Abstract
Due to current challenges in the early detection, less than 40% of individuals diagnosed with hepatocellular carcinoma (HCC) are viable candidates for surgical intervention. Therefore, validating and launching of a novel precise diagnostic approach is essential for early diagnosis. Based on developing evidence using circulating tumor cells and their derivatives, circulating miRNAs, and extracellular vesicles (EVs), liquid biopsy may offer a reliable platform for the HCC’s early diagnosis. Each liquid biopsy analyte may provide significant areas for diagnosis, prognostic assessment, and treatment monitoring of HCC patients depending on its kind, sensitivity, and specificity. The current review addresses potential clinical applications, current research, and future developments for liquid biopsy in HCC management.
Keywords: HCC biomarkers, Liquid biopsy, Circulating tumor DNA, Circulating tumor cells, Extra-cellular vesicles
Introduction
It is estimated that liver cancer cases and deaths will rise by over 50% in the next two decades [1–4]. The incidence of HCC and resulting fatality rates are rapidly rising in the United States [5, 6]. This increase is mainly due to a rise in the number of people afflicted with both alcoholic and non-alcoholic liver diseases, as well as the presence of hepatitis C virus [7, 8].
Patients with various stages of HCC have a wide range of prognoses; hence, routine screening is advised for those who are at risk. In cases of early identification, surgical removal of the affected tissue has a positive outlook, with a 5-year survival rate exceeding 70% [9]. Despite many proposed treatments, no treatments for advanced HCC have been officially authorized [10]. It is imperative to advance early detection methods. A study on liver cancer in Italy revealed that patients who received semi-annual surveillance had a 5-year survival rate of 32.7%, while those who received annual surveillance had a rate of 25.2%. Patients who showed symptoms had the lowest survival rate at 12.2%. These findings highly support the idea that a semi-annual HCC screening and surveillance program might increase survival rates [11]. Early diagnosis is, therefore, essential for managing HCC and offers a variety of therapeutic options, including microwave ablation, hepatectomy, and hepatic transplantation. The US Food and Drug Administration (FDA) has approved lenvatinib and the tyrosine kinase inhibitors, such as sorafenib as the initial treatments for HCC; for patients with serum levels of AFP below 400 ng/mL, cabozantinib, ramucirumab, and regorafenib have been authorized as second-line treatments [12–14]. Nivolumab and pembrolizumab's phase III trials for anti-tumor activity have been unsuccessful in achieving their primary endpoints [15]. Recently, a phase III study combined bevacizumab and atezolizumab and revealed objective response rates of almost 30% in HCC patients [16]. On the other hand, selective internal radiation therapy (SIRT), a systemic approach using tyrosine kinase inhibitors, and transarterial chemoembolization (TACE), all improved survival rates [17]. The patients who receive these treatments typically have a high recurrence rate because they are only effective in the early stages of HCC [18].
With so many systemic therapies available, it is essential to create the right tools to group patients based on how they respond. For early diagnosis, prognostic assessment, molecular stratification, therapy prognosis, and monitoring of tumor progression, a non-invasive longitudinal strategy is necessary in HCC patients. An accessible liquid biopsy-based method has quickly emerged as a favorable diagnostic tool in the cancer research field [19]. The liquid biopsy is comprised of a variety of elements, including circulating tumor cells, their DNA, cell-free DNA, circulating miRNAs, and EVs [17].
Liquid biopsy offers several benefits, including non-invasive detection and characterization, treatment response prediction, toxicity evaluation for non-responders, prevention of discarding highly effective drugs, identification of resistance mechanisms, and optimization of future clinical trial design [20]. In this review, we emphasized the most pertinent molecular data on liquid biopsy applications in HCC treatment and discussed potential novel molecular targets for future research.
Current and potential diagnostic biomarkers
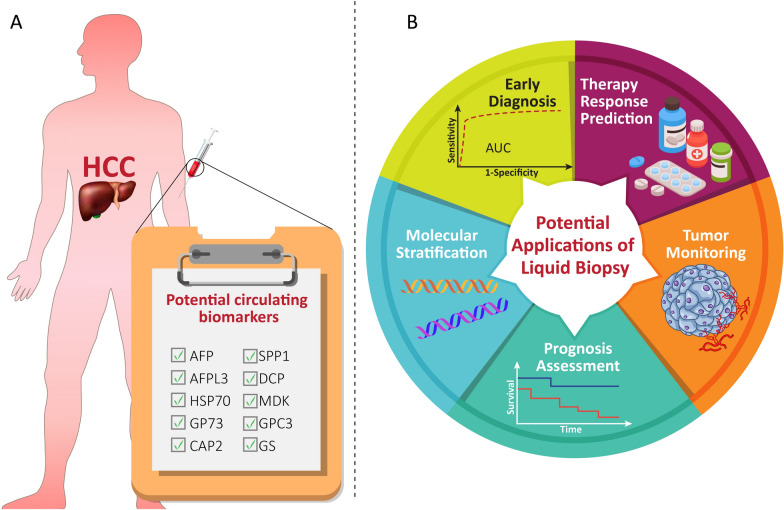
Imaging and serological tests are two key techniques extensively used for HCC screening. The main problem with HCC screening is that these techniques don’t perform well for diagnosis when the tumor is small or in an early stage. Alpha-fetoprotein (AFP) has traditionally been checked in HCC patients as a screening biomarker. AFP, used alongside MRI, CT, and ultrasound scanning, is the primary biomarker in the fifth phase for monitoring HCC [19]. However, AFP assessment is typically ineffective at identifying early HCC cases and has a low sensitivity for diagnosing HCC (47–64%) [21, 22]. Combined AFP measurement and ultrasonography have a 63% sensitivity for early-stage HCC diagnosis [23]. Scientists are developing new indicators that can identify the presence of HCC at an early stage, predict its chances of recovery, and determine the effectiveness of treatment to overcome this limitation. Osteopontin (SPP1), Midkine (MDK), and GALAD are potential options that have not yet been officially approved [24] (Fig. 1A). MDK demonstrates diagnostic potential in early-stage and AFP-negative HCC patients [25, 26]. To identify HCC at an early stage and forecast the probability of patient survival, GALAD uses a statistical algorithm that utilizes age, gender, serum levels of AFP, AFP-L3, and DCP. This model might be an effective tool for following high-risk people. When comparing AFP, AFP-L3, DCP, and GALAD individually, GALAD showed a greater ability to accurately detect HCC in three different cohorts (Germany, Japan, and Hong Kong). This means it had a higher percentage of correctly identifying individuals with and without HCC compared to other biomarkers. GALAD, a recently developed method for monitoring HCC, is currently undergoing rigorous evaluation through clinical trials [27]. Other potential markers for early detection of HCC include glypican-3 (GPC3), heat shock protein 70 (HSP70), glutamine synthetase (GS), cyclase-associated protein 2 (CAP2), and Golgi protein 73 (GP73). However, these markers have not yet been validated through clinical studies [21, 28].
Fig. 1.
Circulating biomarkers and applications of liquid biopsy. A This section represents the conventional and potential circulating biomarkers identified in patients with HCC. B This section demonstrates the potential applications of liquid biopsy as a non-invasive strategy for managing HCC patients. Liquid biopsy offers a promising method for early diagnosis, prediction of therapy response, tumor monitoring, prognosis assessment and, molecular stratification
Emerging biomarkers: liquid biopsies
Liquid biopsies can serve as a non-invasive tool with a wide range of potential applications (Fig. 1B), as they contain a diverse range of materials released from primary or secondary tumors [29]. Recently, a number of liquid biopsy techniques have emerged, aiming to assess biomarkers for HCC and providing useful information through non-intrusive means in order to achieve timely detection [30]. Additionally, for individuals who are not candidates for surgery, non-invasive diagnostic procedures can serve as the foundation for mutation profiling. The association between mutations that were identified in plasma samples and tissue specimens is typically over 70% [31].
Circulating tumor cells (CTCs) as biomarker
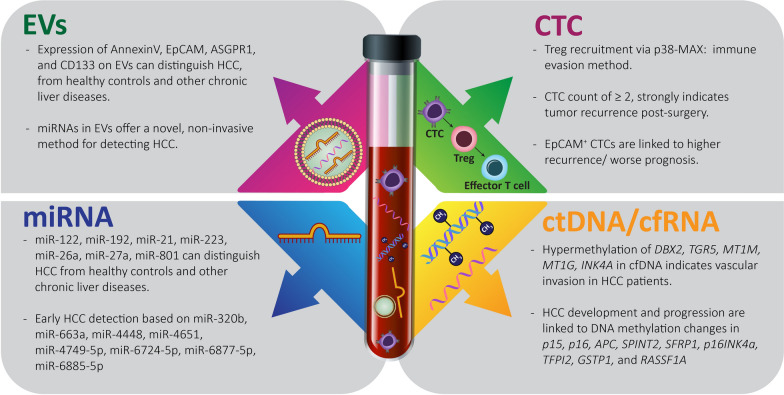
CTCs, a diverse group of cells, hold critical tumor data and provide a more precise representation of tumor heterogeneity compared to a tissue biopsy [32, 33]. The most typical mechanism of HCC metastasis is hematogenous dissemination [34]. To promote better results for HCC patients, further investigations into the fundamental mechanisms involved are imperative. CTCs act as "seeds" for intrahepatic and extrahepatic metastasis (EHM) in HCC, according to several studies [35]. CTCs are highly dependable biological markers that can detect and track early relapses of hepatocellular carcinoma (HCC), as they spread throughout the stages of tumor development. For many years, the study of CTCs has been hindered by their extremely low frequencies in the bloodstream [36]. During dispersion, CTCs are exposed to a number of stressors imposed by circulation microenvironments, such as anoikis, oxygen/nutrient deprivation, immune surveillance, and shear forces. Before effective colonization can occur, the aforementioned challenges must be overcome [37]. Therefore, focusing on CTCs and researching the changes that take place during their hematogenous spread could result in the identification of novel tumor metastatic pathways. During hematogenous transportation, the EMT process in CTCs was dynamically activated [38]. Table 1 summarizes the clinical research on the efficacy of CTCs in HCC cases. CTCs possess the capability to modify their physical characteristics and molecular communication patterns in order to adapt and thrive in the challenging circulatory microenvironment, allowing them to spread and establish themselves in distant regions [39]. The single-cell RNA sequencing (scRNA-seq) findings of CTCs indicated a considerable diversity among both intravascular and intervascular areas. The study compared CTCs from nearby vascular regions to uncover the spatiotemporal changes in gene expression related to immune-evasion signaling and the cell cycle that takes place along the CTCs’ journey through the blood vessels. It was revealed that the stimulation of CTC immune evasion (Fig. 2) is mainly dependent on the activation of regulatory T cells (Tregs) through TGFβ1-p38-MAX signaling [40]. Moreover, to overcome the immune surveillance obstacle, circulating tumor cells (CTCs) have the ability to heighten the levels of immunosuppressive chemokines such as CXCL5 and CCL5, facilitating the recruitment of neutrophils and Tregs, respectively. By modifying the antitumor immune response, these immunosuppressive cells will aid CTC proliferation. CTCs may stimulate adaptive stress response pathways to regain cellular homeostasis. This increases their phenotypic heterogeneity and their capacity to withstand stress. The peripheral CTCs' adaptation-related evolution is explained by the transcriptional diversity found there [41].
Table 1.
List of ongoing clinical studies to evaluate CTCs in HCC patients after different interventions
| Participants | Intervention | Objective-result/status | NCT number | |
|---|---|---|---|---|
| 1 | 57 | TACE | A decrease in CTC count after surgery indicates that postoperative TACE is effective in preventing the recurrence of HCC/Completed | NCT02032368 |
| 2 | 220 | Propofol, Sevoflurane | Recruiting | NCT04601961 |
| 3 | 150 | 1.curative surgery or radiofrequency ablation therapy 2. TACE 3. systemic therapy | Recruiting | NCT01930383 |
| 4 | 184 | FOLFOX4(infusional FU, LV, and OXA) | Recruiting | NCT04521491 |
| 5 | 45 | Radiotherapy | Recruiting | NCT02066974 |
| 6 | 200 | Hepatic resection | Recruiting | NCT04800497 |
| 7 | 100 | Sorafenib | Completed | NCT01481805 |
| 8 | 300 | CTC Capture for early diagnosis and postoperative tumor recurrence monitoring of liver Cancer /Recruiting | NCT04688606 | |
| 9 | 37 | Durvalumab + SBRT | Recruiting | NCT04913480 |
| 10 | 50 | PD-1/ Sorafenib | Recruiting | NCT04152356 |
| 11 | 200 | Recruiting | NCT02036216 | |
| 12 | 256 | TACE/ Epirubicin/ lipiodol | Not yet recruiting | NCT02631499 |
| 13 | 40 | - | Recruiting | NCT04506398 |
| 14 | 20 | IRE & multiple NK immunotherapies | Completed | NCT03008343 |
| 15 | 167 | Sorafenib, RFA or surgery, Everolimus, Lanreotide | Completed | NCT02973204 |
| 16 | 25 | Temsirolimus | Recruiting | NCT01567930 |
| 17 | 18 | COMBIG-DC (ilixadencel) | Completed | NCT01974661 |
Fig. 2.
Novel and emerging biomarkers: EVs, CTCs, miRNAs, and ctDNA/cfRNAs are the known liquid biomarkers. These biomarkers can be detected in circulation, making them useful for non-invasive diagnostic tests. They provide valuable information about the presence and progression of diseases, helping in early detection and personalized treatment strategies
Combinations of various biomarkers, including pAkt and pERK [42], EMT markers (vimentin and twist) [43], Survivin and MAGE-3 [44], Glypican-3, CK, and CK [45], taMPs, Annexin V, ASGPR1 and EpCAM, [46] were used to detect CTC in order to assess the status and prognosis of metastasis, as well as to track the effectiveness of sorafenib. Despite significant advancements in single-cell RNA sequencing (scRNA-seq) of CTCs [47], a substantial knowledge gap remains regarding the plasticity and adaptive mechanisms of CTCs during their dissemination [48]. The effectiveness of CTC-based monitoring for HCC can be improved by integrating it with additional sophisticated techniques, such as high-throughput microfluidic CTC-iChip and RNA-based digital PCR, which can aid in the detection of specific CTC-derived signals. Kalinich et al. demonstrated this method by first isolating CTCs using CTC-iChip technology and then profiling them using RNA-based digital PCR. These findings have the potential to function as gauges indicating the likelihood of HCC being present. The above mentioned report resulted in a predictive value of 83% for negative cases and 86% for positive cases with cirrhosis as an underlying condition [49].
The analysis of CTCs has faced early difficulties [50]. CTCs are challenging to find in the early stages of the illness because they are incredibly uncommon in circulation and their number correlates with tumor volume [51]. Instead of early diagnosis, they are probably more useful for prognosis. A higher number of circulating tumor cells (CTCs) indicates a bleak prognosis, suggesting a heightened chance of cancer recurrence and a decreased likelihood of survival. Following some treatments, particularly surgical therapies, cancer cell dissemination appears to be facilitated. Spreading CTCs is associated with liver manipulation [52]. Ha et al. found that fluctuations in CTC counts after hepatectomy are independently associated with reduced survival rates and increased tumor recurrence [53]. One potential breakthrough in personalized medicine could be achieved by developing methods for isolating live CTCs from patients with HCC and transforming them into three-dimensional structures resembling spheroids. After obtaining and growing CTCs, Zhang et al. looked into how sensitive each person was to oxaliplatin and sorafenib [54]. In the future, various therapeutic candidates may be assessed using xenografts [55–57]. Significant temporal heterogeneity in the morphology of CTCs after anticancer therapy has been found, which is similar to primary tumor’s heterogeneity [38, 58]. However, the anatomically distinct areas of the human circulatory system's CTC spatial heterogeneity have remained elusive. [48].
The CTC population in liver-efferent vessels was remarkably diverse. After they enter the bloodstream, tumor cells engage in interactions with platelets. CTCs are physically shielded and protected from immune cell attacks and blood shear force damage by the formation of tumor cell-platelet microaggregates [59]. It is possible to predict lung and liver metastases by counting CTCs and looking for CTC clusters in hepatic and peripheral veins [60].
When serum AFP was combined with CTC detection, the performance in identifying HCC patients was improved compared to AFP alone [61]. Single-cell mRNAseq in people with HCC led to genome-wide transcriptome profiling for accurate CTC detection, underscoring its possible role in identifying HCC driver genes and its ability to check the heterogeneity of HCC [62]. Recent research has shown that combining mRNA screening and CTC detection can significantly enhance the efficacy of monitoring and diagnosing HCC. For quite some time, therapeutic research in the clinical setting has primarily focused on CTCs. Interestingly, the FDA-approved Cell Search platform has shown a correlation between CTC counts and progression free survival (PFS) and their overall survival (OS) in numerous types of cancer such as prostate, colorectal, and breast [63]. In addition to quantifying CTCs, various characteristics linked to them, including epithelial, mesenchymal, and stem cell identities, as well as CTC clusters, have been thoroughly investigated. These phenotypes were associated with various kinetics and functions [64]. Other studies have shown that higher risk of metastasis is associated with CTCs that have detectable AFP mRNA [65]. Additionally, patients undergoing surgical resection had a lower chance of survival if they had CTCs with aneuploid chromosome 8 [64]. The unique predictive significance of CTC counts was established through the utilization of a combination of biomarkers and the advanced StreamX imaging technology, which can generate detailed images of isolated CTCs at a high resolution [45, 66]. The key components utilized in immune capture techniques to identify circulating tumor cells (CTCs) are human epidermal growth factor receptor (Her2), epithelial cell adhesion molecule (EpCAM), different types of cytokeratins (CK19, CK8, and CK18), and mesenchymal markers such as vimentin and N-cadherin. These markers play a critical role in biological property-based technologies [67]. According to Sun YF et al., the CellSearchTM system (CSS) can identify EpCAM + CTCs in HCC patients. The researchers analyzed blood samples from 123 patients with HCC, both before and after resection. Among these patients, 82 had at least one EpCAM + CTC, while 51 had two or more EpCAM + CTCs. They proposed that detecting two or more CTCs in the preoperative test could strongly predict tumor recurrence in HCC patients with serum AFP levels below 400 ng/mL or a low risk of tumor recurrence [68]. The clinical evidence showed a strong correlation between the amount of EpCAM + CTCs and the levels of AFP in individuals with advanced HCC [69]. CTCs that were positively marked with EpCAM were associated with a more unfavorable outlook and an increased likelihood of tumor recurrence following liver resection [70]. Similarly, Shen et al. showed that an elevated number of EpCAM-positive CTCs following TACE was associated with an unfavorable outlook for individuals with inoperable liver cancer [71]. The presence of EpCAM in only a small portion of HCC tumors implies that EpCAM-based tests may specifically target EpCAM + HCC and may not be reliable in predicting all types of HCC [72]. A recent report indicates that by determining the CTC pERK/pAkt phenotype, one can ascertain the level of sensitivity to sorafenib [42]. Furthermore, the presence of PD-L1-positive CTCs cells results in a notable response from checkpoint inhibitors [73]. The detection of CTCs expressing MAGE3, Survivin, and CEA was linked to a consistent reaction to cryosurgery treatment [44]. Additionally, it was revealed that the presence of pERK + /pAkt or PD-L1 + CTCs in HCC patients was closely associated with the effectiveness of anti-PD-1 treatment or sorafenib [42, 74]. Nonetheless, most of these investigations were limited by their low number of participants and lack of extended future monitoring data [69].
An innovative idea, still in the early experimental stages, involves recovering CTCs and expanding them ex vivo using microfluidic technology. This method aims to facilitate individualized drug susceptibility testing. After receiving FDA approval, medications that target and inhibit tumor growth may undergo standard testing protocols to ensure they are suitable for use. This process identifies and removes ineffective medications against the tumor, particularly those showing resistance in CTCs obtained from patients [75, 76]. Utilizing this cost-effectiveness can contribute to reducing healthcare expenses. Likewise, it may result in the discovery of the most potent individualized cancer treatment and save time for the patient's benefit. Unfortunately, the main barrier to the utility of these strategies is how to prompt a proper CTC expansion. Although there is an experimental protocol to speed up the expansion of CTCs, most protocols still take months [76]. Despite all the advancements, CTCs have not yet gained acceptance as a practical diagnostic tool for HCC.
CTC analysis plays a crucial role in the advancement of tailored medical treatment by providing prompt detection, continuous surveillance, and detailed examination of the tumor. According to the published data, it is not recommended to use CTC assay as an independent method for diagnosis of HCC but may indicate a poor prognosis when combined with complex clinical characteristics [77]. Several matters must be addressed prior to the implementation of CTC analysis in medical environments. The disparity in the detection techniques is one of the main problems. There are many different ways to isolate and detect CTCs, and developing a highly sensitive and targeted technique is very difficult. Thus, it is crucial to validate CTC analysis methods, such as sample preparation, enrichment, and detection. In order to properly assess the effectiveness of CTC detection technologies, it is necessary to conduct large-scale, prospective studies across multiple centers with significant sample sizes and prolonged periods of observation. Despite the fact that, at this time, the application of CTC detection is in a research setting, it is hoped that upcoming technological advancements will make it possible in clinical practice.
Circulating tumor DNA as a molecular biomarker
Cell-free RNA (cfRNA) and cell-free DNA (cfDNA) include circulating tumor RNA (ctRNA) and circulating tumor DNA (ctDNA), respectively. These terms are utilized to characterize nucleic acids that are found circulating freely throughout the body [78]. CfDNA concentrations in healthy people range from 10 to 15 ng/ml [79]. Cell-free DNA can be detected in cells that undergo apoptosis, including myeloid and lymphoid cells. This DNA is in the form of double-stranded fragments that range from 150 to 200 base pairs in length, with a short half-life, lasting less than one hour [80]. Due to programmed cell death and/or active release of DNA from cancerous cells, fragments of tumor DNA that are found in the blood are referred to as circulating tumor DNA (ctDNA) or circulating free DNA (cfDNA) [81], whose allele content ranges from less than 0.01% to more than 60% [82]. The increased quantity of cell death in the form of both apoptotic and necrotic carcinoma cells, which possess a greater level of specificity compared to cfDNA, is principally the underlying cause for the raised levels of ctDNA in the blood and the corresponding quantity in individuals afflicted with tumors. Although the mechanism is unknown, living tumor cells have also been seen to be actively releasing ctDNA [83]. There are various options available for ctDNA testing, which include assays that focus on specific hot spots and variants, as well as more comprehensive next-generation sequencing panels that can analyze mutations in hundreds of genes at once [84]. The same patient's tumor cells and plasma DNA samples should be examined simultaneously for any differential molecular changes that might serve as ctDNA biomarkers. These biomarkers should not be found in healthy individuals [85]. The concentrations of ctDNA can vary from 0.1% to over 90%, which correlates with tumor size and burden. This variability highlights the significance of ctDNA as a non-invasive biomarker for personalized cancer therapy [86]. The attributes of ctDNA are dictated by modifications unique to the tumor, including alterations in methylation patterns, somatic mutations, integration of viral DNA, loss of heterozygosity, and preexisting abnormalities in chromosomes [83]. The detection and evaluation of ctDNA in liquid biopsy-based diagnoses is complex due to the extensive range and inconsistent levels of ctDNA within the background of cfDNA [87]. Because liver cfDNA exhibits a defined size distribution and ends at identified genomic positions, there is no randomness involved in the fragmentation of cfDNA [88]. According to Jiang et al., the composition of ctDNA in blood samples from HCC patients was considerably smaller in comparison to the composition of ctDNA in samples taken from individuals who were in good health [89]. Conducting gene integrity analysis on the size of ctDNA molecules in plasma samples of patients with HCC is a successful method for distinguishing ctDNA from non-tumor-related cfDNA [89]. Moreover, studies have demonstrated that HCC tumor specimens contain circulating cell-free DNA fragments that terminate at specific genomic sites, making them useful as a diagnostic indicator for differentiating individuals with HCC from those without the condition [90]. The assessment of the total amount of ctDNA in the blood sample reflects the progression of cancer and the weight of the tumor [85, 91]. In 2006, Iizuka and colleagues revealed that cfDNA exhibited a sensitivity of 69.2% and a specificity of 93.3%, surpassing the effectiveness of utilizing AFP for detecting HCC patients within a population infected with HCV [92]. In a similar vein, Piciocchi and colleagues discovered that hepatocellular carcinoma (HCC) could be differentiated from individuals with chronic liver disease (CLD) and cirrhosis with a sensitivity of 91%, specificity of 43%, and an area under the curve (AUC) value of 0.69 [93].
The amount of cfDNA in the blood can increase in conditions like cancer, surgery, inflammation, and tissue damage. Two distinct alterations are examined during ctDNA analysis: the quantity of circulating ctDNA and tumor-specific genetic anomalies. A proposed HCC diagnostic index by Yan et al. incorporated cfDNA, age, and AFP, achieving a sensitivity of 87% and a specificity of 100% [85]. Genetic alterations such as copy number variations (CNVs), DNA mutations, gene methylations, and gene fusions are representative instances of the various DNA-related modifications that are encompassed in the term cfDNA, which indicates the condition of DNA within cancerous cells. cfDNA is found in samples of blood, saliva, ascites fluid, pleural effusion, urine, and stool. There is currently disagreement over the source of cfDNA extraction, and many studies prefer plasma as the preferred biological matrix [94]. The amount of cfDNA in plasma is somewhat influenced by the time between sample preparation and analysis, despite the fact that blood cell DNA is less likely to contaminate plasma [95]. It is advised to use distinct tubes for blood collection. When blood processing must be completed within 6 h, EDTA tubes are usually the initial preference, while Streck or CellSave blood collection tubes may be more suitable if the samples require preservation for longer than 6 h [96]. These specifics are essential to the ctDNA extraction procedure and directly impact the method’s stability and precision. Recently, commercial cfDNA testing assays have become available. Instead of an invasive tissue biopsy, these tests could be used to profile a tumor using a plasma sample genetically. Due to the brief amount of time that the plasma cfDNA fragments remain in circulation and the ability to detect variations throughout different regions of a tumor, this technique enabled scientists to analyze cancer genetic alterations in a timely and diverse manner [97]. Notably, elevated levels of cfDNA can also occur in autoimmune and inflammatory circumstances like systemic lupus erythematosus, during pregnancy cirrhosis, chronic hepatitis, rheumatoid arthritis, and following physical activity [98]. The precision of using whole cfDNA as a diagnostic approach may be hindered by its lack of specificity. An astounding investigation revealed that by examining the positioning of nucleosomes, one can accurately identify both the specific cell and original tissue where cfDNA originated from [99] (see Table 2).
Table 2.
List of lnRNA-based biomarkers in early detection, monitoring, and prognosis of HCC
| Name | Biomarker role | Refs. | |
|---|---|---|---|
| 1 | MALAT1 | Regulation of miRNAs (miR-204, miR-143-3p, miR-30a-5p, miR-200a, miR-124-3p, miR-195, and miR-22) involving in EMT | [139] |
| 2 | CTBP | Detection of cirrhotic HCC | [140] |
| 3 | LINC00152, XLOC014172, RP11-160H22.5 + AFP | Differentiating HCC patients from those with cirrhosis, chronic hepatitis, and normal individuals | [141] |
| 4 | Linc00974 | Elevated serum level in HCC patients | [142] |
| 5 | miRNAs miR-21, miR-199, and miR-122 | Upregulation in HCC samples | [145, 146] |
| 6 | miR-145, miR-29a, miR-133a, miR-143, miR-505 miR-29c, and miR-192 | Enhanced accuracy | [151] |
| 7 | miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801 | HCC detection among healthy individuals or cirrhotic patients | [152] |
| 8 | miR-6885-5p, miR-663a, miR-320b, miR-4448, miR-4749-5p, miR-6724-5p, miR-6877-5p, andmiR-4651 | Early stage detection of HCC | [132] |
| 11 | miR-4651 | Poor outcome | [155] |
| 12 | miR-1, miR-122, miR-26a, miR-29a, and miR-223-3p, miR-155, miR-96, and miR-193-5p | Poor outcome | [156] |
| 13 | miR-424-5p and miR-101-3p, miR-128, miR-139-5p, miR-382-5p, and miR-410 | Overall survival | [157] |
| 14 | miR-1246, miR-500 | Predicting early relapse after surgery | [158, 159] |
| 15 | miR-122, miR-26a, miR-2 | Prognostic indicators | [160] |
| 16 | miR-645, miR-200a, miR-122, miR-125b, miR-374b, miR-107, miR-15b, miR-320, and miR-30a | Overall survival | [163] |
For patients with lung cancer, the US FDA has confirmed using ctDNA and an EGFR mutation analysis as a diagnostic test. At present, there is a lack of research on the practical significance of liquid biopsy for advanced HCC [100].
There are two types of ctDNA-based technology platforms: targeted and untargeted. Targeted platforms focus on identifying specific mutations in predetermined genes, like KRAS. Untargeted platforms search the genome for previously unknown genomic changes, such as ones that make tumors resilient to certain targeted treatments. Examples of precise methods developed in recent years for finding single-nucleotide mutations in ctDNA include BEAMing, Capp-Seq, digital PCR, Safe-SeqS, and TAm-Seq. Additionally, whole-genome sequencing (WGS) can detect copy-number alterations and gather more details about tumor samples [101–106]. The discovery of altered genetic sequences in 38.6% of individuals with hepatocellular carcinoma indicates a potential avenue for discovering novel treatment options [107]. Felden et al. conducted an analysis of ctDNA mutations in 121 cases of advanced-stage HCC and suggested that the use of liquid biopsy could serve as a new method for predicting the prognosis of HCC. As per the results of their study, the most commonly mutated regions in the circulating tumor DNA (ctDNA) of advanced HCC patients were TERT (51%), TP53 (32%), CTNNB1 (17%), PTEN (8%), AXIN1, ARID1A, ARID2, KMT2D, BAP1, RB1, KEAP1, and TSC2 (5–10%) [31]. The alteration of TP53 at codon 249 appears to exhibit a distinct level of specificity. Moreover, alterations in DNA methylation at various genes, such as p15 [108], p16 [109], APC [110], SPINT2 [111], SFRP1 [112], p16INK4a [113], TFPI2 [114], GSTP1 [115] and RASSF1A [115], are closely associated with the formation of HCC. A recent analysis investigated genetic alterations and protein markers in multiple forms of stage I cancer through the utilization of a blood-based screening method. The research revealed that the mutation TP53 c.747G > T (p.R249S) was only present in a small percentage (3–4%) of pancreatic and stomach cancer tissue samples, and in 20% of HCC plasma samples. This mutation was not detected in a control group of 812 individuals without any health issues. In the tissue and plasma samples of patients with HCC, mutations in both KRAS and CTNNB1 were discovered at the same time [116]. In addition, individuals diagnosed with Hepatitis B (HBV) exhibited a greater occurrence of ERBB2 gene mutations [117]. Furthermore, it was determined that combining the analysis of cfDNA mutations with protein markers was particularly successful in detecting early-stage HCC among asymptomatic individuals at high risk, such as those who tested positive for HBV surface antigen [117]. According to the results from either tumor biopsies or cfDNA from blood samples, it was found that males with HCV and/or cirrhosis caused by alcohol had the most significant occurrence of TERT promoter mutations. Consequently, identifying cfDNA's TERT promoter mutations could aid in the early detection of HCC among individuals at risk [118].
It is important to note that mutations are more readily identifiable in more severe instances. Out of 7 cases of HCC, nearly all [6] were found to have mutations in their cell-free DNA (cfDNA) if their tumors were > 5 cm or had spread to other areas. Conversely, only 9% of mutations were detected in the cfDNA of individuals with smaller tumors that had not metastasized [118]. The frequency of more commonly occurring mutations among HCC patients can differ among different groups. Among HCC patients in multiple European groups, ARID1A had the highest frequency of mutations in ctDNA [119]. The quantity of somatic mutation locations present in ctDNA serves as an indicator of a genetic modification in the underlying cancer and exhibits a direct relationship with the level of tumor mass in patients with HCC. Patients diagnosed with HCC have a significantly elevated rate of mutated genes (including TERT, CTNNB1, ARID1A, RAS, TP53, and AXIN) detected within their ctDNA [17]. The mentioned ctDNA mutant genes have been associated with numerous crucial signaling pathways. HCC is shaped and advances due to the maintenance of telomeres, activation of the MAPK/RAS pathway, functioning of the p53 signaling pathway, activity of the Wnt-β catenin pathway, and involvement of the SWI/SNF complex-related pathway [120].
The identification of numerous hypermethylated genes as vascular invasion biomarkers included DBX2 [121], TGR5 [122], MT1M, MT1G [122], and INK4A [123], in the cfDNA of patients with HCC. Although hypermethylation at multiple genes has been shown as a potential biomarker to diagnose HCC, there is currently no universally recognized algorithm utilizing this information in different institutions or facilities [36]. Transformations in the methylation pattern of ctDNA take place at the initial stages of cancer progression and have the potential to be reversed, rendering them valuable indicators for prompt cancer detection and the integration of preventive strategies. Moreover, in both pathological and physiological states, the unique methylation pattern of each cell type remains unaltered and exceptionally steadfast [51, 124]. The findings have indicated that in 92% of HCC patients, the simultaneous presence of p15 and p16 methylation was identified [108]. The promoter of the Ras association domain family 1A (RASSF1A) was hypermethylated in almost 90% of HCC patients. A modification in the methylation arrangement of this gene has the capability to differentiate between individuals with HCC and those who are healthy, as well as chronic HCV-positive patients. The predictive accuracy of this distinction is 77.5% for HCC individuals and 72.5% for chronic HCV-positive patients [125]. Therefore, detecting an atypical methylation pattern in the promoter region of ctDNA may serve as a useful indicator for early detection of HCC, specifically in individuals at high risk, including those with smaller tumors [126].
After examining the methylation patterns of p15, p16, and RASSF1A in the serum of 50 HCC patients, it was determined that there is a high predictive accuracy of 89%. This was achieved through 84% sensitivity and 94% specificity, indicating the reliability of the results [109]. An algorithm of the four genes, APC, GSTP1, RASSF1A, and SFRP1, with 92.7% sensitivity and 81.9% specificity, accurately differentiated individuals with HCC from those who were not afflicted, achieving 92.7% accuracy in identifying positive cases and 81.9% accuracy in correctly identifying negative cases [112]. Furthermore, a combination of data from one specific miRNA (miR-203) and three genes exhibiting noticeably elevated levels of methylation (APC, COX2, and RASSF1A) was utilized to construct a forecasting model. The proposed model successfully identified over 75% of patients who were not diagnosed using AFP levels of > 20 ng/mL [127]. The threshold of 73.0 ng/ml effectively distinguishes between the individuals with HCC and those with HCV, achieving a sensitivity of 69.2% and a specificity of 93.3%. Combining cfDNA screening with AFP data has a 50% sensitivity for HCC diagnosis. This can enhance the accuracy of HCC detection [92]. A notable observation is that a panel utilizing multiple parameters, such as age, cfDNA, and AFP, was more effective in diagnosing with a sensitivity of 87% and specificity of 100%, with an AUC of 0.98. This outperformed the use of each parameter individually [85]. Further research is necessary to ascertain the feasibility of utilizing both cfDNA quantification data and somatic mutation analysis as a joint approach for screening HCC. Moreover, the varying employment of detection techniques among studies has resulted in the current debate surrounding the optimal threshold for an elevated level of cfDNA [126]. In contrast, Xu and colleagues discovered that the DNA methylation profiles of HCC tumor DNA were congruent with 10 distinct biomarkers in circulating ctDNA. They suggested creating a new method for predicting diagnoses called the diagnostic algorithm prediction model. This model would utilize cfDNA samples from a significantly large number of 1098 HCC patients, and it would have a higher level of specificity and sensitivity compared to the currently used AFP method. In addition, the model showed a robust correlation with the extent of tumor growth, treatment effectiveness, and disease stage [128].
ctDNA, in addition to timely detection, plays a crucial role in predicting outcomes for HCC. The measurement of cfDNA levels can serve as a distinct indicator for the potential of HCC recurrence and spread to other organs, as it has a significant inverse correlation with both disease-free survival and overall survival [129, 130]. According to the findings of Tokuhisa et al., individuals diagnosed with HCC as a result of HCV infection were more likely to develop metastases and experience a shorter overall survival (OS) if they underwent a liver resection procedure [129]. Based on various evidence, it has been found that patients who have undergone liver transplantation, and liver resection, and have been prescribed sorafenib experience a more negative outcome if they have high levels of cfDNA [130, 131].
Using targeted ctDNA analysis is an effective method for identifying intra-tumoral heterogeneity and can also serve as a prognostic indicator for a negative outcome. The most frequently observed mutations in ctDNA included hotspot variants such as CTNNB1, TP53, and TERT. The extent of diversity within the mutated alleles found in 27 out of 48 patients (56.3%) with HCC ranged significantly, from as low as 0.33% to as high as 23.7% [132]. A separate investigation utilizing the MiSeqTM technique revealed a correlation between the presence of vascular invasion in patients and the presence of ctDNA mutations, ultimately pointing to a decreased duration of disease-free survival [133]. Furthermore, three-year disease-free survival (DFS) was linked to allelic imbalance on chromosome 8p at D8S264 in plasma ctDNA samples. DFS was also found to be correlated with the detection of allelic imbalance at the D8S258 locus and the levels of circulating ctDNA [134]. The constraints of tumor variety can be typically surmounted, and the future outlook for HCC can be determined through continually identifying mutations related to the tumor in ctDNA. Mutations found in the bloodstream are not solely derived from cancer, inherited mutations can also be identified in plasma specimens. The source of the tumors must be determined by analyzing similar tumor samples [126].
Cell-free non-coding RNAs as molecular biomarker
Long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) make up the majority of known groups of RNAs that have been found in circulation as potential disease biomarkers. RNA molecules of varying lengths, with long sequences exceeding 200 base pairs and shorter ones of only 28 base pairs, are present in the group of cell-free/non-coding RNA. These RNAs have an essential role in regulation of gene expression. A vast number of lncRNAs have been associated with cancer progression and can influence gene expression through various mechanisms. These include recruiting regulatory protein complexes, acting as molecular decoys, altering genome structure, and modulating posttranslational modifications [135–137]. HCC demonstrates irregular expression of HULC, MEG3, HOTAIR, HOTTIP, MALAT-1, and MVIH, making them potential biomarkers for the disease [138–140]. MALAT1 is a significant oncogenic lncRNA in HCC that has the ability to sequester a number of miRNAs. MALAT1 can sequester miR-140 and, therefore, enhances VEGF-A expression. Besides, MALAT1 activation causes macrophages to polarize to the M2 state and promotes tumor growth by modifying the tumor microenvironment [141]. MALAT1 controls the miRNAs that can promote cell proliferation, EMT, and invasion, including miR-204, miR-143-3p, miR-30a-5p, miR-200a, miR-124-3p, miR-195, and miR-22 [142]. The lncRNA CTBP exhibited superior specificity and sensitivity when compared to other RNA-based biomarkers in detecting cirrhotic HCC in contrast to healthy individuals [143]. Together with AFP, the levels of LINC00152, XLOC014172, and RP11-160H22.5 in the bloodstream possess the capability to differentiate individuals with HCC from those with cirrhosis, chronic hepatitis, and normal individuals with exceedingly precise accuracy [144]. HCC patients’ serum levels of the long intergenic non-protein coding RNA 974 (Linc00974) were elevated [145].
Seventy or even more miRNAs have been proposed as novel molecular biomarkers [146]. The EV-loaded miRNAs or linked to those certain proteins argonaute 2, nucleomorphin 1, or lipoproteins (LDL, HDL) can be found in circulation [147]. A meta-analysis suggests that the significantly upregulated miRNAs miR-21, miR-199, and miR-122 demonstrate strong potential as dependable biomarkers for diagnosing HCC due to their increased specificity [148, 149].
However, it should be noted that research on circulating miR-21 in HCC has produced conflicting results. Some studies report overexpression of miR-21 [150, 151], and others indicate downregulation in HCC; these discrepancies highlight the complexity of miR-21’s role and the need for further investigation to clarify its function in this type of cancer [132, 152]. A diverse range of panels consisting of numerous circulating miRNAs has been assessed to enhance the accuracy of HCC diagnosis, as the effectiveness of a single miRNA is limited. Research has demonstrated that a group of specific miRNAs (miR-145, miR-29a, miR-133a, miR-143, miR-505, miR-29c, and miR-192) have a greater area under the curve (AUC) compared to using just a single serum AFP when identifying small early-stage HCC tumors and those that are AFP-negative [153]. A distinct set of miRNAs, namely miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801, were utilized by scientists to effectively identify cases of HCC among individuals who were unaffected, had chronic hepatitis B, or had cirrhosis [154]. In a comparable manner, a model computational process utilizing the assessment and identification of eight specific miRNAs (miR-6885-5p, miR-663a, miR-320b, miR-4448, miR-4749-5p, miR-6724-5p, miR-6877-5p, andmiR-4651) effectively identified HCC in its early stages by analyzing serum samples from patients with an accuracy of 97.7% and precision of 94% (Fig. 2) [155].
It remains unclear whether miRNAs are expressed and secreted into the bloodstream by HCC cells, non-HCC cells, or other cells within or beyond the tumor microenvironment. Research has shown that several miRNAs have irregular expression levels in cancer. For example, miR-663a levels were significantly lower in HCC tissue compared to the surrounding non-tumor tissue. This reduction inhibited the proliferation and invasion of HCC cells by targeting the High-mobility group AT-hook 2 (HMGA2) [156]. Furthermore, research revealed that the levels of miR-320b were diminished in multiple types of cancer, such as colon cancer, leading to the activation of c-Myc [157]. In addition, individuals who had HCC and were positive for aflatoxin B1 displayed heightened levels of miR-4651 in their bloodstream, and this rise was often linked to an unfavorable outcome [158]. Furthermore, the levels of miR-1, miR-122, miR-26a, miR-29a, and miR-223-3p were reduced in the blood, whereas miR-155, miR-96, and miR-193-5p were increased and linked to an unfavorable outcome [159]. An investigation analyzed the complete miRNome profile of 116 individuals with HCC, discovering six miRNAs that act as indicators of prognosis. Specifically, reduced levels of miR-424-5p and miR-101-3p, as well as increased levels of miR-128, miR-139-5p, miR-382-5p, and miR-410, were linked to decreased survival rates [160]. An increase in miR-1246 levels, along with a decrease in miR-500 and miR-224 levels in the bloodstream, could serve as potential indicators for predicting early relapse after a surgical procedure [161, 162]. Higher and lower expression of miR-122, miR-26a, and miR-2, respectively, are prognostic indicators in people undergoing radiofrequency ablation [163]. Also some researchers discovered that evaluating miRNAs could help predict how people would react to TACE [164] or sorafenib [165]. The latest study, which examined plasma samples from individuals enrolled in the regorafenib registration trial (RESOURCE), discovered several plasma miRNAs (miR-645, miR-200a, miR-122, miR-125b, miR-374b, miR-107, miR-15b, miR-320, and miR-30a) that were found to correlate with overall survival (OS) [166].
Extra cellular vesicles as an innovative biomarker
EVs are small sacs surrounded by a membrane that are actively released by both non-diseased and cancerous cells. Their dimensions vary between 40 and 150 nm, and they transport a diverse array of cellular components, such as proteins, lipids, and different forms of RNA (mRNA, DNA, and non-coding RNA). Through the process of attaching to different cells, they are able to transfer their contents and subsequently regulate the internal pathways of the recipient cells. Tumor cells release EVs that often reflect the molecular characteristics of the original tumor. EVs can be acquired without the need for invasive procedures and serve as biomarkers in the bloodstream, making them ideal for screening and identifying these specific markers [35]. Moreover, the study conducted by Aydin et al. revealed that reductions in both exosome levels and eGPC3 levels after treatment had correlated more strongly with positive outcomes from locoregional chemotherapy among individuals with HCC waiting for a liver transplant, compared to changes in AFP levels in the blood [167]. Compared to the controls, HCC individuals had increased levels of circulating EVs. In order to make a distinction between HCC from other cancers, cirrhotic patients, and healthy individuals, another subpopulation of extracellular vesicles covered with ASGPR1, AnnexinV, EpCAM, and CD133 was recognized (Fig. 2) [46]. The majority of studies have concentrated on examining the composition of EVs rather than their overall quantity. It has been reported that the diagnostic accuracy of the polymeric immune receptor (PIGR) and 3-binding protein (LG3BP) is higher than AFP alone [168]. EV-derived AFP, GPC3, and hnRNPH1 mRNA hold promising potential as biomarkers for early detection and monitoring of HCC [169, 170]. Elevated levels of CCT8 and CFL1 in blood samples are associated with more advanced tumor stages, evidence of vascular invasion, and decreased rates of overall and disease-free survival. CCT8 and Cofilin-1, two proteins that are highly present in exosomes derived from HCC, have been recognized as promising circulating biomarkers for the prognosis and diagnosis of HCC patients [171]. There is an interesting study report on the various cargos found in the EVs derived from tumor cells [172].
EV microRNAs were examined in multiple studies. According to some studies, AFP and EVs miRNAs have similar diagnostic accuracies [173, 174], while others proved the latter's superiority [169, 175]. Elevated miR-21-5p and miR-144-3p levels were found in EVs, while EV-free serum showed no significant changes. The identification of miRNAs in serum has been greatly improved due to the presence of EVs, thereby highlighting that miRNAs are significantly abundant in serum EVs [173]. In their study, Li and colleagues discovered that there were no changes in the quantity of plasma microparticles measuring 50–400 nM and the amount of miR-144 they contained. However, observed a significant increase of nearly four times in the precursor of miR-144 within the extracellular vesicle pellet. This suggests that compared to serum alone, the detection of miRNAs in EVs is more sensitive [176]. The discovered evidence indicated that the presence of miRNAs within EVs could potentially present a new and non-intrusive method for diagnosing liver cancer. The findings from a research study utilizing miR-21 in serum as a biomarker for HCC demonstrated that individuals with HCC had notably elevated levels of miR-21 in their serum compared to those who were healthy donors [177]. Additionally, Wang and colleagues showed that the levels of miR-21 in extracellular vesicles were markedly higher than those found in the depleted supernatants or complete serum. Furthermore, in comparison to individuals without any medical conditions, subjects diagnosed with hepatocellular carcinoma exhibited substantially elevated expression of miR-21 in exosomes within their serum [178]. A recent investigation discovered that individuals with chronic liver inflammation exhibited elevated levels of miR-21 in their bloodstream compared to those affected by HCC [179].
Few studies have examined EVs as a prognosis biomarker. Most of them were primarily concerned with evaluating EV-derived miRNAs, particularly following surgical treatments [180, 181]. miR-21 was found to be a common miRNA in multiple studies, and high levels of this miRNA have repeatedly been linked to poor survival and increased risk of progression [181]. A worse prognosis was associated with low levels of EV-derived miR-718, miR-125b, miR-638, and miR-320a [182, 183], and high levels of EV-derived miR-665 and miR-10b in other studies [181, 184]. Similarly, AUC values for a 4-miRNAs panel, including miRNA-200a miRNA-21, miRNA-122, and, miRNA-10b, were higher in the diagnosis of HCC than those for AFP. (0.943 vs. 0.826) in a reproducible animal model of HCC [185]. EVs and their content may hold great promise as diagnostic and prognostic biomarkers in HCC patients. Additionally, although miR-148a alone significantly outperformed AFP, patients' early HCC was more easily distinguished from liver cirrhosis when miR-122, miR-148a, and AFP were combined [169]. However, larger prospective studies are needed to establish their function as biomarkers in liquid biopsy. Several challenges and hurdles must be addressed before they can be applied in a clinical setting. One of the primary challenges is the isolation and purification of EVs. Current methods, such as ultracentrifugation, size-exclusion chromatography, and immunoaffinity capture, often result in low yield and purity. Also, the cost of EV-based liquid biopsies is currently high, limiting their accessibility [186]. Moreover, there is a lack of standardized protocols for the characterization of EVs. The heterogeneity of EVs, which vary in size, composition, and origin, complicates their analysis. Standardizing these protocols is crucial for ensuring reproducibility and reliability in clinical settings [172].
Perspective and future directions
Multi-cancer early detection (MCED) approach provides a new horizon for the early detection of a variety of cancers simultaneously [187]. In this context, CancerSEEK was a project through which the detection of eight different cancers was evaluated by taking advantage of mutation analysis and the presence of eight proteins [116]. Notably, the sensitivity of such a test for early-stage liver cancer (stage I) was estimated at 100%. The high sensitivity of the outlined test proposes it as a screening test for early detection of HCC. One of the key issues in reducing cancer mortality is the screening process. The DETECT-A study was a prospective trial that followed participants with false-positive results from the CancerSEEK program. The findings confirmed the reliability of an image-based diagnostic approach, showing that 98% of those with false-positive results remained cancer-free after 3.5 years of follow-up [188]. Another MCED study, OncoSeek, utilized a panel of seven protein tumor markers and AI to detect multiple cancers, including liver cancer. The OncoSeek test showed an overall sensitivity of 51.7% with liver cancer detection sensitivity ranging from 31.1% to 77.6%. This approach significantly reduced false positive results and improved sensitivity to 92.9% [189].
Conclusions
In summary, considering the difficulties in monitoring HCC and determining treatment response, better methods for creating superior biomarkers are crucial. There is growing evidence that suggests the use of liquid biopsies as potential substitutes for tissue biopsy as the diagnostic and prognostic biomarkers in HCC. However, these studies are generally small and lack statistical power to draw firm conclusions about the disease parameters associated with approved detection biomarkers. Liquid biopsies provide a more easily accessible source of molecular data, which might enable noninvasive diagnosis and characterization of the primary tumor. Artificial intelligence (AI) plays a pivotal role in this context by analyzing complex datasets from liquid biopsies, enhancing the accuracy and reliability of biomarker detection. AI algorithms can integrate various clinical and molecular data, improving the sensitivity and specificity of HCC detection and monitoring, ultimately leading to better patient outcomes.
Acknowledgements
We would like to express our sincere gratitude to our colleagues at Royan Institute, Liver Research Group for their support. E.M. would like to acknowledge the support from the National Institute of Biomedical Imaging and Bioengineering (5T32EB009035).
Author contributions
RS and MP drafted manuscript and contributed substantially to this manuscript. HM and MH reviewed the draft and critically contributed to reviewing the manuscript. AZ created the illustrations. MV developed the concept, contributed to drafting and critical review. MV and HM approved the manuscript for submission.
Funding
Not applicable.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hamed Mirzaei, Email: mirzaei-h@kaums.ac.ir, Email: h.mirzaei2002@gmail.com.
Massoud Vosough, Email: masvos@yahoo.com.
References
- 1.Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77(6):1598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chouhan S, Singh S, Athavale D, Ramteke P, Vanuopadath M, Nair BG, et al. Sensitization of hepatocellular carcinoma cells towards doxorubicin and sorafenib is facilitated by glucosedependent alterations in reactive oxygen species, P-glycoprotein and DKK4. J Biosci. 2020;45:1. [PubMed] [Google Scholar]
- 3.Athavale D, Chouhan S, Pandey V, Mayengbam SS, Singh S, Bhat MK. Hepatocellular carcinoma-associated hypercholesterolemia: involvement of proprotein-convertase-subtilisin-kexin type-9 (PCSK9). Cancer Metab. 2018;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chouhan S, Singh S, Athavale D, Ramteke P, Pandey V, Joseph J, et al. Glucose induced activation of canonical Wnt signaling pathway in hepatocellular carcinoma is regulated by DKK4. Sci Rep. 2016;6:27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61(1):191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tapper EB, Parikh ND. Mortality due to cirrhosis and liver cancer in the United States, 1999–2016: observational study. BMJ. 2018;362:k2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D, Li AA, Perumpail BJ, Gadiparthi C, Kim W, Cholankeril G, et al. Changing trends in etiology-based and ethnicity-based annual mortality rates of cirrhosis and hepatocellular carcinoma in the United States. Hepatology. 2019;69(3):1064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thrift AP, El-Serag HB, Kanwal F. Global epidemiology and burden of HCV infection and HCV-related disease. Nat Rev Gastroenterol Hepatol. 2017;14(2):122–32. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7(3):308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shokoohian B, Negahdari B, Aboulkheyr Es H, Abedi-Valugerdi M, Baghaei K, Agarwal T, et al. Advanced therapeutic modalities in hepatocellular carcinoma: novel insights. J Cell Mol Med. 2021;25(18):8602–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cucchetti A, Trevisani F, Pecorelli A, Erroi V, Farinati F, Ciccarese F, et al. Estimation of lead-time bias and its impact on the outcome of surveillance for the early diagnosis of hepatocellular carcinoma. J Hepatol. 2014;61(2):333–41. [DOI] [PubMed] [Google Scholar]
- 12.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–73. [DOI] [PubMed] [Google Scholar]
- 13.Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018;379(1):54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(2):282–96. [DOI] [PubMed] [Google Scholar]
- 15.Finn RS, Ryoo BY, Merle P, Kudo M, Bouattour M, Lim HY, et al. Pembrolizumab as second-line therapy in patients with advanced hepatocellular carcinoma in KEYNOTE-240: a randomized, double-blind, phase III trial. J Clin Oncol. 2020;38(3):193–202. [DOI] [PubMed] [Google Scholar]
- 16.Cheng A-L, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. Updated efficacy and safety data from IMbrave150: atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J Hepatol. 2021. 10.1016/j.jhep.2021.11.030. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Li J, Gassa A, Buchner D, Alakus H, Dong Q, et al. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int J Biol Sci. 2020;16(9):1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Sun H, Wang Z, Cong W, Wang J, Zeng M, et al. Guidelines for the Diagnosis and Treatment of Hepatocellular Carcinoma (2019 Edition). Liver Cancer. 2020;9(6):682–720. [DOI] [PMC free article] [PubMed]
- 20.Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E, Viatour P. Hepatocellular carcinoma: old friends and new tricks. Exp Mol Med. 2020;52(12):1898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo SI, Kim HS, Kim WJ, Shin WG, Kim DJ, Kim KH, et al. Diagnostic value of PIVKA-II and alpha-fetoprotein in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2015;21(13):3928–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154(6):1706-18.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta S, Parikh ND. Biomarker development for hepatocellular carcinoma early detection: current and future perspectives. Hepatic oncology. 2017;4(4):111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thokerunga E, Kisembo P, FangFang H, Zi W, Yu Z, Bongolo CC, et al. Serum midkine for AFP-negative hepatocellular carcinoma diagnosis: a systematic review and meta-analysis. Egypt Liver J. 2023;13(1):25. [Google Scholar]
- 26.Zhu W-W, Guo J-J, Guo L, Jia H-L, Zhu M, Zhang J-B, et al. Evaluation of midkine as a diagnostic serum biomarker in hepatocellular carcinoma. Clin Cancer Res. 2013;19(14):3944–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berhane S, Toyoda H, Tada T, Kumada T, Kagebayashi C, Satomura S, et al. Role of the GALAD and BALAD-2 serologic models in diagnosis of hepatocellular carcinoma and prediction of survival in patients. Clin Gastroenterol Hepatol. 2016;14(6):875-86. e6. [DOI] [PubMed] [Google Scholar]
- 28.Di Tommaso L, Destro A, Seok JY, Balladore E, Terracciano L, Sangiovanni A, et al. The application of markers (HSP70 GPC3 and GS) in liver biopsies is useful for detection of hepatocellular carcinoma. J Hepatol. 2009;50(4):746–54. [DOI] [PubMed] [Google Scholar]
- 29.Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16(9):398–406. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Rong Y, Yi K, Huang L, Chen M, Wang F. Circulating tumor cells in hepatocellular carcinoma: single-cell based analysis, preclinical models, and clinical applications. Theranostics. 2020;10(26):12060–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Felden J, Craig AJ, Garcia-Lezana T, Labgaa I, Haber PK, D’Avola D, et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene. 2021;40(1):140–51. [DOI] [PubMed] [Google Scholar]
- 32.Pantel K, Speicher M. The biology of circulating tumor cells. Oncogene. 2016;35(10):1216–24. [DOI] [PubMed] [Google Scholar]
- 33.Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology. 2021;73(1):422–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poon RT-P, Ng IO-L, Lau C, Yu W-C, Yang Z-F, Fan S-T, et al. Tumor microvessel density as a predictor of recurrence after resection of hepatocellular carcinoma: a prospective study. J Clin Oncol. 2002;20(7):1775–85. [DOI] [PubMed] [Google Scholar]
- 35.Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut. 2018;67(12):2204–12. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Han X, Yu X, Xu Z, Yang G, Liu B, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senft D, Ze’ev AR. Adaptive stress responses during tumor metastasis and dormancy. Trends Cancer. 2016;2(8):429–42. [DOI] [PubMed] [Google Scholar]
- 38.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salehi M, Lavasani ZM, Keshavarz Alikhani H, Shokouhian B, Hassan M, Najimi M, et al. Circulating tumor cells as a promising tool for early detection of hepatocellular carcinoma. Cells. 2023;12(18):2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y-F, Wu L, Liu S-P, Jiang M-M, Hu B, Zhou K-Q, et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat Commun. 2021;12(1):4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells—mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14(3):155–67. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Shi L, Zhang X, Sun B, Yang Y, Ge N, et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget. 2016;7(3):2646–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YM, Xu SC, Li J, Han KQ, Pi HF, Zheng L, et al. Epithelial-mesenchymal transition markers expressed in circulating tumor cells in hepatocellular carcinoma patients with different stages of disease. Cell Death Dis. 2013;4(10):e831-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J, Li Y, Liang S, Zeng J, Liu G, Mu F, et al. Circulating tumour cells as biomarkers for evaluating cryosurgery on unresectable hepatocellular carcinoma. Oncol Rep. 2016;36(4):1845–51. [DOI] [PubMed] [Google Scholar]
- 45.Ogle LF, Orr JG, Willoughby CE, Hutton C, McPherson S, Plummer R, et al. Imagestream detection and characterisation of circulating tumour cells—a liquid biopsy for hepatocellular carcinoma? J Hepatol. 2016;65(2):305–13. [DOI] [PubMed] [Google Scholar]
- 46.Julich-Haertel H, Urban SK, Krawczyk M, Willms A, Jankowski K, Patkowski W, et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J Hepatol. 2017;67(2):282–92. [DOI] [PubMed] [Google Scholar]
- 47.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y-F, Wu L, Liu S-P, Jiang M-M, Hu B, Zhou K-Q, et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat Commun. 2021;12(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalinich M, Bhan I, Kwan TT, Miyamoto DT, Javaid S, LiCausi JA, et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2017;114(5):1123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the cell search system in patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010;2010: 617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, et al. Liquid biopsy in patients with hepatocellular carcinoma: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23(31):5650–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Y, Xue P, Liu S, Zhang L, Guan Q, Zhu J, et al. Metal-based hybrid nanoparticles as radiosensitizers in cancer therapy. Colloid Interface Sci Commun. 2018;23:45–51. [Google Scholar]
- 53.Ha Y, Kim TH, Shim JE, Yoon S, Jun MJ, Cho YH, et al. Circulating tumor cells are associated with poor outcomes in early-stage hepatocellular carcinoma: a prospective study. Hepatol Int. 2019;13(6):726–35. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Zhang X, Zhang J, Sun B, Zheng L, Li J, et al. Microfluidic chip for isolation of viable circulating tumor cells of hepatocellular carcinoma for their culture and drug sensitivity assay. Cancer Biol Ther. 2016;17(11):1177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Wang B, Wu J, Zhang C, Zhou Y, Yang X, et al. Association of preoperative EpCAM circulating tumor cells and peripheral treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer. 2016;16(1):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukherji R, Suguru S, Xiao J, Geng X, Wang H, Noel MS, et al. Success rates and clinicopathologic associations with experimental outcomes of a novel circulating tumor cell (CTC) technology in advanced colon cancer (CC) and pancreatic cancer (PC). Am Soc Clin Oncol. 2023. 10.1200/JCO.2023.41.4_suppl.805. [Google Scholar]
- 57.Liu Y, Zhao W, Hodgson J, Egan M, Cooper Pope CN, Hicks G, et al. CTC-race: single-cell motility assay of circulating tumor cells from metastatic lung cancer patients. ACS Nano. 2024;18(12):8683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2(11):995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strilic B, Offermanns S. Intravascular survival and extravasation of tumor cells. Cancer Cell. 2017;32(3):282–93. [DOI] [PubMed] [Google Scholar]
- 60.Sun YF, Guo W, Xu Y, Shi YH, Gong ZJ, Ji Y, et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin Cancer Res. 2018;24(3):547–59. [DOI] [PubMed] [Google Scholar]
- 61.Guo W, Yang XR, Sun YF, Shen MN, Ma XL, Wu J, et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin Cancer Res. 2014;20(18):4794–805. [DOI] [PubMed] [Google Scholar]
- 62.D’Avola D, Villacorta-Martin C, Martins-Filho SN, Craig A, Labgaa I, von Felden J, et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci Rep. 2018;8(1):11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bauernhofer T, Zenahlik S, Hofmann G, Balic M, Resel M, Pirchmoser R, et al. Association of disease progression and poor overall survival with detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer. Oncol Rep. 2005;13(2):179–84. [PubMed] [Google Scholar]
- 64.Wang L, Li Y, Xu J, Zhang A, Wang X, Tang R, et al. Quantified postsurgical small cell size CTCs and EpCAM(+) circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018;412:99–107. [DOI] [PubMed] [Google Scholar]
- 65.Jin J, Niu X, Zou L, Li L, Li S, Han J, et al. AFP mRNA level in enriched circulating tumor cells from hepatocellular carcinoma patient blood samples is a pivotal predictive marker for metastasis. Cancer Lett. 2016;378(1):33–7. [DOI] [PubMed] [Google Scholar]
- 66.Dent BM, Ogle LF, O’Donnell RL, Hayes N, Malik U, Curtin NJ, et al. High-resolution imaging for the detection and characterisation of circulating tumour cells from patients with oesophageal, hepatocellular, thyroid and ovarian cancers. Int J Cancer. 2016;138(1):206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14(9):623–31. [DOI] [PubMed] [Google Scholar]
- 68.Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, et al. Circulating stem cell–like epithelial cell adhesion molecule–positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57(4):1458–68. [DOI] [PubMed] [Google Scholar]
- 69.Kelley RK, Magbanua MJM, Butler TM, Collisson EA, Hwang J, Sidiropoulos N, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer. 2015;15(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.von Felden J, Schulze K, Krech T, Ewald F, Nashan B, Pantel K, et al. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget. 2017;8(52):89978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen J, Wang WS, Zhu XL, Ni CF. High epithelial cell adhesion molecule-positive circulating tumor cell count predicts poor survival of patients with unresectable hepatocellular carcinoma treated with transcatheter arterial chemoembolization. J Vasc Interv Radiol. 2018;29(12):1678–84. [DOI] [PubMed] [Google Scholar]
- 72.Carr BI. Hepatocellular carcinoma: diagnosis and treatment. Cham: Springer; 2016. [Google Scholar]
- 73.Winograd P, Hou S, Court CM, Lee YT, Chen PJ, Zhu Y, et al. Hepatocellular carcinoma-circulating tumor cells expressing PD-L1 are prognostic and potentially associated with response to checkpoint inhibitors. Hepatol Commun. 2020;4(10):1527–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu X, Gao X-S, Xiong W, Guo W, Han L, Bai Y, et al. Increased programmed death ligand-1 expression predicts poor prognosis in hepatocellular carcinoma patients. Onco Targets Ther. 2016;9:4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345(6193):216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khoo BL, Grenci G, Lim YB, Lee SC, Han J, Lim CT. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat Protoc. 2018;13(1):34–58. [DOI] [PubMed] [Google Scholar]
- 77.Sun C, Liao W, Deng Z, Li E, Feng Q, Lei J, et al. The diagnostic value of assays for circulating tumor cells in hepatocellular carcinoma: a meta-analysis. Medicine. 2017;96(29): e7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40(3):172–86. [DOI] [PubMed] [Google Scholar]
- 79.Leon S, Shapiro B, Sklaroff D, Yaros M. Free DNA in the serum of cancer patients and the effect of therapy. Can Res. 1977;37(3):646–50. [PubMed] [Google Scholar]
- 80.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem. 2010;56(8):1279–86. [DOI] [PubMed] [Google Scholar]
- 81.Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramkissoon LA, Pegram W, Haberberger J, Danziger N, Lesser G, Strowd R, et al. Genomic profiling of circulating tumor DNA from cerebrospinal fluid to guide clinical decision making for patients with primary and metastatic brain tumors. Front Neurol. 2020;11:544680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yan L, Chen Y, Zhou J, Zhao H, Zhang H, Wang G. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis. 2018;67:92–7. [DOI] [PubMed] [Google Scholar]
- 86.Hasenleithner SO, Speicher MR. A clinician’s handbook for using ctDNA throughout the patient journey. Mol Cancer. 2022;21(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379(18):1754–65. [DOI] [PubMed] [Google Scholar]
- 88.Jiang P, Sun K, Tong YK, Cheng SH, Cheng THT, Heung MMS, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci USA. 2018;115(46):E10925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang P, Chan CW, Chan KA, Cheng SH, Wong J, Wong VW-S, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci. 2015;112(11):1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang P, Sun K, Tong YK, Cheng SH, Cheng TH, Heung MM, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci. 2018;115(46):E10925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marchio A, Amougou Atsama M, Béré A, Komas NP, Noah Noah D, Atangana PJA, et al. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin Exp Med. 2018;18(3):421–31. [DOI] [PubMed] [Google Scholar]
- 92.Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, Stark M, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res. 2006;26(6c):4713–9. [PubMed] [Google Scholar]
- 93.Piciocchi M, Cardin R, Vitale A, Vanin V, Giacomin A, Pozzan C, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int. 2013;7(4):1050–7. [DOI] [PubMed] [Google Scholar]
- 94.Martignano F. Cell-free DNA: an overview of sample types and isolation procedures. In: Casadio V, Salvi S, editors. Cell-free DNA as diagnostic markers. New York: Springer; 2019. p. 13–27. [DOI] [PubMed] [Google Scholar]
- 95.Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 2016;14(10):898–908. [DOI] [PubMed] [Google Scholar]
- 96.van Dessel LF, Beije N, Helmijr JC, Vitale SR, Kraan J, Look MP, et al. Application of circulating tumor DNA in prospective clinical oncology trials—standardization of preanalytical conditions. Mol Oncol. 2017;11(3):295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nevel KS, Wilcox JA, Robell LJ, Umemura Y. The utility of liquid biopsy in central nervous system malignancies. Curr Oncol Rep. 2018;20(8):60. [DOI] [PubMed] [Google Scholar]
- 98.Siravegna G, Mussolin B, Venesio T, Marsoni S, Seoane J, Dive C, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol. 2019;30(10):1580–90. [DOI] [PubMed] [Google Scholar]
- 99.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1–2):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Underwood JJ, Quadri RS, Kalva SP, Shah H, Sanjeevaiah AR, Beg MS, et al. Liquid biopsy for cancer: review and implications for the radiologist. Radiology. 2020;294(1):5–17. [DOI] [PubMed] [Google Scholar]
- 101.Murtaza M, Dawson S-J, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–12. [DOI] [PubMed] [Google Scholar]
- 102.Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59(12):1722–31. [DOI] [PubMed] [Google Scholar]
- 103.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci. 2011;108(23):9530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):13668–68. [DOI] [PubMed] [Google Scholar]
- 107.Huang A, Zhao X, Yang X-R, Li F-Q, Zhou X-L, Wu K, et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J Hepatol. 2017;67(2):293–301. [DOI] [PubMed] [Google Scholar]
- 108.Wong IH, Lo YM, Yeo W, Lau WY, Johnson PJ. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res. 2000;6(9):3516–21. [PubMed] [Google Scholar]
- 109.Zhang YJ, Wu HC, Shen J, Ahsan H, Tsai WY, Yang HI, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13(8):2378–84. [DOI] [PubMed] [Google Scholar]
- 110.Iyer P, Zekri AR, Hung CW, Schiefelbein E, Ismail K, Hablas A, et al. Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp Mol Pathol. 2010;88(1):107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Iizuka N, Oka M, Sakaida I, Moribe T, Miura T, Kimura N, et al. Efficient detection of hepatocellular carcinoma by a hybrid blood test of epigenetic and classical protein markers. Clin Chim Acta. 2011;412(1–2):152–8. [DOI] [PubMed] [Google Scholar]
- 112.Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol. 2011;91(3):702–7. [DOI] [PubMed] [Google Scholar]
- 113.Wong IH, Zhang J, Lai PB, Lau WY, Lo YM. Quantitative analysis of tumor-derived methylated p16INK4a sequences in plasma, serum, and blood cells of hepatocellular carcinoma patients. Clin Cancer Res. 2003;9(3):1047–52. [PubMed] [Google Scholar]
- 114.Sun FK, Fan YC, Zhao J, Zhang F, Gao S, Zhao ZH, et al. Detection of TFPI2 methylation in the serum of hepatocellular carcinoma patients. Dig Dis Sci. 2013;58(4):1010–5. [DOI] [PubMed] [Google Scholar]
- 115.Chang H, Yi B, Li L, Zhang HY, Sun F, Dong SQ, et al. Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp Mol Pathol. 2008;85(2):96–100. [DOI] [PubMed] [Google Scholar]
- 116.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaseb AO, Sánchez NS, Sen S, Kelley RK, Tan B, Bocobo AG, et al. Molecular profiling of hepatocellular carcinoma using circulating cell-free DNA. Clin Cancer Res. 2019;25(20):6107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jiao J, Watt GP, Stevenson HL, Calderone TL, Fisher-Hoch SP, Ye Y, et al. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: prevalence and risk factors. Hepatol Commun. 2018;2(6):718–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Howell J, Atkinson SR, Pinato DJ, Knapp S, Ward C, Minisini R, et al. Identification of mutations in circulating cell-free tumour DNA as a biomarker in hepatocellular carcinoma. Eur J Cancer. 2019;116:56–66. [DOI] [PubMed] [Google Scholar]
- 120.Whittaker S, Marais R, Zhu A. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29(36):4989–5005. [DOI] [PubMed] [Google Scholar]
- 121.Zhang P, Wen X, Gu F, Deng X, Li J, Dong J, et al. Methylation profiling of serum DNA from hepatocellular carcinoma patients using an infinium human methylation 450 BeadChip. Hepatol Int. 2013;7(3):893–900. [DOI] [PubMed] [Google Scholar]
- 122.Zhang J, Zhong QJC. Histone deacetylase inhibitors cell death. Cell Mol Life Sci. 2014;71(20):3885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang G, Krocker JD, Kirk JL, Merwat SN, Ju H, Soloway RD, et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin Chem Lab Med. 2014;52(6):899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20(3):274–81. [DOI] [PubMed] [Google Scholar]
- 125.Yeo W, Wong N, Wong WL, Lai PB, Zhong S, Johnson PJ. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver Int. 2005;25(2):266–72. [DOI] [PubMed] [Google Scholar]
- 126.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu C-Y, Chen S-Y, Peng H-L, Kan P-Y, Chang W-C, Yen C-J. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget. 2017;8(4):6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu R-h, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–61. [DOI] [PubMed] [Google Scholar]
- 129.Tokuhisa Y, Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, et al. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br J Cancer. 2007;97(10):1399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ono A, Fujimoto A, Yamamoto Y, Akamatsu S, Hiraga N, Imamura M, et al. Circulating tumor DNA analysis for liver cancers and its usefulness as a liquid biopsy. Cell Mol Gastroenterol Hepatol. 2015;1(5):516–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oh CR, Kong SY, Im HS, Kim HJ, Kim MK, Yoon KA, et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer. 2019;19(1):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang A, Zhang X, Zhou S-L, Cao Y, Huang X-W, Fan J, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Cancer. 2016;7(13):1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liao W, Yang H, Xu H, Wang Y, Ge P, Ren J, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7(26):40481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ren N, Qin L-X, Tu H, Liu Y-K, Zhang B-H, Tang Z-Y. The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2006;132(6):399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3(9):2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dashti F, Mirazimi SMA, Rabiei N, Fathazam R, Rabiei N, Piroozmand H, et al. The role of non-coding RNAs in chemotherapy for gastrointestinal cancers. Mol Ther Nucleic Acids. 2021;26:892–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pourhanifeh MH, Vosough M, Mahjoubin-Tehran M, Hashemipour M, Nejati M, Abbasi-Kolli M, et al. Autophagy-related microRNAs: possible regulatory roles and therapeutic potential in and gastrointestinal cancers. Pharmacol Res. 2020;161: 105133. [DOI] [PubMed] [Google Scholar]
- 138.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29(3):946–50. [DOI] [PubMed] [Google Scholar]
- 140.Lai M-c, Yang Z, Zhou L, Zhu Q-q, Xie H-y, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–6. [DOI] [PubMed] [Google Scholar]
- 141.Hou Z-H, Xu X-W, Fu X-Y, Zhou L-D, Liu S-P, Tan D-M. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol Cell Physiol. 2020;318(3):C649–63. [DOI] [PubMed] [Google Scholar]
- 142.Ghafouri-Fard S, Gholipour M, Hussen BM, Taheri M. The impact of long non-coding RNAs in the pathogenesis of hepatocellular carcinoma. Front Oncol. 2021;11:649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.El-Tawdi AHF, Matboli M, Shehata HH, Tash F, El-Khazragy N, Azazy AE, et al. Evaluation of circulatory RNA-based biomarker panel in hepatocellular carcinoma. Mol Diagn Ther. 2016;20(3):265–77. [DOI] [PubMed] [Google Scholar]
- 144.Yuan W, Sun Y, Liu L, Zhou B, Wang S, Gu D. Circulating LncRNAs serve as diagnostic markers for hepatocellular carcinoma. Cell Physiol Biochem. 2017;44(1):125–32. [DOI] [PubMed] [Google Scholar]
- 145.Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5(12): e1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mocan T, Simão AL, Castro RE, Rodrigues CMP, Słomka A, Wang B, et al. Liquid biopsies in hepatocellular carcinoma: are we winning? J Clin Med. 2020;9(5):1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pezzuto F, Buonaguro L, Buonaguro FM, Tornesello ML. The role of circulating free DNA and microRNA in non-invasive diagnosis of HBV- and HCV-related hepatocellular carcinoma. Int J Mol Sci. 2018;19(4):1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56(1):167–75. [DOI] [PubMed] [Google Scholar]
- 149.Ding Y, Yan JL, Fang AN, Zhou WF, Huang L. Circulating miRNAs as novel diagnostic biomarkers in hepatocellular carcinoma detection: a meta-analysis based on 24 articles. Oncotarget. 2017;8(39):66402–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhou L, Yang ZX, Song WJ, Li QJ, Yang F, Wang DS, et al. MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int J Oncol. 2013;43(2):661–9. [DOI] [PubMed] [Google Scholar]
- 151.Amr KS, Ezzat WM, Elhosary YA, Hegazy AE, Fahim HH, Kamel RR. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene. 2016;575(1):66–70. [DOI] [PubMed] [Google Scholar]
- 152.Zhuang C, Jiang W, Huang D, Xu L, Yang Q, Zheng L, et al. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2016;40(4):386–96. [DOI] [PubMed] [Google Scholar]
- 153.Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16(7):804–15. [DOI] [PubMed] [Google Scholar]
- 154.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29(36):4781–8. [DOI] [PubMed] [Google Scholar]
- 155.Yamamoto Y, Kondo S, Matsuzaki J, Esaki M, Okusaka T, Shimada K, et al. Highly sensitive circulating microRNA panel for accurate detection of hepatocellular carcinoma in patients with liver disease. Hepatol Commun. 2020;4(2):284–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang W, Li J, Guo X, Zhao Y, Yuan X. miR-663a inhibits hepatocellular carcinoma cell proliferation and invasion by targeting HMGA2. Biomed Pharmacother. 2016;81:431–8. [DOI] [PubMed] [Google Scholar]
- 157.Wang H, Cao F, Li X, Miao H, Xing H, et al. LmiR-320b suppresses cell proliferation by targeting c-Myc in human colorectal cancer cells. BMC Cancer. 2015;15:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wu XM, Xi ZF, Liao P, Huang HD, Huang XY, Wang C, et al. Diagnostic and prognostic potential of serum microRNA-4651 for patients with hepatocellular carcinoma related to aflatoxin B1. Oncotarget. 2017;8(46):81235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Loosen SH, Wirtz TH, Roy S, Vucur M, Castoldi M, Schneider AT, et al. Circulating levels of microRNA193a-5p predict outcome in early stage hepatocellular carcinoma. PLoS ONE. 2020;15(9): e0239386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jin Y, Wong YS, Goh BKP, Chan CY, Cheow PC, Chow PKH, et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci Rep. 2019;9(1):10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Han J, Li J, Qian Y, Liu W, Liang J, Huang Z, et al. Identification of plasma miR-148a as a noninvasive biomarker for hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2019;43(5):585–93. [DOI] [PubMed] [Google Scholar]
- 162.Chuma M, Toyoda H, Matsuzaki J, Saito Y, Kumada T, Tada T, et al. Circulating microRNA-1246 as a possible biomarker for early tumor recurrence of hepatocellular carcinoma. Hepatol Res. 2019;49(7):810–22. [DOI] [PubMed] [Google Scholar]
- 163.Cho HJ, Kim JK, Nam JS, Wang HJ, Lee JH, Kim BW, et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis B virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin Biochem. 2015;48(16–17):1073–8. [DOI] [PubMed] [Google Scholar]
- 164.Ali HEA, Emam AA, Zeeneldin AA, Srour R, Tabashy R, El-Desouky ED, et al. Circulating miR-26a, miR-106b, miR-107 and miR-133b stratify hepatocellular carcinoma patients according to their response to transarterial chemoembolization. Clin Biochem. 2019;65:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nishida N, Arizumi T, Hagiwara S, Ida H, Sakurai T, Kudo M. MicroRNAs for the prediction of early response to sorafenib treatment in human hepatocellular carcinoma. Liver Cancer. 2017;6(2):113–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Teufel M, Seidel H, Köchert K, Meinhardt G, Finn RS, Llovet JM, et al. Biomarkers associated with response to regorafenib in patients with hepatocellular carcinoma. Gastroenterology. 2019;156(6):1731–41. [DOI] [PubMed] [Google Scholar]
- 167.Aydin Y, Koksal AR, Thevenot P, Chava S, Heidari Z, Lin D, et al. Experimental validation of novel glypican 3 exosomes for the detection of hepatocellular carcinoma in liver cirrhosis. J Hepatocell Carcinoma. 2021;8:1579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. 2017;66(4):1125–43. [DOI] [PubMed] [Google Scholar]
- 169.Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018;7(5):1670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56(3):479–84. [DOI] [PubMed] [Google Scholar]
- 171.Cho HJ, Baek GO, Yoon MG, Ahn HR, Son JA, Kim SS, et al. Overexpressed proteins in HCC cell-derived exosomes, CCT8, and cofilin-1 are potential biomarkers for patients with HCC. Diagnostics (Basel). 2021;11(7):1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He D, Cui B, Lv H, Lu S, Zhu Y, Cheng Y, et al. Blood-derived extracellular vesicles as a promising liquid biopsy diagnostic tool for early cancer detection. Biomolecules. 2024;14(7):847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pu C, Huang H, Wang Z, Zou W, Lv Y, Zhou Z, et al. Extracellular vesicle-associated mir-21 and mir-144 are markedly elevated in serum of patients with hepatocellular carcinoma. Front Physiol. 2018;9:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang Y, Xi H, Nie X, Zhang P, Lan N, Lu Y, et al. Assessment of miR-212 and other biomarkers in the diagnosis and treatment of HBV-infection-related liver diseases. Curr Drug Metab. 2019;20(10):785–98. [DOI] [PubMed] [Google Scholar]
- 175.Sorop A, Iacob R, Iacob S, Constantinescu D, Chitoiu L, Fertig TE, et al. Plasma small extracellular vesicles derived miR-21-5p and miR-92a-3p as potential biomarkers for hepatocellular carcinoma screening. Front Genet. 2020;11:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Li J, Rohailla S, Gelber N, Rutka J, Sabah N, Gladstone RA, et al. MicroRNA-144 is a circulating effector of remote ischemic preconditioning. Basic Res Cardiol. 2014;109(5):423. [DOI] [PubMed] [Google Scholar]
- 177.Guo X, Lv X, Lv X, Ma Y, Chen L, Chen Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget. 2017;8(27):44050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014: 864894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50(2):136–42. [DOI] [PubMed] [Google Scholar]
- 180.Hao X, Xin R, Dong W. Decreased serum exosomal miR-320a expression is an unfavorable prognostic factor in patients with hepatocellular carcinoma. J Int Med Res. 2020;48(4):300060519896144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ, et al. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics. 2019;9(7):1965–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tao N-N, Ren J-H, Tang H, Ran L-K, Zhou H-Z, Liu B, et al. Deacetylation of Ku70 by SIRT6 attenuates Bax-mediated apoptosis in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;485(4):713–9. [DOI] [PubMed] [Google Scholar]
- 184.Qu Z, Wu J, Wu J, Ji A, Qiang G, Jiang Y, et al. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(46):80666–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu W-h, Ren L-n, Wang X, Wang T, Zhang N, Gao Y, et al. Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J Cancer Res Clin Oncol. 2015;141:1767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nieuwland R, Enciso-Martinez A, Bracht JWP. Clinical applications and challenges in the field of extracellular vesicles. Med Gen. 2023;35(4):251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hackshaw A, Clarke CA, Hartman A-R. New genomic technologies for multi-cancer early detection: rethinking the scope of cancer screening. Cancer Cell. 2022;40(2):109–13. [DOI] [PubMed] [Google Scholar]
- 188.Lennon AM, Buchanan AH, Rego SP, Choudhry OA, Elias PZ, Sadler JR, et al. Outcomes following a false-positive multi-cancer early detection test: results from DETECT-A, the first large, prospective, interventional MCED study. Cancer Prevent Res. 2024. 10.1158/1940-6207.CAPR-23-0451. [DOI] [PubMed] [Google Scholar]
- 189.Luan Y, Zhong G, Li S, Wu W, Liu X, Zhu D, et al. A panel of seven protein tumour markers for effective and affordable multi-cancer early detection by artificial intelligence: a large-scale and multicentre case–control study. EClinicalMedicine. 2023;61:102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.