Abstract
Background
The influence of the glucagon-like peptide-1 receptor (GLP-1R) on bone metabolism is well-established. However, it has been observed that single nucleotide polymorphisms (SNPs) in the GLP-1R gene can partially affect its function. Therefore, this study aims to investigate the association between SNPs in the GLP-1R gene and postmenopausal osteoporosis (PMOP) within the Chinese Han population.
Methods
This study employed a cross-sectional case–control design, recruiting a total of 152 participants, including 76 patients with osteoporosis (OP) (case group) and 76 healthy individuals (control group). Seven tag SNPs of GLP-1R were selected from the National Center of Biotechnology Information and Genome Variation Server. The association between GLP-1R polymorphisms and PMOP risk was assessed using different genetic models and haplotypes, while also exploring SNP-SNP and SNP-environment interactions.
Results
Our results showed that minor alleles A at rs3765468, A at rs3765467 and G at rs4714210 showed significant associations with an increased risk of OP. Individuals with rs3765468 AG-AA genotype and rs3765467 AG-AA genotype exhibited a significantly higher risk of PMOP. Moreover, haplotype analysis revealed a significant association of the GACACA haplotype on PMOP risk (P = 0.033). Additionally, a multiplicative interaction was observed between rs3765468 and rs3765467 that was associated with an increased risk of PMOP (Pinteraction = 0.012).
Conclusions
Specific SNPs in the GLP-1R gene were linked to an increased risk of PMOP. This study improves our understanding of the genetic basis of PMOP in this population and suggests that genetic screening can identify individuals at risk for developing PMOP, enabling early prevention.
Keywords: Glucagon-like peptide-1 receptor, Single-nucleotide polymorphism, Postmenopausal osteoporosis, Haplotype, Bone mineral density
Background
Osteoporosis (OP) is a progressive systemic skeletal disease characterized by low bone mass and microarchitectural deterioration of bone tissue [1], leading to increased susceptibility to fractures, particularly fragility fractures. These fractures are a prevalent condition with significant implications for global healthcare, emphasizing the importance of effective management strategies to minimize morbidity and healthcare expenditures [2]. Postmenopausal osteoporosis (PMOP) is a prevalent age-related metabolic bone disorder. Approximately 10% of the global population is affected by OP [3]. In China, the estimated prevalence of OP in adults aged ≥ 40 years is 20.6% for women [4]. In the management of PMOP or corticosteroid-induced OP, various medications have shown efficacy [5, 6]. Recently, many studies used bone turnover markers to monitor the efficacy and safety of drugs influencing bone turnover and as therapy monitors in PMOP [7, 8]. The anti-OP drugs, in particular, exhibit a positive impact on bone mineral density (BMD) in postmenopausal women with OP, making them a valuable therapeutic option [9]. BMD is a reliable and quantifiable indicator for the gold standard of OP diagnosis. However, BMD demonstrates a significant degree of genetic heritability [10]. Research suggests that genetic factors contribute to 60%−80% of variation in BMDs, with genes accounting for 50%−85% of variation in the peak BMD [11]. Although OP has a strong genetic propensity, the identification of genetic molecular mechanisms associated with OP remains a formidable challenge. To date, genome-wide association studies (GWAS) have identified more than 500 loci that are linked to OP [12]. Moreover, numerous other single nucleotide polymorphisms (SNPs) have been discovered to be correlated with OP [13–15].
Glucagon-like peptide-1 receptor (GLP-1R), a member of the B1 family of G-protein-coupled receptors, is composed of 463 amino acids and exerts its function through specific binding to glucagon-like peptide-1 (GLP-1). Our previous studies have demonstrated that GLP-1R agonists can promote bone formation while inhibiting bone resorption [16–18]. Moreover, Meng et al. reported that activation of GLP-1R could enhance the osteogenic potential of bone marrow stromal cells to ameliorate OP [19]. Additionally, mice lacking GLP-1R expression (GLP-1R−/−) exhibited cortical osteopenia and increased susceptibility to bone fragility due to augmented bone resorption by osteoclasts [20, 21]. Zhang et al. also provided evidence indicating that the A allele at rs2295006 of the GLP-1R gene predicts a decrease in BMDs [22]. These findings underscore a critical role for GLP-1R in regulating bone metabolism, with its genetic variation profoundly impacting BMDs.
However, there is currently a lack of research data regarding the association between the GLP-1R gene and OP. Given that OP is a multifactorial disease influenced by both genetic and environmental factors [23], this study aims to investigate the relationship between GLP-1R SNPs (rs3765468, rs3765467, rs4714210, rs10305420, rs10305445, rs761386, and rs761387), SNP-SNP and SNP-environmental interactions on susceptibility to OP in postmenopausal Chinese Han women. This investigation may provide valuable insights for genetic screening of individuals at risk for OP, enhance our understanding of OP pathogenesis, and establish an underlying genetic basis for developing GLP-1R agonists as novel anti-osteoporotic agents.
Methods
Recruitment of participants
The study recruited a total of 152 postmenopausal Chinese Han females from the Endocrinology Department of the Hebei Medical University Third Hospital between March 2023 and June 2023. Among them, there were 76 patients with OP (the case group) and an equal number of healthy individuals (the control group). The inclusion criteria were as follows: 1) All participants were aged between 45–80 years, with a natural menopause duration of more than one year; 2) The diagnosis of OP in postmenopausal women was based on the T-value according to the criteria established by the World Health Organization [a T-value ≤ − 2.5 standard deviations (SD) indicated OP, while a T-value ≥ − 1.0 SD indicated normal BMD]; 3) None of the participants had previously received any anti-OP treatment.
The exclusion criteria included patients with thyroid and parathyroid disease, autoimmune disease, diabetes mellitus, severe cardiovascular, hepatic, and renal disease, metabolic or hereditary skeletal disorders, malignancy, long-term use of hormones or immunosuppressants, as well as any anti-OP drugs.
The study protocol was approved by the Medical Ethics Committee of the Hebei Medical University Third Hospital [NO.2023–026-1]. The participants were required to provide written informed consent in accordance with the principles outlined in the Declaration of Helsinki prior to their participation in the study.
Laboratory assessment
Demographic data from all participants were collected encompassing nationality, age, menopause onset age, height, weight, smoking and drinking habits, as well as family history of OP. Standard laboratory methods were employed for measuring serum levels of parathyroid hormone (PTH), 25-Hydroxy vitamin D, calcium, and phosphorus. Body mass index (BMI) was calculated using the formula: BMI = weight/height2 (kg/m2).
Measurement of BMDs
The BMDs of all 152 participants at the lumbar spine (L1-L4), femoral neck, and total hip were assessed using a Dual-energy X-ray absorptiometry device (Hologic Discovery A, MA, USA). To ensure rigorous quality control measures, the instrument underwent daily calibration. The coefficient of variation for the BMDs of the lumbar spine, femoral neck and total hip was found to be 1.14%, 1.29%, and 1.80%, respectively.
SNP selection, DNA extraction and genotyping
The tag SNPs were selected based on the databases of the National Center of Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/SNP/) and the Genome Variation Server (GVS) (https://gvs.gs.washington.edu/GVS150/index.jsp). The selection criteria for these SNPs included minor allele frequency (MAF) ≥ 0.01, and linkage disequilibrium (LD) measured by an r2 value ≥ 0.33. After reviewing relevant research reports [24–26], seven SNPs of the GLP-1R gene were identified, namely rs3765468, rs3765467, rs4714210, rs10305420, rs10305445, rs761386, and rs761387.
The genomic DNA was isolated from peripheral blood leukocytes using the Genomic DNA Kit (Tiangen, Beijing, China) following the manufacturer's protocol. Subsequently, we assessed the quality and concentration of the extracted DNA by employing a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Genotyping was performed through the polymerase chain reaction-ligase detection reaction. The primers sequences for polymerase chain reaction were designed by Primer Premier 5 software. Furthermore, the results were further processed using the 3730 XL Gene Sequencer (Applied Biosystems, MA, USA) and analyzed with GeneMarker software version V2.6.3.
Statistical analysis
Statistical analyses were performed using SPSS version 22.0. The normal distribution of variables was assessed through the Kolmogorov–Smirnov test. Variables conforming to normality were presented as mean ± SD, while those deviating from normality were reported as median (interquartile range). Student’s t-test was employed for comparing normally distributed data, whereas the Mann–Whitney U test was used for non-normally distributed data. Qualitative data were expressed as number (%) and compared using the chi-square (χ2) test. Hardy–Weinberg equilibrium (HWE), allele and genotype frequencies were analyzed using the χ2 test (P ≥ 0.05). The online tool SHEsis (http://analysis.bio-x.cn/myAnalysis.php) was employed for LD and haplotype analysis. Binary logistic regression was utilized to examine the associations between SNPs and PMOP. The SNPStats software (https://www.snpstats.net/start.htm.) was employed to investigate SNP-SNP and SNP-environmental interactions. The optimal model selection was based on Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC). The statistical significance was determined at a significance level of P < 0.05.
Results
Baseline characteristics of participants
The baseline characteristics of all participants are presented in Table 1. There were statistically significant differences between the two groups in terms of age, menopausal duration, BMI, and family history of OP (P < 0.05), which were identified as potential influencing factors for PMOP between the case and control groups. No statistically significant differences were observed in terms of other variables (P > 0.05).
Table 1.
Baseline characteristics of the study population
| Variables | Control group (n = 76) | Case group (n = 76) | P Value |
|---|---|---|---|
| Age (Years) | 0.000 | ||
| <50 | 6 (7.89) | 0 (0.00) | |
| 50 – | 36 (47.37) | 18 (23.68) | |
| 60 – | 24 (31.58) | 29 (38.16) | |
| ≥ 70 | 10 (13.16) | 29 (38.16) | |
| Age of menopause (Years) | 0.138 | ||
| <50 | 27 (35.53) | 36 (47.37) | |
| ≥ 50 | 49 (64.47) | 40 (52.63) | |
| Menopausal duration (Years) | 0.000 | ||
| <10 | 42 (55.26) | 14 (18.42) | |
| 10 – | 22 (28.95) | 33 (43.42) | |
| ≥ 20 | 12 (15.79) | 29 (38.16) | |
| BMI (kg/m2) | 0.001 | ||
| <24 | 18 (23.68) | 36 (47.37) | |
| 24– | 32 (42.11) | 30 (39.47) | |
| ≥ 28 | 26 (34.21) | 10 (13.16) | |
| Smoking status (Yes/No) | 2 (2.63)/74 (97.37) | 4 (5.26)/72 (94.74) | 0.681 |
| Drinking status (Yes/No) | 12 (15.79)/64 (84.21) | 9 (11.84)/67 (88.16) | 0.481 |
| Family history of OP (Yes/No) | 15 (19.74)/ 61 (80.26) | 27 (35.53)/ 49 (64.47) | 0.030 |
| FBG (mmol/L) | 5.35 ± 0.59 | 5.44 ± 0.66 | 0.347 |
| PTH (pg/ml) | 52.83 (42.46–66.24) | 49.96 (37.67–63.59) | 0.411 |
| 25(OH)D (ng/ml) | 21.28 (17.07–25.48) | 18.19 (15.82–23.75) | 0.097 |
| Blood calcium (mmol/L) | 2.27 ± 0.11 | 2.26 ± 0.13 | 0.642 |
| Blood phosphorus (mmol/L) | 1.22 ± 0.17 | 1.22 ± 0.18 | 0.887 |
BMI, body mass index; OP, osteoporosis; FBG, fasting blood glucose; PTH, parathyroid hormone; 25 (OH) D, 25-hydroxy vitamin D
Genotype and allele frequencies of GLP-1R SNPs
Among the seven GLP-1R SNPs, rs10305445 was excluded due to its non-conformity with the HWE in the case group, indicating an underrepresentation of this specific GLP-1R SNP in the target population. The remaining six SNPs met both criteria of MAF > 0.01 and HWE. Therefore, our study participants provided a representative sample of the population (Table 2).
Table 2.
The characteristics of seven GLP-1R SNPs
| SNPs | Chr.position | Alleles | MAF in dbSNP | MAF | HWE (P value) | Functions | ||
|---|---|---|---|---|---|---|---|---|
| Control group | Case group | Control group | Case group | |||||
| rs3765468 | 39065817 | G > A | 0.09 | 0.092 | 0.197 | 0.377 | 0.487 | Synonymous-codon |
| rs3765467 | 39065819 | G > A | 0.04 | 0.178 | 0.303 | 0.755 | 0.602 | Nonsynon missense |
| rs4714210 | 39087709 | A > G | 0.24 | 0.211 | 0.355 | 0.799 | 0.481 | Non-coding transcript variant |
| rs10305420 | 39048860 | C > T | 0.17 | 0.138 | 0.184 | 0.597 | 0.748 | Missense |
| rs10305445 | 39058761 | C > T | 0.18 | 0.053 | 0.053 | 0.628 | 0.000 | Intron-variant |
| rs761386 | 39079095 | C > T | 0.16 | 0.151 | 0.105 | 0.120 | 0.305 | Upstream-variant |
| rs761387 | 39078425 | A > G | 0.20 | 0.158 | 0.118 | 0.102 | 0.942 | Non-coding transcript variant |
SNPs, single nucleotide polymorphisms; MAF, minimum allele frequency; dbSNP, the Genetic Variation Database of NCBI; HWE, Hardy-Weinberg equilibrium
The distribution of genotype and allele frequencies for the GLP-1R SNPs is presented in Table 3. Significantly higher genotype frequencies of rs3765468 and rs3765467 G-carrier (GG and GA) were observed in the case group compared to the control group (P = 0.025, 0.035). Additionally, a significantly higher genotype frequency of rs4714210 A-carrier (AA and AG) was found in the case group than in the control group (P = 0.022). Moreover, minor alleles A at rs3765468, A at rs3765467, and G at rs4714210 showed significant associations with an increased risk of OP (P = 0.009, 0.011, and 0.005, respectively). However, no significant differences were observed between genotypes or alleles for other GLP-1R SNPs (rs10305420, rs761386, and rs761387) among both groups (P > 0.05).
Table 3.
Allele and genotype frequencies of GLP-1R SNPs
| SNPs | Genotypes/alleles | Control group [n (%)] | Case group [n(%)] | χ2 | P value | OR (95%CI) |
|---|---|---|---|---|---|---|
| rs3765468 | GG | 62 (81.6) | 48 (63.2) | 6.975 | 0.025 | |
| GA | 14 (18.4) | 26 (34.2) | ||||
| AA | 0 (0.0) | 2 (2.6) | ||||
| G | 138 (90.8) | 122 (80.3) | 6.803 | 0.009 | 2.42 (1.23–4.78) | |
| A | 14 (9.2) | 30 (19.7) | ||||
| rs3765467 | GG | 51 (67.1) | 36 (47.4) | 6.709 | 0.035 | |
| GA | 23 (30.3) | 34 (44.7) | ||||
| AA | 2 (2.6) | 6 (7.9) | ||||
| G | 125 (82.2) | 106 (69.7) | 6.508 | 0.011 | 2.01 (1.17–3.45) | |
| A | 27 (17.8) | 46 (30.3) | ||||
| rs4714210 | AA | 47 (61.8) | 33 (43.4) | 7.642 | 0.022 | |
| AG | 26 (34.2) | 32 (42.1) | ||||
| GG | 3 (3.9) | 11 (14.5) | ||||
| A | 120 (78.9) | 98 (64.5) | 7.848 | 0.005 | 2.07 (1.24–3.45) | |
| G | 32 (21.1) | 54 (35.5) | ||||
| rs10305420 | CC | 57 (75.0) | 51 (67.1) | 1.174 | 0.556 | |
| CT | 17 (22.4) | 22 (28.9) | ||||
| TT | 2 (2.6) | 3 (3.9) | ||||
| C | 131 (86.2) | 124 (81.6) | 1.192 | 0.275 | 1.41 (0.76–2.61) | |
| T | 21 (13.8) | 28 (18.4) | ||||
| rs761386 | CC | 53 (69.7) | 60 (78.9) | 1.690 | 0.194 | |
| CT | 23 (30.3) | 16 (21.1) | ||||
| TT | 0 (0.0) | 0 (0.0) | ||||
| C | 129 (84.9) | 136 (89.5) | 1.441 | 0.230 | 0.66 (0.33–1.31) | |
| T | 23 (15.1) | 16 (10.5) | ||||
| rs761387 | AA | 52 (68.4) | 59 (77.6) | 3.041 | 0.219 | |
| AG | 24 (31.6) | 16 (21.1) | ||||
| GG | 0 (0.0) | 1 (1.3) | ||||
| A | 128 (84.2) | 134 (88.2) | 0.995 | 0.319 | 0.72 (0.37–1.38) | |
| G | 24 (15.8) | 18 (11.8) |
SNPs, single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval
Association of GLP-1R SNPs with the risk of PMOP
The association between GLP-1R SNPs and the risk of PMOP was further investigated using binary logistic regression analysis with different genetic models. In the dominant model, individuals with rs3765468 AG-AA genotype and rs3765467 AG-AA genotype exhibited a significantly higher risk of PMOP compared to those with GG genotype (adjusted P = 0.04, 0.037, respectively). Furthermore, in both the dominant and recessive models, individuals carrying AG-GG and GG genotypes of rs4714210 showed the increased risk of PMOP (P = 0.023, 0.021, respectively), although these differences were not statistically significant after adjusting for confounding factors (P = 0.051, 0.19, respectively). Based on the AIC and BIC values obtained from the analysis, the dominant model was considered to be the optimal model for these three SNPs (Table 4).
Table 4.
Genetic model analysis of the association of GLP-1R SNPs with the risk of PMOP
| SNPs | Genotypes | Control group | Case group | OR (95%CI) | P value | AIC | BIC | OR* (95%CI) | P*value | AIC* | BIC* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3765468 | |||||||||||
| Dominant | GG | 62 (81.6) | 48 (63.2) | 1 | 0.010 | 208.2 | 214.2 | 1 | 0.040 | 178.1 | 208.4 |
| AG-AA | 14 (18.4) | 28 (36.8) | 2.58 (1.23–5.44) | 2.50 (1.02–6.10) | |||||||
| Recessive | GG-AG | 76 (100.0) | 74 (97.4) | 1 | 0.094 | 211.9 | 218 | 1 | 0.061 | 178.8 | 209.1 |
| AA | 0 (0.0) | 2 (2.6) | – | – | |||||||
| Overdominant | GG-AA | 62 (81.6) | 50 (65.8) | 1 | 0.026 | 209.8 | 215.8 | 1 | 0.110 | 179.7 | 210.0 |
| AG | 14 (18.4) | 26 (34.2) | 2.30 (1.09 − 4.87) | 2.07 (0.85 − 5.06) | |||||||
| rs3765467 | |||||||||||
| Dominant | GG | 51 (67.1) | 36 (47.4) | 1 | 0.014 | 208.6 | 214.7 | 1 | 0.037 | 177.0 | 207.2 |
| AG-AA | 25 (32.9) | 40 (52.6) | 2.27 (1.17 − 4.37) | 2.34 (1.04 − 5.22) | |||||||
| Recessive | GG-AG | 74 (97.4) | 70 (92.1) | 1 | 0.140 | 212.5 | 218.6 | 1 | 0.260 | 177.4 | 207.6 |
| AA | 2 (2.6) | 6 (7.9) | 3.17 (0.62 − 16.24) | 3.30 (0.90 − 17.09) | |||||||
| Overdominant | GG-AA | 53 (69.7) | 42 (55.3) | 1 | 0.065 | 211.3 | 217.4 | 1 | 0.250 | 181.0 | 211.2 |
| AG | 23 (30.3) | 34 (44.7) | 1.87 (0.96 − 3.63) | 1.59 (0.72 − 3.51) | |||||||
| rs4714210 | |||||||||||
| Dominant | AA | 47 (61.8) | 33 (43.4) | 1 | 0.023 | 209.2 | 215.1 | 1 | 0.051 | 176.4 | 206.6 |
| AG-GG | 29 (38.2) | 43 (56.6) | 2.11 (1.10 − 4.04) | 2.70 (0.99 − 6.14) | |||||||
| Recessive | AA-AG | 73 (96.0) | 65 (85.5) | 1 | 0.021 | 209.4 | 215.4 | 1 | 0.190 | 176.8 | 207.0 |
| GG | 3 (4.0) | 11 (14.5) | 4.12 (1.10 − 15.41) | 4.47 (0.87 − 16.49) | |||||||
| Overdominant | AA-GG | 50 (65.8) | 44 (57.9) | 1 | 0.320 | 213.7 | 219.8 | 1 | 0.280 | 181.2 | 211.4 |
| AG | 26 (34.2) | 32 (42.1) | 1.40 (0.72 − 2.70) | 1.55 (0.70 − 3.44) |
*Adjusted the age, menopausal duration, family history of OP, and BMI
OR, odds ratio; CI, confidence interval; AIC, Akaike Information Criterion; BIC, Bayesian Information Criterion
Haplotype analysis of GLP-1R SNPs with the risk of PMOP
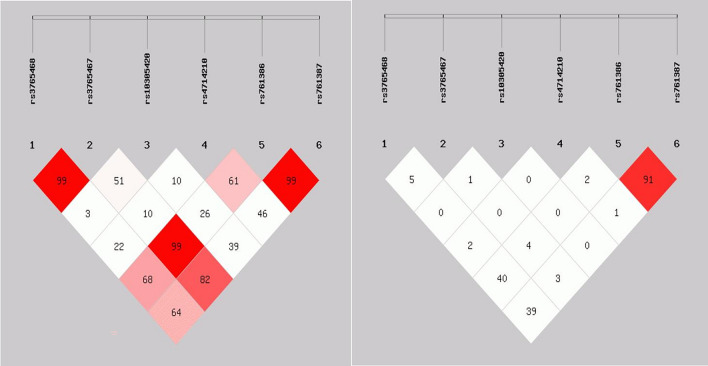
The LD analysis revealed a stronger LD among the six GLP-1R SNPs, indicating that these SNPs were considered a linkage domain (Fig. 1). Haplotype analysis demonstrated that the GGCACA haplotype had the highest frequency and could be selected as the reference haplotype. Furthermore, we observed a significant association between the GACACA haplotype and an increased risk of PMOP, even after adjusting for confounding factors (P = 0.033) (Table 5). Importantly, none of the remaining four haplotypes showed a significant impact on PMOP (P > 0.05) (Table 5).
Fig. 1.
Linkage disequilibrium diagram of the GLP-1R SNPs
Table 5.
Correlation analysis between GLP-1R SNPs haplotypes and PMOP risk (Frequence > 1%)
| Haplotypes | Frequence | OR (95% CI) | P value | |||||
|---|---|---|---|---|---|---|---|---|
| rs3765468 | rs3765467 | rs10305420 | rs4714210 | rs761386 | rs761387 | |||
| G | G | C | A | C | A | 0.327 | 1.00 | Referent |
| G | A | C | A | C | A | 0.155 | 4.24 (1.14 − 15.72) | 0.033 |
| G | G | C | G | C | A | 0.143 | 5.36 (0.98 − 25.33) | 0.055 |
| G | G | T | A | C | A | 0.102 | 2.01 (0.48 − 8.36) | 0.340 |
| A | G | C | A | T | G | 0.069 | 4.64 (0.99 − 20.96) | 0.050 |
| G | A | C | G | C | A | 0.049 | 2.89 (0.45 − 18.43) | 0.260 |
OR are adjusted for age, menopausal duration, family history of OP, and BMI
OR, odds ratio; CI, confidence interval
Association between GLP-1R SNP-SNP and SNP-environment multiplicative interactions and PMOP risk
To further investigate the association between GLP-1R SNP-SNP and SNP-environment interactions and the risk of PMOP, we performed multiplicative interaction analysis using the dominant model (Tables 6 and 7). The analysis revealed a significant multiplicative interaction between rs3765468 and rs3765467 of the GLP-1R gene, which was associated with an increased risk of PMOP (adjusted Pinteraction = 0.012) (Table 6). And Table 7 shows the multiplicative interactions between GLP-1R SNPs and environmental factors in relation to PMOP risk. However, no significant association was observed between the multiplicative interactions of GLP-1R SNP-environment and PMOP risk (Pinteraction > 0.05).
Table 6.
The association between the multiplicative interaction of GLP-1R SNP-SNP and PMOP risk
| SNP*SNP | Control group (n = 76) | Case group (n = 76) | OR (95% CI) | Pinteraction | |
|---|---|---|---|---|---|
| rs3765468*rs3765467 | 0.012 | ||||
| GG | GG | 41 | 12 | 1.00 | |
| AG-AA | GG | 10 | 24 | 7.70 (2.40 − 24.68) | |
| GG | AG-AA | 21 | 36 | 5.90 (2.14 − 16.29) | |
| AG-AA | AG-AA | 4 | 4 | 3.37 (0.64 − 17.81) | |
| rs3765468*rs4714210 | 0.330 | ||||
| GG | AA | 41 | 21 | 1.00 | |
| AG-AA | AA | 6 | 12 | 3.65 (0.90 − 14.71) | |
| GG | AG-GG | 21 | 27 | 3.09 (1.19 − 8.03) | |
| AG-AA | AG-GG | 8 | 16 | 4.46 (1.33 − 14.92) | |
| rs3765467*rs4714210 | 0.950 | ||||
| CC | AA | 32 | 14 | 1.00 | |
| AC-AA | AA | 15 | 19 | 2.36 (0.76 − 7.36) | |
| CC | AG-GG | 20 | 22 | 2.73 (0.94 − 7.95) | |
| AC-AA | AG-GG | 9 | 21 | 6.81 (2.00 − 23.15) |
P value was adjusted by age, menopausal duration, family history of OP, and BMI
SNP, single nucleotide polymorphism; OR, odds ratio, CI, confidence interval
Table 7.
The association between the multiplicative interaction of GLP-1R SNP-environment and PMOP risk
| Age | Menopausal duration | Family history of OP | BMI | |
|---|---|---|---|---|
| rs3765468 | ||||
| P-dominant | 0.160 | 0.400 | 0.310 | 0.550 |
| P-recessive | – | – | 1.000 | 1.000 |
| P-overdominant | 0.360 | 0.260 | 0.310 | 0.450 |
| rs3765467 | ||||
| P-dominant | 0.240 | 0.200 | 0.770 | 0.680 |
| P-recessive | 0.130 | 0.057 | – | 0.120 |
| P-overdominant | 0.600 | 0.290 | 0.770 | 0.540 |
| rs4714210 | ||||
| P-dominant | 0.170 | 0.260 | 0.058 | 0.200 |
| P-recessive | 0.620 | 0.660 | 0.530 | 0.470 |
| P-overdominant | 0.300 | 0.480 | 0.069 | 0.290 |
Adjusted confounders
OP, osteoporosis; BMI, body mass index
Discussion
GLP-1R is widely distributed in various organs, including the pancreas, gastrointestinal tract, bone, brain, and heart. Consequently, it exerts significant effects on glucose regulation, weight loss, bone metabolism, cardiovascular function, and nervous system activity [26]. GLP-1R gene is located on chromosome 6 (chr 6p21) and consists of 13 exons and 12 introns [27]. In order to further elucidate the physiological mechanism of GLP-1R from a genetic perspective, extensive research has been conducted on GLP-1R polymorphisms. SNPs, which are the most common type of DNA sequence variations at the genomic level caused by single nucleotide changes [28], represent the predominant form of human heritable variation. In recent years, numerous studies have investigated the biological effects associated with GLP-1R SNPs.
Continuing this line of investigation, specific GLP-1R SNPs have been implicated in various disease susceptibilities. For instance, the G allele of the GLP-1R rs3765467 variant has been identified as a risk allele for susceptibility to T2DM in the Asian population [29]. The presence of GLP-1R rs3765467 can significantly reduce insulin secretion by pancreatic beta cells and the concentration of AMP in circulation, promoting apoptosis of beta cells [30]. Moreover, there is a significant association between the GLP-1R rs3765467 polymorphism and individual variations in both GLP-1 concentration and blood glucose levels [30]. Meanwhile, GLP-1R rs3765467 (G/A) variant was related to dyslipidemia incidence and might serve as a risk factor for Chinese patients with T2DM [31]. Furthermore, an association between GLP-1R rs4714210 and the risk of coronary artery disease in T2DM patients within the Chinese Han population was identified [32]. GLP-1R rs10305420 is not only associated with inter-individual variations in glycated hemoglobin reduction but also with weight loss. In a study involving 285 obese Chinese patients with T2DM, GLP-1R rs10305420 gene was genotyped after six months of exenatide treatment. The results demonstrated significant reductions in both glycated hemoglobin levels and BMI among patients following the six-month treatment period. Furthermore, consistent correlations were observed between the minor allele T of rs10305420 and reductions in weight and glycated hemoglobin during exenatide therapy. Therefore, GLP-1R rs10305420 holds potential as a genetic biomarker for precision medicine targeting overweight individuals with comorbid T2DM [33]. GLP-1R rs10305420 allele was found to be associated with both HbA1c levels and weight loss in patients treated with exenatide [34]. Additionally, a study revealed that GLP-1R rs10305445 could potentially increase the risk of cardiovascular disease within a five-year period among chronic kidney disease patients undergoing hemodialysis [35]. GLP-1R rs761386 (C/T) polymorphism exhibited an association with obesity in the Chinese population [36]. Moreover, individuals with the GLP-1R rs761386 CT/TT genotype demonstrated elevated glucose levels at 2 h during a 75 g oral glucose tolerance test among patients with poorly controlled T2DM [37]. The G allele of GLP-1R rs761387 was linked to higher levels of both GLP-1 and glucose but not insulin [38]. However, the current research investigating the correlation between GLP-1R SNPs and bone metabolism remains significantly limited.
Studies have demonstrated the presence of GLP-1R on the surface of osteoblast-like MC3T3-E1 cells, osteoclasts and bone marrow stromal cells [16–19]. Meanwhile, GLP-1R agonists, have been shown to stimulate bone formation and inhibit bone resorption through multiple signaling pathways [39, 40]. By binding to GLP-1R and acting on osteoblasts, Liraglutide, a GLP-1R agonists, activated phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) and cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA) signaling pathways, thereby promoting osteogenic differentiation and bone formation [18]. Feng et al. reported that exendin-4 increased the mRNA levels of key factors involved in osteogenic differentiation such as alkaline phosphatase (ALP), collagen-1 (Col-1), osteocalcin (OC) and runt-related transcription factor 2 (Runx2) through mitogen-activated protein kinase (MAPK) activation [41]. Furthermore, liraglutide can suppress osteoclastogenesis through the inhibition of NF-κB and MAPK pathways via GLP-1R [17]. Meng et al. confirmed that activating GLP-1R promoted osteogenic differentiation while inhibiting adipogenic differentiation by regulating the β-catenin signaling pathway [19]. In conclusion, the impact of GLP-1/GLP-1R is pivotal in the regulation of bone metabolism.
The regulation of bone metabolism by GLP-1/GLP-1R, as discussed, plays a crucial role in maintaining bone health and density. This is particularly significant in the context of OP, a condition characterized by thinning bones and decreased bone density, which makes the lumbar spine more vulnerable to degenerative changes. As OP weakens the structural integrity and stability of bones, the lumbar spine becomes increasingly susceptible to injury and degeneration under load. Lumbar degenerative diseases can further aggravate OP by causing disc degeneration and joint degenerative conditions, thereby perpetuating a vicious cycle of decreased bone density and spinal instability. Notably, the importance of addressing OP in surgical contexts, such as lumbar spinal fusion procedures, has been emphasized, with preoperative screening for OP highlighted as a critical step to mitigate complications [42]. In this regard, recent research exploring the impact of targeted mud therapy on serum serotonin levels in patients with lumbar degenerative diseases provides additional insights into potential therapeutic interventions that could modulate biomarkers and influence pathological conditions [43]. Understanding the intricate interplay between bone metabolism regulation by GLP-1/GLP-1R and the pathophysiology of OP, as well as exploring novel therapeutic approaches, is essential for developing comprehensive strategies to manage and prevent lumbar degenerative diseases and their associated complications.
Building on the understanding of the crucial role of GLP-1/GLP-1R in bone metabolism and its implications for OP, our team has delved deeper into the genetic aspects of this relationship. Recently, our team has found that the A allele of GLP-1R rs1042044 was closely associated with an increased risk of PMOP, and this risk was significantly amplified through an SNP-SNP interaction with rs2268641 [44]. Given the multitude of mutation sites and intricate interactions of the GLP-1R gene, we expanded the scope of our research to comprehensively comprehend the genetic mechanism underlying GLP-1R gene mutations and susceptibility to OP. After thorough review of numerous references, we identified seven significant SNPs for in-depth analysis. In this study, we found that individuals carrying rs3765468 AG-AA genotype and rs3765467 AG-AA genotype exhibited a significantly higher risk of PMOP for Chinese Han women. Furthermore, we also observed a significant association between the GACACA haplotype and an increased risk of PMOP, even after adjusting for confounding factors. Notably, there was a significantly multiplicative interaction between rs3765468 and rs3765467 that further contributed to an increased susceptibility to PMOP.
GLP-1R rs3765467 (p.Arg131Gln) is a non-synonymous mutation in which arginine is replaced by glutamine at position 131 on exon 5 of GLP-1R [29]. Therefore, it is hypothesized that the rs3765467 variation may potentially affect the binding efficiency of the ligand to GLP-1R and thus interfere with downstream signal transduction. However, the rs3765468 variant located in exon 4 does not result in any alteration to the amino acid sequence of the translated protein. Interestingly, our study discovered a SNP-SNP multiplicative interaction between rs3765467 and rs3765468. Epistasis, a genetic phenomenon, may involve one gene masking or modifying the phenotype of another gene [45]. The close proximity of rs3765468 to rs3765467 on the same chromosome suggests that it could exert genetic effects through SNP-SNP interactions or strong linkage disequilibrium with other biologically impactful SNPs.
The occurrence of PMOP is influenced by a combination of genetic variations and environmental factors, thereby suggesting that gene-environment interactions may partially contribute to the missing genetic predisposition. Unfortunately, this study did not uncover the relationship between the multiplicative interaction of GLP-1R SNP-environment and PMOP risk. Furthermore, although the GACACA haplotype showed a statistically significant association with an increased risk of PMOP, its specific mechanism remains unknown, necessitating further comprehensive research. Nevertheless, exploring genetic markers at the haplotype level could reveal novel insights into genetic alterations.
In conclusion, specific SNPs of GLP-1R gene were identified to be correlated with increased risk of PMOP for Chinese Han ethnicity. This study contributes to a deeper comprehension of the genetic underpinnings of PMOP among Chinese populations and suggests that genetic screening could serve as a valuable tool for identifying individuals at risk of developing this condition, thereby facilitating early preventive measures.
However, our study has several limitations. Firstly, this study was conducted at a single center and only included participants from a local region, potentially introducing sample selection bias. Secondly, the limited sample size and number of SNPs may restrict the generalizability of our findings. Thus, it is important to consider that other polymorphisms of GLP-1R gene could also influence susceptibility to OP. Thirdly, we solely focused on the clinical aspects of OP. Further studies should explore the functional implications of genetic variations in GLP-1R loci in order to gain a better understanding of its underlying pathophysiological mechanism. Overall, while our finding regarding gene polymorphism are limited in scope, identifying significant SNPs impacting OP and comprehending their cellular and molecular functions will significantly contribute to elucidating the pathophysiological mechanism of OP. If our results are validated by prospective studies, GLP-1R polymorphisms could potentially serve as predictors for PMOP in the Chinese Han population.
Conclusions
Specific SNPs in the GLP-1R gene were linked to an increased risk of PMOP. This study improves our understanding of the genetic basis of PMOP in this population and suggests that genetic screening can identify individuals at risk for developing PMOP, enabling early prevention.
Acknowledgements
We express our gratitude to the participants in this study, as well as the hospital staff who provided invaluable assistance with sample collection. We also thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Abbreviations
- GLP-1R
Glucagon-like peptide-1 receptor
- SNPs
Single nucleotide polymorphisms
- PMOP
Postmenopausal osteoporosis
- OP
Osteoporosis
- BMD
Bone mineral density
- GWAS
Genome-wide association studies
- GLP-1
Glucagon-like peptide-1
- SD
Standard deviations
- PTH
Parathyroid hormone
- BMI
Body mass index
- NCBI
National Center of Biotechnology Information
- GVS
Genome Variation Server
- MAF
Minor allele frequency
- LD
Linkage disequilibrium
- HWE
Hardy-Weinberg equilibrium
- AIC
Akaike Information Criterion
- BIC
Bayesian Information Criterion
- T2DM
Type 2 diabetes mellitus
- HbA1c
Glycated Hemoglobin A1c
- PI3K
Phosphoinositide 3-kinase
- cAMP
Cyclic Adenosine monophosphate
- PKA
Protein kinase A
- ALP
Alkaline phosphatase
- Col-1
Collagen-1
- OC
Osteocalcin
- Runx2
Runt-related transcription factor 2
- MAPK
Mitogen-activated protein kinase
Author contributions
XB performed the experiments and wrote the paper. CL and HL analyzed and interpreted the data. YW and PX revised the manuscript. YL conceived and designed the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (NO.82170892), Project Plan of Medical Science Research of Hebei Province (NO.20221155), Precision Medicine Joint Fund Cultivation Project of Natural Science Foundation of Hebei Province(NO.H2020206314) and Government-Funded Clinical Medical Outstanding Talent Project (NO.ZF2023102).
Data availability
The data from this study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Consent for publication
I certify that this manuscript is a unique submission and is not being considered for publication, in part or in full, with any other source in any medium.
Ethics approval and consent to participate
The study protocol was approved by the Medical Ethics Committee of the Hebei Medical University Third Hospital [NO.2023–026-1]. The participants were required to provide written informed consent in accordance with the principles outlined in the Declaration of Helsinki prior to their participation in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Compston JE, McClung MR, Leslie WD. Osteoporosis Lancet. 2019;393(10169):364–76. 10.1016/S0140-6736(18)32112-3. [DOI] [PubMed] [Google Scholar]
- 2.Migliorini F, Giorgino R, Hildebrand F, Spiezia F, Peretti GM, Alessandri-Bonetti M, et al. Fragility Fractures: Risk Factors and Management in the Elderly. Medicina (Kaunas). 2021;57(10):1119. 10.3390/medicina57101119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu F, Xia W. The epidemiology of osteoporosis, associated fragility fractures, and management gap in China. Arch Osteoporos. 2019;14(1):32. 10.1007/s11657-018-0549-y. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Yu W, Yin X, Cui L, Tang S, Jiang N, et al. Prevalence of Osteoporosis and Fracture in China: The China Osteoporosis Prevalence Study. JAMA Netw Open. 2021;4(8): e2121106. 10.1001/jamanetworkopen.2021.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migliorini F, Colarossi G, Baroncini A, Eschweiler J, Tingart M, Maffulli N. Pharmacological Management of Postmenopausal Osteoporosis: a Level I Evidence Based - Expert Opinion. Expert Rev Clin Pharmacol. 2021;14(1):105–19. 10.1080/17512433.2021.1851192. [DOI] [PubMed] [Google Scholar]
- 6.Migliorini F, Colarossi G, Eschweiler J, Oliva F, Driessen A, Maffulli N. Antiresorptive treatments for corticosteroid-induced osteoporosis: a Bayesian network meta-analysis. Br Med Bull. 2022;143(1):46–56. 10.1093/bmb/ldac017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migliorini F, Maffulli N, Spiezia F, Tingart M, Maria PG, Riccardo G. Biomarkers as therapy monitoring for postmenopausal osteoporosis: a systematic review. J Orthop Surg Res. 2021;16(1):318. 10.1186/s13018-021-02474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migliorini F, Maffulli N, Spiezia F, Peretti GM, Tingart M, Giorgino R. Potential of biomarkers during pharmacological therapy setting for postmenopausal osteoporosis: a systematic review. J Orthop Surg Res. 2021;16(1):351. 10.1186/s13018-021-02497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migliorini F, Maffulli N, Colarossi G, Eschweiler J, Tingart M, Betsch M. Effect of drugs on bone mineral density in postmenopausal osteoporosis: a Bayesian network meta-analysis. J Orthop Surg Res. 2021;16(1):533. 10.1186/s13018-021-02678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang TL, Shen H, Liu A, Dong SS, Zhang L, Deng FY, et al. A road map for understanding molecular and genetic determinants of osteoporosis. Nat Rev Endocrinol. 2020;16(2):91–103. 10.1038/s41574-019-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23(3):303–26. 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 12.Sabik OL, Farber CR. Using GWAS to identify novel therapeutic targets for osteoporosis. Transl Res. 2017;181:15–26. 10.1016/j.trsl.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, Gu Z, He Y, Wang C, Duan J. The effect of SOX4 gene 3’UTR polymorphisms on osteoporosis. J Orthop Surg Res. 2021;16(1):321. 10.1186/s13018-021-02454-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdi S, Binbaz RA, Mohammed AK, Ansari MGA, Wani K, Amer OE, et al. Association of RANKL and OPG Gene Polymorphism in Arab Women with and without Osteoporosis. Genes (Basel). 2021;12(2):200. 10.3390/genes12020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conti V, Russomanno G, Corbi G, Toro G, Simeon V, Filippelli W, et al. A polymorphism at the translation start site of the vitamin D receptor gene is associated with the response to anti-osteoporotic therapy in postmenopausal women from southern Italy. Int J Mol Sci. 2015;16(3):5452–66. 10.3390/ijms16035452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao L, Li SL, Li YK. Liraglutide Promotes the Osteogenic Differentiation in MC3T3-E1 Cells via Regulating the Expression of Smad2/3 Through PI3K/Akt and Wnt/β-Catenin Pathways. DNA Cell Biol. 2018;37(12):1031–43. 10.1089/dna.2018.4397. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Li S, Wang N, Xue P, Li Y. Liraglutide, a glucagon-like peptide-1 receptor agonist, suppresses osteoclastogenesis through the inhibition of NF-κB and MAPK pathways via GLP-1R. Biomed Pharmacother. 2020;130: 110523. 10.1016/j.biopha.2020.110523. [DOI] [PubMed] [Google Scholar]
- 18.Wu X, Li S, Xue P, Li Y. Liraglutide, a glucagon-like peptide-1 receptor agonist, facilitates osteogenic proliferation and differentiation in MC3T3-E1 cells through phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT), extracellular signal-related kinase (ERK)1/2, and cAMP/protein kinase A (PKA) signaling pathways involving β-catenin. Exp Cell Res. 2017;360(2):281–91. 10.1016/j.yexcr.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Meng J, Ma X, Wang N, Jia M, Bi L, Wang Y, et al. Activation of GLP-1 Receptor Promotes Bone Marrow Stromal Cell Osteogenic Differentiation through β-Catenin. Stem Cell Reports. 2016;6(4):579–91. 10.1016/j.stemcr.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada C, Yamada Y, Tsukiyama K, Yamada K, Udagawa N, Takahashi N, et al. The murine glucagon-like peptide-1 receptor is essential for control of bone resorption. Endocrinology. 2008;149(2):574–9. 10.1210/en.2007-1292. [DOI] [PubMed] [Google Scholar]
- 21.Mabilleau G, Mieczkowska A, Irwin N, Flatt PR, Chappard D. Optimal bone mechanical and material properties require a functional glucagon-like peptide-1 receptor. J Endocrinol. 2013;219(1):59–68. 10.1530/JOE-13-0146. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, He J, Sun X, Luo X, Zeng J, He W, Liu X, Feng Bo. Relationship between glucagon-like peptide-1 receptor gene polymorphism and bone mineral density in postmenopausal women in Shanghai. Annals of Palliative Medicine. 2020;9(4):1732–41. 10.21037/apm-19-396. [DOI] [PubMed] [Google Scholar]
- 23.Ralston SH, de Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 2006;20(18):2492–506. 10.1101/gad.1449506. [DOI] [PubMed] [Google Scholar]
- 24.Luo P, Fan Y, Xiong Y, Feng H, Yang Z, Zhang C, et al. Genetic variants of the GLP-1R gene affect the susceptibility and glucose metabolism of gestational diabetes mellitus: a two-center nested case-control study. Diabetol Metab Syndr. 2022;14(1):190. 10.1186/s13098-022-00963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan Z, Du Y, Li R, Zhang S, Xu Y, Zhang X, et al. Association between glucagon-like peptide-1 receptor gene polymorphism and treatment response to GLP1R agonists in Chinese patients with type 2 diabetes: a prospective cohort study. Eur J Clin Pharmacol. 2022;78(5):793–9. 10.1007/s00228-021-03249-z. [DOI] [PubMed] [Google Scholar]
- 26.Michałowska J, Miller-Kasprzak E, Bogdański P. Incretin Hormones in Obesity and Related Cardiometabolic Disorders: The Clinical Perspective. Nutrients. 2021;13(2):351. 10.3390/nu13020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simó R, Hernández C. GLP-1R as a Target for the Treatment of Diabetic Retinopathy: Friend or Foe? Diabetes. 2017;66(6):1453–60. 10.2337/db16-1364. [DOI] [PubMed] [Google Scholar]
- 28.Shi R, Nejad MI, Zhang X, Gu LQ, Gates KS. Generation and Single-Molecule Characterization of a Sequence-Selective Covalent Cross-Link Mediated by Mechlorethamine at a C-C Mismatch in Duplex DNA for Discrimination of a Disease-Relevant Single Nucleotide Polymorphism. Bioconjug Chem. 2018;29(11):3810–6. 10.1021/acs.bioconjchem.8b00663. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki K, Akiyama M, Ishigaki K, Kanai M, Hosoe J, Shojima N, et al. Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nat Genet. 2019;51(3):379–86. 10.1038/s41588-018-0332-4. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Li P, Li R, Yu Z, Sun X, Ji G, et al. GLP1R Single-Nucleotide Polymorphisms rs3765467 and rs10305492 Affect β Cell Insulin Secretory Capacity and Apoptosis Through GLP-1. DNA Cell Biol. 2020;39(9):1700–10. 10.1089/dna.2020.5424. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Yang Z, Ren S, Shen B, Zhang Y, Zong H, et al. Association between GLP-1R gene polymorphism and dyslipidemia in Chinese patients with type 2 diabetes mellitus: A case-control study. Gene. 2023;878: 147589. 10.1016/j.gene.2023.147589. [DOI] [PubMed] [Google Scholar]
- 32.Ma X, Lu R, Gu N, Wei X, Bai G, Zhang J, et al. Polymorphisms in the Glucagon-Like Peptide 1 Receptor (GLP-1R) Gene Are Associated with the Risk of Coronary Artery Disease in Chinese Han Patients with Type 2 Diabetes Mellitus: A Case-Control Study. J Diabetes Res. 2018;2018:1054192. 10.1155/2018/1054192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu MJ, Wang KY, Liu HM, Cao R. GLP1R variant is associated with response to exenatide in overweight Chinese Type 2 diabetes patients. Pharmacogenomics. 2019;20(4):273–7. 10.2217/pgs-2018-0159. [DOI] [PubMed] [Google Scholar]
- 34.Rathmann W, Bongaerts B. Pharmacogenetics of novel glucose-lowering drugs. Diabetologia. 2021;64(6):1201–12. 10.1007/s00125-021-05402-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terranegra A, Arcidiacono T, Macrina L, Brasacchio C, Pivari F, Mingione A, et al. Glucagon-like peptide-1 receptor and sarcoglycan delta genetic variants can affect cardiovascular risk in chronic kidney disease patients under hemodialysis. Clin Kidney J. 2020;13(4):666–73. 10.1093/ckj/sfz182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu T, Liu M, Liu Q, Wang B, Wang M, Qu M, et al. Associations of TCF7L2 rs11196218 (A/G) and GLP-1R rs761386 (C/T) Gene Polymorphisms with Obesity in Chinese Population. Diabetes Metab Syndr Obes. 2021;14:2465–72. 10.2147/DMSO.S310069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin CH, Lee YS, Huang YY, Hsieh SH, Chen ZS, Tsai CN. Polymorphisms of GLP-1 receptor gene and response to GLP-1 analogue in patients with poorly controlled type 2 diabetes. J Diabetes Res. 2015;2015: 176949. 10.1155/2015/176949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorsey-Trevino EG, Kaur V, Mercader JM, Florez JC, Leong A. Association of GLP1R Polymorphisms With the Incretin Response. J Clin Endocrinol Metab. 2022;107(9):2580–8. 10.1210/clinem/dgac374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie B, Chen S, Xu Y, Han W, Hu R, Chen M, et al. The Impact of Glucagon-Like Peptide 1 Receptor Agonists on Bone Metabolism and Its Possible Mechanisms in Osteoporosis Treatment. Front Pharmacol. 2021;12: 697442. 10.3389/fphar.2021.697442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Meng J, Jia M, Bi L, Zhou Y, Wang Y, et al. Exendin-4, a glucagon-like peptide-1 receptor agonist, prevents osteopenia by promoting bone formation and suppressing bone resorption in aged ovariectomized rats. J Bone Miner Res. 2013;28(7):1641–52. 10.1002/jbmr.1898. [DOI] [PubMed] [Google Scholar]
- 41.Feng Y, Su L, Zhong X, Guohong W, Xiao H, Li Y, et al. Exendin-4 promotes proliferation and differentiation of MC3T3-E1 osteoblasts by MAPKs activation. J Mol Endocrinol. 2016;56(3):189–99. 10.1530/JME-15-0264. [DOI] [PubMed] [Google Scholar]
- 42.Filley A, Baldwin A, Ben-Natan AR, Hansen K, Arora A, Xiao A, et al. The influence of osteoporosis on mechanical complications in lumbar fusion surgery: a systematic review. N Am Spine Soc J. 2024;18: 100327. 10.1016/j.xnsj.2024.100327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupu A-A, Oprea D, Obada B, Iliescu DM, Botnarciuc M, Ionescu A-M, Mihailov CI, Iliescu M-G, Caraban BM. Variation of serum serotonin values under specific peloidotherapy in patients with degenerative pathology of the lumbar spine. Balneo and PRM Research Journal. 2023. 10.12680/balneo.2023.588. [Google Scholar]
- 44.Liu C, Bao X, Tian Y, Xue P, Wang Y, Li Y. Polymorphisms in the glucagon-like peptide-1 receptor gene and their interactions on the risk of osteoporosis in postmenopausal Chinese women. PLoS ONE. 2023;18(12): e0295451. 10.1371/journal.pone.0295451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arkin Y, Rahmani E, Kleber ME, Laaksonen R, März W, Halperin E. EPIQ-efficient detection of SNP-SNP epistatic interactions for quantitative traits. Bioinformatics. 2014;30(12):i19-25. 10.1093/bioinformatics/btu261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data from this study are available from the corresponding author upon reasonable request.