Abstract
Background
Cellular prion protein (PrPC) is a widely expressed membrane-anchored glycoprotein, which has been associated with the development and progression of several types of human malignancies, controlling cancer stem cell activity. However, the different molecular mechanisms regulated by PrPC in normal and tumor cells have not been characterized yet.
Methods
To assess the role of PrPC in patient-derived glioblastoma stem cell (GSC)-enriched cultures, we generated cell lines in which PrPC was either overexpressed or down-regulated and investigated, in 2D and 3D cultures, its role in cell proliferation, migration, and invasion. We evaluated the role of PrPC in supporting GSC stemness and the intracellular signaling involved using qRT-PCR, immunocytofluorescence, and Western blot.
Results
Stable PrPC down-regulation leads to a significant reduction of GSC proliferation, migration, and invasiveness. These effects were associated with the inhibition of the expression of stemness genes and overexpression of differentiation markers. At molecular level PrPC down-regulation caused a significant inhibition of Wnt/β-catenin pathway, through a reduced expression of Wnt and Frizzled ligand/receptor subtypes, resulting in the inhibition of β‐catenin transcriptional activity, as demonstrated by the reduced expression of its target genes. The specificity of PrPC in these effects was demonstrated by rescuing the phenotype and the biological activity of PrPC down-regulated GSCs by re-expressing the protein. To get insights into the distinct mechanisms by which PrPC regulates proliferation in GSCs, but not in normal astrocytes, we analyzed structural features of PrPC in glioma stem cells and astrocytes using Western blot and immunofluorescence techniques. Using Pi-PLC, an enzyme that cleaves GPI anchors, we show that, in GSCs, PrP is retained within the plasma membrane in an immature Pro-PrP isoform whereas in astrocytes, it is expressed in its mature PrPC form, anchored on the extracellular face of the plasma membrane.
Conclusions
The persistence of Pro-PrP in GSCs is an altered cellular mechanism responsible of the aberrant, constitutive activation of Wnt/β-catenin pathway, which contributes to glioblastoma malignant features. Thus, the activity of Pro-PrP may represent a targetable vulnerability in glioblastoma cells, offering a novel approach for differentiating and eradicating glioblastoma stem cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03581-1.
Keywords: Glioblastoma stem cells; Prion protein; Pro-prion, Wnt/β-catenin pathway.
Background
Glioblastoma (GBM, WHO grade 4 glioma) is the most aggressive primary brain tumor [1]. The mainstay GBM treatment, comprising maximal surgical resection, radiotherapy, and chemotherapy with temozolomide [2], is non-effective and unavoidably tumors relapse due to the rapid brain invasion. The infiltrative nature of GBM is dependent on the presence of GBM stem cells (GSCs), a small cell population responsible for tumor development, growth, and brain invasion, conferring GBM chemo- and radio-resistance [3, 4]. Thus, GSC eradication is mandatory for effective therapies. However, despite preclinical knowledge of GSC biology and pharmacology greatly improved in recent years, the approved therapeutic approaches did not parallel these advances, and patients’ life expectancy has only marginally increased.
One crucial strategy to eradicate GSCs is the targeting of the intracellular signaling pathways that control stem cell-like features. Noteworthy, cancer stem cells proliferation is dependent on proteins previously considered non-involved in cell cycle progression. Among these, cellular prion protein (PrPC) is crucial to grant GSCs with stemness properties [5–8].
PrPC is an extracellular, cell membrane-anchored glycoprotein whose physiological role is still not completely understood [9, 10]. PrPC is widely expressed in the central nervous system (CNS), but it is also present in peripheral tissues (bone marrow, lymphoid organs, heart, skeletal muscles, lung). A 3D structurally altered PrPC conformer (PrPSc) is the causal agent of prion diseases, a group of fatal and transmissible neurodegenerative diseases (e.g., Creutzfeldt–Jakob disease) [11]. Misfolded PrPSc accumulates within the brain as intra- and extra-cellular amyloid-like aggregates, causing neurotoxicity and astrogliosis and providing these diseases with transmissible features [12–16].
In both CNS and hematopoietic cells, PrPC controls the balance between cell survival and death, and proliferation and differentiation of stem cells [8, 17], suggesting its possible involvement in cancer biology [18]. Indeed, PrPC favors doxorubicin resistance and in vivo invasiveness in colorectal and breast cancers [19–23].
In normal cells, neo-synthesized, immature pro-prion (Pro-PrP) undergoes to endo-proteolytic processing in the endoplasmic reticulum and Golgi, to remove the C-terminal signal peptide, which is substituted by a glycosylphosphatidylinositol (GPI) anchor, and the insertion of one or two N-linked oligosaccharide residues, before being translocated to the plasma membrane, forming the mature GPI-anchored PrPC [24]. This process is altered in some tumors (e.g., pancreatic ductal carcinoma and melanoma) [25], leading to the retention of Pro-PrP as major PrP isoform. Pro-PrP contains the C-terminal GPI anchor peptide, a domain rich in hydrophobic amino acids that causes its retention within plasma membrane. PrPC expression as Pro-PrP occurs in ~ 40% of the pancreatic cancers but not in non-neoplastic pancreatic tissues [26], and, in an experimental setting, it favors the metastatic diffusion of melanoma cells [27].
Although PrPC is highly expressed in CNS cells [28], only few studies reported its expression in GBM [29, 30]. On the basis of the observation that PrPC controls stemness/differentiation equilibrium in normal and tumor cells [31, 32], we previously characterized the role of PrPC in sustaining stem-like features of GSCs, showing that its downregulation inhibits GSC proliferation and self-renewal, and abolishes their tumorigenicity [6].
Studies investigating the molecular pathways controlled by PrPC to induce its biological effects in normal and cancer cells showed its interaction with different partners: CD44 [33], stress-inducible protein 1 [34, 35], laminin [36, 37], Notch [26, 38], and components of the Wnt/β-catenin pathway. In particular, Wnt/β-catenin signaling was identified as a central pathway mediating the effects of PrPC in intestinal epithelial cells, showing an interaction between PrPC and β-catenin/transcription factor 7–like 2 (TCF7L2) complex [39]. Wnt/β-catenin pathway (or canonical Wnt pathway) directs the early phases of brain development, regulating neuronal migration and polarization, axon guidance and dendrite development [40]. Moreover, in cancer stem cell, it controls proliferation, self-renewal, stemness maintenance, and epithelial-to-mesenchymal transition (EMT) [41]. Wnts compose a family of secreted autocrine/paracrine ligands that, upon binding to Frizzled (FZD) receptors, promote β-catenin transcriptional activity preventing its phosphorylation and proteolytic degradation. β-catenin-target genes include c-Myc, cyclin D1, T cell factor 1 (TCF1), matrix metallopeptidase 7 (MMP7), axin2, and CD44, among others, indicating that the maintenance of stem cell properties in normal tissues is one of the major physiological roles of Wnt/β-catenin pathway. In contrast, overactive Wnt/β-catenin pathway is a hallmark of cancer stem cells, controlling tumor development, progression, and invasiveness [42, 43].
Here, we characterized the relationship between PrPC and Wnt/β-catenin pathway in GSCs isolated from 5 human GBMs and stably transfected to overexpress or downregulate PrPC. Moreover, the presence of alterations in PrPC topology (i.e., the persistence of Pro-PrP) was assessed as molecular determinant of the GSC malignant phenotype. We demonstrate that the expression of Pro-PrP is essential to sustain GSC malignancy through the overstimulation of the canonical Wnt pathway.
Materials and methods
Materials
Primary and secondary antibodies are reported in Table S3. R-Spondin-1 was obtained from Sigma-Aldrich (Milano, Italy). "Development-WNT signaling pathway. Part 2 H96 Predesigned 96-well panel for use with SYBR® Green" was purchased from BioRad (Milano, Italy) and used for mRNA array, following manufacturer’s instructions.
Human GBM specimens and establishment of primary cultures
Human GBM specimens were obtained from the Neurosurgery Dept. of IRCCS Ospedale Policlinico San Martino (Genova, Italy), after Institutional Ethical Committee approval (CER Liguria 360/2019) and patients’ informed consent. All samples were diagnosed as primary GBM WHO grade 4 (Table S1). All patients underwent surgery for the first time and had not received prior chemo- or radiotherapy. GBM primary cultures were obtained by mechanical dissociation of specimens followed by filtration through a 40 μm strainer (BD Biosciences, Buccinasco, Italy) to remove aggregates, and cultured as previously described [44].
GSC-enriched culture
GSCs were selected from primary cultures using stem cell-permissive medium [DMEM-F12/Neurobasal (1:1) (Gibco/Thermo Fischer Scientific) supplemented with 1X B27 (Gibco/Thermo Fischer Scientific), 2 mM L-glutamine (EuroClone) 1% penicillin/streptomycin (EuroClone), insulin 15 µg/ml (Sigma-Aldrich) heparin 2 µg/ml (Sigma-Aldrich), 10 ng/ml bFGF and 20 ng/ml EGF (Miltenyi Biotec)]. Cells were grown as monolayer on Matrigel (BD Biosciences), as reported [45]. In these conditions, cells retain main GSC features, as routinely assessed by stem cell marker expression and in vivo tumorigenicity after orthotopic injection of 10,000 neurosphere-derived cells in NOD/SCID mice [6, 44, 46].
Rat astrocyte primary cultures
Sprague–Dawley rat astrocytes were obtained by 7-day old pup cerebral cortices, isolated according to standard procedures [47]. Experimental procedures and animal care complied with the EU Parliament and Council Directive (2010/63/EU) and were approved by the Italian Ministry of Health (prot. 75F11.N.6DX) in accordance with D.M. 116/1992. All efforts were made to minimize animal suffering and to reduce the number of animals used for the experiments.
Gene expression profiling interactive analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn), a web-based data mining platform with large RNA sequencing data from The Cancer Genome Atlas (TCGA) [48] was used to estimate. by Kaplan–Meier survival curves. the effect of PRNP gene expression level on tumor prognosis; low-grade glioma (LGG) and GBM samples were divided into two groups (high and low expression groups) according to the median level of expression of PRNP. Overall survival (OS) and disease-free survival (DFS) were generated, and the hazard ratio (HR) and P-value based on Cox PH Model calculated. P value < 0.05 indicated statistical significance.
RNA sequencing
RNA was extracted from 106 GSCs using Qiagen RNeasy mini kit following manufacturer’s recommendations. RNA Quality was checked with Agilent Tape Station and ranged between RIN 8.6 to 9.9. Library preparation was carried out with Truseq stranded mRNA (Illumina) and sequenced on Illumina platform with 150 cycles paired end flow cells [49]. Up to 80Mreads were obtained for each sample. Reads were aligned with STAR 2.7.5c [50] and counted with RSEM 1.3.2 [51] using GRCh38.p13 as reference genome. RNA-seq data have been deposited at NCBI Geo data set (GSE179356) [52].
Stable silencing and overexpression of PrP in GSCs cultures
GSCs were transfected with hPrP short hairpin RNA (shRNA) Plasmid (Santa Cruz Biotechnology), containing a pool of 3 target-specific plasmids each encoding 19–25 nt (plus hairpin) shRNAs, designed to knock-down gene expression. For each transfection, “solution 1” (1 µg of plasmid DNA resuspended in 10 µl and mixed with 90 Plasmid Transfection Medium) and “solution 2” (4 µl of Plasmid Transfection Reagent with 96 µl Plasmid Transfection Medium) are mixed and incubated for 40 min at room temperature. After cell wash with 2 ml of Transfection Medium, 200 µl Plasmid DNA/ Plasmid Transfection Reagent Complex (Solution 1 + Solution 2) is added dropwise to each well. Cells are incubated for 7 h at 37 °C in a CO2 incubator under normal culture conditions. Following incubation, 1 ml of normal growth medium containing 2 times the normal serum and antibiotics concentration is added, and cells are incubated for additional 18–24 h. Stably transfected PrPC cultures were selected by puromycin (5 µg/ml) [6]. The same procedure was performed with control shRNA plasmids encoding a scrambled shRNA sequence that will not lead to the specific degradation of any known cellular mRNA. Using this protocol, we generated knock-down GSCs (GBM KD) and control GSCs (GBM SCR).
GSCs were also transfected with human PRNP natural open reading frame (ORF) mammalian expression plasmid (Sino Biological Inc.), to stably overexpress PrPC (GBM OV); the same plasmid was used to revert PrPC down-regulation in GBM KD cells (GBM REV). Transfected cells were selected by hygromycin resistance. Differential PrPC expression among the transfected GSC cultures (GBM SCR, KD, OV, and REV) from 5 GBMs, evaluated by qRT-PCR and Western blot, is reported in Supplementary Figs. 1 and 2.
Fig. 1.
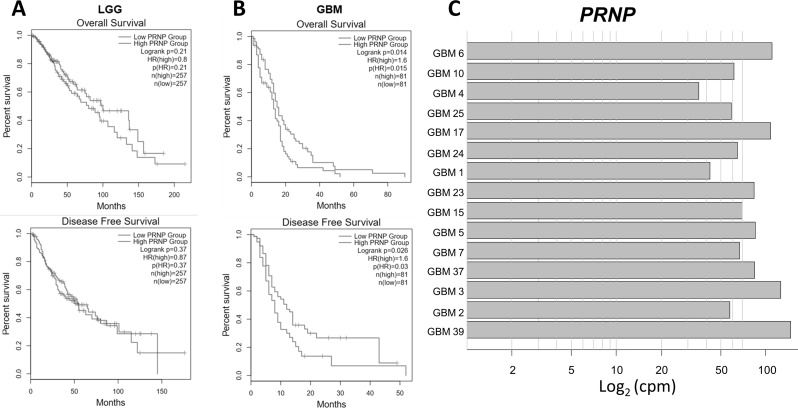
Prognostic significance of PrPC expression in GBM and transcript levels in patient-derived GSC cultures. Kaplan–Meier survival analysis of overall survival and disease-free survival of low-grade gliomas (LGG) (total 514 patients) (A) and GBM (total 162 patients) (B) with high or low expression level of PRNP, using GEPIA (http://gepia2.cancer-pku.cn/#index) in TCGA database (https://portal.gdc.cancer.gov/). A statistically significant difference was observed only in GBM (OS: P = 0.014; PFS: P = 0.026). C) PRNP transcript levels in GSCs isolated from 15 human GBMs. Data was obtained by RNA-seq, and expressed as counts per million reads mapped (cpm)
Fig. 2.
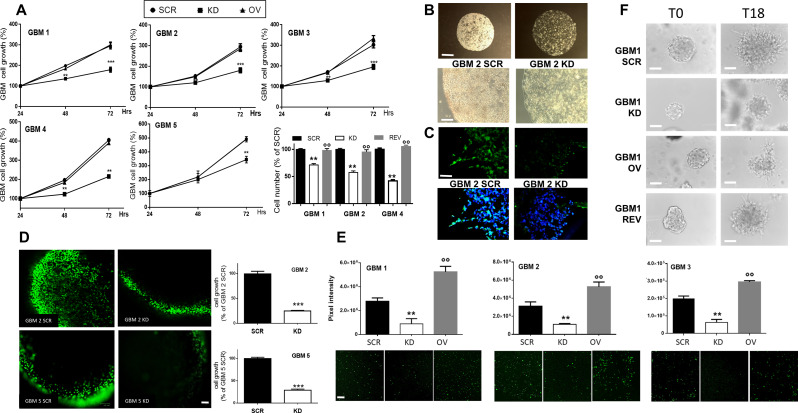
PrPC expression modulates GSC proliferation in 2D and 3D cultures. A) Growth curves of GBM 1–5 GSC cultures, comparing GBM SCR, KD and OV. Proliferation rate was evaluated 24-48-72 h from plating, by MTT assay. Each point represents the mean ± SD (n = 3, each performed in quadruplicate). ** p < 0.01 *** p < 0.001 vs. both GBM SCR and OV (t-test). Histogram represents viable cell number counts, using Trypan bleu exclusion test from GBM 1, 2, and 4 SCR, KD, REV evaluated 24 h after plating. Each bar represents the mean ± SD (n = 3, each performed in triplicate). ** p < 0.01 vs. GBM SCR; °°p < 0.01 vs. GBM KD (ANOVA, Tukey’s post-test). B) Representative images of 3D cultures of GBM2 SCR (left) and KD (right) cells, embedded in Matrigel and allowed to grow for 15 days, showing the cell density and the cell expansion within the spheroids. Upper panels: scale bar = 400 μm, lower panels: scale bar = 200 μm. C) Representative images of PrPC-expressing cells (green) in 3D GBM2 SCR (left) and KD (right) cultures, analyzed by immunofluorescence. Nuclei are counterstained with DAPI (blue). Scale bar = 100 μm. D) Representative images of proliferating cells (green) within GBM2 and GBM5 SCR and KD 3D organoids after 10 days of culture, assessed by 5-EdU labeling. Scale bar = 200 μm. Histograms show the percentage of labelled (proliferating) cells obtained by ImageJ analysis of n = 7 spheroids for each culture; each available plan was analyzed in at least three different areas. ***p < 0.001 vs. GBM SCR (t-test). E) Effects of PrPC downregulation on GSC migration. Lower panels: Representative images of fluorescently labelled SCR, KD, and OV cells from GBM1, 2, and 3 migrated in 18 h towards 10%-containing medium used as chemoattractant. Scale bar = 200 μm. Upper panels: quantification of the fluorescence intensity of migrated cells using ImageJ software. Data represent the mean ± SD (n = 3). ** p < 0.01 vs. respective GBM SCR, °° p < 0.01 vs. respective GBM KD (ANOVA Tukey’s post-test). F) Matrigel invasion ability of GBM1 SCR, KD, OV, and REV. Representative images depict GSC spheroids embedded in Matrigel at time T0 (left column) and at time T18 (right column). Invasion was calculated by ImageJ, counting the cells that leave the spheroid to invade the Matrigel matrix. Quantification is reported in Supplementary Fig. 4B. Scale bar = 100 μm
3D GSC cultures
Three-dimensional spheroids were obtained as previously described [53]. Briefly, 2,500 GBM SCR and KD cells were re-suspended in 20 µL droplets of Matrigel® and cultured in complete stem medium. Growth of spheroids was monitored using a digital camera (Leica ICC50 HD) until dense aggregates are formed within Matrigel droplets. Proliferating cells were labelled using EdU-Detect Pro Cell Proliferation Kit 488 for Imaging (Base Click) following the manufacturer’s protocol. The fluorescent probe 5-Edu (5-ethynyl-2’-deoxyuridine), incorporates into the DNA during the cell cycle S phase, thus labeling the cells in active proliferation. Fluorescence was detected with Zoe® Fluorescent cell imager (BioRad) and intensity analyzed using the ImageJ software [52].
Western blot
Cells were lysed in 20 mM Tris-HCl pH 7.4, 140 mM NaCl, 2 mM EDTA, 2 mM EGTA, 10% glycerol, 1% Igepal, 1 mM dithiothreitol, 1 mM sodium orthovanadate, 1 mM phenylmethylsulphonyl fluoride (all from Sigma-Aldrich), and the ‘‘Complete’’ protease inhibitor cocktail (Roche, Monza, Italy), as reported [54]. Proteins (25 µg) were size-fractionated by 12% SDS-PAGE transferred to a poly-vinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA, USA) and probed with primary antibodies. Horseradish peroxidase-linked anti-rabbit or anti-mouse IgG secondary antibodies were used. Antibody-reactive bands were detected by ECL (GE Healthcare) and quantified using ChemiDoc™ Imaging System (Bio-Rad).
Immunocytofluorescence
Immunofluorescence was performed in cells plated on glass coverslips as previously reported [55]. GSCs were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with normal goat serum and immuno-stained with anti-PrP Saf 32 antibody (SPI-bio, Chennai India). Immunoreactivity was evidenced with anti-mouse Alexa Fluor-488, (Molecular Probes). When specified, DAPI (Sigma-Aldrich) was used to counterstain nuclei and cell membranes, respectively. Slides were photographed with DM2500 microscope equipped with DFC350FX digital camera (Leica).
Cell proliferation assays
MTT assay: Mitochondrial activity, as index of cell viability was evaluated by measuring the reduction of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma-Aldrich] reduction assay. Cells (3000 cells/well) were incubated with MTT (final concentration 0.2 mg/ml) for 2 h. Medium was removed, and the amount of formazan crystals dissolved in DMSO. was determined by measuring absorbance at 570 nm using a BioTek ELx800 plate reading spectrophotometer [55].
Cell count: proliferation rate of GSCs, grown in standard conditions for 3 days, was determined by counting viable cell number by Trypan blue exclusion assay using the automated TC20 Cell Counter (Bio-Rad) cell counter every 24 h [56].
Migration Assay
GSCs were stained with 10µM of the fluorescent dye Vybrant™ CFDA SE (ThermoFisher) for 15 min, and plated at a density of 1 × 104 cells/well in the upper chambers of FluoroBlok™ HTS 96 multiwell (8 μm pore, Corning), Migration was evaluated after 18 h. by confocal microscope (BioRad MRC 1024 ES) counting, in 3 microscopic fields for each condition, cells that have migrated in the lower chambers [57].
3D invasion assay
GSCs were grown without Matrigel in complete medium for 7 days to allow sphere generations. Then, spheres were embedded within 80% Matrigel and 20% culture medium and seeded on the bottom of a 60 mm2 Petri dish. After Matrigel polymerization, culture medium has been added (time T0) and the spheres were incubated for 18 h (T18). In this time interval, being cell proliferation minimal, we can exclude that changes in the proliferation rate in the different GSC cultures may interfere with the invasive behavior.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using the Aurum™ Total RNA Mini Kit (Bio-Rad), according to the manufacturer’s instruction. Reverse transcription was performed with 1 µg of RNA using the iScript cDNA Synthesis Kit (Bio-Rad), and cDNA products were analyzed using the SsoFastTM Eva Green mix (Bio-Rad) on a CFX96 Touch real-time PCR (Bio-Rad). Cycling conditions were set at 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, for 37 cycles. Primer sequences, designed on mature transcripts, are reported in Table S4.
Target gene expression in each sample was normalized based on the levels of RPLP0 and 28 S housekeeping genes. All qRT-PCR runs included negative controls without mRNA templates and cDNA transcription to check reagents for contaminations.
PrPC GPI digestion by Pi-PLC
The capacity of the bacterial enzyme phosphoinositide phospholipase C (PI-PLC) to detach PrPC from plasma membrane was used to discriminate mature PrPC from Pro-PrP in both GSCs and astrocytes [26]. Cells were plated on plastic dishes and glass coverslips and treated for 30 min. at 5 °C with phosphoinositide phospholipase C (PI-PLC) 0.5 enzymatic U/ml in PBS. Immunoblotting was used to compare the amount of PrPC released in the medium with those retained in cell membrane. Immunostaining was conducted to detect the changes of PrPC expression on cell membrane after enzymatic cleavage.
Statistical analysis
Unless otherwise specified, all experiments were repeated three times. Data are reported as mean ± SEM. Statistical analysis (ANOVA followed by post-hoc Tukey’s test and unpaired t-test was performed using Prism version 5.02 software (GraphPad, San Diego, CA, United States). P ≤ 0.05 was considered statistically significant.
Results
PrPC expression in human GBMs and in patient-derived GSC cultures
To assess the significance of PRNP expression in low-grade glioma (LGG) or GBM patients’ survival, we searched the GEPIA database, performing Kaplan-Meier curves for overall survival (OS) and disease-free survival (DFS). In LGG, neither OS nor DFS were correlated to PRNP expression (Fig. 1A) while, in GBM patients high PRNP expression predicted a reduced OS (HR = 1.6, P = 0.015) and DFS (HR = 1.6, P = 0.03) (Fig. 1B).
PRNP expression in human GSCs was then analyzed by RNA sequencing. Using GSC cultures isolated from 15 human GBMs, we show high levels of PrP mRNA in GSCs isolated from all the tumors (Fig. 1C).
Experimental modulation of PrPC expression in GSC cultures
To assess the role of PrPC in GSCs, we developed a cell model in which PrPC was downregulated or overexpressed. To cope with GBM intertumoral variability, we used five GSC cultures isolated from independent patients (GBM 1–5, see Table S1 for patients’ and tumors’ characteristics). Cells from each culture were transfected with a pool of shRNA for PrP to obtain PrP-down-regulated cells (GBM 1–5 KD) or with the human PRNP-ORF mammalian expression plasmid, to induce PrPC overexpression (GBM 1–5 OV). Control cells were generated by transfecting a scrambled DNA sequence (GBM 1–5 SCR), to exclude effects of the transfection procedures. Lastly, the specificity of the observed effects was shown by rescuing GBM KD cells by re-transfection with the human PRNP plasmid, obtaining GBM REV (1, 2, and 4) cells.
PrPC down-regulation, overexpression, and rescue were monitored by qRT-PCR and immunoblot (Figures S1 and S2). A significant, although incomplete, reduction of the expression of PrPC mRNA (Figure S1A and B) and protein (Figure S2A and C) was obtained in all GBM KD cells, while a marked increase occurred in GBM OV cultures. GBM REV displayed higher PrPC levels than GBM SCR cells, reaching similar levels to GBM OV cells.
PrPC controls GSC proliferation in 2D and 3D cultures
To explore the role of PrPC in GSC proliferation, we compared 1-3-day growth curves of GBM 1–5 SCR, KD, and OV, by MTT assays. GBM KD cells showed a slower proliferation rate than GBM SCR cells, independently from the characteristics of the original GBM (Fig. 2A), supporting that PrPC expression is required to sustain GSC growth. On the other hand, PrPC overexpression (GBM OV) did not further increase proliferation rate respect to GBM SCR cells. The role of PrPC in supporting GSC proliferation was confirmed in GBM REV (i.e., GBM KD cells rescued for PrPC expression). Proliferation of GBM REV, measured as viable cell count, regained the levels of the respective SCR cells (Fig. 2A, histogram panel). Finally, to validate the experimental model, we compared GBM 1–2 SCR proliferation to parental wild-type GBM1-2 cultures obtaining superimposable curves, thus demonstrating that the transfection procedure per se did not modify GSC proliferation (Figure S3A).
These data demonstrate that PrPC expression supports GSC proliferation, and that the reduced growth rate observed in GBM KD cells is solely dependent on PrPC down-regulation.
GSC 3D organoids, generated by growing patient-derived GBM stem-like cells in Matrigel scaffolds, represent in vitro models endowed with a better clinical translational impact than 2D cultures [58, 59]. 3D organoids allow the evaluation of the influence of the interactions among tumor cells and between tumor cells and the extracellular matrix (ECM), better reproducing the reciprocal modulation observed in the in vivo setting [52]. Thus, we determined whether PrPC expression affects GSC ability to grow up in 3D structures. GBM 2 and 5, SCR and KD cells were seeded into Matrigel pearls and allowed to grow for 15 days in the culture medium used for 2D cultures (stem cell-permissive medium containing EGF and bFGF). All cultures developed 3D tissue-like structures (Fig. 2B), retaining the starting levels of PrPC expression (Fig. 2C). However, SCR cells formed compact organoids with high cellularity, while GBM KD organoids appeared less populated (Fig. 2B). Indeed, a striking reduction in cell proliferation cell fraction, as assessed by 5-EdU fluorescent labelling, was observed in GBM KD cells as compared to GBM SCR (-76% for GBM 2 KD, and − 72% for GBM 5 KD; Fig. 2D), paralleling the results obtained in monolayer cultures.
PrPC down-regulation impairs GSC migration and invasion
The role of PrPC in the migration of GSCs was evaluated in a transwell assay using fluorescently labeled GBM 1–3 SCR, KD, and OV cells and complete medium as chemoattractant, for 18 h, to prevent that cell proliferation may interfere with the assay (Fig. 2E). Non-migrated cells were scraped off from the top side of the membrane, while migrated cells, present on the underside of the filter, were imaged and quantified by fluorescence microscopy. PrPC downregulation significantly impaired GSC migration in all cultures tested, as compared to the respective SCR cells (GBM 1 KD: -60%; GBM 2 KD: -66%; GBM 3 KD: -71%) (Fig. 2E). However, in contrast to the results obtained in the proliferation assay, PrPC overexpression increased cell migration (+ 80%, + 69%, and + 35% in GBM 1, 2, and 3 OV, vs. respective SCR cells). Similar results were obtained in a 3D invasion test, in which GBM1 spheroids were embedded in Matrigel and, after 18 h, cells leaving the spheroid and invading ECM were imaged by brightfield microscopy and quantified by ImageJ software. GSC KD cells showed reduced invasive ability in comparison with respective SCR and OV cells, but a complete recovery of the invasive behavior was observed in REV cells (Fig. 2F and Figure S3B for quantification).
The expression of PrPC is required for GSC proliferation and survival in the absence of growth factors
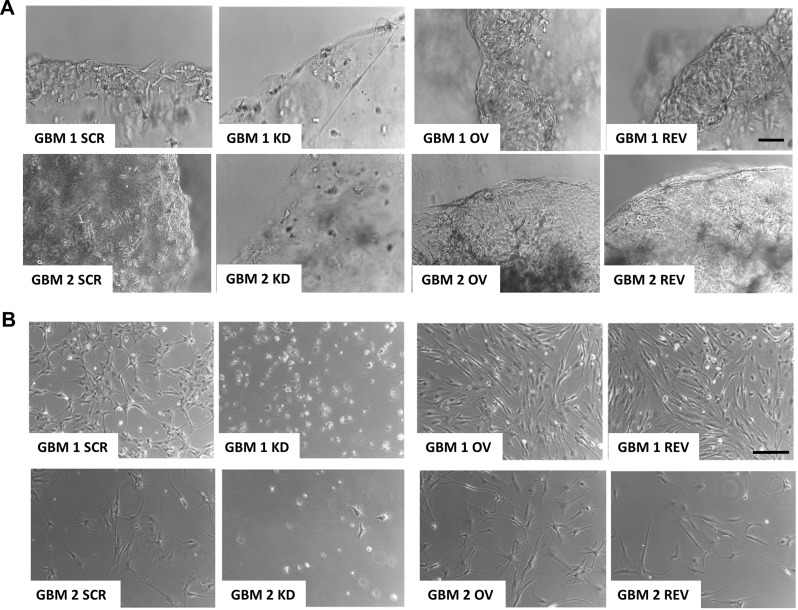
Cancer stem cell-derived organoids can survive in growth factor-free media, and this is considered a feature related to tumorigenic ability and ascribed to the activation of stemness-related pathways, including Wnt/β-catenin [60]. Thus, we verified whether GSC 3D cultures, grown in the absence of growth factors, might unmask PrPC-dependent differences in organoid growth. Therefore, we monitored the role of PrPC in GSC ability to survive into self-assembled 3D organoids. GBM 1 and 2 cells were seeded in Matrigel and let grow in the absence of EGF and bFGF for 15 days (Fig. 3). In these experimental conditions, GBM SCR and OV were viable, continued to proliferate, and retained 3D tissue-like structures. Conversely, GBM KD cells did not survive, and organoids progressively degenerated (Fig. 3A). Importantly, GBM REV organoids survived also in the absence of growth factors, exhibiting cellular morphology and branching interacting with the matrix, like GBM SCR and OV organoids (Fig. 3A). This suggests that PrPC expression might induce specific signaling to support GSC survival and stemness even in the absence of growth factors.
Fig. 3.
PrPC drives GSC survival. Representative images of 3D (A) and 2D (B) GBM1 and GBM2 cell cultures, after 15 days of growth in medium without growth factors. In both culture models, GBM KD failed to survive, leading to 3D organoid and 2D monolayer degeneration; conversely, GBM SCR, OV, and REV were viable and grew in well-organized 3D structures or in 2D monolayers. (A) Scale bar = 100 μm; (B) Scale bar = 50 μm
To further support this hypothesis, we performed the same experiment in 2D cultures. GBM KD cells did not survive when grown for 15 days without growth factors, at odds with all PrPC-expressing GSC cultures, including GBM REV (Fig. 3B).
PrPC expression sustains the stem-like phenotype of GSCs
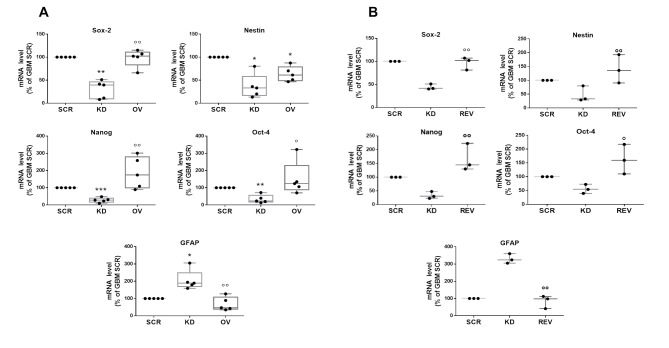
PrPC leverage on GSC stemness was directly investigated by measuring, in the five GSC cultures, the expression of genes related to stemness (i.e., Sox2, Oct-4, Nanog, and nestin) and astrocytic differentiation (glial fibrillary acidic protein, GFAP) by qRT-PCR (Fig. 4A; individual GSC culture data are reported in Table S2). A statistically significant reduction in the expression of stem markers occurred in all five GBM KD cultures, although displaying the expected intertumor variability. Conversely, GBM KD showed an increased GFAP expression as compared to GBM SCR cells. GBM OV cells did not differ from SCR in the expression of all these markers, except for nestin, which, unexpectedly, was slightly down-regulated (Fig. 4A). Sox2 and Oct4 downregulation and GFAP overexpression in KD cells were confirmed by Western Blot in selected GSC cultures (Figure S4). These data show that the expression of PrPC in GSCs favors the retention of a stem-like phenotype, while its downregulation directs cells toward a differentiation pathway.
Fig. 4.
PrPC sustains the stem-like phenotype of GSCs. mRNA levels of stemness (Sox2, nestin, Nanog, Oct-4) and differentiation (GFAP) marker genes in GBM1-5 (dots), evaluated by qRT-PCR. Box-whisker plot represents the mean (line in the center of the box), the 75th, and 25th percentiles (upper and lower ends of boxes) and outliers. A) Comparison of GBM1-5 SCR, KD and OV cells. B) Comparison of GBM1, 2 and 4 SCR, KD, and REV. *p < 0.05, **p < 0.01, ***p < 0.0001 vs. respective GBM SCR; °p < 0.05 °°p < 0.01 vs. respective GBM KD (ANOVA, Tukey’s post-test)
Again, the specificity of PrPC in determining GSC stem/differentiated state, was confirmed by GBM REV cells, in which the alterations in gene expression observed in GBM KD cells were recovered, restoring the stem-like phenotype observed in GBM SCR cells (Fig. 4B and Figure S4).
PrPC expression in GSCs controls Wnt/β-catenin signaling pathway activity
Dysregulation of Wnt/β-catenin signaling pathway, which plays the role of controller of GSC stemness, has been identified in many cancers [42], and PrPC was reported to modulate Wnt/β-catenin pathway in proliferating intestinal epithelial cells [39]. Thus, we hypothesize that PrPC might control GSC stem-like phenotype promoting Wnt/β-catenin signaling.
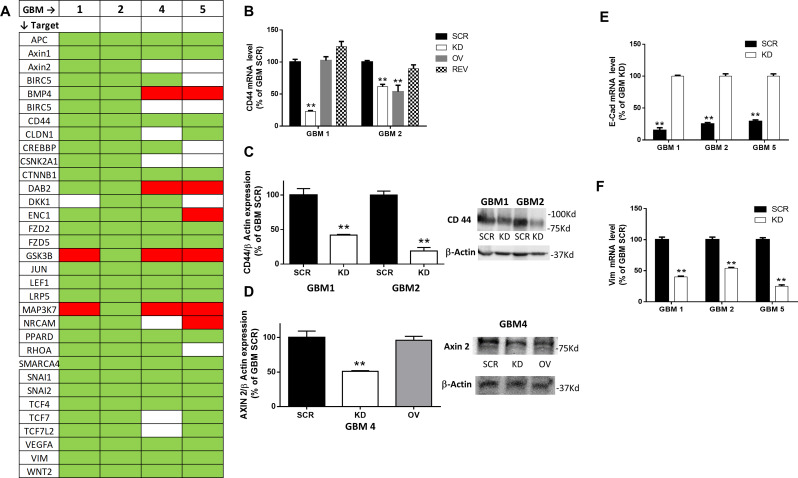
Using a commercial Wnt/β-catenin pathway-related mRNA array, we assessed mRNA expression profiles of different components of the pathway and β-catenin target genes, comparing GBM SCR with GBM KD cells derived from GBM 1, 2, 4, and 5 (Fig. 5A). The results revealed that, with few exceptions, the mRNA for most of the analyzed genes was downregulated in GBM KD cells, with a similar pattern across all GBM tested (Fig. 5A). Thus, the expression of the components of Wnt/β-catenin pathway and of β-catenin target genes are highly dependent on PrPC content.
Fig. 5.
PrPC downregulation affects Wnt/β catenin signaling pathway. A) Heatmap showing changes in mRNA levels of Wnt pathway-related genes in GBM1-2-4-5 KD cells, compared to respective GBM SCR. Data were obtained using a predesigned PCR panel for Wnt signaling pathway. Green = downregulation; red = upregulation; white = unchanged. N = 3. B) CD44 mRNA levels in GBM1 and GBM2 SCR, KD, OV, and REV, analyzed by qRT-PCR. Bars represent the mean ± SD. **p < 0.01 vs. respective GBM SCR (t-test). C) Quantification of CD44 protein expression normalized to β-actin content (left). Bars represent the mean ± SD (n = 3). **p < 0.01 vs. respective GBM SCR (t-test). Representative immunoblot shows the content of CD44 protein in GBM1 and GBM2 SCR and KD (right). D) Quantification of Axin2 protein expression normalized to β-actin content (left). Bars represent the mean ± SD (n = 3). **p < 0.01 vs. respective GBM SCR (t-test). Representative immunoblot shows the content of Axin 2 protein in GBM4 SCR, KD and OV (right). E) E-cadherin mRNA levels in GBM1, 2 and 5 SCR (black bars) and KD (white bars) cells, analyzed by qRT-PCR. Bars represent the mean ± SD. **p < 0.01 vs. respective GBM SCR; °°p < 0.01 vs. respective GBM KD (t-test). F) Vimentin mRNA levels in GBM1, 2 and 5 SCR (black bars) and KD (white bars) cells, analyzed by qRT-PCR. Bars represent the mean SD. **p < 0.01 vs. respective GBM KD (t-test)
In-depth analysis highlighted that all GBM KD cells show a reduced expression of components of canonical Wnt/β-catenin pathway [61], such as Wnt2, FZD2, 5, and low density lipoprotein receptor-related protein 5 (LRP5), representing ligand and receptors activating the pathway, altogether with components of the β-catenin regulatory complex: adenomatous polyposis coli (APC), Axin 1 and 2, and β-catenin itself (CTNNB1). Moreover, negative regulators of the canonical pathway, such as glycogen synthase kinase-3β (GSK3β), which phosphorylates β-catenin and directs it toward ubiquitin-mediated degradation [30] and disabled 2 (Dab2), a stabilizer of the β-catenin degradation complex, are upregulated in GBM KD cells as compared to GBM SCR in 3/4 and 2/4 cultures, respectively.
PrPC down-regulation was also associated with inhibition of β-catenin transcriptional activity, as evidenced by the reduced expression of downstream genes dependent on the activation of the Wnt/β-catenin pathway. In particular, the expression of CD44, SNAIL 1 and 2, vimentin, claudin-1 (CLDN1), c-Jun, lymphoid enhancer-binding factor 1 (LEF1), RHO, SWI/SNF related, matrix associated, actin-dependent regulator of chromatin subfamily A-4 (SMARCA 4), T-cell factor 4 and 7 (TCF4 and TCF7), and vascular endothelial growth factor A (VEGFA) was downregulated in 4/4 GBM KD cultures; mRNAs for ectodermal-neural cortex 1 (ENC1) in 3/4; and casein kinase 2 alpha 1 (CSNK2A1), and survivin (BIRC5) were reduced in 2/4 cultures. Moreover, bone morphogenetic protein 4 (BMP4), involved in the inhibition of GSC proliferation and invasion, promoting their differentiation [62, 63], was upregulated in GBM KD cells.
Array data were validated by qRT-PCR and Western blot (Fig. 5B, C, and D). CD44 and AXIN2 mRNA and protein, chosen as representative genes, showed a reduced expression in GBM KD cells as compared to SCR cells. Moreover, PrPC rescue in GBM REV restored CD44 mRNA content, to levels comparable to GBM SCR (Fig. 5B).
β-catenin plays also a role in the control of the expression of several genes involved in EMT [64]. GBM KD showed downregulation of several EMT-related genes in the mRNA array previously described (Fig. 5A), including snail (SNAI1), slug (SNAI2), twist1 and twist2, and vimentin, while E-cadherin is upregulated. The changes in the expression of the last two genes were also validated in qRT-PCR experiments (Fig. 5E, F).
Overall, these data support the notion that reduced PrP expression impairs β-catenin signaling and target gene expression, EMT ability and, consequently, that PrP contributes to the stem-like, malignant phenotype through the Wnt/β catenin pathway overactivation.
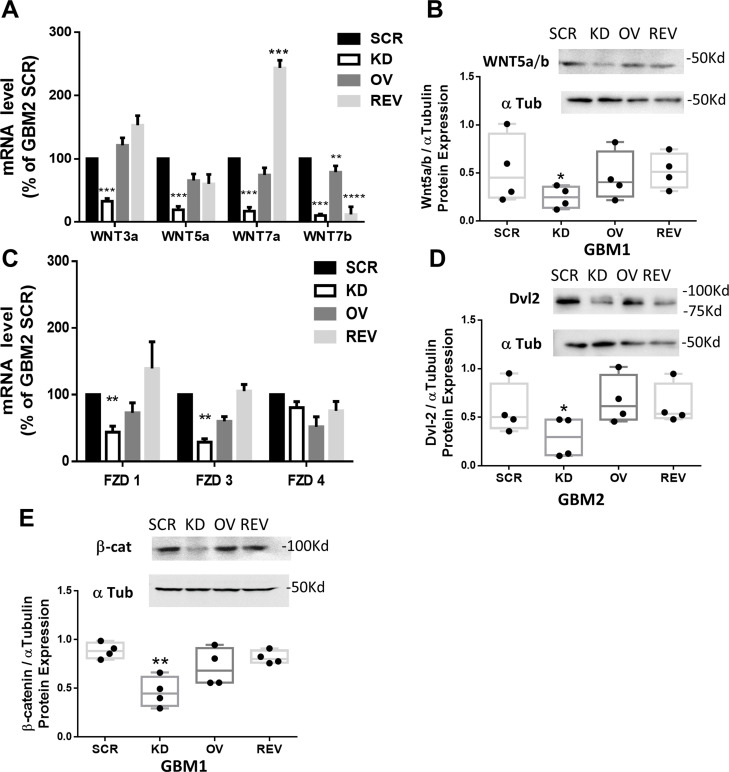
This observation was further confirmed by evaluating mRNA and protein content of the components of the canonical Wnt/β-catenin pathway in GSCs, after modulation of PrP expression. In GBM OV cells the expression of Wnt members was not modified as compared to GBM SCR cells, but, in GBM KD cells, a significant downregulation of Wnt3a, 5a, 7a and 7b isoforms (Fig. 6A, B), and FZD receptor subtypes 1 and 3 (but not FZD4) occurred (Fig. 6C). The reintroduction of PrPC in GBM REV cells significantly rescued Wnt expression (except for Wnt7b), to levels comparable or even higher (Wnt7a) than observed in GBM SCR. Similarly, FZD1 and FZD3 downregulation, observed in GBM KD cells, was reverted by the re-expression of PrPC. The reduction of Wnt5a/b expression in GBM KD cells, and the rescue in GBM REV cells were also confirmed at protein level by immunoblot (Fig. 6B). The expression of Dvl-2, a cytosolic FZD downstream effector, and of the non-phosphorylated (active) β-catenin isoform, were also significantly reduced in GBM KD cells in comparison with respective GBM SCR. Conversely, no significant changes were observed in GBM OV cells as compared to respective GBM SCR, while in GBM REV cells protein content was rescued at levels comparable to GBM SCR, further demonstrating the specificity of the correlation between the phenotype and the downregulation of PrPC (Fig. 6D and E).
Fig. 6.
PrPC downregulation influences the mRNA and protein content of components of the Wnt/β catenin pathway. A) mRNA levels of Wnt subtypes, in GBM2 SCR, KD, OV, and REV cells analyzed by qRT-PCR. Bars represent the mean ± SEM (n = 4). **p < 0.01 vs. respective GBM SCR; ***p < 0.001 vs. respective GBM SCR (t-test). B) Representative Western blot (upper panel) and densitometric quantification (lower panel) of Wnt5a/b protein content in GBM1 SCR, KD, and REV cells, normalized to α-tubulin content. Bars represent the mean ± SD (n = 3). *p < 0.05 vs. GBM SCR (t-test). C) mRNA levels of Frizzled (FZD) subtypes, in GBM2 SCR, KD, OV, and REV analyzed by qRT-PCR. Bars represent the mean ± SEM (n = 4). **p < 0.01 vs. respective GBM SCR (t-test). D) Representative Western blot (upper panel) and densitometric quantification (lower panel) of Dvl2 in GBM2 SCR, KD, and REV cells, normalized to α-tubulin content. Bars represent the mean ± SD (n = 3). *p < 0.05 vs. GBM SCR (t-test). E) Representative Western blot (upper panel) and densitometric quantification (lower panel) of β-catenin in GBM1 SCR, KD, and REV cells, normalized to α-tubulin content. Bars represent the mean ± SD (n = 3). **p < 0.01 vs. GBM SCR (t-test)
Altogether, we propose that PrPC expression sustains the constitutive activation of Wnt/β-catenin pathway, and its down-regulation reduces the expression of several up-stream components of this signaling cascade, impairing β-catenin transcriptional activity. Thus, the dependence of Wnt/β-catenin activity on PrPC expression may represent a molecular link with the reduced stemness observed in GBM KD cells, further corroborating the role of this pathway as primary target of PrPC signaling in GSCs.
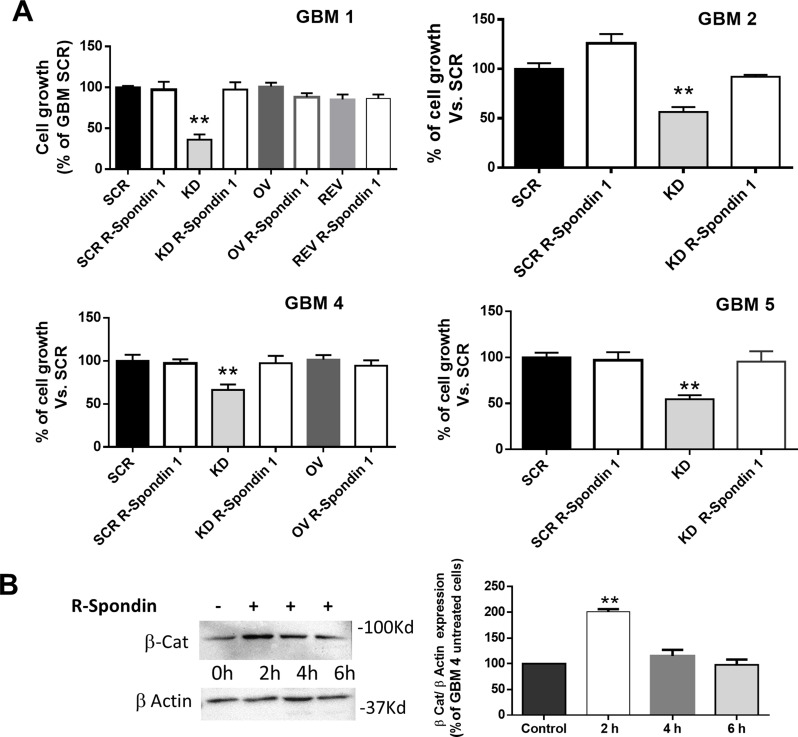
The correlation between PrPC and Wnt/β-catenin pathway in GSC proliferation was assessed treating for 48 h GSC cultures, expressing different PrPC levels, with R-Spondin1 (500ng/ml), a protein able to potentiate β-catenin activity [65], and measuring the proliferation rate by MTT assay. While R-Spondin1 did not change the proliferation of all the GSC cultures expressing PrPC (GBM SCR, OV, and REV), because the Wnt/β-catenin pathway is likely fully active, in GBM KD cells from all the GBM tested, R-Spondin1 treatment increased proliferation rate, to a level comparable to that observed in GBM SCR cells (Fig. 7A).
Fig. 7.
Activation of β-catenin pathway with R-Spondin1 restores proliferation in PrPC-downregulated GSCs. A) Cell proliferation of GBM1, 2, 4, and 5 after 48 h of treatment with R-Spondin1 (500 ng/ml), measured by MTT assay. Bars represent the mean ± SD (n = 3) **p < 0.01 (t-test). B) Left panel: Representative immunoblot of GBM4 KD for non-phosphorylated β-catenin at time 0 (untreated control), and after 2, 4, and 6 h of treatment with R-Spondin 1 (500ng/ml). Right panel: Quantification was carried out by densitometric analysis and normalized to β-actin content. Bars represent the mean ± SD (n = 3). **p < 0.01 vs. untreated GBM KD (t-test)
To confirm that R-Spondin1 effect was directly correlated to β-catenin pathway activation, we measured, in GBM KD, the content of non-phosphorylated (active) β-catenin after R-Spondin1 treatment. The results demonstrated an increased accumulation of active β-catenin starting 2 h after the treatment, thus confirming that R-Spondin1 proliferative effects are related to increased β-catenin transcriptional activity (Fig. 7B).
Wnt/β-catenin activation also relies on growth factor receptor activity leading to Akt and MAPK pathway activation. Since GSCs are grown in the presence of EGF and bFGF, their regulation may affect Wnt/β-catenin activity. To dissect this issue, first we performed immunoblots for the active/phosphorylated forms of Akt and ERK1/2 in GBM SCR, KD, and OV grown in the presence of EGF and bFGF. GSCs, independently from PrPC expression levels, showed similar Akt and ERK1/2 activation, indicating that the reduced proliferation observed in GBM KD cells is not related to changes EGF/bFGF activity (Figure S5A, B). On the other hand, GBM KD proliferation was recovered by R-Spondin1 treatment also in cells grown in a medium devoid of EGF and bFGF (Figure S6). Thus, we conclude that reduced proliferation rate after PrPC downregulation is mainly dependent on the inhibition of Wnt/β-catenin pathway.
GSCs express the immature pro-prion (Pro-PrP) isoform
We demonstrated that the modulation of Wnt/β-catenin pathway is at the basis of the PrPC control of GSC stemness. However, since this effect is initiated by signals starting at membrane level, it is reasonable that the transmembrane Pro-PrP immature form may represent the trigger of the activation of β-catenin. In fact, differently from the mature form, Pro-PrP is a transmembrane protein, which interacts with cytoskeleton elements, as filamin-A [66, 67], and possibly favors the aberrant Wnt/β-catenin signaling contributing to the malignant behavior of GSCs.
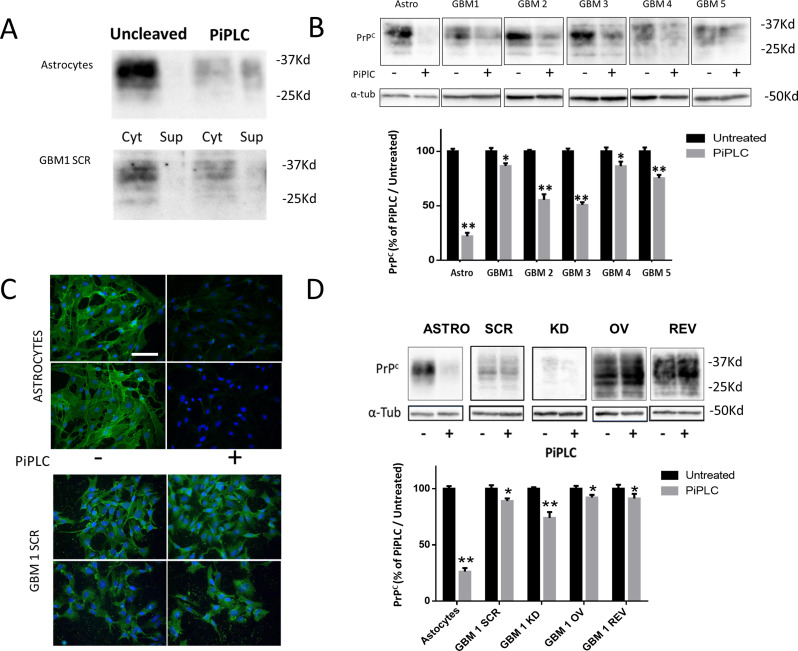
To analyze PrP topology in GSCs and discriminate between the two isoforms, since both PrPC and Pro-PrP isoforms are equally recognized by available anti-PrP antibodies, we treated GSCs with phosphatidylinositol-specific phospholipase C (Pi-PLC). Pi-PLC cleaves GPI anchors causing the release in the culture medium of the extracellularly anchored PrPC, only [26]. Conversely, being Pro-PrP membrane-tethered and devoid of GPI, it is not affected by Pi-PLC cleavage. As experimental control for GSCs, we used primary cultures of rat astrocytes, known to express high concentration of mature PrPC on the plasma membrane external side.
In untreated astrocytes, PrPC is mainly membrane-bound and no immunoreactive PrP was detected in the culture medium (Fig. 8A). After Pi-PLC treatment (0.5 enzymatic units at 5 °C for 30 min) a remarkable reduction of PrPC in the cellular fraction was paralleled by a significant increase of the protein released in the culture medium (Fig. 8A), with the cleavable protein representing about 80% of total PrPC (Fig. 8B). Conversely, in GBM1 SCR cells after Pi-PLC treatment, PrP content was slightly reduced in the cell fraction and only traces were detectable in the medium, indicating that most PrP molecules do not contain cleavable GPI anchor being bona fide Pro-PrP (Fig. 8A). The prevalent Pro-PrP expression was confirmed in all 5 GBM SCR cells, independently from the initial PrPC content (Fig. 8B). Similar results were obtained in immunofluorescence experiments (Fig. 8C), in which PrPC and Pro-PrP were identified by performing the immunostaining under non-permeabilizing conditions, to detect only PrP molecules exposed on the plasma membrane. In control astrocytes, bright PrPC immunoreactivity was detectable on cell surface, but it was almost completely abolished after treatment with Pi-PLC. As a control, to identify intracellular PrP, we performed the same analysis in astrocytes permeabilized before the incubation with the primary antibody. In this experimental setting, we clearly demonstrated that Pi-PLC treatment was able to remove only the extracellular GPI-anchored PrPC, leaving unaltered the intracellular protein (Figure S7). Conversely, no significant reduction in PrP immunofluorescence was observed in GBM SCR cells. Altogether, these data show that, while normal astrocytes mainly express the mature form of PrPC, which is properly anchored on the extracellular side of the plasma membrane through an enzymatically accessible GPI, PrP expressed by GSCs is present in a conformation that hinders GPI anchor to Pi-PLC, likely being Pro-PrP.
Fig. 8.
GSCs contain Pro-PrP, the immature form of PrPC. A) Representative immunoblot of PrPC cellular fraction (Cell) or released in the supernatant (Sup) in control cells (Uncleaved) or following GPI digestion by Pi-PLC (PiPLC) in normal astrocytes and GBM1 SCR cells. B) Representative immunoblot of retained PrPC, following GPI digestion by Pi-PLC in astrocytes and GBM1-5 SCR cells (upper panel). PrPC quantification by densitometric analysis, normalized to α-tubulin, is expressed as percentage of Pi-PLC-treated compared to untreated cells (lower panel). Bars represent the mean ± SD (n = 2). *p < 0.05, **p < 0.01 vs. respective untreated cells (t-test). C) Representative images of immunofluorescence for PrPC (green) showing the profile of membrane PrPC expression in normal astrocytes and GBM1 SCR cells, in control conditions (-) and after digestion for 30 min with Pi-PLC (+). After fixation, cells have been immunoprobed without permeabilization to evidence PrPC expression in plasma membrane only. Nuclei are counterstained with DAPI (blue). Scale bar = 100 µM. D) Representative immunoblot of retained or released PrPC, following GPI digestion with Pi-PLC in astrocytes and GBM1 SCR, KD, OV, and REV cells. PrPC quantification by densitometric analysis, normalized to α-tubulin, is expressed as a percentage of Pi-PLC-treated compared to untreated cells. Bars represent the mean ± SD. *p < 0.05, **p < 0.01 vs. respective untreated cells (t-test)
Moreover, we assessed, by immunoblotting, the membrane topology of PrPC in GBM1 OV and GBM1 REV cells to verify whether the PrP isoform produced in transfected cells retains the features of Pro-PrP. The lack of Pi-PLC-induced PrP cleavage, expressed as percentage of the immunoreactivity observed in mock-treated cells (Fig. 8D) confirmed that also in OV and REV cultures, Pro-PrP is the main isoform expressed.
To show that only Pro-PrP favors cell proliferation (while PrPC has no effects), we evaluated, by 5-EdU labeling, the effects of Pi-PLC treatment on astrocyte proliferation. Serum-starved astrocytes were treated or not with Pi-PLC and then let them grow for 48 h in FBS-containing medium. No difference in cell proliferation was observed under both experimental conditions (Figure S8), confirming that, differently from what observed with Pro-PrP in GSCs, the removal of the membrane GPI-anchored PrPC does not influence cell proliferation.
Discussion
In this study, we identified an intracellular pathway by which PrP controls the stem-like features of GSCs. The possible therapeutic interference with the mechanisms balancing GSC stemness or differentiation represents a relevant goal to develop novel pharmacological approaches to treat GBM. Although high PrPC expression characterizes all brain resident cells, including neurons and astrocytes, possibly involved in neurotrophic effects [28], in GBM this protein acquires an aberrant role, contributing to malignancy and boosting cell proliferation and brain invasion [7, 8]. Thus, we hypothesized that this discrepancy might be dependent on different intracellular signaling activated by PrPC in normal cells and in GSCs. Although PrPC is expressed in most established GBM cell lines [68], its expression in human GBM was never systematically assessed, and only few studies identified PrPC in GBM specimens [30]. We analyzed the available database for gene expression profiling, showing an inverse relationship between PRNP transcript levels and DFS and OS in GBM, but not in LGG, patients. Since GSCs are at the apex of GBM cell heterogeneity, we propose that PrPC could represent a GSC signature. This was confirmed by RNA-seq analysis in 15/15 GSC cultures, which showed a high PRNP expression.
In our study, the direct correlation between PrPC expression levels and GSC malignant phenotype was reproduced in five independently isolated, patient-derived GSC cultures, in which PrPC expression was stably down- or up-regulated. The use of GSC cultures obtained from different patients is particularly relevant to cope with the known intertumor GBM variability. In particular, the observation that similar results were obtained in GSCs derived from different GBM molecular subtypes (i.e., neural and mesenchymal types) further support the notion that the reported data may represent a general GSC feature.
In the presence of reduced PrPC levels, GSCs displayed low proliferation and migration/invasion rates in both 2D and 3D models. Moreover, another peculiar feature of GSCs, the ability to survive in both 2D monolayers and 3D organoids in the absence of growth factors [60], was also dependent on PrPC expression, since GBM KD cells did not survive in these conditions, at odds with GSC cultures in which PrPC was expressed. PrPC down-regulation also leads GSCs to acquire a differentiated phenotype, characterized by increased GFAP expression, and down-regulation of stemness factors (Sox2, Oct-4, Nanog, and nestin), and EMT-related genes, which, conversely, characterize PrPC-expressing GSCs. All these changes were dependent on PrPC expression since a complete biological and phenotypical recovery occurred after re-expression of the protein in GBM KD cells.
Thus, we focused on the identification of the intracellular signaling by which PrPC support GSC stemness. PrPC was reported to regulate different metabolic and signaling pathways in GBM, changing regular biosynthetic processes and facilitating the stem-like cell features. For example, PrPC overexpression was reported to sustain the malignant features of GBM cells through the activation of the PI3K/Akt/mTOR and ERK1/2 pathways, due to the association with the co-chaperone STI1/HOP [31, 69]. Moreover, PrPC controls GBM cell autophagic flux or the vesicle formation and release, two processes associated to tumorigenesis [70, 71].
Although we cannot exclude the participation of different molecular pathways in PrPC modulation of GSC functioning, our study provides direct evidence that alterations in PrPC expression cause the modulation of Wnt/β-catenin signaling. Indeed, Wnt/β-catenin pathway supports stem cell maintenance in the early phase of brain development [72], and increases proliferation, self-renewal, migration, and EMT in normal stem cells [41, 73]. On the other hand, aberrant activation of Wnt pathway leads to development, progression, and invasion of different human tumors, including GBM [74, 75].
We demonstrate that the expression of both components of the β-catenin pathway (Wnt ligands and FZD receptors, intracellular regulators such as Dvl2, and β-catenin itself) and the genes transcribed upon β-catenin activation, are downregulated in GBM KD cells. Importantly, both Wnt5a, β-catenin, and other pathway components analyzed were recovered in GBM REV, confirming the direct relationship between PrPC content and the expression of these proteins. Thus, we propose that, in the absence of PrPC, the reduced Wnt/β-catenin signaling causes the loss of the malignant features of GSCs. This relationship was also confirmed by the pharmacological activation of the β-catenin pathway using R-Spondin1, which was able to rescue β-catenin intracellular content and restore proliferation rate of GBM KD cells to similar levels than GBM SCR cells. R-Spondin1 controls β-catenin activity also in the presence of low concentrations of Wnt ligands interacting with the co-receptor Lgr5, which leads to the activation of the pathway and the modulation of the FZD-Lrp5/6 complex [65]. However, R-Spondin1 was ineffective in GBM SCR, OV, and REV, suggesting that a maximal activation of the pathway was already present in the cells expressing high levels of PrPC. This observation provides a possible molecular explanation for the observed low impact of PrPC overexpression on GSC malignant features.
In several human tumors, PrPC has been detected as an immature isoform, Pro-PrP [18, 27, 67]. To be anchored on the external leaflet of plasma membrane, neosynthesized PrPC requires the removal of the C-terminal peptide and the insertion of a GPI anchor. This step is defective in tumors, and the resulting protein, named Pro-PrP, retains the GPI signal peptide and cannot be anchored to the plasma membrane. Thus, being Pro-PrP a protein inserted within the membrane lipid bilayer [25], we hypothesized that it might interfere with membrane-originating intracellular pathways, altering cell biological activity. Thus, as possible explanation of how PrPC might interfere with Wnt/β-catenin signaling, we first demonstrated that PrPC is predominantly expressed as Pro-PrP also in GBM. Importantly, this unconventional topology of PrPC was retained after its overexpression in GSCs or in the rescuing of KD cells, suggesting that PrPC maturation is equally defective when derived from either endogenous or exogenous sources, and that it may represent a relevant feature of GSCs, as reported in other tumor stem cells [26]. Thus, the presence of Pro-PrP is one of the main determinants of the malignant phenotype of GSCs, likely via the aberrant hyperactivation of the Wnt/β-catenin pathway.
The observation that Pro-PrP is the predominant form in GSCs isolated from different human GBMs, suggests that this might be a general feature of GSCs to sustain the stem-like malignant features of this cell subpopulation. Thus, pharmacological targeting of Pro-PrP may represent a novel strategy to counteract GSC proliferation and invasiveness. Wnt signaling grants a functional cooperation between signaling of growth factors, adhesion protein, and β-catenin transcriptional activity, stabilizing the molecular machinery involved in cell-cell interactions [75]. Thus, Pro-PrP, controlling Wnt signaling, might link these pathways to the cytoskeleton in the adhesion complex.
Conclusions
We report that PrP, causing a sustained hyperactivation of the Wnt/β-catenin pathway, acts as a pivotal regulator of GSC stem-like properties and their malignant phenotype. Thus, the interaction between Pro-PrP and Wnt signaling may represent a novel relevant therapeutic target for GBM. Finally, we provide evidence that the transmembrane protein Pro-PrP is the predominant PrP isoform expressed in GSCs. The persistence of Pro-PrP in GSCs supports cell proliferation and invasiveness, subverting the normal function of mature PrPC which, being expressed at high levels in neurons and astrocytes, mainly sustains the differentiated, quiescent state of the cells. The Pro-PrP expressed by GSCs may interact with Wnt/β-catenin pathway causing its constitutive hyperactivation and enhancing proliferation, migration, and invasion. Thus, Pro-PrP activity may represent a targetable GBM cell vulnerability and a novel differentiating approach for GSC eradication.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
“Not applicable”.
Abbreviations
- BIRC5
survivin
- BMP4
bone morphogenetic protein 4
- CSNK2A1
casein kinase 2 alpha 1
- CLDN1
claudin-1
- CNS
central nervous system
- Dab2
disabled 2
- DFS
disease-free survival
- ENC1
ectodermal-neural cortex 1
- EMT
epithelial-to-mesenchymal transition
- FZD
frizzled receptors
- GBM
glioblastoma
- GPI
glycosylphosphatidylinositol
- GSCs
GBM stem cells
- GSK3β
glycogen synthase kinase-3β
- HR
hazard ratio
- LGG
Low-grade glioma
- LEF1
lymphoid enhancer-binding factor 1
- MMP7
Matrix metallopeptidase 7
- OS
Overall survival
- Pi-PLC
Phosphatidylinositol phospholipase C
- Pro-PrP
Immature pro-prion
- PrPC
Cellular prion protein
- PrP Sc
Prion protein scrapie
- GBM SCR
GBM scrambled
- GBM KD
GBM knock-down
- GBM REV
GBM reverted
- GBM OV
GBM overexpressed
- shRNA
Short hairpin RNA
- SMARCA 4
SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily A member 4
- TCF 1
T-cell factor 1
- VEGFA
Vascular endothelial growth factor A
- VIM
Vimentin
- 5-Edu
5-ethynyl-2’-deoxyuridine
- MTT
[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- APC
adenomatous polyposis coli
Author contributions
AC, ID, MT, and ST performed the experiments; AC, ID, AP, FB, and TF acquired and analyzed the data; AC and TF conceived and designed the study; AC, and TF wrote the manuscript; all authors reviewed and/or revised the manuscript.
Funding
Work supported by #NEXTGENERATIONEU (NGEU) and funded by the Ministry of University and Research (MUR), National Recovery and Resilience Plan (NRRP), project MNESYS (PE0000006) – A Multiscale integrated approach to the study of the nervous system in health and disease (DN. 1553 11.10.2022) to AC, FB; and TF, and by a grant from Italian “Fondazione Giovanni Celeghin Contro i Tumori Cerebrali” (grant 2018) to TF.
Data availability
RNA-seq data were deposited at NCBI Geo data set (GSE179356).
Declarations
Ethics Declaration
Human GBM specimens were obtained from the Neurosurgery Dept. of IRCCS Ospedale Policlinico San Martino (Genova, Italy), after Institutional Ethical Committee approval (CER Liguria 360/2019) and patients’ informed consent.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alessandro Corsaro and Irene Dellacasagrande equally contributed and should both be considered as first authors.
References
- 1.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, et al. Effect of Tumor-Treating Fields Plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a Randomized Clinical Trial. JAMA. 2017;318(23):2306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Florio T, Barbieri F. The status of the art of human malignant glioma management: the promising role of targeting tumor-initiating cells. Drug Discov Today. 2012;17(19–20):1103–10. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 5.Antony H, Wiegmans AP, Wei MQ, Chernoff YO, Khanna KK, Munn AL. Potential roles for prions and protein-only inheritance in cancer. Cancer Metastasis Rev. 2012;31(1–2):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corsaro A, Bajetto A, Thellung S, Begani G, Villa V, Nizzari M, et al. Cellular prion protein controls stem cell-like properties of human glioblastoma tumor-initiating cells. Oncotarget. 2016;7(25):38638–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryskalin L, Busceti CL, Biagioni F, Limanaqi F, Familiari P, Frati A et al. Prion protein in Glioblastoma Multiforme. Int J Mol Sci. 2019;20(20). [DOI] [PMC free article] [PubMed]
- 8.Thellung S, Corsaro A, Bosio AG, Zambito M, Barbieri F, Mazzanti M et al. Emerging role of Cellular prion protein in the maintenance and expansion of glioma stem cells. Cells. 2019;8(11). [DOI] [PMC free article] [PubMed]
- 9.Grimaldi I, Leser FS, Janeiro JM, da Rosa BG, Campanelli AC, Romao L, et al. The multiple functions of PrP(C) in physiological, cancer, and neurodegenerative contexts. J Mol Med (Berl). 2022;100(10):1405–25. [DOI] [PubMed] [Google Scholar]
- 10.Watts JC, Bourkas MEC, Arshad H. The function of the cellular prion protein in health and disease. Acta Neuropathol. 2018;135(2):159–78. [DOI] [PubMed] [Google Scholar]
- 11.Prusiner SB, Prions. Proc Natl Acad Sci U S A. 1998;95(23):13363–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corsaro A, Thellung S, Chiovitti K, Villa V, Simi A, Raggi F, et al. Dual modulation of ERK1/2 and p38 MAP kinase activities induced by minocycline reverses the neurotoxic effects of the prion protein fragment 90–231. Neurotox Res. 2009;15(2):138–54. [DOI] [PubMed] [Google Scholar]
- 13.Corsaro A, Thellung S, Villa V, Nizzari M, Florio T. Role of prion protein aggregation in neurotoxicity. Int J Mol Sci. 2012;13(7):8648–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thellung S, Gatta E, Pellistri F, Corsaro A, Villa V, Vassalli M, et al. Excitotoxicity through NMDA receptors mediates cerebellar granule neuron apoptosis induced by prion protein 90–231 fragment. Neurotox Res. 2013;23(4):301–14. [DOI] [PubMed] [Google Scholar]
- 15.Thellung S, Villa V, Corsaro A, Pellistri F, Venezia V, Russo C, et al. ERK1/2 and p38 MAP kinases control prion protein fragment 90-231-induced astrocyte proliferation and microglia activation. Glia. 2007;55(14):1469–85. [DOI] [PubMed] [Google Scholar]
- 16.Stohr J, Weinmann N, Wille H, Kaimann T, Nagel-Steger L, Birkmann E, et al. Mechanisms of prion protein assembly into amyloid. Proc Natl Acad Sci U S A. 2008;105(7):2409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YJ, Baskakov IV. The cellular form of the prion protein is involved in controlling cell cycle dynamics, self-renewal, and the fate of human embryonic stem cell differentiation. J Neurochem. 2013;124(3):310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Go G, Lee SH. The Cellular prion protein: a promising therapeutic target for Cancer. Int J Mol Sci. 2020;21(23). [DOI] [PMC free article] [PubMed]
- 19.Cheng Y, Tao L, Xu J, Li Q, Yu J, Jin Y, et al. CD44/cellular prion protein interact in multidrug resistant breast cancer cells and correlate with responses to neoadjuvant chemotherapy in breast cancer patients. Mol Carcinog. 2014;53(9):686–97. [DOI] [PubMed] [Google Scholar]
- 20.Chieng CK, Say YH. Cellular prion protein contributes to LS 174T colon cancer cell carcinogenesis by increasing invasiveness and resistance against doxorubicin-induced apoptosis. Tumour Biol. 2015;36(10):8107–20. [DOI] [PubMed] [Google Scholar]
- 21.de Lacerda TC, Costa-Silva B, Giudice FS, Dias MV, de Oliveira GP, Teixeira BL, et al. Prion protein binding to HOP modulates the migration and invasion of colorectal cancer cells. Clin Exp Metastasis. 2016;33(5):441–51. [DOI] [PubMed] [Google Scholar]
- 22.Lim JH, Go G, Lee SH. PrPC regulates the Cancer Stem Cell properties via Interaction with c-Met in Colorectal Cancer cells. Anticancer Res. 2021;41(7):3459–70. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Qian J, Wang F, Ma Z. Cellular prion protein accelerates colorectal cancer metastasis via the Fyn-SP1-SATB1 axis. Oncol Rep. 2012;28(6):2029–34. [DOI] [PubMed] [Google Scholar]
- 24.Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51(2):229–40. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Yu S, Nakamura F, Yin S, Xu J, Petrolla AA, et al. Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J Clin Invest. 2009;119(9):2725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Gao Z, Hu L, Wu G, Yang X, Zhang L, et al. Glycosylphosphatidylinositol Anchor Modification Machinery Deficiency is responsible for the formation of Pro-prion protein (PrP) in BxPC-3 protein and increases Cancer Cell Motility. J Biol Chem. 2016;291(8):3905–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Zhang J, Ke JR, Yu Z, Shi R, Gao SS, et al. Pro-prion, as a membrane adaptor protein for E3 ligase c-Cbl, facilitates the ubiquitination of IGF-1R, promoting melanoma metastasis. Cell Rep. 2022;41(12):111834. [DOI] [PubMed] [Google Scholar]
- 28.McLennan NF, Rennison KA, Bell JE, Ironside JW. In situ hybridization analysis of PrP mRNA in human CNS tissues. Neuropathol Appl Neurobiol. 2001;27(5):373–83. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi Y, Kakeya T, Yamazaki T, Takekida K, Nakamura N, Matsuda H, et al. G1-dependent prion protein expression in human glioblastoma cell line T98G. Biol Pharm Bull. 2002;25(6):728–33. [DOI] [PubMed] [Google Scholar]
- 30.Luo Q, Wang Y, Fan D, Wang S, Wang P, An J. Prion protein expression is correlated with Glioma Grades. Virol Sin. 2020;35(4):490–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iglesia RP, Prado MB, Cruz L, Martins VR, Santos TG, Lopes MH. Engagement of cellular prion protein with the co-chaperone Hsp70/90 organizing protein regulates the proliferation of glioblastoma stem-like cells. Stem Cell Res Ther. 2017;8(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang J, Pan Y, Zhang D, Guo C, Shi Y, Wang J, et al. Cellular prion protein promotes proliferation and G1/S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007;21(9):2247–56. [DOI] [PubMed] [Google Scholar]
- 33.Du L, Rao G, Wang H, Li B, Tian W, Cui J, et al. CD44-positive cancer stem cells expressing cellular prion protein contribute to metastatic capacity in colorectal cancer. Cancer Res. 2013;73(8):2682–94. [DOI] [PubMed] [Google Scholar]
- 34.Santos TG, Lopes MH, Martins VR. Targeting prion protein interactions in cancer. Prion. 2015;9(3):165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanata SM, Lopes MH, Mercadante AF, Hajj GN, Chiarini LB, Nomizo R, et al. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J. 2002;21(13):3307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauczynski S, Nikles D, El-Gogo S, Papy-Garcia D, Rey C, Alban S, et al. The 37-kDa/67-kDa laminin receptor acts as a receptor for infectious prions and is inhibited by polysulfated glycanes. J Infect Dis. 2006;194(5):702–9. [DOI] [PubMed] [Google Scholar]
- 37.Rieger R, Edenhofer F, Lasmezas CI, Weiss S. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat Med. 1997;3(12):1383–8. [DOI] [PubMed] [Google Scholar]
- 38.Martin-Lanneree S, Halliez S, Hirsch TZ, Hernandez-Rapp J, Passet B, Tomkiewicz C, et al. The Cellular prion protein controls Notch Signaling in neural Stem/Progenitor cells. Stem Cells. 2017;35(3):754–65. [DOI] [PubMed] [Google Scholar]
- 39.Besnier LS, Cardot P, Da Rocha B, Simon A, Loew D, Klein C, et al. The cellular prion protein PrPc is a partner of the wnt pathway in intestinal epithelial cells. Mol Biol Cell. 2015;26(18):3313–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chenn A. Wnt/beta-catenin signaling in cerebral cortical development. Organogenesis. 2008;4(2):76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nager M, Bhardwaj D, Canti C, Medina L, Nogues P, Herreros J. beta-catenin signalling in Glioblastoma Multiforme and Glioma-initiating cells. Chemother Res Pract. 2012;2012:192362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–50. [DOI] [PubMed] [Google Scholar]
- 44.Griffero F, Daga A, Marubbi D, Capra MC, Melotti A, Pattarozzi A, et al. Different response of human glioma tumor-initiating cells to epidermal growth factor receptor kinase inhibitors. J Biol Chem. 2009;284(11):7138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gatti M, Pattarozzi A, Bajetto A, Wurth R, Daga A, Fiaschi P, et al. Inhibition of CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human glioblastoma stem-like cells affecting self-renewal activity. Toxicology. 2013;314(2–3):209–20. [DOI] [PubMed] [Google Scholar]
- 46.Carra E, Barbieri F, Marubbi D, Pattarozzi A, Favoni RE, Florio T, et al. Sorafenib selectively depletes human glioblastoma tumor-initiating cells from primary cultures. Cell Cycle. 2013;12(3):491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thellung S, Gatta E, Pellistri F, Villa V, Corsaro A, Nizzari M, et al. Different Molecular mechanisms Mediate Direct or Glia-Dependent prion protein fragment 90–231 neurotoxic effects in cerebellar granule neurons. Neurotox Res. 2017;32(3):381–97. [DOI] [PubMed] [Google Scholar]
- 48.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbieri F, Bajetto A, Dellacasagrande I, Solari A, Wurth R, Fernandez V, et al. Stem-like signatures in human meningioma cells are under the control of CXCL11/CXCL12 chemokine activity. Neuro Oncol. 2023;25(10):1775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, Gingeras TR. Mapping RNA-seq reads with STAR. Curr Protoc Bioinf. 2015;51(11 4):1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbieri F, Bosio AG, Pattarozzi A, Tonelli M, Bajetto A, Verduci I, et al. Chloride intracellular channel 1 activity is not required for glioblastoma development but its inhibition dictates glioma stem cell responsivity to novel biguanide derivatives. J Exp Clin Cancer Res. 2022;41(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, et al. A three-Dimensional Organoid Culture System Derived from Human Glioblastomas recapitulates the hypoxic gradients and Cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pattarozzi A, Carra E, Favoni RE, Wurth R, Marubbi D, Filiberti RA, et al. The inhibition of FGF receptor 1 activity mediates sorafenib antiproliferative effects in human malignant pleural mesothelioma tumor-initiating cells. Stem Cell Res Ther. 2017;8(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thellung S, Scoti B, Corsaro A, Villa V, Nizzari M, Gagliani MC, et al. Pharmacological activation of autophagy favors the clearing of intracellular aggregates of misfolded prion protein peptide to prevent neuronal death. Cell Death Dis. 2018;9(2):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barbieri F, Wurth R, Pattarozzi A, Verduci I, Mazzola C, Cattaneo MG, et al. Inhibition of Chloride Intracellular Channel 1 (CLIC1) as Biguanide Class-Effect to impair human glioblastoma stem cell viability. Front Pharmacol. 2018;9:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vitale RM, Thellung S, Tinto F, Solari A, Gatti M, Nuzzo G, et al. Identification of the hydantoin alkaloids parazoanthines as novel CXCR4 antagonists by computational and in vitro functional characterization. Bioorg Chem. 2020;105:104337. [DOI] [PubMed] [Google Scholar]
- 58.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(1):126–36. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poudel H, Sanford K, Szwedo PK, Pathak R, Ghosh A. Synthetic matrices for Intestinal Organoid Culture: implications for Better Performance. ACS Omega. 2022;7(1):38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Liu Y, Liu B, Wang J, Wei S, Qi Z, et al. A growth factor-free culture system underscores the coordination between wnt and BMP signaling in Lgr5(+) intestinal stem cell maintenance. Cell Discov. 2018;4:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren Q, Chen J, Liu Y. LRP5 and LRP6 in wnt signaling: similarity and divergence. Front Cell Dev Biol. 2021;9:670960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–5. [DOI] [PubMed] [Google Scholar]
- 63.Zhao X, Sun Q, Dou C, Chen Q, Liu B. BMP4 inhibits glioblastoma invasion by promoting E-cadherin and claudin expression. Front Biosci (Landmark Ed). 2019;24(6):1060–70. [DOI] [PubMed] [Google Scholar]
- 64.Loh CY, Chai JY, Tang TF, Wong WF, Sethi G, Shanmugam MK et al. The E-Cadherin and N-Cadherin switch in Epithelial-To-Mesenchymal transition: signaling, therapeutic implications, and challenges. Cells. 2019;8(10). [DOI] [PMC free article] [PubMed]
- 65.Ter Steege EJ, Bakker ERM. The role of R-spondin proteins in cancer biology. Oncogene. 2021;40(47):6469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prado MB, Melo Escobar MI, Alves RN, Coelho BP, Fernandes CFL, Boccacino JM et al. Prion protein at the leading edge: its role in cell motility. Int J Mol Sci. 2020;21(18). [DOI] [PMC free article] [PubMed]
- 67.Sy MS, Li C, Yu S, Xin W. The fatal attraction between pro-prion and filamin A: prion as a marker in human cancers. Biomark Med. 2010;4(3):453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lenzi P, Busceti CL, Lazzeri G, Ferese R, Biagioni F, Salvetti A et al. Autophagy Activation Associates with suppression of prion protein and improved mitochondrial status in Glioblastoma Cells. Cells. 2023;12(2). [DOI] [PMC free article] [PubMed]
- 69.Lopes MH, Santos TG, Rodrigues BR, Queiroz-Hazarbassanov N, Cunha IW, Wasilewska-Sampaio AP, et al. Disruption of prion protein-HOP engagement impairs glioblastoma growth and cognitive decline and improves overall survival. Oncogene. 2015;34(25):3305–14. [DOI] [PubMed] [Google Scholar]
- 70.Armocida D, Busceti CL, Biagioni F, Fornai F, Frati A. The role of Cellular prion protein in Glioma Tumorigenesis could be through the autophagic mechanisms: a narrative review. Int J Mol Sci. 2023;24(2). [DOI] [PMC free article] [PubMed]
- 71.Boccacino JM, Dos Santos Peixoto R, Fernandes CFL, Cangiano G, Sola PR, Coelho BP, et al. Integrated transcriptomics uncovers an enhanced association between the prion protein gene expression and vesicle dynamics signatures in glioblastomas. BMC Cancer. 2024;24(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noelanders R, Vleminckx K. How wnt signaling builds the brain: Bridging Development and Disease. Neuroscientist. 2017;23(3):314–29. [DOI] [PubMed] [Google Scholar]
- 73.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, et al. A role for wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423(6938):409–14. [DOI] [PubMed] [Google Scholar]
- 74.McCord M, Mukouyama YS, Gilbert MR, Jackson S. Targeting WNT signaling for multifaceted glioblastoma therapy. Front Cell Neurosci. 2017;11:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tompa M, Kalovits F, Nagy A, Kalman B. Contribution of the wnt pathway to defining Biology of Glioblastoma. Neuromolecular Med. 2018;20(4):437–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited at NCBI Geo data set (GSE179356).