Abstract
Background
Pure partial trisomy 16q12.1q22.1 is a rare chromosome copy number variant (CNV). The primary clinical phenotypes associated with this syndrome include abnormal facial morphology, global developmental delay (GDD), short stature, and reported predisposing factors for atypical behavior, autism, the development of learning disabilities, and neuropsychiatric disorders. The dosage-sensitive genes associated with partial trisomy are not disclosed preventing to establish a genotype-phenotype correlation.
Methods
We report a case of a Chinese patient diagnosed with GDD and an abnormal facial shape, who was found to have partial trisomy 16 through karyotyping and high-throughput sequencing analysis. Karyotype and CNV tracing analyses were also conducted on the biological parents of the patient to assess for any chromosomal structural abnormalities. Additionally, we included 29 patients with pure partial trisomy 16q, reported in the DECIPHER database and the literature. We and performed a genotype-phenotype correlation analysis.
Results
The proband, a 2-year-old female, was found to have a de novo 21.96 Mb duplication located between 16q12.1q22.1, with no other deletions observed on other chromosomes, indicating a pure partial trisomy of 16q. Through genotype and phenotype analysis of 29 individuals, we found that patients with the duplicated region located at the distal region of 16q may exhibit more severe symptoms than those with duplication at the proximal region; however, no relationship was identified between phenotype and the size of the duplicated segment.
Conclusion
We report, for the first time, a patient with partial trisomy 16q validated by multiple genetic tests, including CNV-seq, whole exome sequencing (WES), and karyotyping. It is speculated that partial trisomy of 16q may be associated with continuous gene duplication. However, functional studies are necessary to identify the causative gene or critical region linked to duplication syndrome of chromosome 16q.
Keywords: Chromosome 16; Duplications; CNV-seq; WES, partial trisomy 16
Introduction
Complete trisomy 16 is a common chromosomal disorder that often results in early fetal abortion, however a few survivors with partial trisomy 16q which arises from the duplication of the long arm of chromosome 16, have been documented [1, 2]. Patients who survive with partial trisomy 16q typically present with multiple organ system involvement, characterized by a core phenotype that includes neurodevelopmental disorders, short stature, and abnormal facial morphology; abnormal heart morphology is observed in approximately half of the patients [3]. Previous studies have identified two trisomy 16 related structural alterations: (1) unbalanced translocations involving chromosome 16 and other chromosomes, and (2) pure partial trisomy 16. Among these, unbalanced translocations are the most prevalent form observed in cases of partial trisomy 16. Chromosome 16 comprises approximately 9.89% of duplicated sequence and features one of the highest levels of segmentally duplicated sequences [4]. This complex structure contributes to chromosomal instability, rendering long arm of chromosome 16 more susceptible to rearrangements via nonallelic homologous recombination (NAHR) with neighboring intrachromosomal segmental duplications [5]. This explains that partial duplication in the 16p11.2 region results from chromosomal rearrangements of parental balance during meiosis, which may also involve varying degrees of deletion in other chromosomes. However, the pathogenic mechanism underlying pure partial duplication of 16q has not been documented. The dose-sensitive genes and the smallest regions of overlap (SROs) associated with partial trisomy 16q remain unclear. Pure partial trisomy 16q is an exceedingly rare condition, with only seven individuals reported in the literature and 21 individuals documented in the DECIPHER database. Nearly 50 years have elapsed since 1975, when the first case of trisomy 16q was identified through karyotype. With the rapid advancements in cellular and molecular genetics, a broader array of genetic tests has been employed to diagnose various genetic disorders. We present for the first time a case of pure partial trisomy 16q diagnosed via karyotyping and high-throughput sequencing (CNV-seq combined with WES). Furthermore, we analyzed and summarized the genotypes and phenotypes of 29 individuals documented in the DECIPHER database, as well as in the established literature and our own study. All reported cases have been confirmed through chromosome karyotyping and/or chromosomal microarray analysis (CMA). The study of pure partial trisomy 16q cases offers a unique opportunity to analyze the phenotypic effects of genomic diseases.
Subjects and methods
The study was approved by the Institutional Ethics Committee of the Sichuan Provincial Maternity and Child Health Care Hospital / Sichuan Provincial Woman’s and Children’s Hospital (protocol code: 202300911-225 and date:2023.09. 11).
Chromosomal karyotype analysis
A total of 0.5 mL of peripheral blood was inoculated in a clean bench, followed by the addition of 5 mL of lymphocyte medium (BIOSCIENCES medium, Israel). The sample was then placed in two independent constant temperature carbon dioxide incubators for a double-line culture over a duration of 72 h under sterile conditions. The harvested cell suspension underwent drop slides and dry slides, followed by staining with Giemsa solution for chromosome scanning and karyotype analysis at the 500-band level. Subsequently, the cells were counted using the MetaSystems Ikaros Chromosome Automated Scanning and Analysis System (ZEISS, Germany). A total of twenty split metaphases were counted, and five karyotypes were analyzed and described in accordance with the International System for Human Cytogenetic or Cytogenomic Nomenclature 2020 (ISCN2020) standard.
Copy number variation sequencing
A total of 2 mL of peripheral blood was extracted, and CNV-seq analysis was performed on the genomic DNA. Genomic DNA was isolated from peripheral white blood cells following the instructions provided with the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). The genomic DNA was fragmented using an ultrasonic interrupter (Covaris-S220, USA) in buffers containing Tris and EDTA. The standard library construction kit RNGS035 (MyGenostics, Beijing, China) was utilized for DNA library preparation. High-throughput sequencing was conducted using the Illumina NovaSeq 500 platform (Illumina, San Diego, California, USA), achieving a total depth of approximately 1X. The results were compared against the human genome reference sequence (GRCh37/hg19) to identify copy number variations (CNVs). Additionally, comparisons were made with several databases, including Online Mendelian Inheritance in Man (OMIM, https://www.omim.org/), the Database of Chromosomal Imbalance and Phenotype in Humans using Ensemble Resources (DECIPHER, https://www.deciphergenomics.org/), the Database of Genomic Variants (DGV, http://dgv.tcag.ca/dgv/app/home), the Clinical Genome Resource (ClinGen, https://www.clinicalgenome.org/), and the PubMed database (https://pubmed.ncbi.nlm.nih.gov/). The pathogenicity of the CNVs was classified according to the joint consensus recommendations of the American College of Medical Genetics and Genomics (ACMG) and ClinGen in 2020 [6].Karyotype and CNV analyses of the biological parents were conducted to assess the presence of structural abnormalities and to further evaluate the risk of recurrence.
Whole exome sequencing
Using the GenCap® whole exome gene capture probe V6.0 (MyGenostics, Beijing, China), the whole exome region of the DNA library was enriched. This was followed by high-throughput sequencing on the Illumina NovaSeq 500 after library construction. The sequencing data were aligned to the GRCh37/hg19 human reference genome using BWA v0.5.9 software. The mean sequencing depth achieved was 160X, with over 99.33% of the target coverage exceeding 20X. The Genome Analysis Toolkit (GATK) software was utilized to analyze single-nucleotide variations (SNVs), as well as small fragment insertions and deletions (InDels). Frequency variations of ≥ 5% were filtered out from the dbSNP 1000 Genomes, ExAC/gnomAD, and ESP6500 databases. Candidate gene variants were compared against The Online Catalog of Human Genes and Genetic Disorders (OMIM), The Human Gene Mutation Database (HGMD), ClinVar, Genomic Variants (DGV) MITOMAP, PubMed, and other relevant databases, along with clinical symptoms. Structural variants (SVs) were detected using Delly version 0.8.6 software, and variants were annotated with table_annovar.pl (annovar_2020). Protein structure prediction softwares, including Provean, SIFT, PolyPhen2, and Mutation Taster, were employed to assess the potential harmfulness of the variations. Pathogenicity was evaluated according to the criteria and guidelines for the interpretation of variations published by the ACMG in 2015 [7].
Results
Phenotype of the patient
The proband, a 2-year-old female, was referred to our hospital due to short stature and global developmental delay. She is the fourth child in her family, with two healthy older brothers and a healthy older sister. Her parents are non-consanguineous. The mother was 34 years old at conception, while the father was 37 years old. Prenatal screening was not completed as planned during the pregnancy. The proband was delivered vaginally at 39 weeks of gestation, with a length of 48 cm (< 25th percentile), a weight of 3.05 kg (< 50th percentile), and a head circumference of 32.5 cm (< 25th percentile), with no history of asphyxia. She experienced postnatal failure to thrive and recurrent respiratory infections. Laboratory analyses, including blood routine tests, blood gas analysis, thyroid hormone levels, and amino and organic acid assessments, were all within normal ranges. Brain magnetic resonance imaging (MRI) indicated delayed myelination. She was evaluated using the Developmental Scale for Children Aged 0–6 Years (WS/T 580–2017), developed by the Chinese National Health and Family Planning Commission, and obtained a developmental quotient of 32. Based on the Chinese guidelines for the diagnosis of Global Developmental Delay, she was subsequently diagnosed with GDD [8]. The proband exhibited characteristic features such as a prominent forehead, microcephaly, abnormal facial shape depressed nasal bridge, long philtrum, high palate and short stature. There was no reported history of sleep disorders or abnormal emotional behavior in the child. The patient’s last visit took place at the age of 3 years. At that time, she had acquired the ability to walk with assistance; however, she had not yet developed verbal communication skills, being able only to pronounce individual words. Physical examination revealed that the proband had a height of 80.5 cm (< 3rd percentile), a weight of 12.3 kg (< 3rd percentile), and a head circumference of 42 cm (< 3rd percentile).
Genetic and bioinformatics analysis
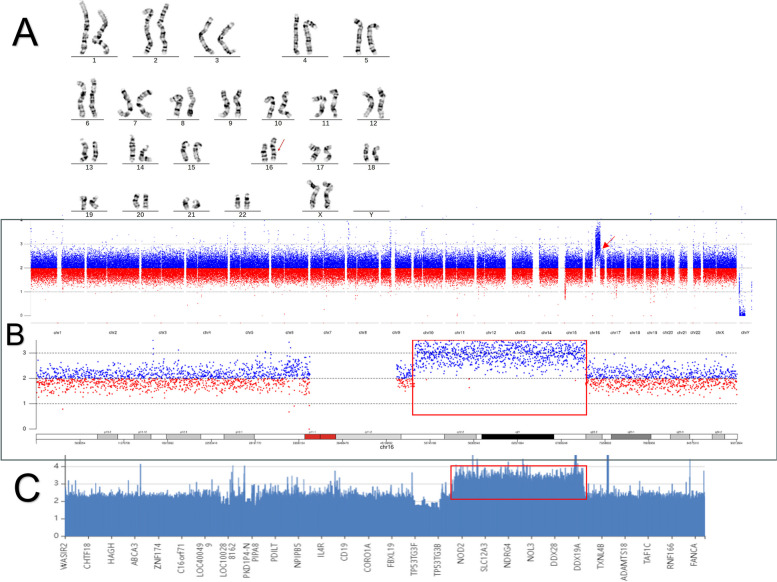
The proband karyotype analysis revealed a 46,XX, dup(16)(q12.1q22.1). The CNV-seq results indicated that the breakpoint of the duplication was located between seq[GRCh37] dup(16)(q12.1q22.1)NC_000016.9:g. 48799549_70756330dup, spanning 21.96 Mb. The WES results indicated that the breakpoint of the duplication was located between seq[GRCh37] dup(16)(q12.1q22.1)NC_000016.9:g.49313314_ 70762759dup, spanning 21.45 Mb (Fig. 1).
Fig. 1.
A karyotype analysis results of the 46,XX,dup(16)(q12.1q22.1) proband: the red arrow indicates chromosome 16 duplicated region; B CNVseq results of the proband: the red framed rectangle and the red arrow indicate the duplicated region; WES results of the proband: the red rectangle and the red arrow point to the duplicated region
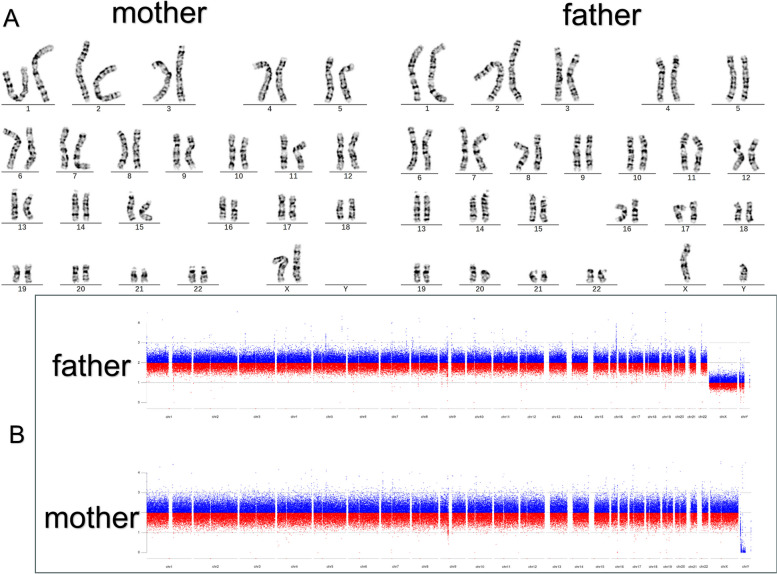
Both the karyotype, CNV-seq and WES results demonstrated that the proband’s parents, brothers and sister did not carry this CNV (Fig. 2).
Fig. 2.
A Karyotype analysis of the proband’s parents (Normal); B CNVseq of the proband’s parents (Normal)
A total of 193 protein-coding genes were identified within the duplicated region, and no records of genomic variants corresponding to this fragment were found in the Database of Genomic Variants (DGV) and DECIPHER databases. According to the CNV classification scoring standard, this variation received a score of 0.90 points, indicating that this region contains more than 50 coding genes, and was classified as a likely pathogenic variation. No genes or genomic regions associated with triplosensitivity (TS) exhibited a TS Score of 3 in this duplication segment. Nine OMIM genes (CTCF, CNOT1, VPS4A, GNAO1, VAC14, RSPRY1, CYLD, IRX5, and ZNF423) were identified with a predicted probability of triplosensitivity (pTriplo) score of ≥ 0.94, according to gnomAD (Table 1).
Table 1.
Genotypes and phenotypes associated with patient’s duplication in the OMIM database
| Gene | CTCF | CNOT1 | VPS4A | GNAO1 | VAC14 | RSPRY1 | CYLD | IRX5 | ZNF423 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OMIM | 604,167 | 604,917 | 609,982 | 139,311 | 604,632 | 616,585 | 605,018 | 606,195 | 604,557 | |||||||
| Cytogenetic | 16q22.1 | 16q21 | 16q22.1 | 16q13 | 16q22.1-q22.2 | 16q13 | 16q12.1 | 16q12.2 | 16q12.1 | |||||||
| Inheritance | AD | AD; | AD | AD | AD; | AD | AR | AR | AD | AR | AD, AR; | AD, AR | ||||
| Phenotype | Intellectual developmental disorder, autosomal dominant 21 | Holoprosencephaly 12, with or without pancreatic agenesis; | Vissers-Bodmer syndrome | CIMDAG syndrome | Developmental and epileptic encephalopathy 17 | Neurodevelopmental disorder with involuntary movements | Striatonigral degeneration, childhood-onset | Spondyloepimetaphyseal dysplasia | Faden-Alkuraya type | Frontotemporal dementia and/or amyotrophic lateral sclerosis 8 | Brooke-Spiegler syndrome | Cylindromatosis, familial | Trichoepithelioma, multiple familial, 1 | Hamamy syndrome | Joubert syndrome 19 | Nephronophthisis 14 |
| Dysfunction of Gene Product | DNA-binding transcription factor; regulating gene expression and chromatin organization | the central scaffolding protein (in)directly binding to all CCR4-NOT partners | critical enzyme regulating endosomal sorting complexes required for transport (ESCRT) function | encodes Gαo, the α subunit of Go; are involved as modulators or transducers in various transmembrane signaling systems | a trimolecular complex that tightly regulates the level of phosphatidylinositol 3,5-bisphosphate [PI (3,5) P2] | encodes a hypothetical RING and SPRY domain-containing protein | a ubiquitin hydrolase; specifically cleaves ‘Lys-63’- and linear ‘Met-1’-linked polyubiquitin chains and is involved in NF-kappa-B activation and TNF-alpha-induced necroptosis | Involved in craniofacial and gonadal development; the regulation of human ventricular depolarization and cardiac electrical conduction | a central role in BMP signaling and olfactory neurogenesis | |||||||
| a | 1 | 1 | 1 | 0.99 | 0.99 | 0.99 | 0.98 | 0.98 | 0.96 | |||||||
agnomAD (v2.1.1) pTriplo, pTriplo scores ≥ 0.94 indicate that the average effect sizes of duplications are as strong as the loss-of-function of genes known to be constrained against protein truncating variants [9]
WES analysis identified two heterozygous single nucleotide variants (SNVs) inherited from the patient’s healthy mother: SETD1A (NM_014712.3) c.1553_1555del (p.Ser518del) and RERE (NM_001042681.2) c.2105 A > G (p.Lys702Arg). The Mendelian mode of inheritance, along with family history and other factors, did not provide a clear explanation for the clinical phenotype observed in the patients. Consequently, the ACMG classified these variants as variants of uncertain significance. Based on the clinical phenotype GDD, abnormal facial shape and the results obtained on the karyotype, CNV-seq and WES., the proband was diagnosed to have a 16q12.1q22.1 duplication.
Discussion
Gains or losses of DNA fragments larger than 1 kb on chromosomes are referred to as copy number variations (CNVs), which are closely associated with neurodevelopmental disorders and various congenital abnormalities in children. Currently, there are numerous explanations for the pathogenesis of the clinical phenotype associated with partial trisomy syndromes. The dosage effect resulting from unbalanced translocations in chromosome function, along with the disruption of expression or coordination of specific genes or gene clusters due to chromosomal breaks and insertions, may influence gene expression to some extent, leading to corresponding phenotypic outcomes [10]. The majority of reported duplications on the long arm of chromosome 16 arise from structural rearrangements during meiosis. This complexity complicates the study of the relationship between genotype and phenotype in this disease [3]. The study of individuals with pure partial trisomy 16q eliminates the interference of additional genetic variants in phenotypic analysis, providing a unique opportunity to examine the phenotypic effects of genomic diseases.
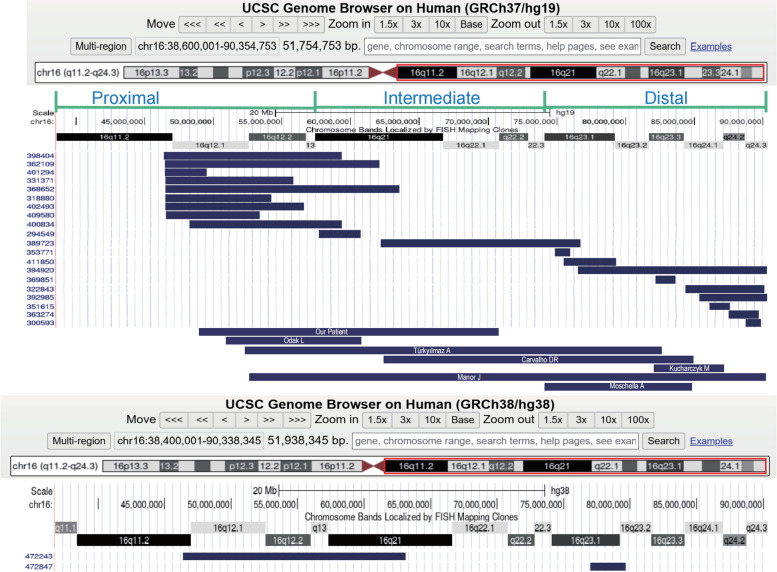
In our study, we conducted an analysis of a total of 29 individuals with overlapping pure partial duplications at 16q, which included 21 cases identified from the DECIPHER database and 7 case reports from the literature (Table 2)(Fig. 3).
Table 2.
The genotype and phenotype of 29 patients with pure partial trisomy 16q
| Serial number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| Coordinates (GRCh37, Hg19) | 46385802_59342499 | 46441545_62089933 | 46464489_49514582 | 46500511_55797192 | 46500741_63537111 | 46505617_54206543 | 46507694_56598191 | 46564557_53360707 | 48242500_59342499 | 57653652_60696431 | 62213235_76769398 | 74915119_76001300 | 75495683_79327457 | 76542500_90294753 | 82184809_83663330 |
| Size | 12.93Mb | 15.65Mb | 3.05Mb | 9.30Mb | 17.04Mb | 7.70Mb | 10.09Mb | 6.80Mb | 11.10Mb | 3.04MB | 14.56Mb | 1.09Mb | 3.83Mb | 13.72Mb | 1.48Mb |
| Regions | 16q11.2q21 | 16q11.2q21 | 16q11.2q12.1 | 16q11.2q12.2 | 16q11.2q21 | 16q11.2q12.2 | 16q11.2q12.2 | 16q11.2q12.2 | 16q12.1q21 | 16q21 | 16q21q23.1 | 16q23.1 | 16q23.1q23.2 | 16q23.1q24.3 | 16q23.3 |
| Abnormal facial shape | + | + | - | - | - | - | + | NA | + | + | + | + | NA | + | NA |
| GDD / ID | + | + | + | - | + | + | NA | + | - | + | + | + | NA | NA | NA |
| Growth abnormality | + | + | - | - | - | - | - | + | + | NA | - | - | NA | NA | NA |
| Behavior problems | |||||||||||||||
| Aggressive | + | - | - | - | + | - | NA | - | - | - | - | NA | NA | - | |
| Hyperactivity | + | - | + | - | - | + | - | NA | - | - | - | - | NA | NA | - |
| Autistic | - | - | - | + | - | - | NA | - | - | - | - | NA | NA | + | |
| Others malformation | |||||||||||||||
| Limbs / musculoskeletal | + | + | - | - | NA | - | - | NA | + | - | + | - | NA | + | NA |
| Hypotonia | + | - | - | - | NA | - | - | NA | - | - | - | - | NA | + | NA |
| Hemiplegia | + | - | - | - | NA | - | - | NA | - | - | - | - | NA | - | NA |
| Cardiovascular | - | + | + | + | NA | - | - | NA | - | - | - | - | NA | - | NA |
| Digestive | NA | + | - | - | NA | - | - | NA | - | - | - | - | NA | - | NA |
| Eye | NA | + | NA | - | NA | - | + | NA | + | - | - | NA | - | NA | |
| Hearing / Dysplastic | - | - | - | - | + | NA | - | - | - | - | NA | - | NA | ||
| Genitourinary | NA | + | NA | - | NA | - | - | NA | - | - | - | - | NA | - | NA |
| Integument | NA | + | NA | - | NA | - | - | NA | - | - | - | - | NA | - | NA |
| Endocrine | NA | - | NA | + | NA | - | - | NA | - | - | - | - | NA | - | NA |
| Macrocephaly | NA | - | NA | + | - | - | + | NA | - | - | - | - | NA | - | NA |
| Craniosynostosis | NA | - | NA | - | + | - | + | NA | - | - | - | - | NA | - | NA |
| Dental structure | NA | - | NA | - | - | + | - | NA | - | + | - | - | NA | - | NA |
| Obesity | NA | - | NA | - | - | - | + | NA | - | - | - | - | NA | - | NA |
| Premature birth | NA | - | NA | - | - | - | - | NA | - | - | - | - | NA | + | NA |
| EEG / Seizure | NA | - | NA | - | - | - | - | NA | + | - | - | - | NA | - | NA |
| Recurrent infections | NA | - | NA | - | - | - | - | NA | - | - | - | - | NA | - | NA |
| References | 398404 | 362109 | 401294 | 331371 | 368652 | 318880 | 402493 | 409580 | 400834 | 294549 | 389723 | 353771 | 411850 | 394920 | 369851 |
| Serial number | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | Serial number | 28 | 29 |
| Coordinates (GRCh37, Hg19) | 84341219_90111263 | 85342500_90294753 | 86106889_87595215 | 87502161_89688617 | 88755341_89897010 | 48799549_70756330 | 50843158_60770795 | 52459169_82285105 | 62586414_84885185 | 84730437_85477564; 85483466_88647238 | NA | 74005024_84546108 | coordinates (GRCh38, Hg38) | 46450037_63178736 | 77045569_79687003 |
| Size | 5.77Mb | 4.95Mb | 1.49Mb | 2.19Mb | 1.14Mb | 21.96Mb | 9.92 Mb | 29.8Mb | 22.3Mb | 747Kb;3.30Mb | 33.5Mb | 10.5 Mb | Size | 16.73 Mb | 2.64Mb |
| Regions | 16q24.1q24.3 | 16q24.1q24.3 | 16q24.1q24.2 | 16q24.2q24.3 | 16q24.3 | 16q12.1q22.1 | 16q12.1q21 | 16q12.1q23.3 | 16q21q24.1 | 16q24.1-q24.3. | 16q12.2q24.3 | 16q22.3q24.1 | Regions | 16q11.2q21 | 16q23.1q23.2 |
| Abnormal facial shape | NA | + | NA | + | NA | + | + | + | + | + | + | + | Abnormal facial shape | NA | NA |
| GDD / ID | NA | NA | NA | + | NA | + | + | + | + | + | + | + | GDD / ID | NA | NA |
| Growth abnormality | NA | NA | NA | - | NA | + | - | + | - | + | + | - | Growth abnormality | NA | NA |
| Behavior problems | Behavior problems | ||||||||||||||
| Aggressive | NA | - | NA | NA | NA | - | + | NA | - | NA | NA | - | Aggressive | NA | NA |
| Hyperactivity | NA | - | NA | NA | NA | - | - | NA | - | NA | NA | - | Hyperactivity | NA | NA |
| Autistic | NA | - | NA | NA | NA | - | - | NA | - | NA | NA | - | Autistic | NA | NA |
| Others malformation | Others Malformation | ||||||||||||||
| Limbs / musculoskeletal | - | + | NA | NA | NA | + | - | + | + | + | - | Limbs / musculoskeletal | NA | NA | |
| Hypotonia | - | - | NA | NA | NA | - | + | - | - | - | - | Hypotonia | NA | NA | |
| Hemiplegia | - | - | NA | NA | NA | - | - | - | - | - | - | Hemiplegia | NA | NA | |
| Cardiovascular | - | + | NA | NA | NA | - | - | + | + | + | - | Cardiovascular | NA | NA | |
| Digestive | - | + | NA | NA | NA | - | - | + | + | - | - | Digestive | NA | NA | |
| Eye | - | - | NA | NA | NA | - | + | - | + | - | Eye | NA | NA | ||
| Hearing / Dysplastic | - | + | NA | NA | NA | + | + | + | + | + | - | - | Hearing / Dysplastic | NA | NA |
| Genitourinary | - | + | NA | NA | NA | - | - | + | + | + | - | Genitourinary | NA | NA | |
| Integument | - | + | NA | NA | NA | - | - | - | - | - | - | Integument | NA | NA | |
| Endocrine | - | - | NA | NA | NA | - | - | + | - | - | - | Endocrine | NA | NA | |
| Macrocephaly | - | - | NA | NA | NA | - | - | - | - | - | - | Macrocephaly | NA | NA | |
| Craniosynostosis | - | - | NA | NA | NA | - | - | - | - | - | - | Craniosynostosis | NA | NA | |
| Dental structure | - | - | NA | NA | NA | - | - | - | - | - | Dental structure | NA | NA | ||
| Obesity | - | - | NA | NA | NA | + | - | - | - | - | + | Obesity | NA | NA | |
| Premature birth | - | - | NA | NA | NA | - | + | - | - | + | + | Premature birth | NA | NA | |
| EEG / Seizure | + | - | NA | NA | NA | - | - | - | + | - | - | - | EEG / Seizure | NA | NA |
| Recurrent infections | - | - | NA | NA | NA | + | + | - | - | - | - | - | Recurrent infections | NA | NA |
| References | 322843 | 392985 | 351615 | 363274 | 300593 | This study | [11] | [12] | [13] | [14] | [15] | [16] | References | 472243 | 472847 |
EEG Electroencephalogram, GDD global developmental delay, ID intellectual disability, NA not applicable, + present, – absent
Fig. 3.
Genomic locations of 29 duplications based on the results obtained in table 2. The annotation is based on GRCh37/hg19 and GRCh38/hg38
With reference to the study by Lonardo F et al. [3] (GRCh37/hg19), we divided the duplications of the 29 carriers into three regions: proximal region [GRCh37](16q11.1q13)NC_000016.9:g. (36600001_57400000); intermediate region [GRCh37](16q21q22.3)NC_000016.9:g.(57400001_74100000); and distal region [GRCh37](16q23.1q24.3) NC_000016.9:g.(74100001_90354753). The duplications among the 29 instances range in size from 1.09 (#12) to 33.5 Mb (#26). 11 individuals (#3, #10, #12, #13, #15, #17, #18, #19, #20, #25, #29) had duplicated segment sizes of less than 5 Mb, while 5 individuals (#4, #6, #8, #22) had duplicated segment sizes between 5 and 10 Mb. Additionally, 9 individuals (#1, #2, #5, #7, #9, #11, #14, #17, #28) had duplicated segment sizes greater than 10 Mb, and 4 individuals (#21, #23, #24, #26) had duplicated segment sizes greater than 22 Mb. Our summary of the 29 individuals revealed that partial trisomic 16q CNVs were concentrated at the proximal end, with 11 individuals (#1 to #9, #27) having proximal duplications, 4 individuals (#10, #11, #21, #24) with proximal-intermediate/interstitial duplications, and 11 individuals (#12-#20, #25, #26, #28) with distal duplications. The most prevalent clinical phenotypes observed were global developmental delay/intellectual disability (GDD/ID) in 20 out of 21 individuals, abnormal facial shape in 14 out of 21, and short stature in 8 out of 19. Two individuals (#23, #26) spanned all three duplication regions. The three individuals with the most severe phenotypes (#17, #25, #26) exhibited multiorgan involvement, affecting systems such as the cardiovascular, digestive, and genitourinary systems, with their duplication regions located at the distal regions. In Table 2, only 3 individuals (#4, #5, #6) lacked phenotype descriptions of GDD or ID, and their mutation coordinates were situated at the proximal regions. This suggests that individuals with distal 16q repeats may experience more severe symptoms than those with proximal duplication, potentially due to a higher density of coding genes in this region (Fig. 3).
The duplication region in our study (#21) completely encompassed the duplication regions observed in cases #10 and #22. The common phenotypes among these cases included GDD and abnormal facial morphology. In contrast, case #22 exhibited additional phenotypes such as aggression and skeletal abnormalities. Four individuals with duplications at the distal 16q2.1 presented with hearing impairment (#23, #24, #25, #26), whereas only case #7 displayed conductive hearing impairment at the proximal end. Furthermore, we noted that the duplication regions and fragment sizes in cases #16 and #17 were similar; however, case #17 presented with a more severe clinical phenotype, thereby underscoring the clinical heterogeneity of this condition. Notably, most patients with larger fragment sizes (#21, #23, #24) did not exhibit more deformities than those with smaller fragment (#4, #6, #8, #22, #17). It is speculated that the phenotype of patients with 16q duplications is more closely reflects the location than the size of the duplication. The only observed duplication-triplication pattern is #25. Additional data are required to substantiate the notion that there are dose-effect genes linked to more severe phenotypes, akin to those seen in MECP2 duplication syndrome, for this complex genomic rearrangement.
We searched the coding genes within the [GRCh37](16q11.1q24.3)NC_000016.9:g.(38600001_90354753) region and identified four neurodevelopment-related genes with pLI and pTriplos scores exceeding 0.99. The GNAO1, CNOT1, and CTCF genes, as documented in OMIM/Morbid, the Gene Curation Coalition, Gene2Phenotype (G2P), and ClinGen, were classified as having definitive pathogenicity, associated with autosomal dominant (AD) inheritance. The AP1G1 gene was also identified as monoallelic by OMIM/Morbid, the Gene Curation Coalition, G2P, and ClinGen, indicating strong evidence of pathogenicity. Among them, the GNAO1 (NM_020988.3) and CNOT1 (NM_016284.5) genes play crucial roles in regulating neurotransmitter release, movement, and neural development. Variants in MYLK3 (NM_182493.3) may be associated with cardiac abnormalities #3, #4, and #5, as the protein encoded by this gene promotes sarcomere formation in cardiomyocytes and enhances cardiomyocyte contractility [17]. Notably, no duplication variants in the aforementioned five genes have been reported to cause disease. Thus, our data do not provide evidence supporting the identification of a possible smallest region of overlap. The various features of trisomy 16q have yet to be attributed to specific genes.
In laboratory settings, CMA and CNV-seq technologies are primarily employed for the analysis of 16q trisomic CNV. The CNV-seq platform generally conducts preliminary screening with a threshold of 100 kb. CNV-seq is influenced by the minimum partition region (Bin) algorithm, which can lead to the detection of CNV fragments that are smaller than their actual size. With advancements in high-throughput sequencing, the costs associated with WES and whole genome sequencing (WGS) have significantly decreased. These methods offer extensive coverage and high efficiency, positioning them as potential preferred diagnostic tests for CNV. These imbalances diminish the sensitivity of WES in specific regions, complicating the accurate assessment of genomic breakpoint locations associated with extensive CNVs, which ultimately limits the ability to determine the true extent of copy number variation [18]. This observation was also evident in our case, where a 0.5 MB difference in CNV duplication size was noted between WES and CNV-seq analyses. In the case we examined, we recommend identification through fluorescent in situ hybridization (FISH) analysis to determine the orientation of proband’s duplicated segments. The proband’s parents, who have three healthy offspring and no plans for additional children, rejected this suggestion. Identifying the critical phenotype-driving genes presents a challenge in the study of pathogenic CNVs. We hypothesize that the pathogenicity associated with a portion of 16q trisomy may be related to continuous gene duplication; however, functional studies are necessary to establish this association.
When a patient presents with global developmental delay/intellectual disability, abnormal facial shape, and involvement of other systems, primary care providers may suspect a genetically related disorder. However, the lack of specialized genetic knowledge and equipment often necessitates a referral to academic medical centers [19]. The uneven distribution of medical resources is a pervasive issue across all regions, and the referral process for patients in remote areas is influenced by various factors, including economic conditions, transportation, and cultural customs. These factors can result in delays in the diagnosis of rare diseases, whether for extended or brief periods. Our patient, who resides in the high mountains and plains of western Sichuan, was diagnosed with global developmental delay in infancy; however, the genetic diagnosis was not confirmed until the age of two. This delay is closely related to the complex referral process for patients. In our medical center, the age at which rare diseases are diagnosed in children living in urban areas is generally younger than in those from remote regions, suggesting that the timing of rare disease diagnosis is influenced by the residential environment. Currently, rare chromosomal disorders, such as trisomy 16q, lack precision medicine and depend on multidisciplinary collaboration to enhance patients’ quality of life. Given the shortage of trained geneticists in developing countries and the limited access clinicians have to assess such patients, relatively efficient and cost-effective approaches to genetic testing are particularly crucial. These approaches not only assist patients in identifying the underlying causes and managing follow-up care but also offer effective genetic counseling for families.
Conclusions
In summary, we report a de novo case of pure partial trisomy 16, which was diagnosed through the mutual verification of karyotype, CNV-seq, and WES. This case enriches the genotypic spectrum of pure partial trisomy 16q. Additionally, we conducted a detailed analysis of 28 cases collected from the literature to raise clinicians’ awareness of this condition. In rare chromosomal diseases, such as partial trisomy 16q, complementary cytogenetic and molecular genetic methods are essential for identifying the effects of copy number loss or gain on the gene dosage map at the DNA sequence level. This understanding is crucial for enhancing clinical genetic counseling. Further research is needed to understand the implications of dosage-sensitive genes associated with partial trisomy of 16q.
Acknowledgements
Thanks to Dr. Heping Fang for reading and reviewing this article. We are grateful to the family for their willing participation and cooperation. Thanks to our family (Mingrong Zhu, Wei Yang, Fenglin Hang, Sha Yang, Jiang Du, Yile Du, Lele Huang) for their company and support for my work.
Authors’ contributions
TD, CA, ZL and ZS collected and integrated data, and wrote the article; ZL and ZS participated in the study’s design and coordination; XJ, HY, FJ and PG interpreted the results of the bone marrow cell smear; WJ and ZL explained the pathology report; ZL, WJ and CA explained the chromosome and WES results; ZS, XF, LG and YL participated in the evaluation of the patient. All authors reviewed the article critically for intellectual content and agreed to the published version of the manuscript.
Funding
This research was funded by Chengdu Bureau of Science and Technology Project (2021-YF05-01658-SN). This study is supported by a grant from the Natural Science Foundation of Sichuan Province (No. 2022NSFSC0784).
Data availability
The datasets generated and/or analysed during the current study are available in the ClinVar repository, VCV003255478.1 - ClinVar - NCBI (nih.gov).
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Ethics Committee of Sichuan Provincial Maternity and Child Health Care Hospital (Protocol 20230911-225 and date of 2023. 09. 11). Written informed consent was obtained from the parents of the patient.
Consent for publication
Written informed consent was obtained from patient’s parents for publication of the details of their medical case and any accompanying images.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lan Zeng and Shuyao Zhu contributed equally to this work.
Contributor Information
Lan Zeng, Email: 277138348@qq.com.
Shuyao Zhu, Email: 330986673@qq.com.
References
- 1.Savary JB, Vasseur F, Manouvrier S, Daudignon A, Lemaire O, Thieuleux M, Poher M, Lequien P, Deminatti MM. Trisomy 16q23?Qter arising from a maternal t(13;16)(p12;q23): case report and evidence of the reciprocal balanced maternal rearrangement by the Ag-NOR technique. Hum Genet. 1991;88(1):115–8. [DOI] [PubMed] [Google Scholar]
- 2.Brisset S, Joly G, Ozilou C, Lapierre JM, Gosset P, LeLorc’h M, Raoul O, Turleau C, Vekemans M, Romana SP. Molecular characterization of partial trisomy 16q24.1-qter: clinical report and review of the literature. Am J Med Genet. 2002;113(4):339–45. [DOI] [PubMed] [Google Scholar]
- 3.Lonardo F, Perone L, Maioli M, Ciavarella M, Ciccone R, Monica MD, Lombardi C, Forino L, Cantalupo G, Masella L, et al. Clinical, cytogenetic and molecular-cytogenetic characterization of a patient with a de novo tandem proximal-intermediate duplication of 16q and review of the literature. Am J Med Genet Part A. 2011;155(4):769–77. [DOI] [PubMed] [Google Scholar]
- 4.Martin J, Han C, Gordon LA, Terry A, Prabhakar S, She X, Xie G, Hellsten U, Chan YM, Altherr M, et al. The sequence and analysis of duplication-rich human chromosome 16. Nature. 2004;432(7020):988–94. [DOI] [PubMed] [Google Scholar]
- 5.Redaelli S, Maitz S, Crosti F, Sala E, Villa N, Spaccini L, Selicorni A, Rigoldi M, Conconi D, Dalprà L, et al. Refining the phenotype of recurrent rearrangements of chromosome 16. Int J Mol Sci. 2019;20(5):1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, Raca G, Ritter DI, South ST, Thorland EC, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Sci. 2020;22(2):245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Sci. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Children′s Health Care Center BCsH. Capital Medical University, National Center for Children′s Health, Center BPSQCaI, Committee CMaCHACBSaBHP, Committee BHCPACaAHDP, Committee MaCHRIAPR, the Subspecialty Group of Developmental and behavioral pediatrics tSoP, Chinese Medical Association: Chinese guideline for the diagnosis of global developmental delay. Chin J Appl Clin Pediatr. 2024;37(9):481489. [Google Scholar]
- 9.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krivega M, Stiefel CM, Storchova Z. Consequences of chromosome gain: a new view on trisomy syndromes. Am J Hum Genet. 2022;109(12):2126–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odak L, Barišić I, Morožin Pohovski L, Riegel M, Schinzel A. Novel duplication on chromosome 16 (q12.1-q21) associated with behavioral disorder, mild cognitive impairment, speech delay, and dysmorphic features: case report. Croatian Med J. 2011;52(3):415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Türkyılmaz A, Yaralı O. A very rare partial trisomy syndrome: De novo duplication of 16q12.1q23.3 in a Turkish girl with developmental delay and facial dysmorphic features. Balkan J Med Genet. 2020;23(1):103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho DR, Moretto ALL, Schneider M, Formigli LM. Clinical features of de novo pure 16q21q24.1 chromosome duplication. Cytogenet Genome Res. 2021;161(3–4):160–6. [DOI] [PubMed] [Google Scholar]
- 14.Kucharczyk M, Kochański A, Jezela-Stanek A, Kugaudo M, Sielska‐Rotblum D, Gutkowska A, Krajewska‐Walasek M. The first case of a patient with de novo partial distal 16q tetrasomy and a data’s review. Am J Med Genet Part A. 2014;164(10):2541–50. [DOI] [PubMed] [Google Scholar]
- 15.Manor J, Dinu D, Azamian MS, Bi W, Darilek S, Lalani SR. A rare description of pure partial trisomy of 16q12.2q24.3 and review of the literature. Am J Med Genet Part A. 2021;185(10):2903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moschella A, Capra AP, Corica D, Pepe G, Di Tommaso S, Sallicandro E, Wasniewska MG, Briuglia S, Aversa T. A novel case of 16q22.3 duplication syndrome in a child with overgrowth: case report and literature review. BMC Med Genom. 2023;16(1):315. [DOI] [PMC free article] [PubMed]
- 17.Hitsumoto T, Tsukamoto O, Matsuoka K, Li J, Liu L, Kuramoto Y, Higo S, Ogawa S, Fujino N, Yoshida S, et al. Restoration of Cardiac myosin light chain kinase ameliorates systolic dysfunction by reducing Superrelaxed myosin. Circulation. 2023;147(25):1902–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lelieveld SH, Spielmann M, Mundlos S, Veltman JA, Gilissen C. Comparison of exome and Genome Sequencing Technologies for the Complete capture of protein-coding regions. Hum Mutat. 2015;36(8):815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen ASA, Berrios CD, Zion TN, Barrett CM, Moore R, Boillat E, Belden B, Farrow EG, Thiffault I, Zuccarelli BD, et al. Genomic answers for kids: toward more equitable access to genomic testing for rare diseases in rural populations. Am J Hum Genet. 2024;111(5):825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the ClinVar repository, VCV003255478.1 - ClinVar - NCBI (nih.gov).