Abstract
Background
Early-maturity cotton varieties have the potential to be cultivated in a wider geographical area, extending as far north as 46 °N in China, and confer to address the issue of competition for land between grain and cotton by reducing their whole growth period (WGP). Therefore, it is of great importance to develop cotton varieties with comprehensive early maturity and high yield following investigating the regulatory mechanism underlying early maturity and identifying early maturity-related genes.
Results
In this study, ‘SCRC19’ and ‘SCRC21’, two excellent cultivars with significantly different WGP, along with their recombinant inbred lines (RILs) consisting of 150 individuals were re-sequenced, yielding 4,092,677 high-quality single nucleotide polymorphisms (SNPs) and 794 bin markers across 26 chromosomes. A genetic map spanning 2213.71 cM was constructed using the 794 bin markers. Based on this map, we identified a total of 78 early maturity-related QTLs, including 12 QTLs for WGP, 4 for SSP, 12 for SFP, 3 for FBP, 11 for NFFB, 8 for NFB, 16 for HNFFB and 12 for PH. Six QTL clusters, each containing more than four traits, were identified. One particular QTL cluster, which had the largest number of QTLs, ranged from 108.5 cM to 109 cM on Dt3, and contained 39 genes. Through functional analysis, we highlighted two early maturity-related candidates of GH_D03G1554 and GH_D03G1541, which were annotated as a BEL1-like homeodomain protein 8 and a homeobox-leucine zipper family protein, respectively.
Conclusions
We have identified a QTL cluster related to six early maturity-associated traits on Dt3. Through annotation of genes from candidate region, we have identified two candidate genes, GH_D03G1554 and GH_D03G1541, whose expression levels in ‘SCRC21’ were significantly higher than those in ‘SCRC19’ at different stages of flower bud development. These candidate genes provide new insights into the study of early-maturity mechanism and offer potential genetic improvement of cotton.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05947-z.
Keywords: Upland cotton, Early maturity, RIL population, QTL cluster, Candidate gene
Background
Cotton is an economically important crop, providing a large amount of natural fiber for the textile industry [1]. Upland cotton (Gossypium hirsutum L.), which is the world’s most widely cultivated cotton, contains early-, middle-, and late- maturing clusters, based on the duration of their whole growth period (WGP). The domestication of WGP for wild Gossypium hirsutum, whose growth period is typically greater than 180 days, initiated at least 5000 years ago [2]. However, early-maturity varieties developed in the past decade have reduced WGP to 100–120 days, such as ‘SCRC19’. The reduction in WGP has greatly expanded planting area of cotton [1]. In addition, early-maturity cotton is suitable to seed directly after wheat or rapeseed harvest, realizing harvesting both cotton and grain in one year, which confers to optimize light and temperature resources and address the land competition between grain and cotton.
Flowering time is a critical factor reflecting WGP [3, 4]. Over the past decade, numerous flowering factors have been identified, such as APETALA1 (AP1), FLOWERING LOCUS T (FT), SHORT VEGETATIVE PHASE (SVP), EARLY FLOWERING MYB PROTEIN (EFM) [5–7]. LEAFY (LFY) and AP1 are key genes in the floral meristem formation. They are activated by some flowering integrators, for example FT, leading to the transition from vegetative growth to reproductive growth [8]. FT protein, which is synthesized in the leaf vasculature and then moved to the shoot apical meristem (SAM) through the phloem, is central for integrating internal and external factors including photoperiod, temperature, gibberellic acid (GA) and plant age [9]. Many genes influence flowering through either positively (CO) or negatively (EFM, SVP) regulating the expression of FT [10]. CONSTANS (CO), whose expression is regulated by the circadian clock, plays a crucial role in plant photoperiod sensing and photoperiodic flowering. In long-day, the CO accumulation of Arabidopsis reaches a threshold, and then activates the expression of FT, resulting in flowering [11]. ASYMMETRIC LEAVES 1 (AS1), which is a vital factor in leaf pattern formation, forms a complex with CO protein and regulates FT expression through directly binding to the FT promoters. Moreover, AS1 positively regulates the expression of GA biosynthesis gene GIBBERELLIN 20-OXIDASE 1 (GA20ox1), whose mutant showed delayed flowering phenotypes regardless of photoperiod [12]. In plants, light clue and other environment factors are integrated to the CO-FT module or the FT integrators and interact with their targets to integrate flowering signaling with plant developmental programs [10].
Genetic map is a valuable tool for quantitative trait locus (QTL) mapping and gene identification of agronomic traits in crops. To date, many genetic maps have been constructed using restriction fragment length polymorphism (RFLP), simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers [13–15]. SNPs are usually used to construct high-density genetic linkage map due to their high rate of polymorphism. The advancements of high-throughput sequencing technology, with reduced cost and shorter time, has greatly accelerated the process of identifying QTLs related to important agronomic traits of cotton by linkage mapping and genome-wide association study (GWAS) [16–23]. For example, Li et al. generated a genome variation map by resequencing 436 cotton accessions and identified abundant significant loci and candidate genes associated with early maturity [1]. Notably, early maturity-related QTLs have predominantly been mapped onto chromosomes Dt3 according to the previous reports [15, 21, 24], and several genes within these QTL regions have been confirmed to be involved in the regulation of early maturity, such as GhAP1 [8], GhEMF2B [25], Ghir_D03G011310 [1]. Despite significant progress in identifying early maturity-related genes, further research is required due to the complexity underlying this trait.
Previously, the two factors of high yield and early maturity were considered direct conflicts [26]. However, ‘SCRC19’, a variety developed by Shandong Academy of Agricultural Sciences (SAAS) in 2005, achieved relatively high yield with a growth period of 105 to 112 days, demonstrating that this variety has great potential for the theoretical study on high-yield, early-maturation crops. In this study, we generated a recombinant inbred line (RIL) population comprising 150 individual lines derived from a cross of early-maturity variety ‘SCRC19’ and medium-maturity variety ‘SCRC21’, aiming to discover the QTLs and candidate genes for early maturity. Our results will contribute to elucidating the genetic mechanism of early maturity and may be useful to explore any potential relationship between WGP and yield.
Materials and methods
Plant materials
The upland cotton cultivars ‘SCRC21’ and ‘SCRC19’ were selected as the parents to generate the F2:8 recombinant inbred lines (RILs) consisting of 150 individuals. ‘SCRC19’ is a commercial short-season cotton variety bred by SAAS with characters of early maturity, disease resistance, high yield and high quality. The WGP of ‘SCRC19’ ranges from 105 to 112 days in the Yellow River basin. ‘SCRC21’ is another excellent commercial cotton variety of SAAS, and has higher plant height, bigger leaves, later flower and longer WGP (126 ~ 133 days in the Yellow River basin) compared to ‘SCRC19’ (Fig. S1). 150 RILs along with their parents were planted in five environments across three locations: Linqing (Shandong Province, 36.80 °N, 115.70 °E) in 2014 and 2015, Heze (Shandong Province, 35.07 °N, 115.57 °E) in 2014 and 2015, Kuitun (Xinjiang Uyghur Autonomous Region, 44.4 °N, 84.9 °E) in 2015. The cotton seeds were hill-sown in a randomized complete block design and the seedlings were kept in a density of approximately 52.5 thousand strains per hectare with row length of 8 m and row spacing of 80 cm and 60 cm in Linqing and Heze; while in Kuitun, a density 240 thousand strains per hectare with row length of 10 m and row spacing of 37.5 cm was used. The planting dates for the five environments were April 26th, April 26th, April 21th, April 21th and April 25th, respectively. Crop management followed local recommendations for the growing area.
Phenotypic measurement and analysis
In this study, the date when 50% of plants reached sprouting, squaring, flowering, boll opening are defined as sprouting, squaring, flowering, boll opening. Eight early maturity-related traits were investigated. PH (plant height from the ground to the main stem tip), NFFB (node number of the first fruiting branch, with the cotyledonary node recorded as zero), HNFFB (height from the ground to the NFFB), were surveyed, as described [24]. WGP, SSP, SFP and FBP were measured as the numbers of days from sowing to boll opening, sprouting to squaring, squaring to flowering, and flowering to boll opening, respectively. For WGP, SSP, SFP, FBP surveys, all of the plants were used, whereas 3 plants from the middle of each row were chosen for PH, NFB (number of fruiting branches), NFFB and HNFFB survey. SAS software was used for the calculation and correlation analysis of phenotypic variation. R software was used for normal distribution test.
Resequencing and genetic map construction
DNA of 150 RILs and their parents was extracted using Plant Genomic DNA Kit from Tiangen Biotech Co., Ltd. (Beijing, China). The concentration and purity of DNA was measured with NanoDrop One Microvolume UV-Vis Spectrophotometer.
Whole genome resequencing was performed in BGI Genomics Co., Ltd. (Shenzhen, China) with a specific process as follows: a small fragment library of about 300 bp in length were constructed using genomic DNA and then were sequenced with BGISEQ-500 sequencing platform to obtain the raw data. High-quality clean data was obtained by removing joint pollution and low-quality reads from raw data using Soapnuke software, then was mapped to the reference genome [27] using BWA (Version: 0.1.17-r1188) software. Reads with the mapping quality (mapQ) greater than 30 were screened for subsequent analysis using samtools software. Population SNP variation was detected using UnifiedGenotypes of GATK4.0 software.
The SNPs were filtered to generate high-quality SNP sites for genetic map construction. The filtering conditions were as follows: (1) in 150 RILs, SNPs with Genotype Quality less than 50 were filtered out; (2) SNPs deletion or heterozygosity between two parents were removed; (3) SNPs with a deletion rate in the offspring samples greater than 20% were filtered out; (4) non-diallelic SNPs were filtered out. Then, the bins were obtained using high-quality SNPs and snpbinner software with the parameters set as the minimum exchange rate of 0.06 and the minimum linkage length of 1 Kb. The genetic map was constructed using the bins through qtl package of R.
QTL analysis
Eight traits of five environments were used for QTL analysis with QTL IciMapping 4.2 software [28]. The parameters were set as follows: the PIN was 0.001; the mapping step was 1.0 cM; and the mapping method of ICIM-ADD, ICIM-EPI, IM-ADD, IM-EPI, were all chosen. The missing phenotype was replaced with the mean value. QTLs with LOD ≥ 2.5 was defined as significant additive QTLs. The method of naming QTLs accorded to Mccouch et al. [29].
Candidate gene identification
The TAIR database (https://www.arabidopsis.org/index.jsp) was used for the annotation of genes within early maturity-related QTL. TBTools software [30] was used for homologous comparison of cotton genes and Arabidopsis genes with parameters of number of threads 2 and e-value 1e− 5. FPKM data of genes, downloaded from CottonFGD (https://cottonfgd.net/) website, were used for the analysis of differently expressed genes.
The heat map was constructed using TBTools software. The protein interaction was predicted through STRING (https://cn.string-db.org/) website with Arabidopsis thaliana as a reference organism.
qRT-PCR analysis
The parental materials of ‘SCRC19’ and ‘SCRC21’ were planted in a greenhouse with a condition of 25℃, 12 h/12 h light/dark in the same day. The squares with a size of 1–2 mm were defined as squares of 1 day post-square (DPS). Squares of 5 DPS, 10 DPS, 15 DPS, 20 DPS represent those 4, 9, 14, 19 days after 1 DPS. Flower bud without bract of ‘SCRC19’ and ‘SCRC21’ at 1 DPS, 5 DPS, 10 DPS, 15 DPS and 20 DPS were collected for qRT-PCR analysis. Total RNA of different samples was extracted using an EASYspin Plus Plant RNA Kit (Aidlab, China), and then converted to cDNA according to the instruction of PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Janpan). The qRT-PCR experiment was performed using SYBR Green Premix Pro TaqHS qPCR Kit (Accurate Biology, China) with three biological repeats and three technical replicates. The endogenous control used in this study was GhActin [17]. 2−ΔΔCt method [31] was used to calculate the relative expression levels of genes in different samples. The primers for qRT-PCR were shown in Table S10.
Results
Phenotypic analysis in parents and RIL population
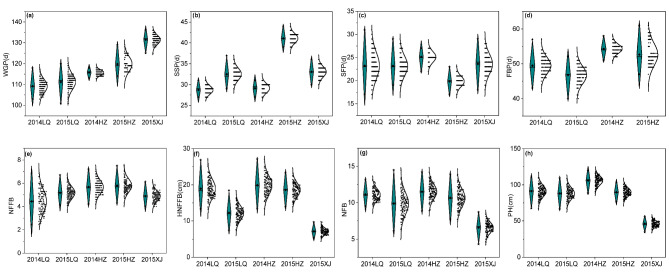
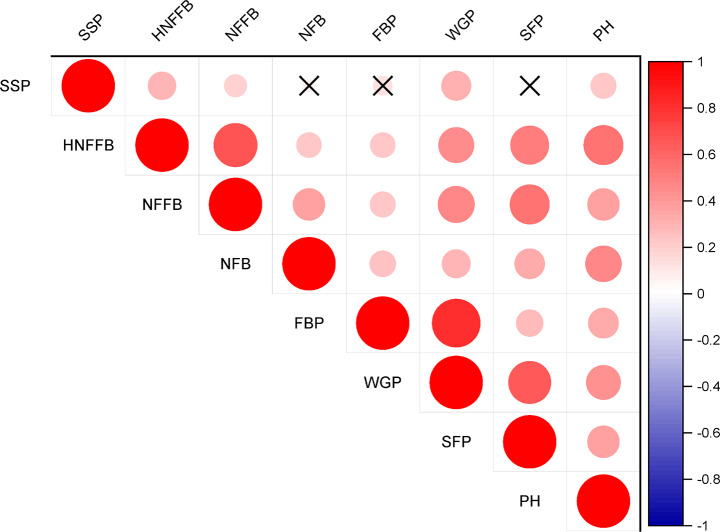
We surveyed eight traits of the parents and RIL population planted in Linqing, Heze in 2014 and 2015 and Xinjiang in 2015. The parents, ‘SCRC21’ and ‘SCRC19’, showed varying degrees in six early maturity-related traits, except SSP and FBP (Table S1). Significant differences (p < 0.05) were observed between ‘SCRC21’ and ‘SCRC19’ for WGP, HNFFB, NFFB, NFB, SFP and PH, with higher phenotypic values found in ‘SCRC21’. Kolmogorov-Smirnov test (K-Stest) was used for normal distribution test of RILs. According to the results, HNFFB, NFFB, NFB and PH in five environments followed an normal distribution with a p-value of > 0.05. And WGP, SSP, SFP and FBP was considered to follow an approximately normal distribution as their fitting curves were approximately normally distributed with a skewness of < 1, although their p-value of K-Stest were less than 0.05. All the eight traits in each environment exhibited a transgressive segregation when compared with their parents (Table S1, Fig. 1). Furthermore, the widely significantly positive correlations were observed among WGP and other early maturity-related traits according to the result of correlation analysis (Table S2, Fig. 2), implying the genetic association of these traits.
Fig. 1.
Phenotypic data in early maturity-related traits in different environments. 2014LQ, 2015LQ, 2014HZ, 2015HZ, 2015XJ represent phenotypic data collected from Linqing in 2014, Linqing in 2015, Heze in 2014, Heze in 2015 and Kuntun, Xinjiang in 2015, respectively; quadrilaterals and stars in the violin plot indicate the mean of the RILs and trait value of their parents, respectively
Fig. 2.
Correlation analysis of eight early-related traits. × indicates that there is no significant correlation between two traits
Genomic resequencing and SNP markers
To construct the genetic map, the RIL population consisting of 150 individuals and their parents were used for resequencing, with an average sequencing depth 35-fold in parents and 7.9-fold in RILs (Table S3). In total, 19,279 million paired-end 150-bp reads were generated, among which high quality bases with Q20 (base calling error at the 0.01 level) account for more than 94% of the total and the average GC (guanine and cytosine) content was 35.2%. Finally, we obtained 79.04 Gb and 82.03 Gb clean reads from ‘SCRC19’ and ‘SCRC21’, respectively, and the clean reads of RIL population ranged from 13.4 Gb to 26.7 Gb. In average, 99.6% of the clean reads were mapped to the reference genome. After filtering by established criteria, a total of 4,092,677 high quality SNPs were obtained in the RIL population and then used to generate bin markers (Table S4), which are groups of consecutive SNPs in the same block for genotyping. Finally, a total of 794 bin markers were generated (Table S5).
Construction and quality assessment of genetic map
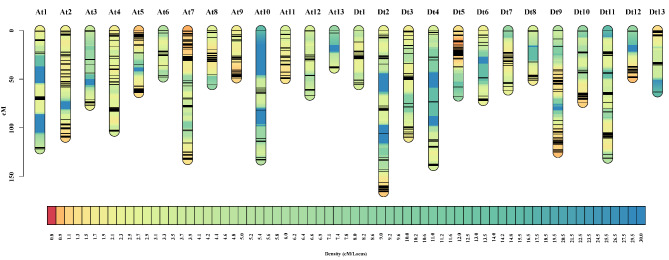
Genetic linkage maps were generated using 794 bin markers, with a total distance of 2213.71 cM and an average inter-bin genetic distance of 2.8 cM (Fig. 3; Table 1). This genetic map covered 26 cotton chromosomes, and the bin number of each chromosome ranged from 8 (At13) to 67 (At7). The longest linkage group was Dt2 of 165.82 cM with 47 bin markers, and the shortest one was At13 of 38.75 cM with 8 bin markers. At10 displayed the largest average inter-bin genetic distance of 7.83 cM, whereas At9 possessed the smallest value at 1.44 cM. Furthermore, for all linkage groups, the average ratio of bin marker intervals (< 5 cM) ranged from 76.5 to 95.0% (Table 1).
Fig. 3.
The genetic map constructed using bin markers
Table 1.
The information of genetic map
| Chromosomes | Map sizes(cM) | Markers | Average distance(cM) | Max gap(cM) | Gap size < 5 (ratio) |
|---|---|---|---|---|---|
| At1 | 121.667 | 26 | 4.680 | 48.776 | 0.846 |
| Dt1 | 55.758 | 22 | 2.534 | 21.513 | 0.864 |
| At2 | 109.773 | 52 | 2.111 | 32.814 | 0.942 |
| Dt2 | 165.823 | 47 | 3.528 | 43.054 | 0.851 |
| At3 | 76.933 | 17 | 4.525 | 28.562 | 0.824 |
| Dt3 | 109.686 | 38 | 2.886 | 24.194 | 0.842 |
| At4 | 103.634 | 44 | 2.355 | 23.321 | 0.864 |
| Dt4 | 138.885 | 24 | 5.787 | 42.930 | 0.833 |
| At5 | 63.911 | 35 | 1.826 | 22.202 | 0.914 |
| Dt5 | 67.487 | 43 | 1.569 | 20.415 | 0.907 |
| At6 | 47.928 | 15 | 3.195 | 18.315 | 0.800 |
| Dt6 | 72.073 | 17 | 4.240 | 32.910 | 0.882 |
| At7 | 132.796 | 67 | 1.982 | 23.603 | 0.896 |
| Dt7 | 61.318 | 27 | 2.271 | 21.682 | 0.926 |
| At8 | 55.765 | 27 | 2.065 | 13.057 | 0.889 |
| Dt8 | 51.167 | 16 | 3.198 | 31.164 | 0.875 |
| At9 | 48.802 | 34 | 1.435 | 15.451 | 0.941 |
| Dt9 | 125.424 | 58 | 2.162 | 29.763 | 0.914 |
| At10 | 133.083 | 17 | 7.828 | 59.056 | 0.765 |
| Dt10 | 73.952 | 28 | 2.641 | 20.912 | 0.893 |
| At11 | 49.625 | 28 | 1.772 | 9.092 | 0.893 |
| Dt11 | 131.262 | 42 | 3.125 | 21.579 | 0.833 |
| At12 | 66.599 | 20 | 3.330 | 24.019 | 0.850 |
| Dt12 | 48.452 | 20 | 2.423 | 35.337 | 0.950 |
| At13 | 38.745 | 8 | 4.843 | 34.411 | 0.875 |
| Dt13 | 63.161 | 22 | 2.871 | 27.465 | 0.909 |
| Mean | 85.143 | 30.538 | 2.788 | 27.907 | 0.876 |
| Total | 2,213.710 | 794 | --- | 59.056 | --- |
To assess the quality of the genetic map, collinearity analysis of each linkage group with cotton genome was performed. The spearman coefficient of 26 linkage groups ranged from 0.754 to 0.985, and was higher than 0.916 in average. What was more, among the 26 groups, 19 groups demonstrated a spearman coefficient higher than 0.9 (Table S6), indicating a strong collinearity between the genetic map and the cotton reference genome. These results suggested that the genetic map is suitable for QTL analysis.
QTL mapping according to phenotypic assessment of RILs
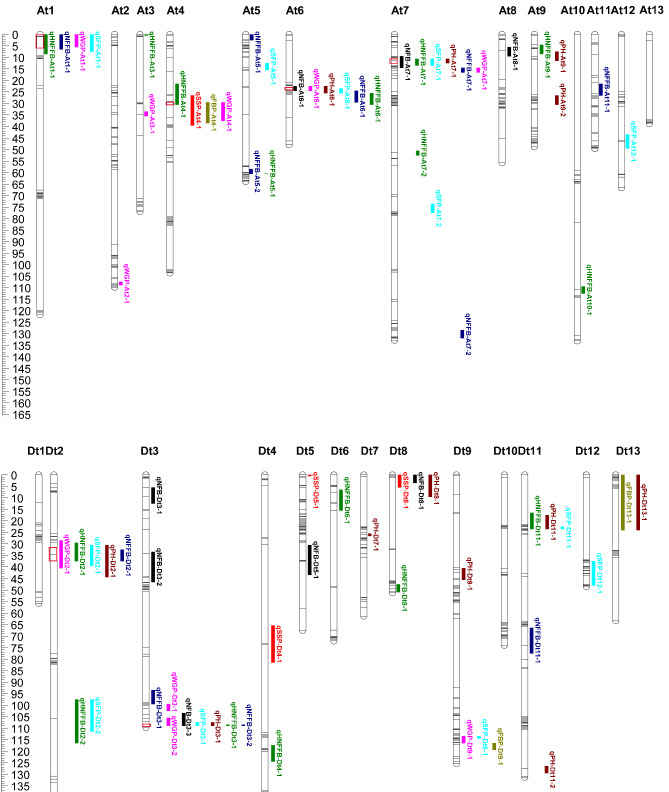
A total of 78 QTLs related to eight traits were detected using QTL IciMapping software (Fig. 4, Table S7). These QTLs, including 12 for WGP, 4 for SSP, 12 for SFP, 3 for FBP, 11 for NFFB, 8 for NFB, 16 for HNFFB and 12 for PH, were distributed on 24 chromosomes, i.e., all chromosomes except At13 and Dt1. The LOD value of all the QTLs ranged from 2.51 to 7.75, with 0.88–13.7% phenotypic variation explained (PVE) by each QTL. Furthermore, At7, Dt2 and Dt3 had most QTLs with a number of 9, 9, 10. The QTLs of WGP, which we cared about most, were distributed on At1, At2, At3, At4, At7, Dt2, Dt3, Dt9; and the QTL numbers were 3, 2, 1 on Dt2, Dt3 and the other chromosomes, respectively. We further compared the QTLs in the present study with early maturity-related QTLs reported previously. As a result, six QTLs were reported in previous QTL linkage studies, such as qPH-Dt9-1 [16, 18]. Besides, three QTLs overlapped with the region enriched for early maturity-associated SNPs and QTLs [1].
Fig. 4.
QTLs for early maturity-related traits. Red boxes mark QTL clusters containing at least four traits
Six QTL clusters, which are defined as specific region on a chromosome contained QTLs of various traits by Said et al. [32], were identified on At1, At4, At6, At7, Dt2, Dt3 (Fig. 4). All these QTL clusters contained QTLs related to more than 4 traits, for example WGP, SFP, NFFB, HNFFB on At1 cluster. The range of clusters were 0-8.5 cM on At1, 21.5–39.5 cM on At6, 9.5–16.5 cM on At7, 28.5–44.5 cM on Dt2 and 108.5–109 cM on Dt3.
Functional annotation of candidate genes in QTL clusters
1081 genes within the six QTL clusters were discovered, including 851 on At1, 4 on At4, 125 on At6, 61 on At7, 1 on Dt2 and 39 on Dt3. To understand the function of these genes, their homologs in Arabidopsis were identified using Arabidopsis Genome Database, and the annotation information of 924 genes was generated (Table S8). Moreover, we performed GO enrichment and KEGG analyses of the 1081 genes and found these genes were enriched in GO terms of “recognition of pollen” and “ADP binding”, which were in the biological process and molecular function category, respectively (Table S9). No gene-enriched pathways were identified through KEGG analyses.
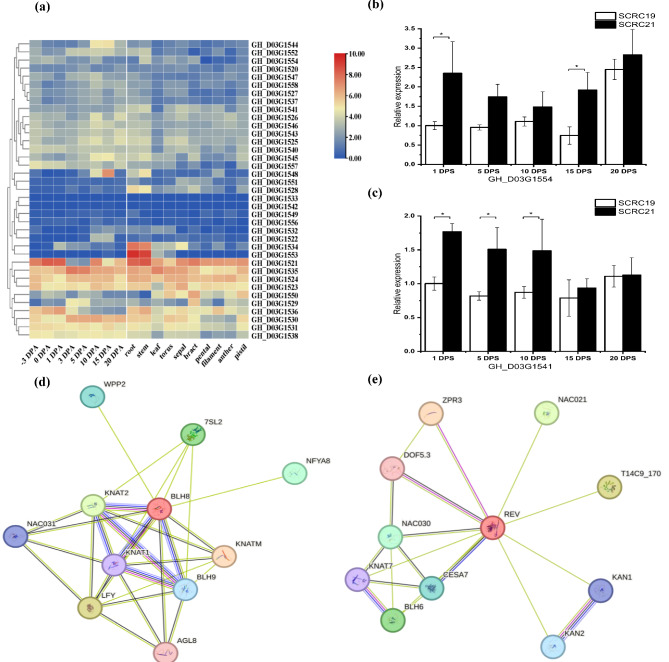
It is reported that chromosome Dt3 is rich in early maturity-related QTLs [1]. In this study, the QTL cluster on Dt3 contained six QTLs of WGP (qWGP-Dt3-2), NFB (qNFB-Dt3-3), SFP (qSFP-Dt3-1), PH (qPH-Dt3-1), HNFFB (qHNFFB-Dt3-1), NFFB (qNFFB-Dt3-2), exhibiting the largest number of QTLs. So, we chose QTL clusters on Dt3 as a candidate region for further study. This cluster ranged from 108.5 to 109 cM, corresponding a physical location of 48,043,162 − 48,682,975 bp, and consisted of 39 genes. Among them, 36 genes were annotated by their homologs in Arabidopsis (Table S8). We found that some genes were involved in regulation of maintaining the normal function of flowers, such as GH_D03G1554, and some genes were related to the phytohormone regulation, such as GH_D03G1544 (annotated as auxin response factor 10). Moreover, we analyzed the expression levels of these genes according to the previously published RNA-seq data. We found that expression levels of the same gene and the expression pattern of different genes all varied at different developmental stages after anthesis (Fig. 5a). For example, GH_D03G1535 was highly expressed before and after anthesis, with a remarkable increase at 3 DPA (day post anthesis) and 5 DPA; whereas the expression level of GH_D03G1548 was low, expect at 10 DPA and 15 DPA. For the expression level in tissues, some genes were highly expressed in all the tissues, such as GH_D03G1521, GH_D03G1535, GH_D03G1524, whereas some genes exhibited tissue-specific expression, such as GH_D03G1534 and GH_D03G1553, whose expression level in root and stem were significantly higher than that in other tissues.
Fig. 5.
Gene expression and protein interaction prediction of genes in QTL cluster on Dt3. (a) The expression of genes from QTL cluster on Dt3. (b) Relative expression of GH_D03G1554 at different stages of flower bud; (c) Relative expression of GH_D03G1541 at different stages of flower bud. 1 DPS, 5 DPS, 10 DPS, 15 DPS and 20 DPS indicate 1, 5, 10, 15 and 20 days post flower bud, respectively. * Indicates significant difference (Diff.) at p = 0.05. The error bars indicate the standard deviation of three biological replicates. (d) Protein interaction prediction of GH_D03G1554; (e) Protein interaction prediction of GH_D03G1541
According to functional annotation in Arabidopsis and protein interaction prediction of 39 genes, two candidate genes were selected for further analysis. GH_D03G1554, annotated as BEL1-like homeodomain protein 8, is homologous to AT2G27990, which plays a key role in the process of plant flowering [33]. Moreover, this gene was predicted to interact with protein of LFY and AGL8 (AGAMOUS-LIKE 8) (Fig. 5d), which are key genes in regulation of plant flowering. GH_D03G1541, another candidate gene, was annotated as homeobox-leucine zipper family protein and homologous to REVOLUTA (AT5G60690) of Arabidopsis. REVOLUTA encoded a homeobox-leucine zipper family protein / lipid-binding START domain-containing protein, which functions as a Class III homeodomain-leucine zipper (HD-ZIPIII) transcription factor regulating the normal growth of floral meristems and the primary shoot apical meristem [34]. What was more, REVOLUTA was predicted to interact with ZPR3 (Fig. 5e), a small ZIP protein functioning as a competitive inhibitor of HD-ZIPIII proteins in regulating shoot apical meristem (SAM) development [35].
To further verify the function of GH_D03G1554 and GH_D03G1541, qRT-PCR was performed using the flower bud from ‘SCRC19’ and ‘SCRC21’ at five developmental stages as follows: 1 DPS, 5 DPS, 10 DPS, 15 DPS and 20 DPS. As shown in Fig. 5, GH_D03G1554 exhibited the highest expression level at 20 DPS in both ‘SCRC19’ and ‘SCRC21’ and its relative expression in ‘SCRC21’ was significantly higher than that in ‘SCRC19’ at 1 DPS and 15 DPS (Fig. 5b). As for GH_D03G1541, the expression in ‘SCRC21’ was significantly high compared with ‘SCRC19’ at 1 DPS, 5 DPS and 10 DPS (Fig. 5c), implying this gene may function at the early stage of flower bud development.
Discussion
The BLH family is involved in flowering regulation
BLH, a subfamily of TALE transcription factor (TF) family, is characterized by three-amino-acid-loop-extensions and the conserved “PYP” (proline-tyrosine-proline) amino acid sequence in the homeodomain (HD), as well as a conserved SKY domain and a BELL domain in the N-terminal, upstream of the HD domain [36]. It is reported that BLH TFs are widely involved in hormone response, stress response and plant development, including the development of meristem, flowering, morphogenesis [36]. In Arabidopsis, 13 BLH TFs are identified, and some of them, for example PNF (POUND-FOOLISH), are found involved in flower development and necessary for inflorescence formation [37–40]. PNY (PENNYWISE) and PNF are both paralogous BEL1-like homeobox genes. The loss of PNY function had no apparent aberrant phenotype, however pnf pny double mutants resulted in abnormal flowering phenotype, implying the potential redundant function of PNY and PNF genes [33]. PNY and PNF also play an important role in long day (LD) - inductive FT-mediated flowering. In pny pnf mutants, the expression of LFY and AP1 are substantially reduced in shoot apices after floral induction, indicating that the floral-promoting gene LFY acts downstream of PNY and PNF. Additionally, overexpression of FT in pny pnf mutants results in terminal flower phenotype, while ectopic expression of FT in wild type Arabidopsis promotes early flowering, suggesting flower formation mediated by FT requires the function of PNY and PNF [38]. What is more, PNY and PNF are associated with the initiation and maintenance of SAM and floral meristem [41–43]. It is reported that PNY and PNF repress BLADE-ON PETIOLE1/2 (BOP1/2) and its transcriptional targets ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA6 (KANT6) to maintain meristem integrity and flowering. Transcriptome analysis of BOP1-overexpressing plants reveals a potential mechanism of microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL)-microRNA172 module by which repression of lateral organ boundary gene by PNY-PNF for flowering [44]. In Gossypium hirsutum, there are 50 BLH genes, among which GhBLH5-A05, GhBLH6-A13, GhBLH7-D06 are involved in drought stress, secondary cell wall biosynthesis, virus response and hormone signaling, respectively [4, 45, 46]. In the present study, we identified a QTL associated with multiple early maturity-related traits on chromosome Dt3, from which two candidate genes of GH_D03G1554 and GH_D03G1541 were highlighted. GH_D03G1554, which is homologous with AtPNF, is differently expressed in flower bud development, and may interact with LFY and AGL8, implying it may be a crucial gene responsible for the difference of WGP between ‘SCRC19’ and ‘SCRC21’.
The function of REVOLUTA in SAM development
The transition from vegetative to reproductive development is necessary for flowering. Two signaling events of SAM development of maintaining stem cell identity and governing the initiation of lateral organs from the flanks of the SAM is closely associated with this progress. In the signaling networks of SAM development, class III homeodomain-leucine zipper (HD-ZIP III) protein is one of the most extensively studied transcription factors [35]. Previous study had reported that HD-ZIPIII gene REVOLUTA (REV) was necessary to promote normal growth of paraclade meristems, floral meristems and the primary shoot apical meristem [34, 47]. In addition, fil rev double mutants of Arabidopsis gave rise to dramatically enhanced flowerless pedicel phenotype compared with filamentous flower (fil) mutants, suggesting an important role of REV in the early step of flower formation [48]. Although REV was reported to function in multiple progresses, including gynoecium development and maintenance of shoot meristem stem cells, through interacting with network of auxin signal [49, 50], but research on REV of cotton was still limited so far. In this study, GH_D03G1541, a candidate gene associated with early maturity trait, is found homologous with REV gene in Arabidopsis (AT5G60690). qRT-PCR analysis indicated that the expression of GH_D03G1541 in ‘SCRC21’ was significantly high compared to ‘SCRC19’ at 1 DPS, 5 DPS and 10 DPS, implying this gene may function at the early stage of flower bud development. While, more experiments are needed to confirm this theory.
Genetic map constructed with bin markers
SNPs have been widely employed in the construction of high-density genetic map because their highly abundance. While constructing genetic map using a great deal of SNPs is a huge challenge for computers. Bin refers to the block of chromosomes in which recombination does not occur in a separate population. In a medium-size RIL population, which contains hundreds of thousands of SNPs for biparental populations, there will probably exits a few thousands genomic bins [51]. Therefore, it will greatly improve the efficiency and accuracy of genetic map construction to use SNP-based bin markers instead of SNP markers. Si et al. constructed a bin genetic linkage map covering 5057 cM using 6303 high-confidence bin markers through re-sequencing 249 individuals from an interspecific F2 population (TM-1 and Hai7124) [16]. In this study, we constructed bin maps of 2213.71 cM using 794 high-confidence bin markers, which were generated from the RIL population including 150 individuals. The number of bin markers and length of bin map are obviously less than the previous reported one (6303 bin markers and 5057 cM), probably because the parents of RIL population in this study are closely related in the pedigree. Specifically, ‘SCRC19’ and ‘SCRC21’ were generated through crossbreeding of ‘Simian 3’ with different maternal varieties followed by multi-generation selection breeding.
Identification of candidate genes associated with traits related to early maturity
Early-maturity cotton is characterized by a dwarf and compact plant architecture, short height of the first fruit branch, a short growth period, and concentrated boll forming [4, 15]. Early maturity is a complex agronomic trait closely related to many quantitative traits, which is supported by the results of correlation analysis of phenotypic traits in our study. Here, we identified 78 QTLs associated with eight traits and found that 9 of them have been reported. Based on these QTLs, six QTL clusters were detected and genes of these clusters were annotated. GH_A01G0120, which was identified from QTL cluster of At1, is homologous with Gh14-3-3 with an identify of 93%, suggesting that there will be a similar function of GH_A01G0120 and Gh14-3-3. 14-3-3 is a key component of a hexameric florigen activation complex, which will activate the downstream floral genes to regulate flowering transition [52]. It has reported that the ectopic overexpression of Gh14-3-3 in Arabidopisis delayed flowering [53]. GH_A07G0875, is annotated as “K-box region and MADS-box transcription factor family protein” and closest homologous with GH_D07G0876 (GhCAL). Overexpression of GhCAL in Arabidopsis has shown to promote flowering, while silencing it in cotton delayed flowering. GhCAL may be involved in a new flowering regulatory pathway through interaction with GhAP-A04 and GhAGL6-D09 to regulate the expression of GhAP-A04 and GhAGL6-D09 [54]. The QTL cluster on chromosome Dt3 was chosen for further analysis because of its association with the most phenotypic traits. In the QTL cluster, GH_D03G1554 and GH_D03G1541 were identified as candidate genes for the reason of the important roles of their homologous genes (AtPNF and AtREV) in flower formation and SAM development, respectively [34, 38, 43, 44, 47]. Although an association between these genes and flowering can be established based on the previous reports, there is currently no direct evidence linking these genes to early maturity. Further experimentations are needed to verify the functions of these genes.
Conclusions
We constructed a genetic linkage map covering 26 chromosomes with a length of 2213.71 cM using 150 RILs and their parents (‘SCRC19’ and ‘SCRC21’). Based on the map, we identified 78 early maturity-related QTLs and 6 QTL clusters. Two candidate genes, GH_D03G1541 and GH_D03G1554, were screened from 39 genes in the Dt3 QTL cluster. The two genes were homologous to AtPNF and AtREV respectively, which were reported to involve flower forming and SAM development, and their expression levels in ‘SCRC21’ were significantly higher than those in ‘SCRC19’ at different stages of flower bud development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
D.Y. and K.F. produced the RIL population. D.Y., K.F., S.G., D.B., W.Z. and F.D. measured early maturity-related traits. G.L. performed the qRT-PCR experiment and tested the expression of candidate genes. X.D. constructed the genetic map and performed QTL analysis. W.X. analyzed all the results and wrote the manuscript. H.Z. designed the experiment and revised the manuscript. All authors have read and approved the final manuscript version.
Funding
This work was supported by Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2023F06); Modern Agricultural Industrial Technology System of Shandong province (SDAIT-03-02); the Key Research and Development Program of Shandong Province (Agricultural Seed Improvement Project, 2023LZGC007).
Data availability
The raw data of RIL individuals and their parents have been deposited in the NCBI Sequence Read Archive under accession number PRJNA1100634.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongsheng Deng, Email: 163dasheng@163.com.
Zongfu Han, Email: zongfuhan@126.com.
References
- 1.Li L, Zhang C, Huang J, Liu Q, Wei H, Wang H, Liu G, Gu L, Yu S. Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L). Plant Biotechnol J. 2020;19(1):109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith CE, Stephens SG. Critical identification of Mexican archaeological cotton remains. Econ Bot. 1971;25(2):160–8. [Google Scholar]
- 3.Zhang L, Zhang F, Zhou X, Poh T, Xie L, Shen J, Yang L, Song S, Yu H, Chen Y. The tetratricopeptide repeat protein OsTPR075 promotes heading by regulating florigen transport in rice. Plant Cell. 2022;34(10):3632–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H, Chen Y, Liu J, Wang Z, Li F, Ge X. Recent advances and future perspectives in early-maturing cotton research. New Phytol. 2022;237(4):1100–14. [DOI] [PubMed] [Google Scholar]
- 5.Khan M, Ai X, Zhang J. Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip Rev RNA. 2014;5(3):347–59. [DOI] [PubMed] [Google Scholar]
- 6.Blümel M, Dally N, Jung C. Flowering time regulation in crops—what did we learn from Arabidopsis? Curr Opin Biotechnol. 2015;32:121–9. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Hua C, Kiang JX, Shen L. A dephosphorylation-dependent molecular switch for FT repression mediates flowering in Arabidopsis. Plant Commun. 2024;5(3):100779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Liu J, Xie X, Wang J, Ma Q, Chen P, Yang D, Ma X, Hao F, Su J. GhAP1-D3 positively regulates flowering time and early maturity with no yield and fiber quality penalties in upland cotton. J Integr Plant Biol. 2023;65(4):985–1002. [DOI] [PubMed] [Google Scholar]
- 9.Song YH, Ito S, Imaizumi TJTPS. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18(10):575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freytes S, Canelo M, Cerdán P. Regulation of Flowering Time: when and where? Curr Opin Plant Biol. 2021;63:102049. [DOI] [PubMed] [Google Scholar]
- 11.Johansson M, Staiger D. Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot. 2015;66(3):719–30. [DOI] [PubMed] [Google Scholar]
- 12.Song YH, Lee I, Lee SY, Imaizumi T, Hong JC. Constans and asymmetric leaves 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J. 2011;69(2):332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Shuwen L, Qianqian G, Xiang Y, Biao C, Caiping Z, Tianzhen, Baoliang Z. Mapping of genes for flower-related traits and QTLs for flowering time in an interspecific population of Gossypium hirsutum × G. Darwinii. J Genet. 2016;95(1):5. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Pei W, Ma Q, Geng Y, Liu G, Liu J, Cui Y, Zhang X, Wu M, Li X, et al. QTL analysis and candidate gene identification for plant height in cotton based on an interspecific backcross inbred line population of Gossypium hirsutum × Gossypium barbadense. Theor Appl Genet. 2019;132(9):2663–76. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Zhao S, Su J, Fan S, Pang C, Wei H, Wang H, Gu L, Zhang C, Liu G, et al. High-density genetic linkage map construction by F2 populations and QTL analysis of early-maturity traits in upland cotton (Gossypium hirsutum L). PLoS ONE. 2017;12(8):e0182918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si Z, Jin S, Chen J, Wang S, Fang L, Zhu X, Zhang T, Hu Y. Construction of a high-density genetic map and identification of QTLs related to agronomic and physiological traits in an interspecific (Gossypium hirsutum × Gossypium barbadense) F2 population. BMC Genomics. 2022;23(1):307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Jia X, Guo X, Wei H, Zhang M, Wu A, Cheng S, Cheng X, Yu S, Wang H. QTL and candidate gene identification of the node of the first fruiting branch (NFFB) by QTL-seq in upland cotton (Gossypium hirsutum L). BMC Genomics. 2021;22(1):882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R, Xiao X, Gong J, Li J, Zhang Z, Liu A, Lu Q, Shang H, Shi Y, Ge Q, et al. QTL mapping for plant height and fruit branch number based on RIL population of upland cotton. J Cotton Res. 2020;3(1):5. [Google Scholar]
- 19.Li C, Wang Y, Ai N, Li Y, Song J. A genome-wide association study of early-maturation traits in upland cotton based on the CottonSNP80K array. J Integr Plant Biol. 2018;60(10):970–85. [DOI] [PubMed] [Google Scholar]
- 20.Guo Y, McCarty JC, Jenkins JN, Saha S. QTLs for node of first fruiting branch in a cross of an upland cotton, Gossypium hirsutum L., cultivar with primitive accession Texas 701. Euphytica. 2007;163(1):113–22. [Google Scholar]
- 21.Su J, Pang C, Wei H, Li L, Liang B, Wang C, Song M, Wang H, Zhao S, Jia X, et al. Identification of favorable SNP alleles and candidate genes for traits related to early maturity via GWAS in upland cotton. BMC Genomics. 2016;17(1):687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Z, Li L, Tang M, Liu Q, Ji Z, Sun D, Liu G, Zhao S, Huang C, Zhang Y, et al. Detection of stable Elite haplotypes and potential candidate genes of Boll Weight Across Multiple Environments via GWAS in Upland Cotton. Front Plant Sci. 2022;13(13):929168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Hu Y, Wang Y, Zhao S, You Y, Liu R, Wang J, Yan M, Zhao F, Huang J, et al. Identification of novel candidate loci and genes for seed vigor-related traits in upland cotton (Gossypium hirsutum L.) via GWAS. Front Plant Sci. 2023;14:1254365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia X, Pang C, Wei H, Wang H, Ma Q, Yang J, Cheng S, Su J, Fan S, Song M, et al. High-density linkage map construction and QTL analysis for earliness-related traits in Gossypium hirsutum L. BMC Genomics. 2016;17(1):909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Q, Qu Z, Wang X, Qiao K, Mangi N, Fan S. EMBRYONIC FLOWER2B, coming from a stable QTL, represses the floral transition in cotton. Int J Biol Macromol. 2020;163:1087–96. [DOI] [PubMed] [Google Scholar]
- 26.Yu H, Shao N, Li J. An unprecedented one-arrow-two-hawks strategy achieves high yield with early flowering in rice. Mol Plant. 2022;15(9):1412–4. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–48. [DOI] [PubMed] [Google Scholar]
- 28.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015;3(3):269–83. [Google Scholar]
- 29.Mccouch S, Cho Y, Yano M, Paul E, Blinstrub M, Morishima H, Mccouch S, Cho Y, Paul E, Morishima H. Report on QTL nomenclature. Nat Genet. 1997;14:11–3. [Google Scholar]
- 30.Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y, et al. TBtools-II: a one for all, all for one bioinformatics platform for biological big-data mining. Mol Plant. 2023;16(11):1733–42. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 – ∆∆CT method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 32.Said JI, Lin Z, Zhang X, Song M, Zhang J. A comprehensive meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genomics. 2013;14(1):776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith HM, Campbell BC, Hake S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr Biol. 2004;14(9):812–7. [DOI] [PubMed] [Google Scholar]
- 34.Talbert P, Adler H, Parks D, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121(9):2723–35. [DOI] [PubMed] [Google Scholar]
- 35.Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell. 2008;20(4):920–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niu X, Fu D. The roles of BLH Transcription Factors in Plant Development and Environmental Response. Int J Mol Sci. 2022;23(7):3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee K, Brocchieri L, Bürglin TR. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol. 2009;26(12):2775–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanrar S, Bhattacharya M, Arthur B, Courtier J, Smith H. Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 2008;54(5):924–37. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, You S, Taylor-Teeples M, Li WL, Schuetz M, Brady SM, Douglas CJ. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell. 2014;26(12):4843–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Zhang X, Ju H, Chen J, Wang S, Wang H, Zhao Y, Chang Y. Ovate family protein1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem Biophys Res Commun. 2016;470(3):492–7. [DOI] [PubMed] [Google Scholar]
- 41.Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009;58(4):641–54. [DOI] [PubMed] [Google Scholar]
- 42.Ung N, Lal S, Smith HMS. The role of PENNYWISE and POUND-FOOLISH in the maintenance of the shoot apical Meristem in Arabidopsis. Plant Physiol. 2011;156(2):605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith HM, Ung N, Lal S, Courtier J. Specification of reproductive meristems requires the combined function of SHOOT MERISTEMLESS and floral integrators FLOWERING LOCUS T and FD during Arabidopsis inflorescence development. J Exp Bot. 2011;62(2):583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan M, Ragni L, Tabb P, Salasini BC, Chatfield S, Datla R, Lock J, Kuai X, Després C, Proveniers M, et al. Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for Meristem Maintenance and flowering in Arabidopsis. Plant Physiol. 2015;169(3):2166–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Q, Wang N, Hao P, Sun H, Wang C, Ma L, Wang H, Zhang X, Wei H, Yu S. Genome-wide identification and characterization of TALE superfamily genes in cotton reveals their functions in regulating secondary cell wall biosynthesis. BMC Plant Biol. 2019;19(1):432. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Ma Q, Wang N, Ma L, Lu J, Wang H, Wang C, Yu S, Wei H. The cotton BEL1-Like transcription factor GhBLH7-D06 negatively regulates the Defense response against Verticillium Dahliae. Int J Mol Sci. 2020;21(19):7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25(2):223–36. [DOI] [PubMed] [Google Scholar]
- 48.Chen Q, Atkinson A, Otsuga D, Christensen T, Reynolds L, Drews GN. The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development. 1999;126(12):2715–26. [DOI] [PubMed] [Google Scholar]
- 49.Nole-Wilson S, Azhakanandam S, Franks RG. Polar auxin transport together with aintegumenta and revoluta coordinate early Arabidopsis gynoecium development. Dev Biol. 2010;346(2):181–95. [DOI] [PubMed] [Google Scholar]
- 50.Roodbarkelari F, Du F, Truernit E, Laux T. ZLL/AGO10 maintains shoot meristem stem cells during Arabidopsis embryogenesis by down-regulating ARF2-mediated auxin response. BMC Biol. 2015;13:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edwards CE, Ewers BE, Weinig C. Genotypic variation in biomass allocation in response to field drought has a greater affect on yield than gas exchange or phenology. BMC Plant Biol. 2016;16(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Sci (New York NY). 2005;309(5737):1052–6. [DOI] [PubMed] [Google Scholar]
- 53.Liu H, Huang X, Ma B, Zhang T, Sang N, Zhuo L, Zhu J. Components and functional diversification of Florigen Activation complexes in Cotton. Plant Cell Physiol. 2021;62(10):1542–55. [DOI] [PubMed] [Google Scholar]
- 54.Cheng S, Chen P, Su Z, Ma L, Hao P, Zhang J, Ma Q, Liu G, Liu J, Wang H, et al. High-resolution temporal dynamic transcriptome landscape reveals a GhCAL‐mediated flowering regulatory pathway in cotton (Gossypium hirsutum L). Plant Biotechnol J. 2020;19(1):153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of RIL individuals and their parents have been deposited in the NCBI Sequence Read Archive under accession number PRJNA1100634.