Abstract
Previous work indicated that the implantation and pregnancy rates of women with endometriosis are lower than those of healthy women during in-vitro fertilisation and embryonic transfer. And there are numerous microRNAs (miRNAs) in human uterine luminal fluid (ULF), some of which are associated with early preimplantation development of embryos. In our study, we sought to determine whether miRNAs in the ULF are differentially expressed between women with and without endometriosis and to uncover the association of miRNAs with the development potential of blastocysts. The implantation and clinical pregnancy rates significantly decreased in women with endometriosis than in those without endometriosis. Notably, hsa-miR-145-5p was upregulated in ULF samples from women with endometriosis (fold change > 2, false discovery rate < 0.001). Moreover, the ratios of mouse/human early embryos that developed into blastocyst-staged embryos (P = 0.0065 and P = 0.0098, respectively) were significantly affected via miR-145-5p upregulation in mouse/human early embryos. Notch signalling pathway components had abnormal expression levels in the mouse/human blastocyst-stage embryos in the miR-145-5p mimic-enriched extracellular vesicles (EVs) group. In conclusions, our study revealed that human extracellular vesicle-derived miRNAs in ULF impacted the developmental potential of blastocysts in women with endometriosis. Moreover, the upregulation of miR-145-5p-enriched EVs in mouse and human embryos negatively affected blastocyst development by suppressing the expression of components of the NOTCH signalling pathway, which may contribute to elucidate the cause of infertility in women with endometriosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13048-024-01579-x.
Keywords: hsa-miR-145-5p, Uterine luminal fluid, Blastocyst development, Endometriosis, Extracellular vesicle, Embryo
Introduction
Endometriosis reportedly causes chronic pelvic pain, is highly prevalent among women of reproductive age [1], and affects 20–50% of infertile women [2]. The implantation and pregnancy rates of women with endometriosis are lower than those of healthy women during in-vitro fertilisation (IVF) and embryonic transfer [3, 4]. During embryo implantation, the blastocyst interacts with and regulates the endometrium, and uterine luminal fluid (ULF) secreted by the endometrial epithelium nurtures the embryo, which is significant for early preimplantation development of embryos [5].
MicroRNAs (miRNAs) are secreted by cells and incorporated into extracellular vesicles (EVs) or associated with proteins that protect them from RNase degradation, endowing them with a long half-life [6]. MiRNAs in EVs, the main molecules playing a regulatory role in EVs, have a primarily gene-silencing function in receptor cells [7]. The presence of miRNAs in the ULF of humans has been identified in several studies [5, 8]. ULF, a viscous fluid secreted by the endometrial glands into the uterine cavity, nurtures the embryo and provides a unique microenvironment in which the first bidirectional dialogue between the maternal endometrium and embryo occurs during the window of implantation (WOI) [9]. As an effective means of intercellular communication, EVs released by the endometrial epithelium into the uterine cavity are involved in the transfer of signalling proteins, miRNAs, and mRNAs to either the embryo or adjacent endometrium. The transfer of materials affects endometrial receptivity and preimplantation embryonic development [8]. EV-derived uterine miRNAs isolated from cows with endometritis reportedly impede early embryonic development [10], and maternal miRNAs secreted into the ULF act as transcriptomic regulators of preimplantation embryos [5]. However, until now, no studies have analysed miRNA expression profile in the ULF of women with endometriosis and the effects of uterine fluid miRNAs on the development of embryos under endometriosis conditions.
This study explored the miRNA expression profile of ULF from women with endometriosis relative to that from women with tubal factor infertility, and quantitative reverse transcription polymerase chain reaction (qRT‒PCR) was used to identify the differential expression of miRNAs associated with endometriosis. Moreover, we investigated the effects and molecular mechanisms of these miRNAs in the development of blastocysts by co-culturing miRNA mimic-EVs with hatching embryos. This research may contribute to elucidating the cause of infertility in women with endometriosis.
Materials and methods
Patients
Thirty women with endometriosis and thirty women with tubal factor infertility were included in this study at Shanghai Ji Ai Genetics and IVF Institute, affiliated with Fudan University, from March 2018 to May 2019. All the women underwent laparoscopic surgical examination of the abdominal cavity and complete excision of endometriotic tissue. The disease was diagnosed clinically or by ultrasonography and verified by surgical findings and postoperative pathological examination. In addition, laparoscopic examination of the abdominal cavity excluded the presence of any other pelvic pathology that could potentially confound the data observed. There were 24 women with stage III and six women with stage IV disease (all women were diagnosed with tubal endometriosis via pathological biopsy). The women in the control group all underwent surgery for laparoscopic tubal sterilisation, and the absence of endometriosis was verified after a surgical examination of the abdominal cavity. Women who repeatedly had high baseline levels of serum follicle-stimulating hormone (FSH) (> 15–20 IU/l), a seriously deformed uterus, or any other active infections were excluded from this study.
ULF sample preparation
The participants with regular menstrual cycles of 25–33 days were selected for collection of ULF samples. ULF samples were obtained from all the patients using a Frydman catheter (Prodimed; Neuilly-en-Thelle, France) during the patient’s implantation window, based on the date of the last menstruation. None of the women received hormonal treatment in the 3 months preceding the biopsy and ULF collection. Briefly, a speculum was inserted into the subject while in the lithotomy position. Subsequently, the cervix was cleansed, and the Frydman catheter was gently introduced to aspirate in 20–50 µL of endometrial secretion as described [5]. ULF samples were stored at − 80 °C until they were used for RNA extraction.
Ovarian stimulation and human oocyte collection
This study involved 52 patients enrolled in the assisted reproduction programme at the Shanghai Ji Ai Genetics and IVF Institute, affiliated with Fudan University. In total, 106 MI oocytes were obtained from 52 consenting couples. And all 106 MI oocytes have matured. The patients were stimulated with GnRH agonists (Ferring Pharmaceuticals, Switzerland) and recombinant FSH (Gonal F, Merck-Serono, Geneva, Switzerland). Human chorionic gonadotropin (hCG; Profasi, Merck-Serono) was injected when there was at least one 18-mm follicle and three or more 16-mm follicles. Ultrasonography-directed oocyte retrieval was performed 36 h after hCG administration. After 2–4 h of incubation, the cumulus masses of the oocytes were removed with a sharp needle and treated with 0.1% hyaluronidase in Dulbecco’s phosphate-buffered saline (DPBS) (w/v) (Irvine Scientific, Santa Ana, CA, USA) in preparation for ICSI. Only MI oocytes without the first polar body (PB) were used in this study. MI oocytes were cultured in a fertilisation medium (Vitrolife, Sweden) supplemented with 10% HSA in an incubator at 37 °C in 6% CO2 in the air for 5–7 h until they became MII oocytes [11]. Oocytes were then fertilised using ICSI and incubated in the fertilisation medium. Normal fertilisation was assessed and confirmed by the presence of two pronuclei and a second PB at 16–18 h after insemination.
Collection of mouse embryos
Female B6D2F1 mice, aged 6–8 weeks, were superovulated using 5 IU of pregnant mare serum gonadotropin (Ningbo Second Hormone Factory, Ningbo, China) followed by 5 IU of hCG after 48 h (Ningbo Second Hormone Factory). The vaginal plug was checked the day following mating. The day when a vaginal plug was observed was considered day 1 of pregnancy. On day 1.5 of pregnancy, the mice were euthanised by cervical dislocation, and embryos were flushed from the oviduct with phosphate buffered saline (PBS; Life Technologies, Grand Island, NY, USA) using a 30-gauge blunt needle (code procedure 2015/VSC/PEA/00048). The embryos were then flushed four times in M2 medium, incubated at 37 °C in 5% CO2 and 95% humidified air, and used for co-culture experiments and transcriptomic assays.
RNA isolation
RNA extraction was performed using a method described in an earlier study [12]. The miRNeasy Kit (QIAGEN, Hilden, Germany) was used to isolate and purify miRNAs according to the manufacturer’s protocol. Briefly, uterine fluid was gently centrifuged to remove cellular debris and blood. The supernatant was stored at 4 °C, and the mucus within the sample was retrieved and suspended in 1 mL of PBS (pH 7.6; Life Technologies). Subsequently, 500 µL of ULF supernatant from each patient was transferred to an Axygen™ centrifuge tube (Corning, Tewksbury, MA, USA) and thoroughly mixed with 700 µL of QIAzol Lysis Reagent (QIAGEN). After 5 min of incubation at 24 °C, 140 µL of chloroform was added to the mixture, and the mixture was vigorously vortexed. Subsequently, the RNA pellet was collected by centrifugation at 3865 ×g for 30 min at 4 °C. The aqueous phase was carefully transferred to a new tube, and a 1.5 volume of absolute ethyl alcohol was added. The RNA pellet was then placed in an RNA-binding column and washed twice. Finally, the pellet was dissolved in 30 µL of nuclease-free H2O.
miRNA analysis and profiling
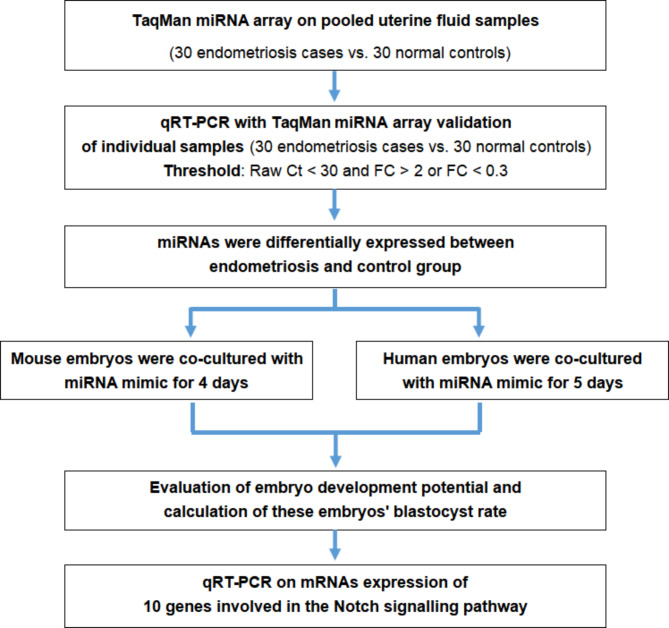
The study design is presented in Fig. 1. Thirty ULF samples from patients with endometriosis were classified as the endometriosis group and 30 samples from control subjects were classified as the non-endometriosis group. Briefly, 30 ng of RNA was initially reverse-transcribed using Megaplex RT Primers Pools A and B and then preamplified with Megaplex Preamp Primers Pools A and B. Subsequently, 900 µL of the preamplified product was loaded on a TaqMan Array Human MicroRNA Card and run on an Applied Biosystems 7900HT thermocycler following the manufacturer’s protocol. The cards contained assays for 766 mature miRNAs in Sanger miRBase version 18.0. Subsequently, miRNA profiling was performed with TaqMan Array Human MicroRNA Cards A and B v3.0 (Applied Biosystems). The analysis was performed following a previous study [12]. Finally, detailed data analysis was performed using the Real-Time Statminer software package (Applied Biosystems).
Fig. 1.
Flow chart of the experimental design
MiRNA validation
To validate the miRNA arrays, we initially measured the expression levels of the candidate miRNAs in each uterine fluid sample in the two groups (30 samples from the endometriosis group and 30 samples from the control group) by qRT‒PCR with a TaqMan miRNA assay. Furthermore, to identify and verify differentially expressed miRNAs correlated with blastocyst development potential, we selected candidate miRNAs in the endometriosis group with Raw Ct (miRNA) < 30 to exclude miRNAs with low expression levels. These miRNAs were selected for subsequent verification analysis and had the highest relative expression quantities. Second, the expression levels were normalised based on an internal reference: U6 snRNA [13]. Finally, the relative expression levels were calculated as 2−ΔCt where ΔCT = Raw Ct (miRNA)-Raw Ct (U6).
Isolation of EVs from ULF
Briefly, ULF samples were diluted in PBS (Life Technologies), vigorously vortexed, and filtered using a 0.22-µm syringe filter (Pall). The filtrates were centrifuged at 300 × g for 10 min to remove whole cells. The supernatant was subjected to a second centrifugation at 2000 × g for 10 min to remove dead cells and then centrifuged again at 10,000 × g for 30 min to remove cell debris. The supernatants were refiltered with a 0.22-µm syringe filter (Pall) and ultracentrifuged at 120,000 × g for 70 min. The supernatants were discarded, and the EVs from ULF (ULF-EVs) pellets were gently washed once with 200 µL of PBS to remove residual extract buffer, resuspended in 20 µL of PBS, and stored at − 80 °C. The size and purity of the isolated ULF-EVs were determined using a nanoparticle tracking analyser (ZetaView PMX 120, Particle Metrix, Germany). Western blotting analysis of the expression of the EV-specific markers CD63 (Abcam, Cambridge, UK), CD81 (Abcam) and CD9 (Abcam) and the negative control marker calnexin (Abcam) was performed, as described [14].
hEEC cultures and EV isolation
Endometrial biopsy samples were obtained from healthy donors in the luteal phase who underwent endometrial biopsy (ages 23–39 years). The endometrial samples were minced into small pieces < 1 mm and then digested in trypsin (Difco, BD Biosciences, MD, USA). Human endometrial epithelial cells (hEECs) were grown from isolated endometrial glands and purified as previously described [15]. This cell type was cultured and grown to confluence in steroid-depleted medium: 75% DMEM (GIBCO, Grand Island, NY, USA) and 25% MCDB-105 (Sigma, St. Louis, MO, USA) containing antibiotics and supplemented with 10% charcoal-dextran-treated FBS (HyClone, Logan, UT) and 5 µg/mL insulin (Sigma). The homogeneity of cultures was determined by analysing the morphological characteristics and verified by immunocytochemical localisation of cytokeratin, vimentin, and CD68. When the cultures reached confluence, they were washed with DMEM (Sigma‒Aldrich) to remove FBS-contaminated EVs and cultured in DMEM (Sigma‒Aldrich). After 48 h, the conditioned medium was collected, and EVs were isolated, as described in the section above.
Western blotting analysis
Antibodies against human CD63, CD9, CD81 (System Biosciences), and calnexin (Enzo Life Sciences) were used for western blotting. First, proteins were extracted from the EVs secreted from primary hEEC cultures with radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with protease and phosphatase inhibitors (Sigma‒Aldrich). The concentration of proteins in the extract was measured using a BCA Kit (Thermo Fisher Scientific, CA, USA). Subsequently, the protein extracts were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Merck Millipore, Germany). First, the membranes were incubated overnight with primary target antibodies diluted 1:1000 in 5% BSA. Second, the membranes were washed three times with 1% PBS-T and incubated with secondary anti-mouse and anti-rabbit antibodies (System Biosciences) at a concentration of 1:20,000. Third, specific proteins were measured using the SuperSignal West Femto Chemiluminescence kit (Thermo Fisher Scientific).
Transmission electron microscopy
Briefly, EVs in suspension, isolated from ULF, were mixed with equal volume of 4% Paraformaldehyde solution for fixation and absorbed by the discharged electron microscope grid. Then, the uranyl acetate was added to the copper grid to precipitate for 1 min, and the floating liquid was absorbed with filter paper. The sample was dried at room temperature for a few minutes before transmission electron microscopy (TEM) (Jeol Korea) imaging was performed at 100 kV.
Embryos co-cultured with miR-145-5p mimic-enriched EVs
Mouse embryos
After fertilisation, the early mouse embryos were successively washed with G-MOPSplus medium (Vitrolife) and G-1 medium (Vitrolife) three times and then cultured in humidified air with 6% CO2 at 37 °C for 48 h. The activated embryos were then randomly divided into two groups. hEECs were transfected with miR-145-5p mimic or pre-miR scramble using Lipofectamine 2000 (Thermo Fisher Scientific). After transfection, the transfection medium was replaced by fresh MEM supplemented with 1% penicillin/streptomycin, 1% l-glutamine, and 10% EV-depleted FBS (Thermo Fisher Scientific). After 48 h of culture, the spent medium was collected for EV extraction. miR-145-5p mimic-enriched EVs were extracted from the spent medium with the Total Exosome Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The miR-145-5p mimic levels in the EVs were determined by reverse transcription qPCR (RT‒qPCR) (Thermo Fisher Scientific). Embryos can take up free and EV-associated miRNAs from uterine fluid [5, 10]. Therefore, to select the most suitable concentration of miR-145-5p mimic EVs, day-1.5 embryos were cultured for 72 h with 50, 100, and 200 µg/mL mimic hsa-miR-145-5p mimic-enriched EVs. To validate the success of the internalization process, a RT‒qPCR was performed in both miR-145-5p mimic-treated and control embryos. In this study, embryos were treated with EVs at a physiological dose of 100 µg/mL. Once the working concentration (100 µg/mL) was selected, the day-1.5 embryos (1–2 embryos per 50 µL droplet) were incubated continuously for 72 h, and their blastocyst rate was calculated.
Human embryos
The human embryos were also randomly divided into two groups. Day 2 human embryos (n = 9) were cultured for 96 h with 50, 100, and 200 µg/ml miR-145-5p mimic EVs to select the most suitable concentration of miR-145-5p mimic-enriched EVs. Once the working concentration (100 µg/mL) was selected, the embryos (1–2 embryos per 50 µL droplet) were continuously cultured for 96 h. Subsequently, embryonic development was evaluated at the blastocyst stages.
Expression levels of NOTCH signalling pathway genes in the miR-145-5p mimic co-cultured and control groups
We collected mouse and human embryos after 72–96 h of co-culture with miRNA-145 mimic-EVs (human embryos: n = 48; mouse embryos: n = 180) or NC mimic-EVs (human embryos: n = 45; mouse embryos: n = 98). The miRNeasy Micro Kit (QIAGEN) was used for the isolation and purification of RNA from the embryos according to the manufacturer’s instructions [16]. The expression levels of 10 target genes (NOTCH1, NOTCH2, NOTCH3, NOTCH4, DLL1, DLL4, JAGGED1, JAGGED2, HES1, and HES2) that encode proteins that are members or regulators of the NOTCH signalling pathway in human/mouse blastocyst-stage embryos were measured by qRT‒PCR. The measurements were then compared between the miR-145-5p mimic co-cultured and control groups. Before PCR, whole transcriptome amplification (TaKaRa, Dalian, China) was performed because the quantity of RNA was limited owing to the small number of embryos. We performed three biological triplicates for each sample.
Statistical analysis
All the experiments were repeated at least three times for each group. The data are presented as the mean ± SEM or mean ± SD. Student’s t-test and chi-square test was used to assess the differences. All the statistical analyses were performed using SPSS (version 19.0; SPSS Inc., Chicago, IL, USA). Differences were considered significant when P < 0.05.
Results
Clinical and medical characteristics of study participants
The baseline demographics and clinical characteristics of participants with and without endometriosis are presented in Table 1. No differences were observed between the two groups in age, body mass index (BMI), years of infertility, hormone levels, the proportion of IVF/ICSI, endometrial thickness, embryo transfer cycles, embryo stage, thawed embryo survival rates and the number of transferred embryos. However, the embryo implantation and clinical pregnancy rates in frozen embryo transfer (FET) cycles were significantly higher in women in the control group than in those in the endometriosis group (all P < 0.01, Table 1). The flow chart of the study design is shown in Fig. 1.
Table 1.
Clinical characteristics of infertile patients with endometriosis and controls in FET cycles
| Variable | Controls (n = 30) |
Endometriosis (n = 30) |
P values |
|---|---|---|---|
| Female age (years) | 34.2 ± 6.1 | 33.5 ± 5.3 | NS |
| BMI (kg/m2) | 22.3 ± 2.4 | 21.6 ± 2.6 | NS |
| Duration of infertility (years) | 4.5 ± 3.1 | 4.9 ± 3.7 | NS |
| Basal FSH (mIU/mL) | 7.7 ± 2.3 | 8.5 ± 3.2 | NS |
| AMH (ng/mL) | 4.31 ± 2.38 | 3.89 ± 1.62 | NS |
| Oestradiol (pg/mL) | 31. 28 ± 8.83 | 34.91 ± 9.75 | NS |
| Progesterone (ng/mL) | 0.83 ± 0.47 | 0.76 ± 0.46 | NS |
| LH (IU/L) | 4.03 ± 1.06 | 4.39 ± 1.57 | NS |
| Proportion of IVF (%) | 18 (60.0) | 19 (63.3) | NS |
| Proportion of ICSI (%) | 12 (40.0) | 11 (36.7) | NS |
| Endometrial thickness (mm) | 10.3 ± 2.4 | 9.8 ± 2.5 | NS |
| Embryo transfer cycles (%) | |||
| Natural cycles (NC) | 33.3% (10/30) | 30.0% (9/30) | NS |
| HRT cycles | 66.7% (20/30) | 70.0% (21/30) | NS |
| Embryo stage (%) | |||
| cleavage | 13.9% (5/36) | 15.4% (6/39) | NS |
| blastocyst | 86.1% (31/36) | 84.6% (33/39) | NS |
| Thawed embryo survival rates (%) | 100% (36/36) | 100% (39/39) | NS |
| No. of transferred embryos | 1.2 ± 0.6 | 1.3 ± 0.7 | NS |
| Implantation rate (%) | 36.1 (13/36) | 23.1 (9/39) | < 0.01 |
| Clinical pregnancy rate (%) | 63.3 (19/30) | 36.7 (11/30) | < 0.01 |
The controls were healthy patients without endometriosis undergoing treatment for tubal factor infertility. FET, Frozen embryo transfer. NS, no significant difference. P values: Unpaired t-test. BMI, body mass index; FSH, follicle-stimulating hormone; IVF, in vitro fertilisation; ICSI, intracytoplasmic sperm injection; HRT, hormone replacement therapy
ULF miRNA profiles and the identification of differentially expressed miRNAs in women with and without endometriosis
The finding that the embryo implantation rate was higher in the controls than in patients with endometriosis demonstrated that differentially expressed miRNAs between the patients with endometriosis and controls might play a role in blastocyst development. MiRNAs with Raw Ct < 30 are considered as highly expressive [17, 18]. Thus, to identify and verify differentially expressed miRNAs correlated with blastocyst development potential, we selected candidate miRNAs in the endometriosis group with Raw Ct (miRNA) < 30 to exclude miRNAs with low expression levels. MiRNAs with high expression levels (Raw Ct < 30) are listed in Supplementary Table 1. As presented in Table 2, 8 miRNAs were upregulated and 9 miRNAs were downregulated in the endometriosis group. These miRNAs were selected for subsequent verification analysis and had the highest relative expression quantities. The differences between the endometriosis and control groups were based on the miRNA profiling results.
Table 2.
MicroRNAs with high expression levels, identified by miRNA array, between uterine fluid samples from control patients and patients with endometriosis
| MicroRNAs | Endometriosis | Controls | FC | ||||
|---|---|---|---|---|---|---|---|
| Raw Ct | ΔCt | RQ | Raw Ct | ΔCt | RQ | ||
| hsa-miR-145-5p | 23.0494 | 4.7841 | 36.2948 | 26.1280 | 7.2737 | 6.4625 | 5.6162 |
| hsa-miR-720 | 26.7546 | 8.4893 | 2.7827 | 29.7631 | 10.9088 | 0.5201 | 5.3503 |
| hsa-miR-106b | 25.2551 | 6.9898 | 7.8678 | 28.8026 | 9.9483 | 1.0122 | 7.7729 |
| hsa-miR-143 | 22.2295 | 3.9642 | 64.0722 | 25.9785 | 7.1242 | 7.1681 | 8.9385 |
| hsa-miR-708 | 22.8780 | 4.6127 | 40.8724 | 26.6919 | 7.8376 | 4.3717 | 9.3493 |
| hsa-miR-486 | 23.5726 | 5.6287 | 5.4628 | 28.1158 | 3.2579 | 67.3218 | 3.7251 |
| hsa-miR-191 | 24.9304 | 6.6651 | 9.8539 | 27.0046 | 8.1503 | 3.5198 | 2.7996 |
| hsa-miR-302b | 26.6159 | 8.3506 | 3.0635 | 29.0073 | 10.1530 | 0.8783 | 3.4880 |
| hsa-miR-21 | 26.2200 | 7.9547 | 4.0308 | 24.8751 | 6.0208 | 15.4013 | 0.2617 |
| hsa-miR-146a | 21.3511 | 3.0858 | 117.7844 | 20.1410 | 1.2867 | 409.8875 | 0.2874 |
| hsa-miR-378 | 29.5752 | 11.3099 | 0.3939 | 23.8004 | 4.9461 | 32.4396 | 0.0121 |
| hsa-miR-126 | 26.2015 | 7.9362 | 4.0828 | 24.1393 | 5.2850 | 25.6482 | 0.1592 |
| hsa-miR-663 | 25.9662 | 7.7009 | 4.8063 | 21.7182 | 2.8639 | 137.3663 | 0.0350 |
| hsa-miR-451 | 24.7608 | 6.4955 | 11.0828 | 22.7280 | 3.8737 | 68.2182 | 0.1625 |
| hsa-miR-3600 | 27.0926 | 8.8273 | 2.2015 | 21.9041 | 3.0498 | 120.7588 | 0.0182 |
| hsa-miR-125a | 28.6388 | 10.3735 | 0.7538 | 26.9847 | 8.1304 | 3.5687 | 0.2112 |
| hsa-miR-205 | 25.8876 | 7.6223 | 5.0752 | 24.0209 | 5.1666 | 27.8419 | 0.1823 |
Internal reference: snRNA U6. ΔCT = Raw Ct (miRNA)-Raw Ct(U6). Inclusion criteria: Raw Ct < 30. Fold Change (FC) = RQ (Endometriosis)/RQ (Controls), FC > 2 or FC < 0.3, RQ, relative quantity, RQ = 2 −ΔCt *1000
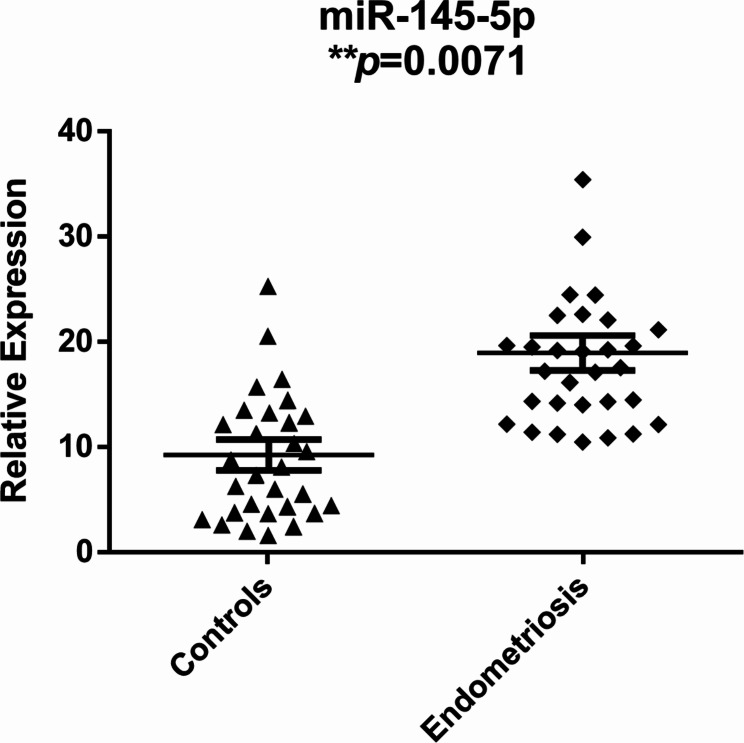
The expression levels of these candidate miRNAs in each uterine fluid sample of the endometriosis and control groups were measured by qRT‒PCR with a TaqMan® miRNA assay. Among the 17 candidate miRNAs, miR-145-5p had significantly higher expression in the endometriosis group than in the control group (P = 0.0071) (Fig. 2). No crucial differences were observed in the expression levels of the remaining miRNAs between the two groups (Supplementary Fig. 1).
Fig. 2.
Relative expression levels of hsa-miR-145-5p in the endometriosis and control groups. Scatter plots depicting the relative expression levels of hsa-miR-145-5p between the two groups during the window of implantation (WOI), unpaired t-test. *P < 0.05; **P < 0.01. Mean ± SD shown by bars
Effects of miR-145-5p mimic-enriched EVs on the development of mouse blastocyst
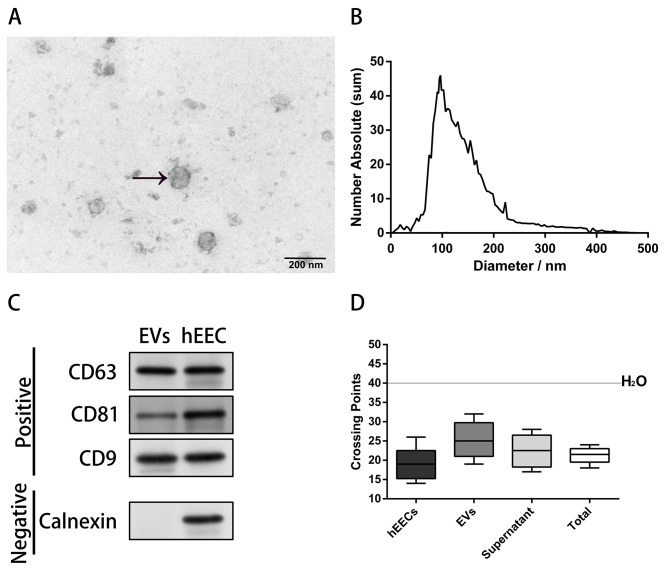
miR-145-5p in the EVs isolated from ULF had also significantly higher expression in the endometriosis group than in the control group (P = 0.0087) (Fig. S3). Maternal endometrial miRNA, secreted by the human endometrium, could be taken up by preimplantation embryo; therefore, we tested the possibility that hsa-miR-145-5p of maternal origin might impede mouse blastocyst development. The following observations supported this possibility. First, the TEM showed that the EVs purified from human ULF consisted of circular vesicular membranes, 50–250 nm in diameter (Fig. 3A). And the nanoparticle tracking analysis revealed that the EVs in the ULF had a mean size of 93.6 nm (Fig. 3B). Second, the western blotting analysis revealed that the EVs from ULF was positive for the three exosomal tetraspanins CD63, CD81, and CD9 and negative for calnexin [8] (Fig. 3C). Third, mouse ULF contains miRNA-carrying CD63 + EVs, which blastocysts can internalise [14]. Among potential miRNAs, hsa-miR-145-5p was identified in hEECs, EVs extracted from primary hEEC cultures, supernatants, and total supernatants without depletion of EVs (Fig. 3D). The control and miR-145-5p mimic-enriched EVs were extracted from the spent medium of hEECs transfected with miR-145-5p mimic or pre-miR scramble. To further explore the function of miR-145-5p in influencing early embryonic development potential, mouse blastocyst development was evaluated by co-incubation with control or miR-145-5p mimic-enriched EVs for 72 h.
Fig. 3.
Characterization of the isolated extracellular vesicles (EVs) from human uterine luminal fluid. A Representative image of transmission electron microscopy (TEM) images showing morphology of isolated EVs purified from human uterine luminal fluid. B Nanoparticle tracking analysis by ZetaView PMX120 indicated that the mean diameter of the EVs was 93.6 nm. C Western blotting analysis suggested that the EVs from uterine luminal fluid expressed three positive markers (CD9, CD81, and CD63) but not one negative marker (calnexin) (Fig. 3C includes cropped blot images, and the original blot image can be obtained from Figure S2). D Crossing points (y-axis) for hsa-miR-145-5p in cells, EVs extracted from primary hEEC cultures, supernatants, and total supernatants without depletion of EVs. The box indicates the first, median, and third quartiles, and the error bars indicate the minimum and maximum values. hEECs, human endometrial epithelial cells
As shown in Fig. 4, mouse blastocyst development was significantly affected by treatment with miR-145-5p mimic-enriched EVs during the 72-h co-culture of day 1.5 embryos. The proportions of embryos that developed into blastocyst-stage embryos were 14.13% ± 1.79% in the miR-145-5p mimic group (n = 180), 45.25% ± 3.79% in the scramble-EVs group (n = 162), and 46.31% ± 4.23% in the untreated control group (n = 205) (P = 0.0065).
Fig. 4.
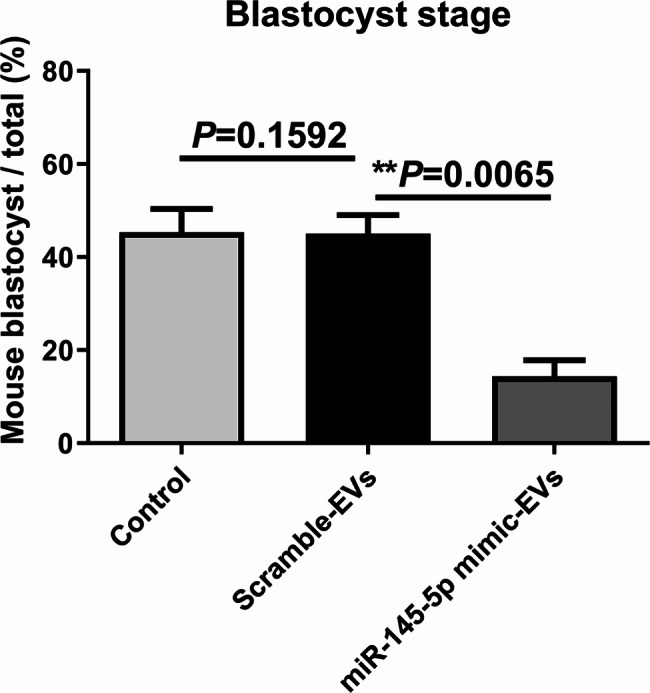
Effects of miR-145-5p mimic-enriched EVs on in vitro development of mouse IVF embryos. Mouse IVF embryos co-incubated with miR-145-5p mimic-enriched EVs or scramble miRNA EVs on day 1.5 of culture. The continuing culture was conducted for 72 h, and the development rate of the blastocyst was calculated. The proportions of day-1.5 embryos that developed to the blastocyst stage in the untreated control (n = 205), scramble-EVs (n = 162), and miR-145-5p mimic groups (n = 180) were 46.31% ± 4.23%, 45.25% ± 3.79%, and 14.13% ± 1.79%, respectively. All the experiments were repeated at least three times. Chi-square test, *P < 0.05; **P < 0.01; ***P < 0.001. EVs, extracellular vesicles
Effects of miR-145-5p mimic-enriched EVs on the expression levels of Notch components in mouse blastocyst
To demonstrate the functional relevance of the maternal tissue–embryo communication mechanism, we investigated the effects of endometrial miRNA uptake on the expression levels of Notch pathway components in blastocyst-stage embryos.
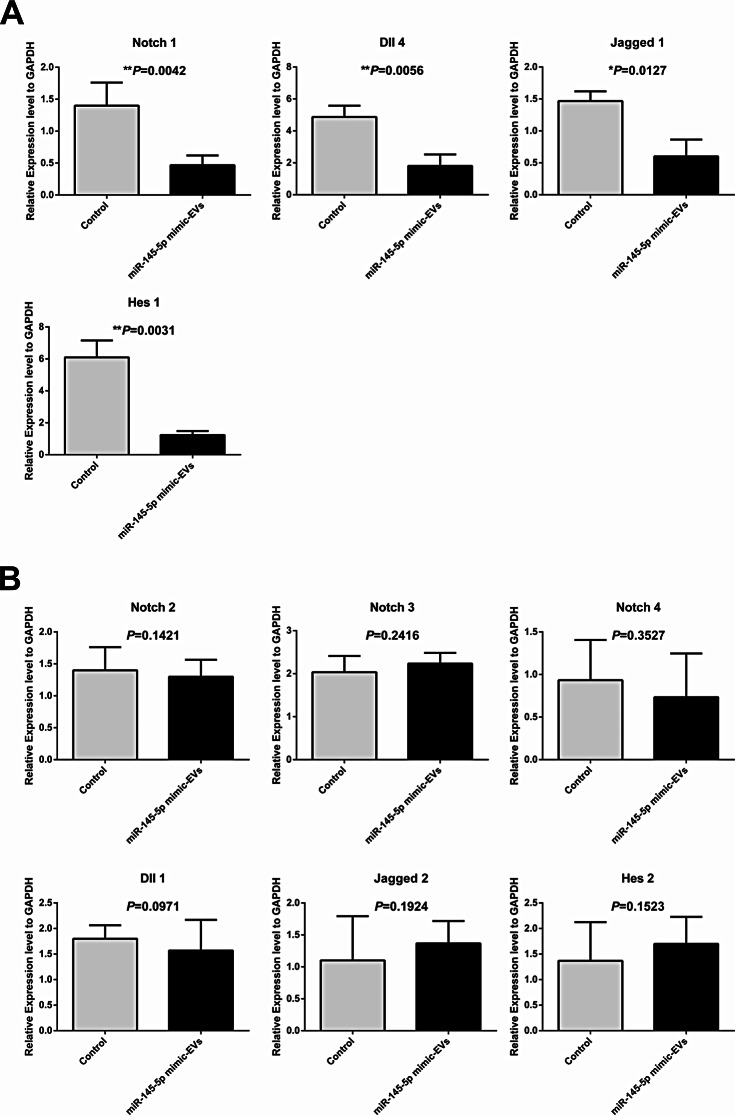
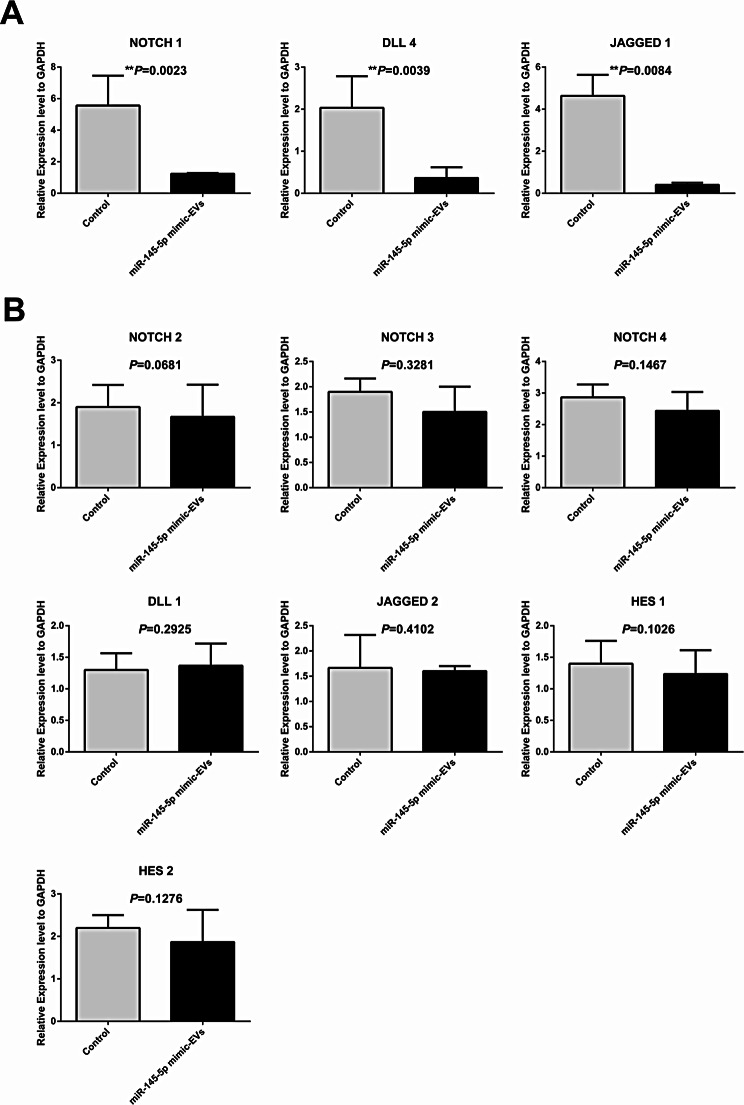
After the co-culture of miR-145-5p mimic-enriched or scramble miRNA EVs with day-1.5 mouse embryos for 72 h, mRNA expression profiles of 10 genes related to the Notch signalling pathway were measured by qRT-PCR in the miR-145-5p mimic (n = 173) and control (n = 119) groups. The results showed that four predicted genes, namely, Notch 1, Dll 4, Jagged 1, and Hes 1, exhibited markedly decreased expression in mouse blastocyst-stage embryos in the miR-145-5p mimic group than in those in the control group (P < 0.01, Fig. 5A). Another six genes (Notch 2, Notch 3, Notch 4, Dll 1, Jagged 2, and Hes 2) had no significant difference between the two groups (P > 0.05, Fig. 5B).
Fig. 5.
Effects of miR-145-5p mimic-enriched EVs on the expression levels of Notch pathway components in mouse blastocyst-stage embryos. Mouse IVF embryos co-incubated with miR-145-5p mimic-enriched EVs or scramble miRNA EVs on day 1.5 of culture. The continuing culture was conducted for 72 h, and the mRNA expression profiles of 10 genes related to the Notch signalling pathway were measured by qRT-PCR. The histograms present four genes that encode proteins that are members or regulators of the Notch signalling pathway with markedly different expression levels (A) and six genes that encode proteins that are members or regulators of the Notch signalling pathway with nonsignificant differences in expression (B) between the miR-145-5p mimic (n = 173) and control (n = 119) groups. All the experiments were repeated at least three times. Unpaired t-test; *P < 0.05; **P < 0.01; ***P < 0.001. EVs, extracellular vesicles
Effects of miR-145-5p mimic-enriched EVs on the development of human blastocyst
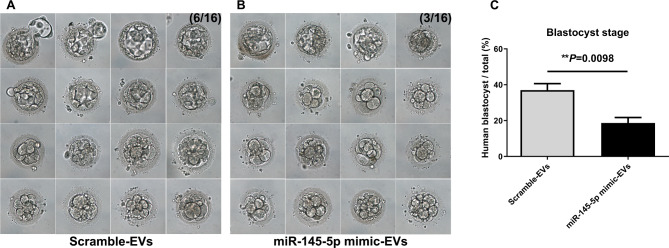
Human blastocyst development in the miR-145-5p mimic group was also significantly influenced (Fig. 6). After the co-cultivation of miR-145-5p mimic-enriched or scramble miRNA EVs with day-2 human embryos for 96 h, photographs of the blastocysts were captured; the images are shown in Fig. 6A and B. The proportions of day-2 embryos that developed into the blastocyst stage were significantly lower (18.75% ± 1.46% vs. 37.5% ± 3.95%, P = 0.0098; Fig. 6C) in the miR-145-5p mimic group (n = 48) than in the scramble-EVs group (n = 45).
Fig. 6.
Effects of miR-145-5p mimic-enriched EVs on in vitro development of human IVF embryos. Human IVF embryos co-incubated with miR-145-5p mimic-enriched EVs or scramble miRNA EVs on day 2 of culture. The continuing culture was conducted for 96 h, and the development rate of the blastocyst was calculated. A Representative image of scramble miRNA EVs added to IVF embryos for 96 h. B Representative image of miR-145-5p mimic-enriched EVs added to IVF embryos for 96 h. C Proportions of embryos that developed into the blastocyst stage in the scramble-EVs (n = 45) and miR-145-5p mimic-EVs (n = 48) groups were 37.5% ± 3.95% and 18.75% ± 1.46%, respectively. All the experiments were repeated at least three times. Chi-square test, *P < 0.05; **P < 0.01; ***P < 0.001. EVs, extracellular vesicles
Effects of miR-145-5p mimic-enriched EVs on the expression levels of Notch components in human blastocysts
After the co-incubation of miR-145-5p mimic-enriched or scramble miRNA EVs with day-2 human embryos for 96 h, mRNA expression profiles of 10 genes related to the Notch signalling pathway were measured by qRT‒PCR in the miR-145-5p mimic and control groups. As with the mouse samples, the results from the human samples also demonstrated that the three predicted genes, namely, NOTCH 1, DLL 4, and JAGGED 1, were significantly downregulated in human blastocyst-stage embryos in the miR-145-5p mimic group than in those in the control group (P < 0.01, Fig. 7A). The expression of the other seven genes (NOTCH 2, NOTCH 3, NOTCH 4, DLL 1, JAGGED 2, HES 1, and HES 2) was not markedly different between the miR-145-5p mimic (n = 48) and control (n = 45) groups (P > 0.05, Fig. 7B).
Fig. 7.
Effects of miR-145-5p mimic-enriched EVs on the expression levels of NOTCH pathway components in human blastocyst-stage embryos. Human IVF embryos co-incubated with miR-145-5p mimic-enriched EVs or scramble miRNA EVs on day 2 of culture. The continuing culture was conducted for 96 h, and the mRNA expression profiles of 10 genes related to the NOTCH signalling pathway were measured by qRT-PCR. The histograms present three crucially differentially expressed genes that encode proteins that are members or regulators of the NOTCH signalling pathway (A) and seven genes that encode proteins that are members or regulators of the Notch signalling pathway with nonsignificant expression differences (B) between the miR-145-5p mimic (n = 48) and control (n = 45) groups. All the experiments were repeated at least three times. Unpaired t-test; *P < 0.05; **P < 0.01; ***P < 0.001. EVs, extracellular vesicles
Discussion
In our studies, the clinical data demonstrate that the implantation rate and clinical pregnancy rate were significantly decreased in patients with endometriosis. MiRNA microarray technology was used to search for miRNAs with aberrant expression in the ULF from patients with endometriosis compared with patients without endometriosis during the WOI. qRT‒PCR validated the finding that hsa-miR-145-5p had significantly different expression levels in the endometriosis group. Subsequently, our in vitro study demonstrated that miR-145-5p–enriched EVs significantly impacted blastocyst development potential. Functional analysis of mouse/human embryos and blastocysts revealed that the abnormal expression of NOTCH signalling pathway-related genes might negatively affect blastocyst-stage embryo development. To the best of our knowledge, this is the first quantitative assessment of ULF miRNA expression that could explain the cause of infertility in women with endometriosis.
Accumulated evidence suggests that the ULF serves as the ‘uterine milk’ that nurtures the embryo during implantation and beyond [19]. ULF contains a rich array of nutrients, proteins, glycodelin, lipids, cytokines, and other molecules. Studies have demonstrated that the constituents of ULF may affect embryo outgrowth and other functionalities during implantation in women with endometriosis [20]. Related studies have demonstrated that the EVs released by the endometrial epithelium into the ULF is involved in the transfer of signalling proteins, mRNAs, and miRNAs to either the trophectodermal of the blastocyst or adjacent endometrial epithelial cells. This transfer of materials influences endometrial receptivity, implantation, and embryonic development [8, 10]. The presence of miRNAs in human ULF was first described by Ng et al. (2013). However, no studies have analysed the relationship between miRNAs–enriched EVs in ULF patients with endometriosis and blastocyst development potential. Therefore, in our study, we hypothesised that miRNAs in the ULF-EVs of women with endometriosis correlated with blastocyst development potential.
In our study, we observed significantly high expression of miR-145-5p in the ULF of patients with endometriosis than in healthy women, which is consistent with earlier studies that suggested that miR-145-5p expression was substantially increased in the eutopic and ectopic endometria of patients with endometriosis [21]. miR-145-5p mimic oligonucleotides in EVs were used to upregulate its expression in mouse/human zygotes to assess the role of miR-145-5p in early embryonic development. This study indicated that the blastocyst outgrowth rate of mouse/human zygotes was significantly affected by the upregulation of miR-145-5p, which further demonstrated that miR-145-5p-enriched EVs have a substantial impact on blastocyst-stage embryonic development potential. Furthermore, consistent with the above observation, decreasing miRNA-145 expression significantly improves the development of bovine somatic cell nuclear transfer embryos in vitro [22], and increased miR-145-5p expression strongly represses pluripotency in human embryonic stem cells [23].
Some reports have indicated that miRNA function is repressed in oocytes [24, 25]; however, others have reported an association between altered miRNA expression levels and oocyte maturation, blastocyst formation, and preimplantation embryo development [17, 26, 27]. This evidence demonstrates that maternal miRNAs are crucial for oocyte and early embryogenesis. Furthermore, miRNAs play critical roles in several signalling pathways. There is evidence to suggest that miR-145-5p participates in regulating multiple signalling pathways, including Notch signalling [28], Wnt/β-catenin signalling [29], and MAPK signalling pathways [30]. The Notch signalling pathway, which is highly conserved among invertebrates and vertebrates, has been implicated as the main regulator of cellular differentiation and proliferation in many adult and embryonic scenarios [31].
Furthermore, several findings have suggested that the components of the Notch signalling pathway are involved in mammalian ovarian folliculogenesis [32] and preimplantation embryogenesis [33–35], including early embryonic development, trophectoderm specification in mouse blastocysts, blastocyst hatching, and embryo implantation. In addition, Notch signalling pathway could be regulated by miR-145-5p [28]. Altogether, these observations demonstrate that the Notch signalling pathway might affect early preimplantation embryogenesis, and we postulate that this may be altered upon miR-145-5p upregulation.
Therefore, we tested 10 genes related to the Notch signalling pathway in treated mouse and human blastocyst-stage embryos. Notch 1, Dll 4, Jagged 1, and Hes 1 were markedly decreased in mouse blastocyst-stage embryos in the miR-145-5p mimic group, and NOTCH 1, DLL 4, and JAGGED 1 were also significantly downregulated in human blastocyst-stage embryos in the miR-145-5p mimic group. Thus, these findings together indicate the suppression of the NOTCH signalling pathway. Furthermore, among the four differentially expressed genes, NOTCH 1, DLL 4, and HES 1 are significant in early embryogenesis, blastocyst development, and hatching of blastocysts [33, 34, 36]. NOTCH 1 plays an essential role in proper early embryonic development owing to its role in the specification of the trophectoderm lineage and the regulation of Notch 1 intracellular domain expression in the mouse blastocyst [34, 35]. Knockdown of maternal and zygotic NOTCH 1 significantly reduces the developmental competence of blastocyst-stage embryos and decreases the cell number of blastocysts [34]. These results indicate that NOTCH 1 downregulation after the co-culture of zygotes with miR-145-5p mimic-EVs may reduce the developmental potential of mouse/human zygotes to the blastocyst stage, thereby decreasing the cell number of blastocysts and inhibiting the specification of the trophectoderm lineage.
The endometrial epithelium releases ULF-EVs containing proteins, mRNAs, and miRNAs that are considered to effectively communicate between the endometrium and blastocysts [8]. miR-145-5p-containing EVs reduced the developmental potential of blastocysts in women with endometriosis, and dysregulation of endometrial miRNAs is observed in subfertile women. Endometriosis-related miRNAs may be abnormally expressed in endometrial-derived EVs and can inhibit the development of preimplantation embryos, contributing to nonsynchronous development between the blastocysts and the endometrium. However, further findings are required to inquire into this possibility.
In conclusions, we discovered that human uterus-derived miRNAs in EVs affected the developmental potential of blastocysts in women with endometriosis. In particular, the upregulation of miR-145-5p in EVs in mouse and human embryos negatively affected blastocyst development by suppressing the expression of components of the NOTCH signalling pathway. However, this study has several limitations. First, the human MII oocytes used in this study were derived from MI oocytes without their first PB, which were exposed to gonadotrophic stimulation; however, failure in maturing in vivo might constitute a sign of their low quality; this should be considered when drawing conclusions from the data. Second, further research should also pay more attention to the mechanism of association of the upregulation of miR-145-5p in the ULF with the blastocyst development potential and the roles of miR-145-5p on blastocyst development. This research may aid in elucidating that the upregulation of miR-145-5p in ULF-EVs may cause infertility in patients with endometriosis.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the ART team of the Shanghai JiAi Genetics and IVF institute for their support in this study.
Abbreviations
- IVF
in vitro fertilization
- ULF
uterine luminal fluid
- EVs
extracellular vesicles
- ULF-EV
extra cellular vesicle from ULF
- MI
metaphase-I
- WOI
window of implantation
- FET
frozen embryo transfer
- hEECs
human endometrial epithelial cells
- qRT‒PCR
quantitative reverse transcription polymerase chain reaction
- ICSI
intracytoplasmic sperm injection
Author contributions
XL and WJ analysed the data and wrote the article. XL and JF arranged the experiments. XL, YX, RG, RQ, YZ, ZL, and WZ collected the uterine luminal fluid and analysed it by miRNA microarrays. JF, YS, and XS conceived the study, obtained funding, and edited the paper. All the authors critically reviewed and approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFC2700100) and Shanghai Medical Public Service Platform Fund (No. SY202211RUE01).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study adhered to the tenets of the Declaration of Helsinki. The studies involving human participants and animal experiments were reviewed and approved by the Institutional Review Committee of Fudan University, and all methods were carried out in accordance with its guidelines and regulations. Written informed consent for study participation was obtained from all subjects. And this study was also carried out in compliance with the ARRIVE guidelines for the reporting of animal experiments.
Consent for publication
Consent for publication was obtained from each patient.
Competing interests
The authors declare no competing interests.
Capsule
miR-145-5p is upregulated in uterine luminal fluid in women with endometriosis. Upregulation of miR-145-5p-enriched extracellular vesicles negatively affected mouse and human blastocyst development by suppressing the NOTCH signalling pathway and may be one of the causes for compromised fertility in women with endometriosis.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiong Li and Jing Fu joint first authors.
Contributor Information
Yijuan Sun, Email: yijuansun2019@163.com.
Xiaoxi Sun, Email: steven30l9@hotmail.com.
References
- 1.Morotti M, Vincent K, Becker CM. Mechanisms of pain in endometriosis. Eur J Obstet Gynecol Reprod Biol. 2017;209:8–13. [DOI] [PubMed] [Google Scholar]
- 2.Mahutte NG, Arici A. New advances in the understanding of endometriosis related infertility. J Reproduct Immunol. 2002;55(1–2):73–83. [DOI] [PubMed] [Google Scholar]
- 3.Evans MB, Decherney AH. Fertility and Endometriosis. Clin Obstet Gynecol. 2017;60(3):497–502. [DOI] [PubMed] [Google Scholar]
- 4.Leone Roberti Maggiore U, Ferrero S, Mangili G, Bergamini A, Inversetti A, Giorgione V, Viganò P, Candiani M. A systematic review on endometriosis during pregnancy: diagnosis, misdiagnosis, complications and outcomes. Hum Reprod Update. 2016;22(1):70–103. [DOI] [PubMed] [Google Scholar]
- 5.Vilella F, Moreno-Moya JM, Balaguer N, Grasso A, Herrero M, Martínez S, Marcilla A, Simón C. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development. 2015;142(18):3210–21. [DOI] [PubMed] [Google Scholar]
- 6.Yoshizawa JM, Wong DT. Salivary microRNAs and oral cancer detection. Methods Mol Biol. 2013;936:313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato M, Suzuki T, Kawano M, Tamura M. Circulating osteocyte-derived exosomes contain miRNAs which are enriched in exosomes from MLO-Y4 cells. Biomed Rep. 2017;6(2):223–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng YH, Rome S, Jalabert A, Forterre A, Singh H, Hincks CL, Salamonsen LA. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE. 2013;8(3):e58502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thouas GA, Dominguez F, Green MP, Vilella F, Simon C, Gardner DK. Soluble ligands and their receptors in human embryo development and implantation. Endocr Rev. 2015;36(1):92–130. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Tian F, Chen C, Feng Y, Sheng X, Guo Y, Ni H. Exosome-derived uterine microRNAs isolated from cows with endometritis impede blastocyst development. Reprod Biol. 2019;19(2):204–9. [DOI] [PubMed] [Google Scholar]
- 11.Yoon SY, Eum JH, Lee JE, Lee HC, Kim YS, Han JE, Won HJ, Park SH, Shim SH, Lee WS, et al. Recombinant human phospholipase C zeta 1 induces intracellular calcium oscillations and oocyte activation in mouse and human oocytes. Hum Reprod. 2012;27(6):1768–80. [DOI] [PubMed] [Google Scholar]
- 12.Sang Q, Yao Z, Wang H, Feng R, Wang H, Zhao X, Xing Q, Jin L, He L, Wu L, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;98(7):3068–79. [DOI] [PubMed] [Google Scholar]
- 13.Rosenbluth EM, Shelton DN, Wells LM, Sparks AE, Van Voorhis BJ. Human embryos secrete microRNAs into culture media–a potential biomarker for implantation. Fertil Steril. 2014;101(5):1493–500. [DOI] [PubMed] [Google Scholar]
- 14.Niu Z, Pang RTK, Liu W, Li Q, Cheng R, Yeung WSB. Polymer-based precipitation preserves biological activities of extracellular vesicles from an endometrial cell line. PLoS ONE. 2017;12(10):e0186534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simo´n C, Piquette GN, Frances A, El-Danasouri I, Polan ML. The effect of interleukin-1 beta (IL-1) on the regulation of IL-1 receptor type I and IL-1 beta messenger ribonucleic acid (mRNA) levels and protein expression in cultured human endometrial stromal and glandular cells. J Clin Endocrinol Metab. 1994;78(3):675–82. [DOI] [PubMed] [Google Scholar]
- 16.Stigliani S, Moretti S, Anserini P, Casciano I, Venturini PL, Scaruffi P. Storage time does not modify the gene expression profile of cryopreserved human metaphase II oocytes. Hum Reprod. 2015;30(11):2519–26. [DOI] [PubMed] [Google Scholar]
- 17.Feng R, Sang Q, Zhu Y, Fu W, Liu M, Xu Y, Shi H, Xu Y, Qu R, Chai R, et al. MiRNA-320 in the human follicular fluid is associated with embryo quality in vivo and affects mouse embryonic development in vitro. Sci Rep. 2015;5:8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu J, Qu RG, Zhang YJ, Gu RH, Li X, Sun YJ, Wang L, Sang Q, Sun XX. Screening of miRNAs in human follicular fluid reveals an inverse relationship between microRNA-663b expression and blastocyst formation. Reprod Biomed Online. 2018;37(1):25–32. [DOI] [PubMed] [Google Scholar]
- 19.Khalil A, Jauniaux E, Cooper D, Harrington K. Pulse wave analysis in normal pregnancy: a prospective longitudinal study. PLoS ONE. 2009;4(7):e6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang NX, Li XL. The Complicated Effects of Extracellular Vesicles and Their Cargos on Embryo Implantation. Front Endocrinol (Lausanne). 2021;12:681266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei S, Xu H, Kuang Y. Systematic enrichment analysis of microRNA expression profiling studies in endometriosis. Iran J Basic Med Sci. 2015;18(5):423–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Xiong Y, Wang F, Liu X, Gao Y, Wang Y, Zhang Y, Jin Y. MicroRNA-145 Inhibitor Significantly Improves the Development of Bovine Somatic Cell Nuclear Transfer Embryos In Vitro. Cell Reprogram. 2016;18(4):230–6. [DOI] [PubMed] [Google Scholar]
- 23.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–58. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20(3):265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20(3):271–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dehghan Z, Mohammadi-Yeganeh S, Salehi M. MiRNA-155 regulates cumulus cells function, oocyte maturation, and blastocyst formation. Biol Reprod. 2020;103(3):548–59. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Zhang W, Fu J, Xu Y, Gu R, Qu R, Li L, Sun Y, Sun X. MicroRNA-451 is downregulated in the follicular fluid of women with endometriosis and influences mouse and human embryonic potential. Reprod Biol Endocrinol. 2019;17(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei B, Liu YS, Guan HX. MicroRNA-145-5p attenuates high glucose-induced apoptosis by targeting the Notch signaling pathway in podocytes. Exp Ther Med. 2020;19(3):1915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Yu X, Yang L. MiR-145 inhibits cervical cancer progression and metastasis by targeting WNT2B by Wnt/β-catenin pathway. Int J Clin Exp Pathol. 2019;12(10):3740–51. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Zhang S, Cheng G, Mei C, Li S, Zhang W, Junjvlieke Z, Zan L. MiR-145 reduces the activity of PI3K/Akt and MAPK signaling pathways and inhibits adipogenesis in bovine preadipocytes. Genomics. 2020;112(4):2688–94. [DOI] [PubMed] [Google Scholar]
- 31.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138(17):3593–612. [DOI] [PubMed] [Google Scholar]
- 32.Vanorny DA, Mayo KE. The role of Notch signaling in the mammalian ovary. Reproduction. 2017;153(6):R187–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batista MR, Diniz P, Torres A, Murta D, Lopes-da-Costa L, Silva E. Notch signaling in mouse blastocyst development and hatching. BMC Dev Biol. 2020;20(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Shi Y, Dang Y, Luo L, Hu B, Wang S, Wang H, Zhang K. NOTCH signaling pathway is required for bovine early embryonic development. Biol Reprod. 2021;105(2):332–44. [DOI] [PubMed] [Google Scholar]
- 35.Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Cañon S, Sasaki H, Hadjantonakis AK, de la Pompa JL, et al. Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell. 2014;30(4):410–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batista MR, Diniz P, Murta D, Torres A, Lopes-da-Costa L, Silva E. Balanced Notch-Wnt signaling interplay is required for mouse embryo and fetal development. Reproduction. 2021;161(4):385–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.