Abstract
Background
Methods for the extraction of DNA from filamentous fungi are frequently laborious and time consuming because most of the available protocols include maceration in liquid nitrogen after the mycelium has been grown in a liquid culture. This paper describes a new method to replace those steps, which involves the growth of the mycelium on cellophane disks overlaid on solid medium and the use of glass beads for cell wall disruption.
Results
Extractions carried out by this method provided approximately 2 μg of total DNA per cellophane disk for the filamentous fungus Trichoderma reesei. To assess the DNA's quality, we made a PCR (Polymerase Chain Reaction) amplification of a gene introduced by a transformation in this fungus's genome (hph gene), with successful results. We also confirmed the quality of the DNA by the use of Southern blotting to analyze the presence of the same gene, which was easily detected, resulting in a sharply defined and strong band.
Conclusions
The use of this method enabled us to obtain pure DNA from Trichoderma reesei, dispensing with the laborious and time-consuming steps involved in most protocols. The DNA obtained was found to be suitable for PCR and Southern blot analyses. Another advantage of this method is the fact that several samples can be processed simultaneously, growing the fungus on multiple well cell culture plates. In addition, the absence of maceration also reduces sample handling, minimizing the risks of contamination, a particularly important factor in work involving PCR.
Background
Many methods for DNA extraction from fungi have been described in the past few years [1-6]. The methods applied to filamentous fungi, such as Trichoderma reesei, are often laborious and time consuming. This is usually because most of the available protocols include the growth of mycelium in a liquid culture, followed by maceration in liquid nitrogen. Although some methods do not involve maceration in liquid nitrogen, they are still time consuming and require special columns [4,5]. On the other hand, some methods recover DNA suitable exclusively for PCR (Polymerase Chain Reaction) [7,8] or use specific apparatii and matrices [9]. In addition to being lengthy, the current methods also involve the handling of large amounts of samples and glassware, which is inconvenient when handling a large number of samples. In this paper, we discuss the development of a much simpler method that precludes liquid cultures and maceration. The method involves the growth of spores on small cellophane disks placed on a solid medium, ressuspension of the grown pellicle in an adequate buffer, cell disruption using glass beads, and subsequent DNA purification either by classical phenol-chloroform extraction followed by DNA precipitation or by a simple and fast direct precipitation.
Results
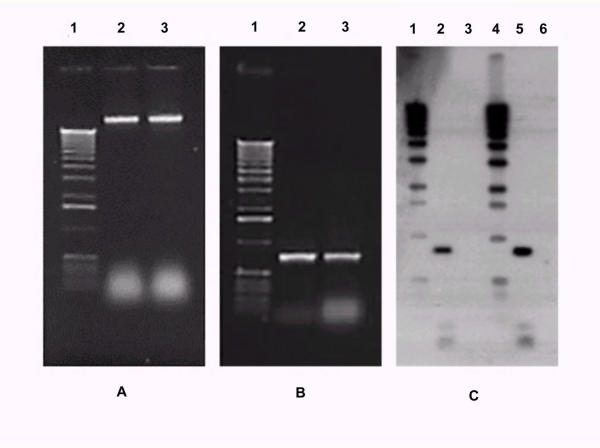
Extractions made by this method provided approximately 2 μg of DNA per cellophane disk. The use of 2 disks resulted in an increment of the quantity obtained, yielding around 3 μg (data not shown). On agarose gels, the DNA appeared as good quality, with high molecular weight and no apparent smear (Fig. 1A). To confirm the DNA's purity and its use for basic molecular analysis, we made a PCR amplification of a fragment of the hph gene present in the fungus. The amplification produced a single band of approximately 750 bp specific to hph gene (Fig. 1B). To substantiate the quality and use for restriction enzyme digestion, a Southern Blot analysis was made using an hph specific probe after the DNA had been digested with EcoRI and BamHI. The autoradiography showed a sharp band with the expected size of approximately 800 bp (Fig. 1C).
Figure 1.
DNA quality analysis. A – Agarose gel electrophoresis of DNA sample purified by the cellophane disk miniprep method. Lane 1 (1 Kb ladder), lane 2–3, genomic DNA from Trichoderma reesei transformants 1 and 2, respectively. B – Amplified hph fragment from DNAs prepared by the cellophane-base method. Agarose gel electrophoresis of PCR samples. Lane 1 (1 Kb ladder), lane 2–3, 750 pb PCR products from transformant 1 and 2, respectively. C – Detection of hph gene in DNA prepared from transformants of Trichoderma reesei. The DNA obtained by the cellophane method was BamHI and EcoRI restricted and subjected to Southern Blot, using an α-32PdATP labeled fragment of the hph gene as a probe. Lane 1 and 4 (1 Kb DNA ladder, probed against itself), lane 2 and 5, restricted DNA from hph Trichoderma reesei transformant 1 and 2, respectively; 3 and 6, DNA from untransformed Trichoderma reesei (control).
Discussion
The method presented in this paper dispenses with the laborious and time-consuming steps of most protocols by using newly germinated spores grown on cellophane. This procedure apparently results in cell walls that are softer than those produced in liquid culture and that are easily broken by glass beads and phenol-chloroform. The use of young spores was a rate-limiting step, since older spores did not produce good results (data not shown). The amount and quality of the DNA obtained by this procedure were suitable for PCR and Southern Blot analyses. One of the advantages of this procedure is that several samples can be simultaneously processed, growing the fungus on 12 well cell culture plates. No more than 2 hours were needed to process 12 or more samples. Another advantage is that the absence of maceration also reduces sample handling, minimizing contamination risks between samples. This is particularly important in work involving amplification by PCR. A similar method using growth of mycelium on cellophane for DNA extraction from Podospora anserina and Ascobolus immersus was previously described [10]. That method used 9-cm (diameter) Petri dishes, long time of growing (2–4 days) and the mycelium needed to be recovered by scraping using a spatula. We avoid this procedure in our protocol using smaller disks. This simple modification can make the method faster and safety, specially when working with pathogenic fungi. On the other hand, we have presented an alternative for phenol-chloroform extraction, avoiding this hazardous step. These procedures were successfully extended to recover DNA from Aspergillus phoenicis and Neurospora crassa (data not shown) and we believe it can easily be adapted to other filamentous fungi, although the growth time on the cellophane disks may require adjustments.
Conclusions
The method described herein is efficient to prepare DNA from the filamentous fungus Trichoderma reesei, resulting in DNA sufficiently pure for PCR and Southern Blot analyses.
Material and Methods
Fungal strain and culture conditions
The Trichoderma reesei strain QM9414 (ATCC number 26921) was transformed for Hygromycin B resistance, as described in [11]. Five-day-old spores of two transformants (TR1 and TR2) were harvested from Potato Dextrose Agar (PDA) slants with 3 ml of sterile water, resulting in a dense spore suspension (109-1010 spores/ml). 40 μl of this suspension was placed on a small 2,2 cm cellophane disk (BIO-RAD#165-0963) sterilized by autoclaving in water and placed on the surface of a solid medium (Potato Dextrose Agar). The culture was then left overnight to grow for 14 hours at 28°C. When a large number of samples was analyzed we grown the fungus on 12 wells cell culture plates.
DNA Purification
Following the fungal growth, the cellophane disk was placed in a 1,5 ml microcentrifuge tube containing 500 μl of a ressuspension buffer (50 mM Tris-HCl, pH 7.5; 10 mM EDTA; 1% SDS). After vigorous whirling, the mycelia pellicle grown on the cellophane disk was readily detached and the cellophane disk removed from the tube. A small volume (200 μl, measured in the tube) of glass beads (Sigma) was added to the tube, which was then vigorously mixed in a vortex for 10 seconds and left to rest for 15 seconds on ice. This step was repeated twice, after which 400 μl of phenol-chloroform was added. The tube was placed in a shaker in a horizontal position and shaken at 200 RPM (37°C) for 15 minutes. Immediately after this procedure, the tube was centrifuged at 15.000 g for 5 minutes, the top aqueous phase (~400 μl) transferred to a new tube and the DNA precipitated by the addition of a 1/10 volume (~40 μl) of 3 M Sodium Acetate, pH 5.8 and 1 ml of cold absolute ethanol. The tube was placed in a freezer at -20°C for 30 minutes and then centrifuged at 15.000 g for 15 minutes, after which the pellet was washed once with 1 ml of ethanol 70% and then with absolute ethanol. After being dried, the DNA pellet was ressuspended in 20 μl of TE-RNAse (20 μg/ml) and quantified spectrophotometrically or by comparison with standards (High Mass DNA ladder – Invitrogen) in a 1% agarose gel. Alternatively and safety, the phenol-chloroform extraction can be suppressed by a direct precipitation after the glass beads step. To the tube still containing the glass beads was added 150 μl of potassium acetate, pH 4.8 (made of 60 ml of 5 M potassium acetate, 11.5 ml of glacial acetic acid, and 28.5 ml of milli-Q water). The tube was vortexed briefly and spun at 15.000 g for 2 min. After transferring the supernatant to a new 1.5 ml tube, 600 μl of isopropanol was added. After mixing by inversion the tube was centrifuged at 15.000 g for 10 min. The resultant DNA pellet was washed with 500 μl of 70% ethanol, air dried and ressuspended in 20 μl of TE-RNAse (20 μg/ml).
PCR
The Polymerase Chain Reaction was accomplished according to a modified Saiki et al. protocol [12], using the purified DNAs as templates. The PCR was carried out using internal primers to amplify a 750 pb fragment of the hph gene (resistance to Hygromicin B)[13] present in the Trichoderma reesei transformants used in this work. PCRs were made in 200 μl tubes containing 100 ng of DNA, 10 pmoles of each primer (Forward CCTGAACTCACCGCGACGTCT and Reverse CTCCGGATGCCTCCGCTCGAAGT), 1× PCR buffer (Invitrogen), 1.5 mM MgCl2, 1 U of Taq DNA polymerase (Invitrogen) and Milli-Q water to 25 μl. The reaction was carried out in an MJ Research thermocycler programmed for 30 cycles of 94°C 1 min, 55°C 1 min and 72°C 1.5 min, with 4-min initial and final steps.
Southern blotting
Southern blotting was performed according to Sambrook et al.[14]. Two μg of DNA were subjected to total digestion using 10 U of the restriction enzymes BamHI and EcoRI for 3 hours. DNA from a non-transformed Trichoderma reesei was included as a control. The digested samples were separated in an 1% agarose gel (TBE buffer) together with a 1 Kb ladder (Invitrogen). The gel's DNA was then transferred to a nylon membrane (Hybon N+ – Amersham Biosciences) which was first hybridized to an α-32PdATP labeled 1 Kb DNA ladder probe (Invitrogen), washed according to the manufacturer's recommendations and rehybridized with an α-32PdATP labeled PCR fragment of the hph gene. Both probes were labeled using the "Ready-to-Go DNA labelling Kit" (Amersham Pharmacia Biotech). After hybridization and washing, the membrane was exposed to X-Ray film (Kodak X-Omat) for 6 hours. The hph probe detected a fragment of approximately 0.8 Kbp.
Acknowledgments
Acknowlegments
The authors thank Dr. João Atílio Jorge, from the Department of Biology, USP-Ribeirão Preto, for providing Aspergillus phoenicis and Neurospora crassa spores. This research work was supported by FAPESP (Grant 96/11018-0). Rodrigo Alexandre Panepucci and Ana Maria Tortella Baião are supported by a CAPES fellowship. Alexandre Cassago are supported by a CNPq fellowship.
Contributor Information
Alexandre Cassago, Email: casslex@rocketmail.com.
Rodrigo Alexandre Panepucci, Email: prap@iris.ufscar.br.
Ana Maria Tortella Baião, Email: amtb@eudoramail.com.
Flavio Henrique-Silva, Email: dfhs@power.ufscar.br.
References
- Specht CA, DiRusso CC, Novotny CP, Ullrich RC. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal Biochem. 1982;119:158–163. doi: 10.1016/0003-2697(82)90680-7. [DOI] [PubMed] [Google Scholar]
- de Graaff L, van den BH, Visser J. Isolation and transformation of the pyruvate kinase gene of Aspergillus nidulans. Curr Genet. 1988;13:315–321. doi: 10.1007/BF00424425. [DOI] [PubMed] [Google Scholar]
- Bainbridge BW, Spreadbury CL, Scalise FG, Cohen J. Improved methods for the preparation of high molecular weight DNA from large and small scale cultures of filamentous fungi. FEMS Microbiol Lett. 1990;54:113–117. doi: 10.1016/0378-1097(90)90267-T. [DOI] [PubMed] [Google Scholar]
- Challen MP, Moore AJ, Martinez-Carrera D. Facile extraction and purification of filamentous fungal DNA. Biotechniques. 1995;18:975–978. [PubMed] [Google Scholar]
- Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985. pp. 17–20.
- Nijs M, Nabben L, Wernars K. Isolation of Fusarium DNA for molecular analysis with and without mechanical cell disruption. Journal of Microbiological Methods. 1996;27:13–17. doi: 10.1016/0167-7012(96)00920-7. [DOI] [Google Scholar]
- Liu D, Coloe S, Baird R, Pedersen J. Rapid Mini-preparation of fungal DNA for PCR. Journal of Clinical Microbiology. 2000;38:471. doi: 10.1128/jcm.38.1.471-471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller FMC, Werner KE, Kasai M, Francesconi A, Chanock SJ, Walsh TJ. Rapid Extraction of Genomic DNA from Medically Important Yeasts and Filamentous Fungi by High-Speed Cell Disruption. Journal of Clinical Microbiology. 1998;36:1625–1629. doi: 10.1128/jcm.36.6.1625-1629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier G, Silar P. Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Current Genetics. 1994;25:122–123. doi: 10.1007/BF00309536. [DOI] [PubMed] [Google Scholar]
- Pentilla M, Nevalainen H, Ratto M, Salminen E, Knowles J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 1987;61:155–164. doi: 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A laboratory manual. Second. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]