to use EG as the main carbon source (Orth et al., 2010, 2011). Furthermore, almost all reports reviewed here indicate that when EG or GA are used as sole carbon sources, there is often a need to supplement with additional carbon sources like glycerol (Wagner et al., 2023), amino acid mixtures like CSM (Panda et al., 2021, 2023), or even complex media (Frazão et al., 2023; Lu et al., 2019; Pandit et al., 2021; Yan et al., 2024) to support adequate microbial growth and more efficient EG assimilation. Moving forward, a thorough investigation of the metabolic pathways and regulatory mechanisms of E. coli during EG and GA assimilation is crucial. These discoveries may help create chassis strains and fermentation conditions that are more economically feasible and efficient.
Secondly, EG (Panda et al., 2021) and its intermediates, including GA (Benov and Fridovich, 1998; Frazão et al., 2023; Wagner et al., 2023), glycolate (Yan et al., 2024) and the reactive aldehyde glyoxylate (Schada von Borzyskowski et al., 2023), have been shown to inhibit growth and pose toxicity challenges to E. coli cells. This underscores the need of a tight process control of the EG assimilating pathway to minimize intermediate accumulation. The same effect is verified with P. putida, where high EG concentration lead to decreased growth (Franden et al., 2018; Li et al., 2019). Moreover, the introduction of heterologous pathways for EG degradation may impose metabolic burden on E. coli, impacting cellular growth, viability, and productivity (Silva et al., 2012). Strategies to mitigate this burden include fine-tuning gene expression, optimizing metabolic flux distribution and genomic integration of the heterologous pathway. Balancing metabolic load and developing robust strains with tolerance to intermediate toxicity will help enhance substrate utilization, product synthesis, and overall efficiency and sustainability of EG biodegradation processes.
Lastly, because of the intricacy of enzyme reactions, substrate specificity, and cofactor requirements, optimizing metabolic pathways for EG biodegradation also poses a considerable challenge. Enhancing EG assimilation will require the development of engineered enzyme variants with increased substrate affinity, cofactor specificity, and catalytic efficiency. Enzymes that will benefit from these approaches include FucO or Gox0313 enzymes for EG oxidation to GA (discussed in Section 5), and other enzymes highlighted throughout Section 4.3 such as ACPS in the SACA pathway (Lu et al., 2019) or FsaAL107Y/A129G and PcTadH in the DHB production pathway (Frazão et al., 2023). Computational modelling methodologies, directed evolution methods, and rational design strategies are useful tools that speed up enzyme optimization pathway engineering. Also, several of the enzymes participating in the pathways covered in this review depend on cofactors such as ATP and NAD(P)+/NAD(P)H to function, highlighting the importance of a controlled intracellular availability of these cofactors and the need to use optimized cofactor regeneration strategies (Bachosz et al., 2023; Sun et al., 2023). Achieving energy and redox balance is crucial not only to support biomass growth, but also to fine tune the functionality and productivity of cellular metabolism and EG assimilating pathways.
The future of utilizing E. coli strains for EG assimilation and PET valorisation in a circular economy context is promising but comes with several challenges and associated areas require further research. One key area that has also been gaining traction is the microbial assimilation of the other PET monomer, TPA. Recent studies have shown that microorganisms such as E. coli and Pseudomonas species, though not able to naturally assimilate TPA, can be engineered with heterologous TPA degrading pathways from other naturally TPA using species, such as the one from Comamonas sp. (Sasoh et al., 2006). Thus, enabling simultaneous TPA assimilation and production of value-added compounds. Examples include the production of gallic acid, pyrogallol, catechol, muconic acid, and vanillic acid (Kim et al., 2019), of 2-pyrone-4,6-dicarboxylic acid, a promising monomer used in biodegradable polymers synthesis (Kang et al., 2020), and of vanillin (Sadler and Wallace, 2021), from TPA using E. coli strains. These advancements provide new avenues for the full conversion of PET waste. Moreover, the first steps in exploring integrated bioprocesses that handle the entire cycle from PET biodegradation, by producing and secreting PET hydrolases, to the assimilation of both EG and TPA, are already started to be taken. Two recent studies using P. putida have explored this concept with modest results (Brandenberg et al., 2022; Liu et al., 2022). These reports specifically highlight the strictly dependency of these systems with the primary activity of PET hydrolases whose activity parameters still need to be improved to match those for optimal bacterial growth. In fact, PET biodegradation still has inferior performance compared to chemical depolymerization (Uekert et al., 2023), which also highlights the need for further research in this area towards PET hydrolases improvement. Nevertheless, these holistic bioprocess approaches have tremendous potential for achieving sustainable and economically viable solutions for PET waste management. By leveraging the metabolic capabilities of microorganisms, it is possible to create open-loop systems where PET waste is converted into valuable products, thus supporting the principles of a circular economy. Future research should prioritize a meticulous understanding of thermodynamic limitations and optimization of these bioprocesses to enhance the robustness and efficiency of microbial chassis. Thus, integrating advanced ME techniques will be essential to improve the overall yield and productivity of the desired end-products.
7. Conclusion
PET biodegradation and assimilation represents a promising avenue with enormous potential for sustainable plastic waste management. This allows the sustainable conversion of PET into valuable NGFs such as EG, thereby alleviating the dependency on other commonly used feedstocks like glucose. Recent advancements in enzyme engineering and microbial biotechnology are paving the way towards enhancing the efficiency of PET biological depolymerization, although careful and complex optimization of PET hydrolases and metabolic pathways is still needed to reach a fully efficient bioprocess. By integrating this process into a circular economy framework, E. coli can be further engineered to utilize EG as a primary carbon source, channelling it into central metabolic pathways to produce value-added products. Continued research and development in optimizing both PET biodegradation and EG assimilation in E. coli will be critical for maximizing the efficiency and viability of this bioconversion process. In conclusion, by harnessing the power of biotechnology, synthetic biology, and systems biology, researchers will be able to unlock the full potential of EG as a renewable microbial feedstock and pave the way for a more sustainable and resource-efficient future.
CRediT authorship contribution statement
Alexandra Balola: Writing – original draft, Investigation, Conceptualization. Sofia Ferreira: Writing – review & editing, Supervision, Conceptualization. Isabel Rocha: Writing – review & editing, Validation, Supervision, Funding acquisition.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used a natural language processing tool (ChatGPT) in order to polish and improve the comprehension of some sections of the manuscript. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Funding sources
This work was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of a Ph.D. Grant (grant number 2020.07984.BD).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The author is an Editorial Board Member/Editor-in-Chief/Associate Editor/Guest Editor for Metabolic Engineering Communications and was not involved in the editorial review or the decision to publish this article.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Data availability
No data was used for the research described in the article.
References
- Achilias D.S., Karayannidis G.P. The chemical recycling of PET in the framework of sustainable development. Water Air Soil Pollut. Focus. 2004;4:385–396. doi: 10.1023/B:WAFO.0000044812.47185.0f. [DOI] [Google Scholar]
- Ashiuchi M., Misono H. Biochemical evidence that Escherichia coli hyi (orf b0508, gip) gene encodes hydroxypyruvate isomerase. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1999;1435:153–159. doi: 10.1016/S0167-4838(99)00216-2. [DOI] [PubMed] [Google Scholar]
- Ashworth J.M., Kornberg H.L. The role of isocitrate lyase in Escherichia coli. Biochim. Biophys. Acta (BBA) - Spec. Sect. Enzymol. Subj. 1964;89:383–384. doi: 10.1016/0926-6569(64)90237-8. [DOI] [PubMed] [Google Scholar]
- Aslan S., Noor E., Benito Vaquerizo S., Lindner S.N., Bar-Even A. Design and engineering of E. coli metabolic sensor strains with a wide sensitivity range for glycerate. Metab. Eng. 2020;57:96–109. doi: 10.1016/j.ymben.2019.09.002. [DOI] [PubMed] [Google Scholar]
- ATSDR Who is at risk of exposure to ethylene glycol? 2022. https://www.atsdr.cdc.gov/csem/ethylene-propylene-glycol/risk_of_exposure.html [WWW Document]. URL.
- Babilas P., Knie U., Abels C. Cosmetic and dermatologic use of alpha hydroxy acids. JDDG J. der Dtsch. Dermatologischen Gesellschaft. 2012;10:488–491. doi: 10.1111/j.1610-0387.2012.07939.x. [DOI] [PubMed] [Google Scholar]
- Bachosz K., Zdarta J., Bilal M., Meyer A.S., Jesionowski T. Enzymatic cofactor regeneration systems: a new perspective on efficiency assessment. Sci. Total Environ. 2023;868 doi: 10.1016/j.scitotenv.2023.161630. [DOI] [PubMed] [Google Scholar]
- Baldomà L., Aguilar J. Involvement of lactaldehyde dehydrogenase in several metabolic pathways of Escherichia coli K12. J. Biol. Chem. 1987;262:13991–13996. doi: 10.1016/S0021-9258(18)47893-3. [DOI] [PubMed] [Google Scholar]
- Bartkus J.M., Mortlock R.P. Isolation of a mutation resulting in constitutive synthesis of L-fucose catabolic enzymes. J. Bacteriol. 1986;165:710–714. doi: 10.1128/jb.165.3.710-714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch O., Hagemann M., Bauwe H. Only plant‐type (GLYK) glycerate kinases produce D‐glycerate 3‐phosphate. FEBS Lett. 2008;582:3025–3028. doi: 10.1016/j.febslet.2008.07.038. [DOI] [PubMed] [Google Scholar]
- Benavides Fernández C.D., Guzmán Castillo M.P., Quijano Pérez S.A., Carvajal Rodríguez L.V. Microbial degradation of polyethylene terephthalate: a systematic review. SN Appl. Sci. 2022;4:263. doi: 10.1007/s42452-022-05143-4. [DOI] [Google Scholar]
- Benov L., Fridovich I. Superoxide dependence of the toxicity of short chain sugars. J. Biol. Chem. 1998;273:25741–25744. doi: 10.1074/jbc.273.40.25741. [DOI] [PubMed] [Google Scholar]
- Benyathiar P., Kumar P., Carpenter G., Brace J., Mishra D.K. Polyethylene terephthalate (PET) bottle-to-bottle recycling for the beverage industry: a review. Polymers. 2022;14:2366. doi: 10.3390/polym14122366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blikstad C., Widersten M. Functional characterization of a stereospecific diol dehydrogenase, FucO, from Escherichia coli: substrate specificity, pH dependence, kinetic isotope effects and influence of solvent viscosity. J. Mol. Catal. B Enzym. 2010;66:148–155. doi: 10.1016/j.molcatb.2010.04.010. [DOI] [Google Scholar]
- Boronat A., Aguilar J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 1979;140:320–326. doi: 10.1128/jb.140.2.320-326.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat A., Caballero E., Aguilar J. Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J. Bacteriol. 1983;153:134–139. doi: 10.1128/jb.153.1.134-139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberg O.F., Schubert O.T., Kruglyak L. Towards synthetic PETtrophy: engineering Pseudomonas putida for concurrent polyethylene terephthalate (PET) monomer metabolism and PET hydrolase expression. Microb. Cell Factories. 2022;21:119. doi: 10.1186/s12934-022-01849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero E., Baldomá L., Ros J., Boronat A., Aguilar J. Identification of lactaldehyde dehydrogenase and glycolaldehyde dehydrogenase as functions of the same protein in Escherichia coli. J. Biol. Chem. 1983;258:7788–7792. doi: 10.1016/S0021-9258(18)32248-8. [DOI] [PubMed] [Google Scholar]
- Cabiscol E., Aguilar J., Ros J. Metal-catalyzed oxidation of Fe2+ dehydrogenases. Consensus target sequence between propanediol oxidoreductase of Escherichia coli and alcohol dehydrogenase II of Zymomonas mobilis. J. Biol. Chem. 1994;269:6592–6597. doi: 10.1016/S0021-9258(17)37413-6. [DOI] [PubMed] [Google Scholar]
- Cabiscol E., Badia J., Baldoma L., Hidalgo E., Aguilar J., Ros J. Inactivation of propanediol oxidoreductase of Escherichia coli by metal-catalyzed oxidation. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1992;1118:155–160. doi: 10.1016/0167-4838(92)90144-3. [DOI] [PubMed] [Google Scholar]
- Cabulong R.B., Bañares A.B., Nisola G.M., Lee W.-K., Chung W.-J. Enhanced glycolic acid yield through xylose and cellobiose utilization by metabolically engineered Escherichia coli. Bioproc. Biosyst. Eng. 2021;44:1081–1091. doi: 10.1007/s00449-020-02502-6. [DOI] [PubMed] [Google Scholar]
- Cahn J.K.B., Baumschlager A., Brinkmann-Chen S., Arnold F.H. Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes. Protein Eng. Des. Sel. 2015;29 doi: 10.1093/protein/gzv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbios Carbios launches industrial demonstration plant for its unique enzymatic recycling technology - carbios. 2021. https://www.carbios.com/en/carbios-annonce-le-demarrage-de-son-demonstrateur-industriel-exploitant-sa-technologie-unique-de-recyclage-enzymatique-c-zyme/
- Chang Y.Y., Wang A.Y., Cronan J.E. Molecular cloning, DNA sequencing, and biochemical analyses of Escherichia coli glyoxylate carboligase. An enzyme of the acetohydroxy acid synthase-pyruvate oxidase family. J. Biol. Chem. 1993;268:3911–3919. doi: 10.1016/S0021-9258(18)53559-6. [DOI] [PubMed] [Google Scholar]
- Chen L., Tang C., Davey K., Zheng Y., Jiao Y., Qiao S.-Z. Spatial-confinement induced electroreduction of CO and CO2 to diols on densely-arrayed Cu nanopyramids. Chem. Sci. 2021;12:8079–8087. doi: 10.1039/D1SC01694F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-M., Zhu Y., Lin E.C.C. The organization of the fuc regulon specifying l-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol. Gen. Genet. MGG. 1987;210:331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- Chen Y.M., Lu Z., Lin E.C. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on L-1,2-propanediol. J. Bacteriol. 1989;171:6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child J., Willetts A. Microbial metabolism of aliphatic glycols bacterial metabolism of ethylene glycol. Biochim. Biophys. Acta Gen. Subj. 1978;538:316–327. doi: 10.1016/0304-4165(78)90359-8. [DOI] [PubMed] [Google Scholar]
- Cho J.S., Kim G.B., Eun H., Moon C.W., Lee S.Y. Designing microbial cell factories for the production of chemicals. JACS Au. 2022;2:1781–1799. doi: 10.1021/jacsau.2c00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocks G.T., Aguilar J., Lin E.C.C. Evolution of L-1,2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J. Bacteriol. 1974;118:83–88. doi: 10.1128/jb.118.1.83-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley C.S., Cascio D., Sawaya M.R., Kopstein J.S., Bobik T.A., Yeates T.O. Structural insight into the mechanisms of transport across the Salmonella enterica pdu microcompartment shell. J. Biol. Chem. 2010;285:37838–37846. doi: 10.1074/jbc.M110.160580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon G.H., Kornberg H.L., Lund P. Purification and properties of malate synthetase. Biochim. Biophys. Acta. 1960;41:217–233. doi: 10.1016/0006-3002(60)90004-4. [DOI] [PubMed] [Google Scholar]
- Fang H., Li D., Kang J., Jiang P., Sun J., Zhang D. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12. Nat. Commun. 2018;9:4917. doi: 10.1038/s41467-018-07412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher E.L., Payne W.J. Bacterial utilization of ether glycols. Appl. Microbiol. 1962;10:542–547. doi: 10.1128/am.10.6.542-547.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franden M.A., Jayakody L.N., Li W.-J., Wagner N.J., Cleveland N.S., Michener W.E., Hauer B., Blank L.M., Wierckx N., Klebensberger J., Beckham G.T. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018;48:197–207. doi: 10.1016/j.ymben.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Frazão C.J.R., Trichez D., Serrano-Bataille H., Dagkesamanskaia A., Topham C.M., Walther T., François J.M. Construction of a synthetic pathway for the production of 1,3-propanediol from glucose. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-48091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazão C.J.R., Wagner N., Rabe K., Walther T. Construction of a synthetic metabolic pathway for biosynthesis of 2,4-dihydroxybutyric acid from ethylene glycol. Nat. Commun. 2023;14:1931. doi: 10.1038/s41467-023-37558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredenberg S., Wahlgren M., Reslow M., Axelsson A. The mechanisms of drug release in poly(lactic-co-glycolic acid)-based drug delivery systems—a review. Int. J. Pharm. (Amst.) 2011;415:34–52. doi: 10.1016/j.ijpharm.2011.05.049. [DOI] [PubMed] [Google Scholar]
- GlobalData Ethylene glycol market capacity and capital expenditure forecasts to 2028. 2024. https://www.globaldata.com/store/report/ethylene-glycol-market-analysis/?utm_source=hs_automation&utm_medium=email&utm_campaign=GD-RS-Request.Sample.Pages&utm_content=72896524 [WWW Document]. URL.
- Gonzalez C.F., Taber W.A., Zeitoun M.A. Biodegradation of ethylene glycol by a salt-requiring bacterium. Appl. Microbiol. 1972;24:911–919. doi: 10.1128/am.24.6.911-919.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotto A., Kornberg H. The metabolism of C2 compounds in micro-organisms. Preparation and properties of crystalline tartronic semialdehyde reductase. Biochem. J. 1961;81:273–284. doi: 10.1042/bj0810273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A., Lahm V., Connock T., Lugal L. 2022. How Circular Is PET? [Google Scholar]
- Güner S., Wegat V., Pick A., Sieber V. Design of a synthetic enzyme cascade for the in vitro fixation of a C1 carbon source to a functional C4 sugar. Green Chem. 2021;23:6583–6590. doi: 10.1039/D1GC02226A. [DOI] [Google Scholar]
- Guo D., Pan H., Li X. Metabolic engineering of Escherichia coli for production of biodiesel from fatty alcohols and acetyl-CoA. Appl. Microbiol. Biotechnol. 2015;99:7805–7812. doi: 10.1007/S00253-015-6809-5. [DOI] [PubMed] [Google Scholar]
- Gupta N.K., Vennesland B. Glyoxylate carboligase of Escherichia coli: a flavoprotein. J. Biol. Chem. 1964;239:3787–3789. doi: 10.1016/S0021-9258(18)91205-6. [DOI] [PubMed] [Google Scholar]
- Hachisuka S. ichi, Chong J.F., Fujiwara T., Takayama A., Kawakami Y., Yoshida S. Ethylene glycol metabolism in the poly(ethylene terephthalate)-degrading bacterium Ideonella sakaiensis. Appl. Microbiol. Biotechnol. 2022;106:7867–7878. doi: 10.1007/S00253-022-12244-Y. [DOI] [PubMed] [Google Scholar]
- Hartmanis M.G.N., Stadtman T.C. Diol metabolism and diol dehydratase in Clostridium glycolicum. Arch. Biochem. Biophys. 1986;245:144–152. doi: 10.1016/0003-9861(86)90198-0. [DOI] [PubMed] [Google Scholar]
- Höhmann S., Briol T.A., Ihle N., Frick O., Schmid A., Bühler B. Glycolate as alternative carbon source for Escherichia coli. J. Biotechnol. 2024;381:76–85. doi: 10.1016/j.jbiotec.2024.01.001. [DOI] [PubMed] [Google Scholar]
- Jin J., Tan T., Wang H., Su G. The expression of spinach glycolate oxidase (GO) in E. coli and the application of GO in the production of glyoxylic acid. Mol. Biotechnol. 2003;25:207–214. doi: 10.1385/MB:25:3:207. [DOI] [PubMed] [Google Scholar]
- Kalikar R.G., Deshpande R.S., Chandalia S.B. Synthesis of vanillin and 4‐hydroxybenzaldehyde by a reaction scheme involving condensation of phenols with glyoxylic acid. J. Chem. Technol. Biotechnol. 1986;36:38–46. doi: 10.1002/jctb.280360107. [DOI] [Google Scholar]
- Kandasamy S., Samudrala S.P., Bhattacharya S. The route towards sustainable production of ethylene glycol from a renewable resource, biodiesel waste: a review. Catal. Sci. Technol. 2019;9:567–577. doi: 10.1039/C8CY02035C. [DOI] [Google Scholar]
- Kang M.J., Kim H.T., Lee M.W., Kim K.A., Khang T.U., Song H.M., Park S.J., Joo J.C., Cha H.G. A chemo-microbial hybrid process for the production of 2-pyrone-4,6-dicarboxylic acid as a promising bioplastic monomer from PET waste. Green Chem. 2020;22:3461–3469. doi: 10.1039/d0gc00007h. [DOI] [Google Scholar]
- Kataoka M., Sasaki M., Hidalgo A.-R.G.D., Nakano M., Shimizu S. Glycolic acid production using ethylene glycol-oxidizing microorganisms. Biosci. Biotechnol. Biochem. 2001;65:2265–2270. doi: 10.1271/bbb.65.2265. [DOI] [PubMed] [Google Scholar]
- Khairul Anuar N.F.S., Huyop F., Ur-Rehman G., Abdullah F., Normi Y.M., Sabullah M.K., Abdul Wahab R. An overview into polyethylene terephthalate (PET) hydrolases and efforts in tailoring enzymes for improved plastic degradation. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232012644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O., Tawfik D.S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Kim H.T., Kim J.K., Cha H.G., Kang M.J., Lee H.S., Khang T.U., Yun E.J., Lee D.-H., Song B.K., Park S.J., Joo J.C., Kim K.H. Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain. Chem. Eng. 2019;7:19396–19406. doi: 10.1021/acssuschemeng.9b03908. [DOI] [Google Scholar]
- Kornberg H.L., Morris J.G. β-Hydroxyaspartate pathway: a new route for biosyntheses from glyoxylate. Nature. 1963;197:456–457. doi: 10.1038/197456a0. [DOI] [PubMed] [Google Scholar]
- Kosiorowska K.E., Moreno A.D., Iglesias R., Leluk K., Mirończuk A.M. Production of PETase by engineered Yarrowia lipolytica for efficient poly(ethylene terephthalate) biodegradation. Sci. Total Environ. 2022;846 doi: 10.1016/j.scitotenv.2022.157358. [DOI] [PubMed] [Google Scholar]
- Kudo H., Ono S., Abe K., Matsuda M., Hasunuma T., Nishizawa T., Asayama M., Nishihara H., Chohnan S. Enhanced supply of acetyl-CoA by exogenous pantothenate kinase promotes synthesis of poly(3-hydroxybutyrate) Microb. Cell Factories. 2023;22:1–13. doi: 10.1186/S12934-023-02083-5/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.G., Roth J.R. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J. Bacteriol. 1995;177:6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Palsson B.Ø. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, l-1,2-propanediol. Appl. Environ. Microbiol. 2010;76:4158–4168. doi: 10.1128/AEM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Kim H.U., Chae T.U., Cho J.S., Kim J.W., Shin J.H., Kim D.I., Ko Y.-S., Jang W.D., Jang Y.-S. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2019;2:18–33. doi: 10.1038/s41929-018-0212-4. [DOI] [Google Scholar]
- Li F., Thevenon A., Rosas-Hernández A., Wang Z., Li Y., Gabardo C.M., Ozden A., Dinh C.T., Li J., Wang Y., Edwards J.P., Xu Y., McCallum C., Tao L., Liang Z.-Q., Luo M., Wang X., Li H., O'Brien C.P., Tan C.-S., Nam D.-H., Quintero-Bermudez R., Zhuang T.-T., Li Y.C., Han Z., Britt R.D., Sinton D., Agapie T., Peters J.C., Sargent E.H. Molecular tuning of CO2-to-ethylene conversion. Nature. 2020;577:509–513. doi: 10.1038/s41586-019-1782-2. [DOI] [PubMed] [Google Scholar]
- Li W., Jayakody L.N., Franden M.A., Wehrmann M., Daun T., Hauer B., Blank L.M., Beckham G.T., Klebensberger J., Wierckx N. Laboratory evolution reveals the metabolic and regulatory basis of ethylene glycol metabolism by Pseudomonas putida KT2440. Environ. Microbiol. 2019;21:3669–3682. doi: 10.1111/1462-2920.14703. [DOI] [PubMed] [Google Scholar]
- Liu M., Ding Y., Chen H., Zhao Z., Liu H., Xian M., Zhao G. Improving the production of acetyl-CoA-derived chemicals in Escherichia coli BL21(DE3) through iclR and arcA deletion. BMC Microbiol. 2017;17:10. doi: 10.1186/s12866-016-0913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Zheng Y., Yuan Y., Zhang T., Li Q., Liang Q., Su T., Qi Q. Valorization of polyethylene terephthalate to muconic acid by engineering Pseudomonas putida. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms231910997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J.M. Glycolate oxidoreductase in Escherichia coli. Biochim. Biophys. Acta Bioenerg. 1972;267:227–237. doi: 10.1016/0005-2728(72)90111-9. [DOI] [PubMed] [Google Scholar]
- Lu H., Diaz D.J., Czarnecki N.J., Zhu C., Kim W., Shroff R., Acosta D.J., Alexander B.R., Cole H.O., Zhang Y., Lynd N.A., Ellington A.D., Alper H.S. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature. 2022;604:662–667. doi: 10.1038/s41586-022-04599-z. [DOI] [PubMed] [Google Scholar]
- Lu X., Liu Yuwan, Yang Y., Wang S., Wang Q., Wang X., Yan Z., Cheng J., Liu C., Yang X., Luo H., Yang S., Gou J., Ye L., Lu L., Zhang Z., Guo Y., Nie Y., Lin J., Li S., Tian C., Cai T., Zhuo B., Ma H., Wang W., Ma Y., Liu Yongjun, Li Y., Jiang H. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Nat. Commun. 2019;10:1378. doi: 10.1038/s41467-019-09095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Cabiscol E., Obradors N., Tamarit J., Ros J., Aguilar J., Lin E.C.C. Evolution of an Escherichia coli protein with increased resistance to oxidative stress. J. Biol. Chem. 1998;273:8308–8316. doi: 10.1074/jbc.273.14.8308. [DOI] [PubMed] [Google Scholar]
- Ma X., Gözaydın G., Yang H., Ning W., Han X., Poon N.Y., Liang H., Yan N., Zhou K. Upcycling chitin-containing waste into organonitrogen chemicals via an integrated process. Proc. Natl. Acad. Sci. USA. 2020;117:7719–7728. doi: 10.1073/pnas.1919862117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S.R., Nunn W.D. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J. Bacteriol. 1982;149:173–180. doi: 10.1128/jb.149.1.173-180.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., Zhang C., Meng T., Wang H., Hu X., Xiao Z., Wang C., Liu J. Effect and mechanism of aluminum(III) for guaiacol–glyoxylic acid condensation reaction in vanillin production. ACS Omega. 2020;5:24526–24536. doi: 10.1021/acsomega.0c03003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Yuan Q., Yang X., Liu P., Cheng Y., Luo J., Liu H., Yao Y., Sun H., Cai T., Ma H. Non-natural aldol reactions enable the design and construction of novel one-carbon assimilation pathways in vitro. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.677596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk J.M., Charusanti P., Aziz R.K., Lerman J.A., Premyodhin N., Orth J.D., Feist A.M., Palsson B.Ø. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc. Natl. Acad. Sci. USA. 2013;110:20338–20343. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montella C., Bellsolell L., Pérez-Luque R., Badía J., Baldoma L., Coll M., Aguilar J. Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli. J. Bacteriol. 2005;187:4957–4966. doi: 10.1128/JB.187.14.4957-4966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mückschel B., Simon O., Klebensberger J., Graf N., Rosche B., Altenbuchner J., Pfannstiel J., Huber A., Hauer B. Ethylene glycol metabolism by Pseudomonas putida. Appl. Environ. Microbiol. 2012;78:8531–8539. doi: 10.1128/AEM.02062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Geographic Great pacific garbage Patch [WWW document] Natl. Geogr. Mag. 2024 https://education.nationalgeographic.org/resource/great-pacific-garbage-patch/ URL. [Google Scholar]
- Ning P., Yang G., Hu L., Sun J., Shi L., Zhou Y., Wang Z., Yang J. Recent advances in the valorization of plant biomass. Biotechnol. Biofuels. 2021;14:102. doi: 10.1186/s13068-021-01949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njau R.K., Herndon C.A., Hawes J.W. Novel β-hydroxyacid dehydrogenases in Escherichia coli and Haemophilus influenzae. J. Biol. Chem. 2000;275:38780–38786. doi: 10.1074/jbc.M007432200. [DOI] [PubMed] [Google Scholar]
- Noor E., Bar-Even A., Flamholz A., Reznik E., Liebermeister W., Milo R. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez M.F., Pellicer M.T., Badia J., Aguilar J., Baldoma L. Biochemical characterization of the 2-ketoacid reductases encoded by ycdW and yiaE genes in Escherichia coli. Biochem. J. 2001;354:707. doi: 10.1042/0264-6021:3540707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradors N., Cabiscol E., Aguilar J., Ros J. Site‐directed mutagenesis studies of the metal‐binding center of the iron‐dependent propanediol oxidoreductase from Escherichia coli. Eur. J. Biochem. 1998;258:207–213. doi: 10.1046/j.1432-1327.1998.2580207.x. [DOI] [PubMed] [Google Scholar]
- OECD Annual plastic waste by disposal method, World, 2000 to 2019. 2023. https://ourworldindata.org/grapher/plastic-fate [WWW Document]. “Recycled” Process. by Our World Data LAb. OECD, “Global Plast. Outlook - Plast. waste by Reg. end-of-life fate” [original data]. URL.
- Ornston L.N., Ornston M.K. Regulation of glyoxylate metabolism in Escherichia coli K-12. J. Bacteriol. 1969;98:1098–1108. doi: 10.1128/jb.98.3.1098-1108.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Conrad T.M., Na J., Lerman J.A., Nam H., Feist A.M., Palsson B. A comprehensive genome-scale reconstruction of Escherichia coli metabolism-2011. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.O. What is flux balance analysis? Nat. Biotechnol. 2010 doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S., Fung V.Y.K., Zhou J.F.J., Liang H., Zhou K. Improving ethylene glycol utilization in Escherichia coli fermentation. Biochem. Eng. J. 2021;168 doi: 10.1016/j.bej.2021.107957. [DOI] [Google Scholar]
- Panda S., Zhou J.F.J., Feigis M., Harrison E., Ma X., Fung Kin Yuen V., Mahadevan R., Zhou K. Engineering Escherichia coli to produce aromatic chemicals from ethylene glycol. Metab. Eng. 2023;79:38–48. doi: 10.1016/j.ymben.2023.06.012. [DOI] [PubMed] [Google Scholar]
- Pandit A.V., Harrison E., Mahadevan R. Engineering Escherichia coli for the utilization of ethylene glycol. Microb. Cell Factories. 2021;20:22. doi: 10.1186/s12934-021-01509-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit A.V., Srinivasan S., Mahadevan R. Redesigning metabolism based on orthogonality principles. Nat. Commun. 2017;8 doi: 10.1038/ncomms15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J., Zheng M., Wang A., Zhang T. Catalytic hydrogenation of corn stalk to ethylene glycol and 1,2-propylene glycol. Ind. Eng. Chem. Res. 2011;50:6601–6608. doi: 10.1021/ie102505y. [DOI] [Google Scholar]
- Pellicer M.T., Badía J., Aguilar J., Baldomà L. Glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J. Bacteriol. 1996;178:2051–2059. doi: 10.1128/jb.178.7.2051-2059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phue J.-N., Shiloach J. Transcription levels of key metabolic genes are the cause for different glucose utilization pathways in E. coli B (BL21) and E. coli K (JM109) J. Biotechnol. 2004;109:21–30. doi: 10.1016/j.jbiotec.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Plastics Europe Plastics – the facts 2022. 2022. https://plasticseurope.org/knowledge-hub/plastics-the-facts-2022/
- Rebsdat S., Mayer D. Ullmann's Encyclopedia of Industrial Chemistry. Wiley; 2000. Ethylene glycol; pp. 803–814. [DOI] [Google Scholar]
- Rodríguez‐Zavala J.S., Allali‐Hassani A., Weiner H. Characterization of E. coli tetrameric aldehyde dehydrogenases with atypical properties compared to other aldehyde dehydrogenases. Protein Sci. 2006;15:1387–1396. doi: 10.1110/ps.052039606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J.C., Wallace S. Microbial synthesis of vanillin from waste poly(ethylene terephthalate) Green Chem. 2021;23:4665–4672. doi: 10.1039/d1gc00931a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim I., González-García S., Feijoo G., Moreira M.T. Assessing the environmental sustainability of glucose from wheat as a fermentation feedstock. J. Environ. Manag. 2019;247:323–332. doi: 10.1016/j.jenvman.2019.06.016. [DOI] [PubMed] [Google Scholar]
- Sallal A.-K.J., Nimer N.A. The intracellular localization of glycolate oxidoreductase in Escherichia coli. FEBS Lett. 1989;258:277–280. doi: 10.1016/0014-5793(89)81673-4. [DOI] [PubMed] [Google Scholar]
- Salusjärvi L., Havukainen S., Koivistoinen O., Toivari M. Biotechnological production of glycolic acid and ethylene glycol: current state and perspectives. Appl. Microbiol. Biotechnol. 2019;103:2525–2535. doi: 10.1007/s00253-019-09640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasoh M., Masai E., Ishibashi S., Hara H., Kamimura N., Miyauchi K., Fukuda M. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 2006;72:1825–1832. doi: 10.1128/AEM.72.3.1825-1832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satapathy A., Gadge S.T., Bhanage B.M. Synthesis of ethylene glycol from syngas via oxidative double carbonylation of ethanol to diethyl oxalate and its subsequent hydrogenation. ACS Omega. 2018;3:11097–11103. doi: 10.1021/acsomega.8b01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad A., Wagner H., Wilhelm C. Optimising biotechnological glycolate production in Chlamydomonas reinhardtii by improving carbon allocation towards the product. Chem. Eng. J. 2023;459 doi: 10.1016/j.cej.2023.141432. [DOI] [Google Scholar]
- Schada von Borzyskowski L., Schulz-Mirbach H., Troncoso Castellanos M., Severi F., Gómez-Coronado P.A., Paczia N., Glatter T., Bar-Even A., Lindner S.N., Erb T.J. Implementation of the β-hydroxyaspartate cycle increases growth performance of Pseudomonas putida on the PET monomer ethylene glycol. Metab. Eng. 2023;76:97–109. doi: 10.1016/j.ymben.2023.01.011. [DOI] [PubMed] [Google Scholar]
- Schada von Borzyskowski L., Severi F., Krüger K., Hermann L., Gilardet A., Sippel F., Pommerenke B., Claus P., Cortina N.S., Glatter T., Zauner S., Zarzycki J., Fuchs B.M., Bremer E., Maier U.G., Amann R.I., Erb T.J. Marine Proteobacteria metabolize glycolate via the β-hydroxyaspartate cycle. Nature. 2019;575:500–504. doi: 10.1038/s41586-019-1748-4. [DOI] [PubMed] [Google Scholar]
- Scheffen M., Marchal D.G., Beneyton T., Schuller S.K., Klose M., Diehl C., Lehmann J., Pfister P., Carrillo M., He H., Aslan S., Cortina N.S., Claus P., Bollschweiler D., Baret J.-C., Schuller J.M., Zarzycki J., Bar-Even A., Erb T.J. A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation. Nat. Catal. 2021;4:105–115. doi: 10.1038/s41929-020-00557-y. [DOI] [Google Scholar]
- Shimizu T., Suzuki K., Inui M. A mycofactocin-associated dehydrogenase is essential for ethylene glycol metabolism by Rhodococcus jostii RHA1. Appl. Microbiol. Biotechnol. 2024;108:1–11. doi: 10.1007/s00253-023-12966-7. [DOI] [PubMed] [Google Scholar]
- Silva F., Queiroz J.A., Domingues F.C. Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 2012;30:691–708. doi: 10.1016/j.biotechadv.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Sridhar S., Zavarise A., Kiema T.-R., Dalwani S., Eriksson T., Hajee Y., Reddy Enugala T., Wierenga R.K., Widersten M. Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity. IUCrJ. 2023;10:437–447. doi: 10.1107/S205225252300444X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhara S., Wu T.T., Chused T.M., Lin E.C.C. Ferrous-activated nicotinamide adenine dinucleotide-linked dehydrogenase from a mutant of Escherichia coli capable of growth on 1,2-propanediol. J. Bacteriol. 1969;98:87–95. doi: 10.1128/jb.98.1.87-95.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples C.A., Williams J.B., Craig G.R., Roberts K.M. Fate, effects and potential environmental risks of ethylene glycol: a review. Chemosphere. 2001;43:377–383. doi: 10.1016/S0045-6535(00)00148-X. [DOI] [PubMed] [Google Scholar]
- Sui B., Wang T., Fang J., Hou Z., Shu T., Lu Z., Liu F., Zhu Y. Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1265139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhang T., Lu B., Li X., Jiang L. Application of cofactors in the regulation of microbial metabolism: a state of the art review. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1145784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szappanos B., Fritzemeier J., Csörgő B., Lázár V., Lu X., Fekete G., Bálint B., Herczeg R., Nagy I., Notebaart R.A., Lercher M.J., Pál C., Papp B. Adaptive evolution of complex innovations through stepwise metabolic niche expansion. Nat. Commun. 2016;7 doi: 10.1038/ncomms11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura J., Ono A., Sugano Y., Huang C., Nishizawa H., Mikoshiba S. Electrochemical reduction of CO2 to ethylene glycol on imidazolium ion-terminated self-assembly monolayer-modified Au electrodes in an aqueous solution. Phys. Chem. Chem. Phys. 2015;17:26072–26078. doi: 10.1039/C5CP03028E. [DOI] [PubMed] [Google Scholar]
- Tiso T., Winter B., Wei R., Hee J., de Witt J., Wierckx N., Quicker P., Bornscheuer U.T., Bardow A., Nogales J., Blank L.M. The metabolic potential of plastics as biotechnological carbon sources – review and targets for the future. Metab. Eng. 2022;71:77–98. doi: 10.1016/j.ymben.2021.12.006. [DOI] [PubMed] [Google Scholar]
- Toraya T., Honda S., Fukui S. Fermentation of 1,2-propanediol and 1,2-ethanediol by some genera of enterobacteriaceae , involving coenzyme B12 -dependent diol dehydratase. J. Bacteriol. 1979;139:39–47. doi: 10.1128/jb.139.1.39-47.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier V., Topham C.M., Gilles A., David B., Folgoas C., Moya-Leclair E., Kamionka E., Desrousseaux M.-L., Texier H., Gavalda S., Cot M., Guémard E., Dalibey M., Nomme J., Cioci G., Barbe S., Chateau M., André I., Duquesne S., Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580:216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Trifunović D., Schuchmann K., Müller V. Ethylene glycol metabolism in the acetogen Acetobacterium woodii. J. Bacteriol. 2016;198:1058–1065. doi: 10.1128/JB.00942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau D.L., Edlich-Muth C., Zarzycki J., Scheffen M., Goldsmith M., Khersonsky O., Avizemer Z., Fleishman S.J., Cotton C.A.R., Erb T.J., Tawfik D.S., Bar-Even A. Design and in vitro realization of carbon-conserving photorespiration. Proc. Natl. Acad. Sci. USA. 2018;115:E11455–E11464. doi: 10.1073/pnas.1812605115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubb C. Feeding our future food system: can we scale glucose production? 2023. https://synthesis.capital/insights/can-we-scale-glucose-production [WWW Document]. Synth. Cap. URL.
- Uekert T., Singh A., DesVeaux J.S., Ghosh T., Bhatt A., Yadav G., Afzal S., Walzberg J., Knauer K.M., Nicholson S.R., Beckham G.T., Carpenter A.C. Technical, economic, and environmental comparison of closed-loop recycling technologies for common plastics. ACS Sustain. Chem. Eng. 2023;11:965–978. doi: 10.1021/acssuschemeng.2c05497. [DOI] [Google Scholar]
- Wagner N., Bade F., Straube E., Rabe K., Frazão C.J.R., Walther T. In vivo implementation of a synthetic metabolic pathway for the carbon-conserving conversion of glycolaldehyde to acetyl-CoA. Front. Bioeng. Biotechnol. 2023;11:1–16. doi: 10.3389/fbioe.2023.1125544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T., Topham C.M., Irague R., Auriol C., Baylac A., Cordier H., Dressaire C., Lozano-Huguet L., Tarrat N., Martineau N., Stodel M., Malbert Y., Maestracci M., Huet R., André I., Remaud-Siméon M., François J.M. Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor 2,4-dihydroxybutyric acid. Nat. Commun. 2017;8 doi: 10.1038/ncomms15828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrmann M., Billard P., Martin-Meriadec A., Zegeye A., Klebensberger J. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio. 2017;8 doi: 10.1128/mBio.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A.Z., Clare R., Mand T.D., Pardo I., Ramirez K.J., Haugen S.J., Bratti F., Dexter G.N., Elmore J.R., Huenemann J.D., Peabody G.L., Johnson C.W., Rorrer N.A., Salvachúa D., Guss A.M., Beckham G.T. Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 2021;67:250–261. doi: 10.1016/j.ymben.2021.07.005. [DOI] [PubMed] [Google Scholar]
- Wiegant W.M., De Bont J.A.M. A new route for ethylene glycol metabolism in Mycobacterium E44. Microbiology. 1980;120:325–331. doi: 10.1099/00221287-120-2-325. [DOI] [Google Scholar]
- Willetts A. Bacterial metabolism of ethylene glycol. Biochim. Biophys. Acta Gen. Subj. 1981;677:194–199. doi: 10.1016/0304-4165(81)90085-4. [DOI] [PubMed] [Google Scholar]
- Wong M.K., Lock S.S.M., Chan Y.H., Yeoh S.J., Tan I.S. Towards sustainable production of bio-based ethylene glycol: progress, perspective and challenges in catalytic conversion and purification. Chem. Eng. J. 2023;468 doi: 10.1016/j.cej.2023.143699. [DOI] [Google Scholar]
- Yan W., Qi X., Cao Z., Yao M., Ding M., Yuan Y. Biotransformation of ethylene glycol by engineered Escherichia coli. Synth. Syst. Biotechnol. 2024;9:531–539. doi: 10.1016/j.synbio.2024.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-P., Zhong G.-F., Lin J.-P., Mao D.-B., Wei D.-Z. Pyrroloquinoline quinone biosynthesis in Escherichia coli through expression of the Gluconobacter oxydans pqqABCDE gene cluster. J. Ind. Microbiol. Biotechnol. 2010;37:575–580. doi: 10.1007/s10295-010-0703-z. [DOI] [PubMed] [Google Scholar]
- Yang X., Yuan Q., Luo H., Li F., Mao Y., Zhao X., Du J., Li P., Ju X., Zheng Y., Chen Y., Liu Y., Jiang H., Yao Y., Ma H., Ma Y. Systematic design and in vitro validation of novel one-carbon assimilation pathways. Metab. Eng. 2019;56:142–153. doi: 10.1016/j.ymben.2019.09.001. [DOI] [PubMed] [Google Scholar]
- Yue H., Zhao Y., Ma X., Gong J. Ethylene glycol: properties, synthesis, and applications. Chem. Soc. Rev. 2012;41:4218. doi: 10.1039/c2cs15359a. [DOI] [PubMed] [Google Scholar]
- Zavarise A., Sridhar S., Kiema T., Wierenga R.K., Widersten M. Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics. FEBS J. 2023;290:465–481. doi: 10.1111/febs.16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelcbuch L., Razo-Mejia M., Herz E., Yahav S., Antonovsky N., Kroytoro H., Milo R., Bar-Even A. An in vivo metabolic approach for deciphering the product specificity of glycerate kinase proves that both E. coli's glycerate kinases generate 2-phosphoglycerate. PLoS One. 2015;10 doi: 10.1371/journal.pone.0122957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ottenheim C., Weingarten M., Ji L. Microbial utilization of next-generation feedstocks for the biomanufacturing of value-added chemicals and food ingredients. Front. Bioeng. Biotechnol. 2022;10:1–22. doi: 10.3389/fbioe.2022.874612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang B., Lin J., Wei D. Oxidation of ethylene glycol to glycolaldehyde using a highly selective alcohol dehydrogenase from Gluconobacter oxydans. J. Mol. Catal. B Enzym. 2015;112:69–75. doi: 10.1016/j.molcatb.2014.12.006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.