Abstract
The pathophysiological mechanism involving the proteolytic processing of amyloid precursor protein (APP) and the generation of amyloid plaques is of significant interest in research on Alzheimer's disease (AD). The increasing significance of the downstream AD-related pathophysiological mechanisms has sparked research interest in other products of the APP processing cascades, including the APP intracellular domain (AICD). The potential importance of AICD in various cellular processes in the central nervous system has been established through the identification of its interactors. The interaction between AICD and its physiological binding partners is implicated in cellular events including regulation of transcriptional activity, cytoskeletal dynamics, neuronal growth, APP processing and cellular apoptosis. On the contrary, AICD is also implicated in neurodegeneration, which is a potential outcome of the functional fluctuation of AICD-mediated neuronal processes within the neuronal network. In this review, we summarize the neuronal functions and pathological manifestations of the dynamic AICD interaction network.
Keywords: APP intracellular domain, neurodevelopment, protein-protein interaction
Introduction to amyloid precursor protein and its intracellular domain
Over the last century, research in Alzheimer's disease (AD) has accumulated a large body of data corroborating the amyloid cascade hypothesis (ACH). ACH posits the extracellular deposition of amyloid-β (Aβ) peptides as the causal agent of AD pathogenesis [1]. Aβ is derived from aberrant proteolysis of the amyloid precursor protein (APP) by β- and γ-secretase sequentially. γ-Secretase-mediated APP cleavage simultaneously releases APP intracellular domain (AICD), a 6-kDa protein fragment, into the cytosol. APP and its closely related mammalian homologues, namely APP-like proteins 1 and 2 (APLP1 and APLP2), are transmembrane proteins which share a conserved structure: a large N-terminal ectoplasmic domain, a single membrane-spanning domain and a small C-terminal cytoplasmic domain [2,3].
APP is a type I transmembrane protein, and such transmembrane proteins, like Notch and low-density lipoprotein receptor protein family (LRP), often function as a cell surface receptor to detect and transduce extracellular signals. For instance, LRP-5 and -6 are involved in Wnt signaling as they are co-receptors to Wnt ligands, that regulate Wnt-mediated cell proliferation[4–6]. Similarly, APP has recently been revealed as a conversed Wnt receptor. CRD of APP on the N-terminal has shown to be the receptor for Wnt ligands, including Wnt5 and Wnt3a, which regulate the expression of APP itself, and also regulate Wnt-mediated neurite development [7]. The extracellular domain of APP is also associated in cell-cell adhesion and synaptic adhesion. X-ray structure confirms the parallel and antiparallel dimerization of APP via its E2 domain [8]. Further research has then revealed the role of APP cis- and trans-dimerization in intercellular adhesion [9]. In addition, since all APP family members are located at pre- and post-synapses, the dimerization of APPs at synapses has been shown to be important for synaptic differentiation and maintenance [10].
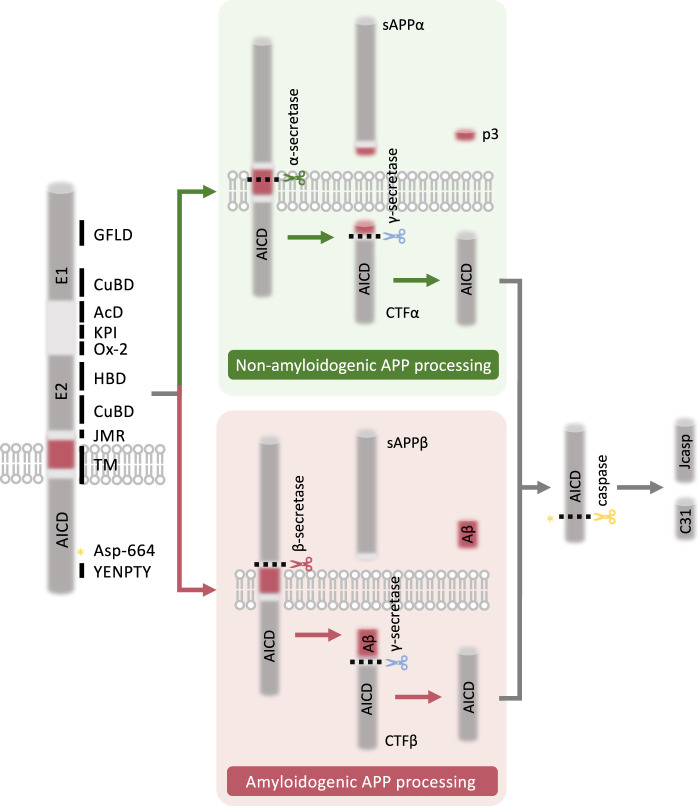
There are eight APP isoforms derived from the alternative splicing of the APP gene. APP695, APP751 and APP770 are the three major mammalian isoforms, with APP695 being the major form expressed in the neurons [11,12]. N-terminally, the ectoplasmic domain of APP consists of E1 and E2 domains connected by an acidic domain (AcD). The ectodomain of APP isoforms also contains an Ox-2 antigen domain (Ox-2) in APP770 and an kunitz protease inhibitor (KPI) domain, which is present in both APP751 and APP770. The E1 domain is composed of a growth-factor-like domain (GFLD), a cysteine-rich domain (CRD) and a copper-binding domain (CuBD) while the E2 domain contains a heparin-binding domain (HBD) and a CuBD. The center of APP is a single-spanning transmembrane helix (TM) connected to the N-terminal ectodomain via the juxtamembrane region (JMR). AICD in the cytosolic domain possesses a highly conserved tyrosine–glutamate–asparagine–proline–threonine–tyrosine (YENPTY) motif through evolution (Figure 1).
Figure 1. Schematic representation of APP structure and processing.
The ectoplasmic domain of APP constitutes E1 and E2 domains, which are connected by a AcD. The JMR connects the ectodomain to the center TM. E1 domain consists of a GFLD and a CuBD while E2 domain contains a HBD and a CuBD. Ox-2 and KPI domain are also present in the ectodomain of APP isoforms. In the cysolic domain, AICD possesses the YENPTY motif. Regulated intramembrane proteolysis of APP occurs in two distinct cleavage pathways, non-amyloidogenic and amyloidogenic processing. In non-amyloidogenic pathway, APP is cleaved within the Aβ-domain by α-secretase, releasing sAPPα and membrane-bound CTFα. Cleavage of CTFα by γ-secretase further secrets p3 fragment and AICD. In amyloidogenic pathway, APP is first cleaved by β-secretase, which releases sAPPβ and CTFβ. CTFβ is further processed by γ-secretase to release AICD and neurotoxic Aβ. AICD can be further cleaved by caspases at Asp-664, releasing two cytotoxic peptides, C31 and Jcasp. AcD, acidic domain; AICD, amyloid precursor protein intracellular domain; APP, amyloid precursor protein; Aβ, amyloid-β; CTFα, C-terminal fragment α; CTFβ, C-terminal fragment β; CuBD, copper-binding domain; E1, E1 domain; E2, E2 domain; GFLD, growth-factor-like domain; HBD, heparin-binding domain; JMR, juxtamembrane region; KPI, kunitz protease inhibitor domain; Ox-2, Ox-2 antigen domain; p3, p3 fragment; sAPPα, secreted APPα; sAPPβ, secreted APPβ; TM, transmembrane helix; YENPTY, tyrosine–glutamate–asparagine–proline–threonine–tyrosine domain.
Regulated intramembrane proteolysis (RIP) of APP family proteins generates secreted and cytosolic peptides that are associated with distinct functions [2]. APP can be cleaved into two mutually exclusive processing pathways, termed non-amyloidogenic and amyloidogenic pathways (Figure 1). During non-amyloidogenic processing of APP, APP is cleaved by α-secretase to release a soluble ectodomain fragment called secreted APPα (sAPPα) and membrane-bound C-terminal fragment α (CTFα). CTFα is then further processed by γ-secretase to release a non-toxic p3 peptide and AICD [13]. In amyloidogenic processing, APP is subsequently cleaved by β-secretase and γ-secretase to release secreted APPβ (sAPPβ) and membrane-bound C-terminal fragment β (CTFβ) which is further cleaved by γ-secretase to secret AICD and neurotoxic Aβ [13]. AICD is the common fragment released from both non-amyloidogenic and amyloidogenic pathways under physiological conditions. However, the amyloidogenic pathway is more prominent in AD, resulting in increased production of Aβ and AICD, which may lead to pathological consequences. Of note, AICD can be further cleaved by caspases at Asp664, releasing two cytotoxic peptides, C31 and Jcasp (Figure 1). Increasing evidence reveals the biological significance of the aforementioned intracellular cleavage products of APP, particularly AICD. Sequence alignment of AICD proteins produced by APP family members across different species demonstrates the protein contains an absolute conserved YENPTY motif [14]. This motif is recognized by AICD interacting proteins (AIP) which contain a phosphotyrosine-binding (PTB) domain. To note, PTB domains play important roles in cell fate determination, protein targeting and trafficking and tyrosine-kinase modulated signaling cascades. FE65 is one of the first identified AIP which interacts with the YENPTY motif via its PTB domain. The following AIPs were then discovered as they interact with AICD in a similar manner, such as JNK-interacting protein 1 (JIP1), Disabled-1 (Dab1), GULP PTB domain containing engulfment adaptor 1 (GULP1), Numb and X11 family [15–18]. Several interactions between AICD and AIPs have been reported to affect the production of Aβ (Table 1). For example, overexpression of X11α or X11β in AD mouse models expressing APP Swedish mutation (APPswe) remarkably attenuates Aβ plaque deposition [19,20]. Conversely, models deficient of X11β or both X11β and X11α exhibit enhanced level of Aβ in the hippocampus [20,21]. Further studies suggested that the interaction between X11 and APP through AICD facilitates the translocation of APP out of detergent-resistant membrane, where β-secretase is active, and hence prevent amyloidogenic cleavage of APP [21]. This demonstrates the importances of the overall role of APP and its cleavage products, including AICD, in neurodegenerative diseases. Most AIPs are adaptor proteins that lack catalytic domains to possess a direct functional role, however, they all composed of diverse protein-protein interacting domains that allow them to bind with different interacting partners to participate in different cellular mechanisms, both AICD-dependently or -independently.
Table 1. List of AIPs which affect Aβ production.
| AIPs | Aβ production ↑ ↓ | Proposed mechanism | References |
|---|---|---|---|
| Caveolin-3 | ↑ | Up-regulate β-secretase-dependent APP cleavage through inactivating G-protein | [26] |
| FE65 | ↑ | Regulate APP trafficking | [27–29] |
| GULP1 | ↑ | Reduce APP expression on the cell surface | [30,31] |
| JIP1 | ↓ | Transport and retain phospho-APP at neurite | [32,33] |
| LRP | ↑ | Elevate APP internalization | [34] |
| Dab1 | ↑ | Increase surface APP expression | [35] |
| X11 | ↓ | Translocate APP out of detergent-resistant Membrane (DRM) to suppress amyloidogenic processing | [21,36] |
| Numb | ↑ | Regulate the sorting of APP into recycling vesicles and decrease the level of cell surface APP | [37] |
| sorLA/LR11 | ↓ | Reduce surface APP expression by retaining APP in Golgi apparatus and attenuate BACE-dependent APP cleavage | [38] |
| CHF5074 | ↓ | Reduces the interaction between APP and PS1, lowering γ-cleavage | [39] |
| Binds to AICD and reduces its translocation to nucleus | [40] |
Physiologically, Aβ is a comparatively minor metabolic product of APP as the majority of APP is processed by α-secretase within the Aβ sequence, thereby prohibiting the production of Aβ. Nevertheless, a shift toward β-secretase processing of APP would increase the production of Aβ, which may contribute to AD pathogenesis, as reviewed elsewhere [22,23]. Notably, there is a significant increase of AICD in the brain of APPswe, a model of the amyloid pathology [24]. Furthermore, in the hippocampus of AD patients, AICD is accumulated in Hirano bodies in dystrophic neurites [25]. Such observation may suggest that AICD also contributes to AD pathogenesis.
Physiological roles of AICD
Gene transcription
AICD is constitutively located in the nucleus [41] where it serves as a transcription factor. Thus, it is suspected that AICD-mediated nuclear signaling can be regulated by the binding of AIP to its evolutionarily conserved regions. Indeed, AICD is reported to form a transcriptional regulatory complex with FE65 and the transcriptional coregulator Tat interactive protein 60 kDa (Tip60), which facilitates AICD-mediated transactivation signaling [42,43]. Moreover, AICD has been found to regulate various gene expression and thereby mediate different cellular processes (Table 2). In particular, AICD controls the expression of neuronal PAS domain Protein 4 (NPAS4), which incidentally controls the release of synaptic gamma-aminobutyric acid (GABA) [44]. AICD also regulates GluN2B expression, which affects synaptic N-methyl-d-aspartate receptor heterodimer assembly and, as a result, regulates excitatory neurotransmission [45].
Table 2. List of AICD-regulated genes in the nervous system.
| Gene | Gene expression ↑ ↓ | Physiological function | References |
|---|---|---|---|
| APP | ↑ | Neurodevelopment | [46] |
| α2-actin | ↑ | Actin dynamics | [47] |
| BACE | ↑ | APP processing | [46] |
| CHOP | ↑ | Apoptosis | [48] |
| Cyclins B1 and D1 | ↑ | Cell cycle activation | [49] |
| GluN2B | ↑ | Regulates neurotransmission | [45] |
| GSK3β | ↑ | Apoptosis, tau phosphorylation | [46] |
| KAI1 | ↑ | Control cell motility and adhesion, apoptosis | [46] |
| NPAS4 | ↑ | Regulate GABA release | [44] |
| Ptch1 | ↑ | Regulate the proliferation of NPCs | [50] |
| p53 | ↑ | Apoptosis | [51] |
| Tip60 | ↑ | Histone acetyltransferase | [46] |
| Transgelin | ↑ | Actin dynamics | [47] |
| Tropomyosin | ↑ | Actin dynamics | [47] |
| VGLUT2 | ↑ | Regulate vesicular uptake of glutamate and synaptic transmission | [52] |
Functions in neural networks
AICD is also implicated in differential functions in neurons. AICD is required during axonal transport. In transgenic Drosophila, overexpression of APP, but not the APP mutant lacking AICD, leads to defects in axonal transport. AICD mediates the interaction between APP and kinesin-I by directly binding to the kinesin-1 light chain. Furthermore, the axonal transport of APP to neuromuscular junctions (NMJs) is dependent on the interaction between AICD and kinesin-1 [53]. Additionally, the physiological roles of AICD in modulating the central nervous system (CNS) have been investigated in vivo in transgenic mouse models FeCγ12 and FeCγ25, both overexpressing AICD and FE65. Abnormal spiking events associated with enhanced seizure susceptibility are observed in these models when compared with mice overexpressing FE65 only [54]. Therefore, it is suggested that overexpression of AICD disrupts the neuronal circuit [54]. Furthermore, these models also demonstrate significantly increased activity of the kinase glycogen synthase kinase-3β (GSK-3β) [24]. The up-regulated activity of GSK-3β is associated with an increase phosphorylation of its downstream substrates which include collapsin responsive mediator protein–2 (CRMP2) protein, a key component of axonal guidance [24], and tau, which bind and stablise microtubule [24,55]. Aberrant phosphorylation of CRMP2 and tau disrupts their binding to microtubule [56,57]. Furthermore, AICD exerts its roles in cytoskeleton organization and dynamics by inducing the expression of genes encoding various regulators of actin dynamics, including transgelin and α2-actin [47]. While down-regulation of α2-actin in neural stem cells (NSCs) impairs NSC migration [58], transgelin knockout mice show cognitive impairments with spatial learning and memory deficits [59]. The above findings implicate proper expression of AICD is essential for the normal functions of neural networks.
Endoplasmic reticulum-mitochondrial signaling
AICD plays a role in maintaining endoplasmic reticulum (ER) calcium storage and mitochondrial functions. Cells lacking AICD (H4-AICD-VSV (Dox)) has shown to reduce ER calcium storage, which increases cytosolic calcium level, and subsequently reduces ATP production level and causes hyperpolarization of the inner mitochondrial membrane potential, indicating the physiological role of AICD in maintaining ER and mitochondrial functions [60]. AICD is shown to regulate the expression and transcription of phosphatase and Phosphatase and tensin homolog–induced kinase 1 (Pink-1) that modulates the turnover of mitochondria via mitophagy and control mitochondrial dynamics by reducing the level of dynamin-like protein 1 and mitofusin 2 [61]. Moreover, AICD also regulates the transcription of coiled-coil-helix-coiled-coil-helix domain containing 6 (CHCHD6), a core component of the multi-subunit protein complex which maintains mitochondrial homeostasis and cristae morphology [62].
AICD and its interactors in neurodevelopment
The aforementioned findings reveal the physiological significance of AICD. As stated above, AICD interacts with different proteins. Therefore, it is possible that some of the functions of AICD are contributed by recruiting different interactors. In the following, we will summarize the roles of AICD and its interactors in both neurodevelopment and neurodegeneration.
Neurogenesis
During neurogenesis, fully differentiated neurons and glia are generated from neural precursor cells (NPCs) in the embryonic brain. In the adult brain, neurogenesis persists in specific regions, like the hippocampus and the subventricular zone. Multiple lines of evidence suggest the role of AICD in neurogenesis, including regulation of NPCs proliferation and transcription. APP has been recognized as a negative modulator of neurogenesis as NPCs of App−/− knockout mice show significant elevated neurogenesis [63]. The effect of APP on neurogenesis has been shown to be associated with AICD. In an AD mouse model carrying the Swedish and Indiana familial AD mutations, platelet-derived growth factor B-chain promoter-driven APP transgenic mice (PDAPP), a mutation that prevents caspase cleavage of AICD to produce C31 peptide leads to a reduction in hippocampal neuronal precursor proliferation when compared with PDAPP mice without the mutation [64]. Furthermore, inhibited caspase cleavage of AICD may also interfere with the intermolecular interactions that involve AICD. For adult neurogenesis, transgenic mouse models FeCγ12 co-expressing AICD and FE65 demonstrates reduction in proliferation adult hippocampal progenitor cells (HPC). However, such reduction in HPC proliferation is not observed in Fe27 transgenic models which overexpress solely FE65. This implicates the role of AICD in suppressing adult neurogenesis in HPC [65]. Additionally, overexpression of AICD can reverse the aberrant neurogenesis in transient axonal glycoprotein (TAG1) knockout mice. Conversely, AICD with a mutated FE65-binding YENPTY motif cannot reverse such effect of TAG1 knockout on neurogenesis, suggesting that both AICD and FE65 participate in neurogenesis [63].
As mentioned earlier, AICD is reported to form a transcriptional regulatory complex with FE65 and Tip60 [42,43]. During neurogenesis, this tripartite complex regulates genes that are implicated in neurogenesis, including stathmin [66]. Stathmin is a neuronal protein that is associated with adult neurogenesis [67]. During embryonic development, AICD acts as a negative regulator of Wnt signaling, which mediates NPCs proliferation and differentiation, through its interaction with GSK3β, a regulator of Wnt signaling activation [68]. The AICD/GSK3β interaction stimulates the kinase activity GSK3β, resulting in an inhibitory effect on Wnt signaling and NPCs proliferation. Moreover, AICD facilitates the functions of nerve growth factor (NGF) in inducing the expression of cyclin-dependent kinase (CDK) inhibitors, including that of p15, p16 and p21, thereby, suppressing NPCs proliferation [68].
Neuronal migration and positioning
The organization of neurons into differential layers of the cerebral cortex during neuronal migration and positioning is essential for the establishment of the CNS. In the embryonic brain, the absence of APP leads to migration defect of NPCs into the cortical plate [69]. AICD interactor Dab1 is an adaptor protein implicated in neuronal migration in the developing cerebral cortex[70,71]. The interaction between full-length APP and Dab1 through AICD is crucial as a mutation in the AICD YENPTY motif and knockdown of Dab1 diminishes the stimulatory effect of APP on neuronal migration into the cortical plate [15,69]. Further elucidation reveals the involvement of disrupted-in-schizophrenia 1 (DISC1) in APP-Dab1 pathway in neuronal migration. Knockdown of APP disrupts the subcellular localization of DISC1 at the centrosome, which is required for proper cortical cell migration into the cortical plate [72]. Furthermore, the overexpression of DISC1 successfully rescues the migration defects caused by the knockdown of APP and Dab1, respectively, but not vice versa [72]. The effect of DISC1 on APP/Dab1 pathway maybe contributed by its interaction with APP-Dab1 complex [72].
The role of APP during neuronal positioning has been suggested by App−/−/Aplp1−/−/Aplp2/−triple knockout mice which exhibit cortical dysplasias, a characteristic of defective neuronal positioning [73]. The function of APP in this process is likely contributed, at least in part, by its interaction with FE65 through its AICD as such neuronal defects resemble those observed in FE65−/−/FE65 Like 1 (FE65L1)−/− double knockout mice [74]. Expression of APP and FE65 significantly up-regulates cell movement mediated by APP, suggesting the role of APP-FE65 complex in cell motility [75]. In fact, APP and FE65 may recruit regulators of the cytoskeleton, like β1-integrin and mammalian homolog of Drosophila Enabled (Mena) [75]. Moreover, the APP/FE65 complex colocalises with Mena in β1-integrins at focal complexes in mobile membrane compartments, implicating the role of the interaction between full-length APP and FE65 through AICD in processes related to integrin-based adhesion and migration [75].
Neurite outgrowth and axonal guidance
During the establishment of the neuronal circuitry, neurite outgrowth generates projections from newly differentiated neurons to form axons and dendrites. Axonal guidance then ensures that axons of newly differentiated neurons are extended to reach their synaptic targets under the effect of guidance cues. AICD is a promoter of neurite outgrowth by acting as a Wnt antagonist [68]. AICD enhances the effect of NGF-dependent induction of neurite outgrowth by attenuating Wnt3a signaling which suppresses the effect of NGF physiologically [68]. The role of APP family proteins in axonal guidance has been revealed as defects in midline crossing of projection axons in App−/−/Aplp2−/− double-knockout mice has been observed [76]. Moreover, binding of Slit2, a glycoprotein that implicates in axonal guidance, to the E1 domain of APP has been shown to mediate APP/FE65/p21-activated kinase 1 (PAK1) multimeric complex formation, and resulted in enhanced axon projection [76]. As PAK1 is a regulator of actin dynamics, the above finding further supports the role of APP/AICD and FE65 in axonal guidance. However, further investigations are required to elucidate the mechanism by which this multiprotein complex activates PAK1. Of note, FE65 concurrently interacts with ADP-ribosylation factor 6 (ARF6) and the Rac1- guanine nucleotide exchange factor complex, Engulfment and Cell Motility 1/dedicator of cytokinesis 1 to stimulate Rac1 [77,78]. Further investigations are still underway to determine the exact implications of these complexes in cytoskeleton remodeling.
Synaptogenesis
The assembly of synapses between neighboring nerves or between muscles facilitates the rapid transmission of signals. App−/−/Aplp2−/− double-knockout mice develop aberrant nerve terminal sprouting with a reduction of synaptic vesicles at the presynaptic terminals, implicating their roles in synaptogenesis [79]. AICD is indispensable for the generation of synaptic connections. The phenotype comprising of synaptic loss observed in the PDAPP AD mouse model could be rescued by an APP mutation that abolishes the generation of C31 from AICD by caspase cleavage at Asp-664 (Figure 1) [64]. It is suggested that AICD is essential for maintaining normal synaptic functions which are impaired in AD [64]. The effect of AICD for inducing axonal arborization is suggested to be mediated by its interaction with Dab1, an adaptor protein for Abl tyrosine kinase, a key regulator of synaptic plasticity, which also interacts with full-length APP through AICD [80,81]. Additionally, App−/−/Aplp2−/− double-knockout mice exhibit multiple NMJ defects, including aberrant nerve terminal sprouting, diffused synaptic distribution and abnormal apposition of presynaptic vesicle proteins and postsynaptic acetylcholine receptors, suggesting the regulatory roles of APP and APLP2 during NMJ development [79,82]. The role of AICD during NMJ formation is further demonstrated in Aplp2−/− knockout expressing secreted sAPPα, whereby sAPPα is insufficient to compensate for the NMJ deficits observed in models deficient of APLP2 [83]. Noteworthy, the interaction between AICD and FE65 may be relevant to the physiological role of APP on neuromuscular synaptogenesis, as similar NMJ defects are also seen in a mouse expressing an APP mutant, that lack the last 15-amino acid of APP, on an Aplp2−/− background [84]. The FE65 interacting YENPTY motif is located within the deleted region. Moreover, compound knockout of APLP2 with either FE65 or FE65L1 intensifies the severity of NMJ impairments [85]. The above findings support the role of AICD-FE65 in NMJ formation.
Additionally, the interaction between Drosophila homologue APP-like (APPL) and Drosophila X11 (dX11) through AICD is crucial for synaptogenesis as the presynaptic expression of mutant APPL or dX11 lacking the critical YENPTY sequence and PTB domain significantly attenuated synaptic formation [86]. It has been reported that APP forms a macromolecular protein complex that induces presynaptic differentiation and the formation of the presynaptic active zone by coupling with X11 and calcium/calmodulin-dependent serine protein kinase (Cask) through the YENPTY motif of APP [87].
AICD and its interactors in neurodegeneration
Neurodegeneration refers to the progressive loss of neurons in the CNS. As neurons are generally post-mitotic, the loss of neurons often implicates the irreversible development of neurodegenerative diseases, such as AD. There are several biochemical pathways that contribute to neuronal death, including the disrupted ER-mitochondria signaling [88], accumulation of neurotoxic peptides [89,90] and the propagation of excitotoxicity [91,92]. These biochemical pathways often lead to the initiation of an apoptotic cascade and eventually cause progressive neuronal death. Since the proposal of ACH, APP and Aβ have become the centers of AD research. As an inevitable product of APP processing, AICD also exerts an important role in neurodegeneration. AICD levels in AD patients are significantly higher than that in non-demented controls [55]. Specifically, AICD has been shown to modulate neuron-specific apoptosis [55,93,94]. Overexpression of AICD in transgenic mice displays AD-like abnormalities, including tau hyperphosphorylation and intracellular aggregation of neurofibrillary tangles, as the activity of GSK-3β, which contributes to hyperphosphorylation of tau, has been elevated [24,55]. Overexpression of AICD also increases the age-dependent loss of hippocampal neurons by inducing neuronal susceptibility to excitotoxic stimuli [55]. Amyloidogenic processing of APP is more pronounced in AD, resulting in increased production of amyloidogenic APP cleavage fragments. Since the overexpression of APP and its cleavage products is implicated in various processes that lead to neurodegeneration, this highlights the importance of the overall role of APP and its cleavage products, including AICD, in neurodegenerative diseases.
Neuronal apoptosis
AICD controls apoptotic pathways by modulating expressions of various proteins through transcriptional regulations. For example, AICD interacts with p53 mRNA and promotes the expression of p53 and its shorter isoform p44, resulting in the activation of p53/p44-mediated proapoptotic signaling pathways. This results in an enhancement of caspase activity [95] and tau hyperphosphorylation in neurons [96]. Recent studies demonstrate that AICD induces ER-stress by enhancing nuclear retention of FoxO3a, a transcription factor involved in neuronal ageing and cell death [97]. During ER-stress-induced apoptosis, AICD also up-regulates C/EBP homologous protein (CHOP) expression by binding to the CHOP promotor and potentiates ER-stress-induced apoptosis [48,98]. Similarly, it also regulates mitochondria homeostasis by up-regulating the transcription of PINK1, which encodes Pink-1, a mitochondrial kinase [61]. Since AICD does not consist of a nuclear localization sequence, it is suggested that AICD translocates into the nucleus to exert the above transactivation functions by forming the AICD-FE65-Tip60 complex [48,95,98,99]. AICD-FE65 also complexes with the transcription factor CP2/LSF/LBP1 [90,100]. This complex induces the transcription and expression of human GSK3β [90,101]. It is found that the knock down of neuronal GSK3β can reverse capspase 3 activation-induced cell death, indicating GSK3β activation can induce neuronal cell death by promoting caspase 3 activation [102].
Besides acting as a transcription factor for proapoptotic proteins, AICD has been found to participate in inducing apoptosis via regulating protein-protein interaction. For example, it is proposed that the phosphorylation of AICD Thr668 weakens AICD-FE65 interaction which releases FE65 for the binding with Bloom syndrome protein (BLM) in the nucleus. Such FE65-BLM complex in nucleus facilitates the re-initiation of DNA replication [103], a process known to associate with apoptosis [104,105]. Furthermore, FE65 can interact with teashirt3 to suppress AICD-FE65 mediated transcriptional activity, yet enhance the inhibition of caspase 4 expression in a FE65-teashirt3 complex-mediated manner [106]. As caspase 4 contributes to both ER-stress-induced apoptosis [107] and innate inflammation response [108], it is suspected that FE65 may participate in neuronal apoptosis by regulating the expression and activity of caspase 4. PTB-containing JIP1 is another AICD interactor predominantly expressed in neural tissues [32]. However, unlike FE65, plenty of JIP1-mediated pathways have been reported to be dependent on the JNK activation pathway. Under UV stress, Aβ exposure and hypoxia, the decrease in JIP1 expression causes neurons to be more prone to JNK-mediated apoptosis [109]. In addition, in response to cytokine, JIP1 is shown to co-operate with Islet-brain 1 (IB1) to modulate apoptosis as an elevated expression of JIP1 and IB1 protects cells from apoptosis [110]. Altogether, JIP1 has an anti-apoptotic potential that can be considered as a neuroprotective factor. Interestingly, JIP1 mediates the phosphorylation of APP and AICD at Thr668 via JNK kinase activity [111]. The phosphorylation of AICD Thr668 increases AICD-induced apoptosis and increases Aβ production [112,113], suggesting that JIP1 promotes apoptosis by facilitating AICD phosphorylation.
Neuroinflammation and neurodegeneration
In addition to intracellular pro-apoptotic signaling pathways, progressive neuronal loss can also be caused by aberrant neuroinflammation. Neuroinflammation is generally considered a neuroprotective mechanism, as it helps eliminate pathogens in the CNS and facilitates the repair of neurons from extracellular insults. However, chronic neuroinflammation has emerged as a major risk factor for age-related neurodegenerative disorders, including AD and Parkinson's disease [114,115]. In AD, the expression levels of pro-inflammatory cytokines, such as interleukins-1 (IL-1), IL-6, IL-8, and tumor necrosis factor α (TNFα), are significantly up-regulated, indicating an inflamed brain environment [116,117]. Activated microglia, the primary macrophages in the CNS, are responsible for releasing these cytokines as the primary source of local inflammatory response in the AD brain [116,118]. Emerging evidence has indicated that APP processing and the AICD may play a role in neuroinflammation as APP is expressed and can be processed in microglia and astrocytes [119]. Inhibiting APP processing in macrophages has been shown to suppress their immunological activities [120]. Similarly, the expression of AICD in transgenic mice has been found to positively induce neuroinflammation [65]. Moreover, the anti-neuroinflammatory drug 1-(3′,4′-dichloro-2-fluoro[1,1′-biphenyl]-4-yl) cyclopropanecarboxylic acid (CHF5074) can effectively decrease neuroinflammation by limiting the expression of pro-inflammatory elements [121]. Interestingly, CHF5074 can interact with AICD fragment and decrease its transcriptional activity and transcriptional output of AICD-regulated genes, like KAI1, by reducing nuclear translocation of AICD [40,122]. These further suggest the potential role of AICD in neuroinflammation. In the cerebrospinal fluid samples of AD patients, several AICD associating proteins, including fibrinogens and peroxiredoxin, which are important for neuroinflammation, have been found to be increased in expression [123–125]. Although the precise mechanisms by which AICD can affect neuroinflammation remain to be elucidated, these findings raise the intriguing speculation that AICD may regulate microglia activation and the expression of pro-inflammatory proteins.
As mentioned, AIPs have been shown to promote amyloidogenic processing of APP to increase Aβ as well as AICD generation (Table 1). Therefore, these interactors participate in neuroinflammation by triggering the production of AICD. Additionally, AICD-AIP complex may contribute neuroinflammation by alternative mechanism as some of these AIPs have been shown to participate in neuroinflammation. For example, GULP1 contributes to the ATP-binding cassette transporter A1 (ABCA1) pathway and regulates astrocytic phagocytosis during ischemic injury [126]. FE65-teashirt3 complex also regulating caspase 4 expression [106], in which caspase 4 is required for the activation of inflammasomes via activating caspase 1 [127]. Nevertheless, the above findings implicate the association of AICD and its interactors in neuroinflammation.
Dysregulation of ER-Mitochondrial signaling
Increase in oxidative stress and mitochondrial dysfunction are recognized as the early events in the progression of sporadic AD [128,129]. In recent years, the proposal of the mitochondrial cascade hypothesis (MCH) believes that mitochondrial dysfunction is the primary cause of AD, as mitochondrial dysfunction causes energetic and/or metabolic impairment and directly leads to neuronal loss. Therapeutic approaches to restoring mitochondrial function have also been shown to effectively slow down the progression of the disease. For example, physical exercise can enhance mitochondrial function and serve a neuroprotective role against AD [130]. A plant-based antioxidant, resveratrol, also shows neuroprotective effects as it protects mitochondria by decreasing mitochondrial oxidative stress and increasing mitochondrial biogenesis [131]. In the APP/PS mice model, the administration and delivery of resveratrol to the brain can significantly improve memory function [132]. Despite MCH suggesting APP processing and amyloid production are the results of mitochondrial dysfunction, in fact, AICD, as the product of APP processing, can in turn cause mitochondrial dysfunction.
As mentioned, the nuclear signaling of AICD is often associated with FE65-mediated nuclear translocation. AICD-FE65 is responsible to act on increasing the expression of α2-actin and transgelin that destabilize actin fibers in neurons [47]. Since mitochondrial function is highly sensitive to actin dynamics, it is reported that AICD-FE65-mediated actin dynamic leads to a significant decrease in mitochondrial membrane potential and ATP production [133]. FE65 also interacts with Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase 2 (SERCA2). As SERCA2 is an important protein for cellular calcium homeostasis, it is reported that FE65-SERCA2 interaction plays an important role in determining cell viability under ER stress by affecting intracellular calcium homeostasis [134]. Notably, it is also reported that the level of cytosolic calcium was elevated in presenilin double knockout mice, suggesting a disrupted SERCA activity [135]. As presenilin is essential for APP γ-cleavage, it is suggested that the absence of AICD can disrupt SERCA-dependent calcium signaling [136]. It is proposed that the interaction of AICD and FE65 may further complex with SERCA2 to regulate ER calcium homeostasis [134]. Numb depletion increases Rho-associated coiled-coil containing protein kinase 1 (ROCK1) activation that enhances dynamin-related protein 1 translocation to mitochondria, consequently promoting mitochondria fragmentation [137]. Intriguingly, ROCK1 is able to interact with and phosphorylate APP at Ser655 to up-regulate amyloidogenic processing [138], which suggests a role of Numb in regulating mitochondrial homeostasis via ROCK-1 induced APP processing.
Additionally, increasing findings suggest that the disruption of ER-mitochondria signaling in AD [139–141]. The communication between ER and mitochondria is essential in regulating numerous cellular events, including calcium and lipid metabolism and UPR response [142–144]. In post-mortem AD brains, the protein expressions of vesicle-associated membrane protein-associated protein B (VAPB) and protein tyrosine phosphatase interacting protein-51 (PTPIP51) are found to be decreased in cortex, indicating the disruption of ER-mitochondria contact in AD brain [139]. Using cell model of AD, the localization of CTFβ increases in mitochondrial-associated ER membranes (MAM), resulting in altering the lipid composition of MAM, ultimately causing mitochondrial dysfunction [145]. Another study also demonstrated the accumulation of CTFβ in ER induces the uptake of cholesterol and cholesterol trafficking from plasma membrane to MAM, reveal a new role of CTFβ in cholesterol disturbances in AD [140]. γ-Secretase activating protein (GSAP) plays a crucial role in regulating γ-secretase activity and has recently been found to participate in mitochondrial function by forming a GSAP-FE65-APP ternary complex. It is proposed that FE65, GSAP and APP-CTR colocalize in MAM. In AD condition, FE65 recruits GSAP to dephosphorylate APP, inducing Aβ production in MAM, which impairs the lipid homeostasis of ER and mitochondria [146].
Future perspectives
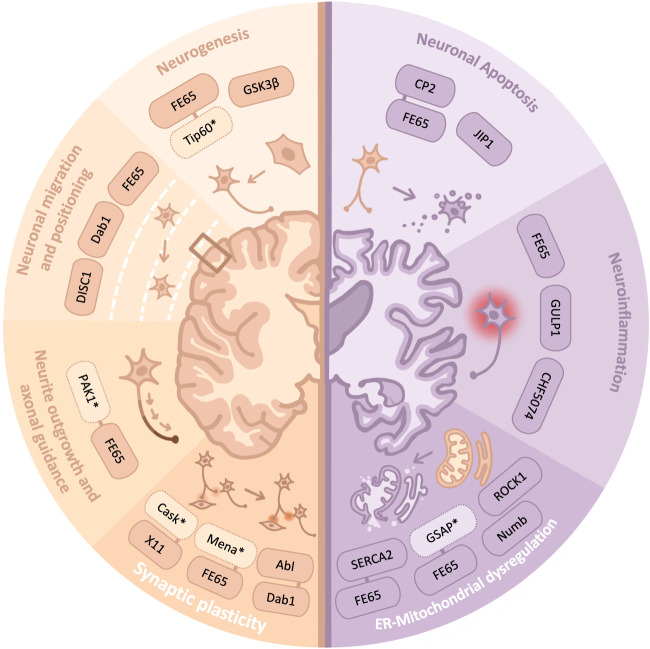
The APP interactome play essential physiological roles during the establishment and maintenance of the neuronal system [147]. In the present review, we have highlighted the indispensable role of AICD in regulating neuronal function, and its contribution to neurodevelopment and neurodegeneration through the interaction between AICD and its binding partners (Figure 2). Over the past decades, the identification of AICD binding partners has aided in understanding the complexity of the roles of AICD in various cellular pathways and has provided us hints of how important APP processing is for neuronal functions from early age, during neurodevelopment, to adulthood neuronal maintenance. Therefore, a comprehensive illustration of the AICD interactome helps to decipher the pathophysiology of AD and suggest druggable targets.
Figure 2. Summary of AICD interactors implicated in neurodevelopment and neurodegeneration.
AICD interactors that are associated with different stages of neurodevelopment (orange) and neurodegeneration (purple) are illustrated. TAG1, transient axonal glycoprotein; Tip60, Tat interactive protein 60 kDa; Dab1, Disabled-1; DISC1, disrupted-in-schizophrenia 1; PAK1, p21-activated kinase 1; Cask, calcium/calmodulin-dependent serine protein kinase; Abl, Abl tyrosine kinase; JIP1, JNK-interacting protein 1; CHF5074, 1-(30,40-dichloro-2-fluoro[1,10-biphenyl]-4-yl) cyclopropanecarboxylic acid; GULP1, GULP PTB domain containing engulfment adaptor 1; SERCA2, Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase 2.
The functional redundancy of APP family proteins during neurodevelopment is demonstrated by single-, double- and triple-knockout mouse mutants. While single-knockout of APPs only leads to subtle phenotypes in mice, double- and triple-knockout of APPs show lethal early postnatally [73,148,149]. The functional compensation between APPs could be due to the overlapping interactome which may stimulate similar signaling pathways. Although ALPL1 and APLP2 lack the Aβ domain, the APPs share a high degree of structural homology and similar presenilin-dependent RIP proteolytic processing as substrates of γ-secretase, which releases the intracellular domains (ICDs) of sizes ∼6 kDa [150]. AICD, APLP1 and APLP2 ICDs (AL1ICD and AL2ICD) are highly similar and possess a conserved structure consisting of motifs, including the caspase cleavage site and YENPTY domain.
The protein-protein network of the AICD interactome co-ordinates cellular functions of AICD as signals received are relayed through the concerted and synchronized actions of functional components of the proteome. It is therefore important to investigate the contribution of the spatio-temporal dynamics of these interactions in the cellular signaling pathways when characterizing the AICD interactome. For example, AICD, AL1ICD and AL2ICD possess differences in cellular localization and nuclear signaling capabilities. While both AICD and AL2ICD bind to FE65 and translocate into the nucleus where they form nuclear AFT complexes, FE65 interacts with the APLP1 holoprotein, resulting in retention of FE65 and AL1ICD in in the extranuclear regions and decreased nuclear AFT complex formation [151]. In addition, full-length APLP1 exhibits slower turnover than APP and APLP2, which may account for a lower availability of AL1ICD and thus reduced ARF complex formation [151]. The differences in the compartmentalization and affinity between ICDs of APPs and their interactors can result in differential levels of downstream signaling. Comparative investigations of the interactions between AICD, AL1ICD and AL2ICD and their shared binding partners using cryo-electron tomography can enable in situ structural studies in unperturbed cellular environments at 3–4 nm resolution, visualizing the localization of macromolecules in cells [152].
The interactions between AICD and its binding partners are dependent on their states of phosphorylation. On AICD, differential phosphorylation statuses of Tyr682 and Thr668 influence the composition of the APP interactome [153–155]. In AD brains, there is an increase in phosphorylation of Tyr682 and Thr668 residues on AICD [156,157], suggesting the implications of phosphorylation and the altered AICD interactome in AD pathogenesis. Furthermore, phosphorylation statuses of AICD, AL1ICD and AL2ICD have varying effects in shaping the ICDs interactome. Phosphorylation of AICD interactors also contributes regulatory mechanisms of signaling pathways involving AICD. For example, the phosphorylation status of FE65 at Ser288 and Ser610 modulate FE65-APP transcriptional activity and APP processing mediated by FE65, respectively [158,159]. Nevertheless, the dynamics of the interactome network created by phosphorylation and dephosphorylation of specific residues of AICD and its interactors as well as the precise effects of these interactions are yet to be further elucidated.
Overlapping features are observed in both neurodevelopmental and neurodegenerative disorders. For instance, AICD, as we have summarized, possesses dual roles in both neurodevelopment and degeneration. Similarly, GSK3 signaling is associated with both neurodevelopmental and degenerative function. GSK3 cycles between on/off states through phosphorylation at different sites whereas Ser9 and Ser21 are considered inactive sites and Tyr216 is considered an active site. LiCl is an FDA-approved drug that manipulates this molecular switch to inactivate GSK3 and to reduce tauopathy and Aβ production [160,161]. Aforementioned, AICD changes its localization and participates in different cellular pathways via interacting with different binding partners. Therefore, manipulating AICD interacting dynamic using small-molecule compounds and peptides have become new therapeutic strategies to treat AD. For example, CHF5074 is an anti-inflammatory compound that targets AICD and reduces its nuclear translocation and transcriptional activity through its interaction with AICD [40]. However, it is to be further investigated whether the neuroprotective abilities of CHF5074 in lowering brain plaque burden and reducing neuroinflammation could be explained by its targeting against AICD activities [122]. Peptide targeting APP-X11 interaction at AICD has also been reported to successfully reduces Aβ levels in neurons by attenuating the interaction [162]. FE65 is known to up-regulate amyloidogenic APP processing through its interaction with AICD. The blockage of FE65-AICD interaction is therefore considered as a promising strategy to decrease Aβ secretion. For example, pentapeptide P33, which interacts with the N-terminal WW domain of FE65, decreases Aβ production and improves learning ability and memory functions in AD animal model by inhibiting FE65-AICD interaction [163]. Additionally, X11 proteins interacts with AICD and in turns inhibits amyloidogenic processing. Brandimarti et al. [164] has introduced gPTB9TM chimeric protein, which is comprised by a N-terminal GFP protein, followed by the PTB domain of X11 protein and transmembrane domain of US9 at the C-terminus. The transmembrane domain of this chimeric protein allows it to localize closely to APP and anchors the PTB domain adjacent to AICD. The expression of gPTB9TM drives the interaction of AICD and the X11-derived PTB domain, resulting in the decrease in amyloidogenic processing [164]. Despite plenty of AICD-targeted therapies aim on attenuating amyloidogenic APP processing, it is worth noting that AICD also holds the positive roles in promoting neurogenesis. With the emerging trend of neuro-regeneration therapies, it is possible that AICD can be a new target to trigger neuro-regeneration.
Perspectives
Research into AICD and its interactome extends our understanding of their roles in both neurodevelopment and neurodegeneration.
AICD interacts with different interactors to form functional complexes to regulate wide range of processes within the nervous system.
The manipulation of the interaction between AICD and its interactors can be the drug target for tackling and ameliorating neurodegenerative pathology.
Abbreviations
- APP
amyloid precursor protein
- ABCA1
ATP-binding cassette transporter A1
- AcD
acidic domain
- ACH
amyloid cascade hypothesis
- AD
Alzheimer's disease
- AICD
APP intracellular domain
- AIP
AICD interacting proteins
- AL1ICD
APLP1 ICDs
- AL2ICD
APLP2 ICDs
- APLP
APP-like protein
- APPL
APP-like
- APPswe
APP Swedish mutation
- ARF6
ADP-ribosylation factor 6
- Aβ
amyloid-β
- BACE
beta-secretase 1
- BLM
Bloom syndrome protein
- Cask
calcium/calmodulin-dependent serine protein kinase
- CDK
cyclin-dependent kinase
- CHCHD6
coiled-coil-helix-coiled-coil-helix domain containing 6
- CHF5074
1-(3′,4′-dichloro-2-fluoro[1,1′-biphenyl]-4-yl) cyclopropanecarboxylicacid
- CHOP
C/EBP homologous protein
- CNS
central nervous system
- CRD
cysteine-rich domain
- CRMP2
collapsin responsive mediator protein–2
- CTFα
C-terminal fragment α
- CTFβ
C-terminal fragment β
- CuBD
copper-binding domain
- Dab1
Disabled-1
- DISC1
disrupted-in-schizophrenia 1
- dX11
Drosophila X11
- ER
endoplasmic reticulum
- FE65L1
FE65 Like 1
- FoxO3a
Transcription factor forkhead box O3 protein
- GABA
gamma-aminobutyric acid
- GFLD
growth-factor-like domain
- GSAP
γ-Secretase activating protein
- GSK3β
glycogen synthase kinase-3β
- GULP1
GULP PTB domain containing engulfment adaptor 1
- HBD
heparin-binding domain
- IB1
Islet-brain 1
- IL
Interleukins
- JIP1
JNK-interacting protein 1
- JMR
Juxtamembrane region
- KPI
Kunitz protease inhibitor
- LRP
lipoprotein receptor protein family
- MCH
mitochondrial cascade hypothesis
- Mena
Mammalian enabled
- NGF
nerve growth factor
- NMJ
neuromuscular junction
- NPAS4
neuronal PAS domain Protein 4
- NPCs
neural precursor cells
- Ox-2
Ox-2 antigen domain
- PAK1
p21-activated kinase 1
- PDAPP
promoter-driven APP transgenic mice
- Pink-1
phosphatase and tensin homolog–induced kinase 1
- PTB
phosphotyrosine-binding
- Ptch1
Patched1
- RIP
regulated intramembrane proteolysis
- ROCK1
Rho-associated coiled-coil containing protein kinase 1
- sAPPα
secreted APPα
- sAPPβ
secreted APPβ
- SERCA2
Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase 2
- TAG1
transient axonal glycoprotein
- Tip60
tat interactive protein 60kDa
- TM
transmembrane helix
- TNFα
tumor necrosis factor α
- VGLUT2
vesicular glutamate transporter 2
- YENPTY
tyrosine–glutamate–asparagine–proline–threonine–tyrosine
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by funds from the Research Grants Council Hong Kong, Health and Medical Research Fund (Hong Kong), CUHK direct grant scheme, United College endowment fund and the TUYF Charitable Trust.
Open Access
Open access for this article was enabled by the participation of Chinese University of Hong Kong in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with JULAC.
Author Contribution
L.L.H.N., J.C. and K.F.L. wrote the manuscript.
References
- 1.Hardy, J.A. and Higgins, G.A. (1992) Alzheimer's disease: the amyloid cascade hypothesis. Science 256, 184–185 10.1126/science.1566067 [DOI] [PubMed] [Google Scholar]
- 2.Jacobsen, K.T. and Iverfeldt, K. (2009) Amyloid precursor protein and its homologues: a family of proteolysis-dependent receptors. Cell. Mol. Life Sci. 66, 2299–2318 10.1007/s00018-009-0020-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gralle, M. and Ferreira, S.T. (2007) Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog. Neurobiol. 82, 11–32 10.1016/j.pneurobio.2007.02.001 [DOI] [PubMed] [Google Scholar]
- 4.Goel, S., Chin, E.N., Fakhraldeen, S.A., Berry, S.M., Beebe, D.J. and Alexander, C.M. (2012) Both LRP5 and LRP6 receptors are required to respond to physiological Wnt ligands in mammary epithelial cells and fibroblasts. J. Biol. Chem. 287, 16454–16466 10.1074/jbc.M112.362137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh, H.D., Ma, J.X. and Takahashi, Y. (2021) Distinct roles of LRP5 and LRP6 in Wnt signaling regulation in the retina. Biochem. Biophys. Res. Commun. 545, 8–13 10.1016/j.bbrc.2021.01.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao, J., Wang, J., Liu, B., Pan, W., Farr, III, G.H., Flynn, C.et al. (2001) Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7, 801–809 10.1016/S1097-2765(01)00224-6 [DOI] [PubMed] [Google Scholar]
- 7.Liu, T., Zhang, T., Nicolas, M., Boussicault, L., Rice, H., Soldano, A.et al. (2021) The amyloid precursor protein is a conserved Wnt receptor. Elife 10, e69199 10.7554/eLife.69199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, Y. and Ha, Y. (2004) The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol. Cell 15, 343–353 10.1016/j.molcel.2004.06.037 [DOI] [PubMed] [Google Scholar]
- 9.Soba, P., Eggert, S., Wagner, K., Zentgraf, H., Siehl, K., Kreger, S.et al. (2005) Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J. 24, 3624–3634 10.1038/sj.emboj.7600824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilling, S., Mehr, A., Ludewig, S., Stephan, J., Zimmermann, M., August, A.et al. (2017) APLP1 is a synaptic cell adhesion molecule, supporting maintenance of dendritic spines and basal synaptic transmission. J. Neurosci. 37, 5345–5365 10.1523/JNEUROSCI.1875-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandbrink, R., Masters, C.L. and Beyreuther, K. (1996) APP gene family. Alternative splicing generates functionally related isoforms. Ann. N. Y. Acad. Sci. 777, 281–287 10.1111/j.1749-6632.1996.tb34433.x [DOI] [PubMed] [Google Scholar]
- 12.Kang, J., Lemaire, H.G., Unterbeck, A., Salbaum, J.M., Masters, C.L., Grzeschik, K.H.et al. (1987) The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature 325, 733–736 10.1038/325733a0 [DOI] [PubMed] [Google Scholar]
- 13.Salbaum, J.M. and Ruddle, F.H. (1994) Embryonic expression pattern of amyloid protein precursor suggests a role in differentiation of specific subsets of neurons. J. Exp. Zool. 269, 116–127 10.1002/jez.1402690205 [DOI] [PubMed] [Google Scholar]
- 14.King, G.D., Perez, R.G., Steinhilb, M.L., Gaut, J.R. and Turner, R.S. (2003) X11alpha modulates secretory and endocytic trafficking and metabolism of amyloid precursor protein: mutational analysis of the YENPTY sequence. Neuroscience 120, 143–154 10.1016/S0306-4522(03)00284-7 [DOI] [PubMed] [Google Scholar]
- 15.Homayouni, R., Rice, D.S., Sheldon, M. and Curran, T. (1999) Disabled-1 binds to the cytoplasmic domain of amyloid precursor-like protein 1. J. Neurosci. 19, 7507–7515 10.1523/JNEUROSCI.19-17-07507.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan, K.S., Kuti, M. and Zhou, M.M. (2002) PTB or not PTB – that is the question. FEBS Lett. 513, 67–70 10.1016/S0014-5793(01)03305-1 [DOI] [PubMed] [Google Scholar]
- 17.Rogelj, B., Mitchell, J.C., Miller, C.C. and McLoughlin, D.M. (2006) The X11/Mint family of adaptor proteins. Brain Res. Rev. 52, 305–315 10.1016/j.brainresrev.2006.04.005 [DOI] [PubMed] [Google Scholar]
- 18.McLoughlin, D.M. and Miller, C.C. (2008) The FE65 proteins and Alzheimer's disease. J. Neurosci. Res. 86, 744–754 10.1002/jnr.21532 [DOI] [PubMed] [Google Scholar]
- 19.Lee, J.H., Lau, K.F., Perkinton, M.S., Standen, C.L., Shemilt, S.J., Mercken, L.et al. (2003) The neuronal adaptor protein X11alpha reduces Abeta levels in the brains of Alzheimer's APPswe Tg2576 transgenic mice. J. Biol. Chem. 278, 47025–47029 10.1074/jbc.M300503200 [DOI] [PubMed] [Google Scholar]
- 20.Lee, J.H., Lau, K.F., Perkinton, M.S., Standen, C.L., Rogelj, B., Falinska, A.et al. (2004) The neuronal adaptor protein X11beta reduces amyloid beta-protein levels and amyloid plaque formation in the brains of transgenic mice. J. Biol. Chem. 279, 49099–49104 10.1074/jbc.M405602200 [DOI] [PubMed] [Google Scholar]
- 21.Saito, Y., Sano, Y., Vassar, R., Gandy, S., Nakaya, T., Yamamoto, T.et al. (2008) X11 proteins regulate the translocation of amyloid beta-protein precursor (APP) into detergent-resistant membrane and suppress the amyloidogenic cleavage of APP by beta-site-cleaving enzyme in brain. J. Biol. Chem. 283, 35763–35771 10.1074/jbc.M801353200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sehar, U., Rawat, P., Reddy, A.P., Kopel, J. and Reddy, P.H. (2022) Amyloid beta in aging and Alzheimer's disease. Int. J. Mol. Sci. 23, 12924 10.3390/ijms232112924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadigh-Eteghad, S., Sabermarouf, B., Majdi, A., Talebi, M., Farhoudi, M. and Mahmoudi, J. (2015) Amyloid-beta: a crucial factor in Alzheimer's disease. Med. Princ. Pract. 24, 1–10 10.1159/000369101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan, K.A. and Pimplikar, S.W. (2005) Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J. Cell Biol. 171, 327–335 10.1083/jcb.200505078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz, D.G., Wang, D. and Greenberg, B.D. (1993) Hirano bodies accumulate C-terminal sequences of beta-amyloid precursor protein (beta-APP) epitopes. J. Neuropathol. Exp. Neurol. 52, 14–21 10.1097/00005072-199301000-00003 [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama, K., Trapp, B.D., Ikezu, T., Ransohoff, R.M., Tomita, T., Iwatsubo, T.et al. (1999) Caveolin-3 upregulation activates beta-secretase-mediated cleavage of the amyloid precursor protein in Alzheimer's disease. J. Neurosci. 19, 6538–6548 10.1523/JNEUROSCI.19-15-06538.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabo, S.L., Lanier, L.M., Ikin, A.F., Khorkova, O., Sahasrabudhe, S., Greengard, P.et al. (1999) Regulation of beta-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J. Biol. Chem. 274, 7952–7957 10.1074/jbc.274.12.7952 [DOI] [PubMed] [Google Scholar]
- 28.Lee, Y.S., Chow, W.N.V. and Lau, K.F. (2017) Phosphorylation of FE65 at threonine 579 by GSK3beta stimulates amyloid precursor protein processing. Sci. Rep. 7, 12456 10.1038/s41598-017-12334-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumanis, S.B., Chamberlain, K.A., Jin Sohn, Y., Jin Lee, Y., Guenette, S.Y., Suzuki, T.et al. (2012) FE65 as a link between VLDLR and APP to regulate their trafficking and processing. Mol. Neurodegener. 7, 9 10.1186/1750-1326-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao, Y., Perkinton, M.S., Chan, W.W., Chan, H.Y., Miller, C.C. and Lau, K.F. (2011) GULP1 is a novel APP-interacting protein that alters APP processing. Biochem. J. 436, 631–639 10.1042/BJ20110145 [DOI] [PubMed] [Google Scholar]
- 31.Beyer, A.S., von Einem, B., Schwanzar, D., Keller, I.E.. Hellrung, A., Thal, D.R.et al. (2012) Engulfment adapter PTB domain containing 1 interacts with and affects processing of the amyloid-beta precursor protein. Neurobiol. Aging 33, 732–743 10.1016/j.neurobiolaging.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 32.Scheinfeld, M.H., Roncarati, R., Vito, P., Lopez, P.A., Abdallah, M. and D'Adamio, L. (2002) Jun NH2-terminal kinase (JNK) interacting protein 1 (JIP1) binds the cytoplasmic domain of the Alzheimer's beta-amyloid precursor protein (APP). J. Biol. Chem. 277, 3767–3775 10.1074/jbc.M108357200 [DOI] [PubMed] [Google Scholar]
- 33.Muresan, Z. and Muresan, V. (2005) Coordinated transport of phosphorylated amyloid-beta precursor protein and c-Jun NH2-terminal kinase-interacting protein-1. J. Cell Biol. 171, 615–625 10.1083/jcb.200502043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pietrzik, C.U., Busse, T., Merriam, D.E., Weggen, S. and Koo, E.H. (2002) The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 21, 5691–5700 10.1093/emboj/cdf568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisiadou, L. and Efthimiopoulos, S. (2007) Expression of mDab1 promotes the stability and processing of amyloid precursor protein and this effect is counteracted by X11alpha. Neurobiol. Aging 28, 377–388 10.1016/j.neurobiolaging.2005.12.015 [DOI] [PubMed] [Google Scholar]
- 36.Miller, C.C., McLoughlin, D.M., Lau, K.F., Tennant, M.E. and Rogelj, B. (2006) The X11 proteins, Abeta production and Alzheimer's disease. Trends Neurosci. 29, 280–285 10.1016/j.tins.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 37.Kyriazis, G.A., Wei, Z., Vandermey, M., Jo, D.G., Xin, O., Mattson, M.P.et al. (2008) Numb endocytic adapter proteins regulate the transport and processing of the amyloid precursor protein in an isoform-dependent manner: implications for Alzheimer disease pathogenesis. J. Biol. Chem. 283, 25492–25502 10.1074/jbc.M802072200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spoelgen, R., von Arnim, C.A., Thomas, A.V., Peltan, I.D., Koker, M., Deng, A.et al. (2006) Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J. Neurosci. 26, 418–428 10.1523/JNEUROSCI.3882-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pizzi, M., Lanzillotta, A., Imbimbo, B.P., Hutter-Paier, B., Villetti, G., Facchinetti, F.et al. (2009) P3-270: CHF5074 reduces brain β-amyloid burden and hyperphosphorylated tau in a mouse model of Alzheimer's disease. Alzheimers Dement. 5, P422 10.1016/j.jalz.2009.04.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Branca, C., Sarnico, I., Ruotolo, R., Lanzillotta, A., Viscomi, A.R., Benarese, M.et al. (2014) Pharmacological targeting of the beta-amyloid precursor protein intracellular domain. Sci. Rep. 4, 4618 10.1038/srep04618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimberly, W.T., Zheng, J.B., Guenette, S.Y. and Selkoe, D.J. (2001) The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J. Biol. Chem. 276, 40288–40292 10.1074/jbc.C100447200 [DOI] [PubMed] [Google Scholar]
- 42.Cao, X. and Sudhof, T.C. (2001) A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 10.1126/science.1058783 [DOI] [PubMed] [Google Scholar]
- 43.Probst, S., Kruger, M., Kagi, L., Thoni, S., Schuppli, D., Nitsch, R.M.et al. (2020) Fe65 is the sole member of its family that mediates transcription regulated by the amyloid precursor protein. J. Cell Sci. 133, jcs242917. 10.1242/jcs.242917 [DOI] [PubMed] [Google Scholar]
- 44.Opsomer, R., Contino, S., Perrin, F., Gualdani, R., Tasiaux, B., Doyen, P.et al. (2020) Amyloid precursor protein (APP) controls the expression of the transcriptional activator neuronal PAS domain protein 4 (NPAS4) and synaptic GABA release. eNeuro 7, ENEURO.0322-19.2020 10.1523/ENEURO.0322-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pousinha, P.A., Mouska, X., Raymond, E.F., Gwizdek, C., Dhib, G., Poupon, G.et al. (2017) Physiological and pathophysiological control of synaptic GluN2B-NMDA receptors by the C-terminal domain of amyloid precursor protein. Elife 6, e25659. 10.7554/eLife.25659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Rotz, R.C., Kohli, B.M., Bosset, J., Meier, M., Suzuki, T., Nitsch, R.M.et al. (2004) The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J. Cell Sci. 117, 4435–4448 10.1242/jcs.01323 [DOI] [PubMed] [Google Scholar]
- 47.Muller, T., Concannon, C.G., Ward, M.W., Walsh, C.M., Tirniceriu, A.L., Tribl, F.et al. (2007) Modulation of gene expression and cytoskeletal dynamics by the amyloid precursor protein intracellular domain (AICD). Mol. Biol. Cell 18, 201–210 10.1091/mbc.e06-04-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi, K., Niidome, T., Akaike, A., Kihara, T. and Sugimoto, H. (2009) Amyloid precursor protein promotes endoplasmic reticulum stress-induced cell death via C/EBP homologous protein-mediated pathway. J. Neurochem. 109, 1324–1337 10.1111/j.1471-4159.2009.06067.x [DOI] [PubMed] [Google Scholar]
- 49.Ahn, K.W., Joo, Y., Choi, Y., Kim, M., Lee, S.H., Cha, S.H.et al. (2008) Swedish amyloid precursor protein mutation increases cell cycle-related proteins in vitro and in vivo. J. Neurosci. Res. 86, 2476–2487 10.1002/jnr.21690 [DOI] [PubMed] [Google Scholar]
- 50.Trazzi, S., Mitrugno, V.M., Valli, E., Fuchs, C., Rizzi, S., Guidi, S.et al. (2011) APP-dependent up-regulation of Ptch1 underlies proliferation impairment of neural precursors in Down syndrome. Hum. Mol. Genet. 20, 1560–1573 10.1093/hmg/ddr033 [DOI] [PubMed] [Google Scholar]
- 51.Ozaki, T., Li, Y., Kikuchi, H., Tomita, T., Iwatsubo, T. and Nakagawara, A. (2006) The intracellular domain of the amyloid precursor protein (AICD) enhances the p53-mediated apoptosis. Biochem. Biophys. Res. Commun. 351, 57–63 10.1016/j.bbrc.2006.09.162 [DOI] [PubMed] [Google Scholar]
- 52.Schrenk-Siemens, K., Perez-Alcala, S., Richter, J., Lacroix, E., Rahuel, J., Korte, M.et al. (2008) Embryonic stem cell-derived neurons as a cellular system to study gene function: lack of amyloid precursor proteins APP and APLP2 leads to defective synaptic transmission. Stem Cells 26, 2153–2163 10.1634/stemcells.2008-0010 [DOI] [PubMed] [Google Scholar]
- 53.Gunawardena, S. and Goldstein, L.S. (2001) Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron 32, 389–401 10.1016/S0896-6273(01)00496-2 [DOI] [PubMed] [Google Scholar]
- 54.Vogt, D.L., Thomas, D., Galvan, V., Bredesen, D.E., Lamb, B.T. and Pimplikar, S.W. (2011) Abnormal neuronal networks and seizure susceptibility in mice overexpressing the APP intracellular domain. Neurobiol. Aging 32, 1725–1729 10.1016/j.neurobiolaging.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghosal, K., Vogt, D.L., Liang, M., Shen, Y., Lamb, B.T. and Pimplikar, S.W. (2009) Alzheimer's disease-like pathological features in transgenic mice expressing the APP intracellular domain. Proc. Natl Acad. Sci. U.S.A. 106, 18367–18372 10.1073/pnas.0907652106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arimura, N., Menager, C., Kawano, Y., Yoshimura, T., Kawabata, S., Hattori, A.et al. (2005) Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol. Cell. Biol. 25, 9973–9984 10.1128/MCB.25.22.9973-9984.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho, J.H. and Johnson, G.V. (2003) Glycogen synthase kinase 3beta phosphorylates tau at both primed and unprimed sites. Differential impact on microtubule binding. J. Biol. Chem. 278, 187–193 10.1074/jbc.M206236200 [DOI] [PubMed] [Google Scholar]
- 58.Zhang, J., Jiang, X., Zhang, C., Zhong, J., Fang, X., Li, H.et al. (2020) Actin alpha 2 (ACTA2) downregulation inhibits neural stem cell migration through rho GTPase activation. Stem Cells Int. 2020, 4764012 10.1155/2020/4764012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, X., Liu, X.Q., Liu, X.L., Wang, X., Zhang, W.D., Huang, X.F.et al. (2023) SM22alpha deletion contributes to neurocognitive impairment in mice through modulating vascular smooth muscle cell phenotypes. Int. J. Mol. Sci. 24, 7117 10.3390/ijms24087117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamid, R., Kilger, E., Willem, M., Vassallo, N., Kostka, M., Bornhovd, C.et al. (2007) Amyloid precursor protein intracellular domain modulates cellular calcium homeostasis and ATP content. J. Neurochem. 102, 1264–1275 10.1111/j.1471-4159.2007.04627.x [DOI] [PubMed] [Google Scholar]
- 61.Goiran, T., Duplan, E., Chami, M., Bourgeois, A., El Manaa, W., Rouland, L.et al. (2018) beta-Amyloid precursor protein intracellular domain controls mitochondrial function by modulating phosphatase and tensin homolog-induced kinase 1 transcription in cells and in Alzheimer mice models. Biol. Psychiatry 83, 416–427 10.1016/j.biopsych.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 62.Shang, Y., Sun, X., Chen, X., Wang, Q., Wang, E.J., Miller, E.et al. (2022) A CHCHD6-APP axis connects amyloid and mitochondrial pathology in Alzheimer's disease. Acta Neuropathol. 144, 911–938 10.1007/s00401-022-02499-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma, Q.H., Futagawa, T., Yang, W.L., Jiang, X.D., Zeng, L., Takeda, Y.et al. (2008) A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat. Cell Biol. 10, 283–294 10.1038/ncb1690 [DOI] [PubMed] [Google Scholar]
- 64.Galvan, V., Gorostiza, O.F., Banwait, S., Ataie, M., Logvinova, A.V., Sitaraman, S.et al. (2006) Reversal of Alzheimer's-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc. Natl Acad. Sci. U.S.A. 103, 7130–7135 10.1073/pnas.0509695103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghosal, K., Stathopoulos, A. and Pimplikar, S.W. (2010) APP intracellular domain impairs adult neurogenesis in transgenic mice by inducing neuroinflammation. PLoS One 5, e11866 10.1371/journal.pone.0011866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muller, T., Schrotter, A., Loosse, C., Pfeiffer, K., Theiss, C., Kauth, M.et al. (2013) A ternary complex consisting of AICD, FE65, and TIP60 down-regulates Stathmin1. Biochim. Biophys. Acta 1834, 387–394 10.1016/j.bbapap.2012.07.017 [DOI] [PubMed] [Google Scholar]
- 67.Jin, K., Mao, X.O., Cottrell, B., Schilling, B., Xie, L., Row, R.H.et al. (2004) Proteomic and immunochemical characterization of a role for stathmin in adult neurogenesis. FASEB J. 18, 287–299 10.1096/fj.03-0973com [DOI] [PubMed] [Google Scholar]
- 68.Zhou, F., Gong, K., Song, B., Ma, T., van Laar, T., Gong, Y.et al. (2012) The APP intracellular domain (AICD) inhibits Wnt signalling and promotes neurite outgrowth. Biochim. Biophys. Acta 1823, 1233–1241 10.1016/j.bbamcr.2012.05.011 [DOI] [PubMed] [Google Scholar]
- 69.Young-Pearse, T.L., Bai, J., Chang, R., Zheng, J.B., LoTurco, J.J. and Selkoe, D.J. (2007) A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J. Neurosci. 27, 14459–14469 10.1523/JNEUROSCI.4701-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ware, M.L., Fox, J.W., Gonzalez, J.L., Davis, N.M., Lambert de Rouvroit, C., Russo, C.J.et al. (1997) Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron 19, 239–249 10.1016/S0896-6273(00)80936-8 [DOI] [PubMed] [Google Scholar]
- 71.Olson, E.C., Kim, S. and Walsh, C.A. (2006) Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression. J. Neurosci. 26, 1767–1775 10.1523/JNEUROSCI.3000-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Young-Pearse, T.L., Suth, S., Luth, E.S., Sawa, A. and Selkoe, D.J. (2010) Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J. Neurosci. 30, 10431–10440 10.1523/JNEUROSCI.1445-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herms, J., Anliker, B., Heber, S., Ring, S., Fuhrmann, M., Kretzschmar, H.et al. (2004) Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. EMBO J. 23, 4106–4115 10.1038/sj.emboj.7600390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guenette, S., Chang, Y., Hiesberger, T., Richardson, J.A., Eckman, C.B., Eckman, E.A.et al. (2006) Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. EMBO J. 25, 420–431 10.1038/sj.emboj.7600926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sabo, S.L., Ikin, A.F., Buxbaum, J.D. and Greengard, P. (2001) The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J. Cell Biol. 153, 1403–1414 10.1083/jcb.153.7.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, B., Li, H., Mutlu, S.A., Bowser, D.A., Moore, M.J., Wang, M.C.et al. (2017) The amyloid precursor protein is a conserved receptor for slit to mediate axon guidance. eNeuro 4, ENEURO.0185-17.2017 10.1523/ENEURO.0185-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li, W., Tam, K.M.V., Chan, W.W.R., Koon, A.C., Ngo, J.C.K., Chan, H.Y.E.et al. (2018) Neuronal adaptor FE65 stimulates Rac1-mediated neurite outgrowth by recruiting and activating ELMO1. J. Biol. Chem. 293, 7674–7688 10.1074/jbc.RA117.000505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan, W.W.R., Li, W., Chang, R.C.C. and Lau, K.F. (2020) ARF6-Rac1 signaling-mediated neurite outgrowth is potentiated by the neuronal adaptor FE65 through orchestrating ARF6 and ELMO1. FASEB J. 34, 16397–16413 10.1096/fj.202001703R [DOI] [PubMed] [Google Scholar]
- 79.Wang, P., Yang, G., Mosier, D.R., Chang, P., Zaidi, T., Gong, Y.D.et al. (2005) Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J. Neurosci. 25, 1219–1225 10.1523/JNEUROSCI.4660-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leyssen, M., Ayaz, D., Hebert, S.S., Reeve, S., De Strooper, B. and Hassan, B.A. (2005) Amyloid precursor protein promotes post-developmental neurite arborization in the Drosophila brain. EMBO J. 24, 2944–2955 10.1038/sj.emboj.7600757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gutierrez, D.A., Chandia-Cristi, A., Yanez, M.J., Zanlungo, S. and Alvarez, A.R. (2023) c-Abl kinase at the crossroads of healthy synaptic remodeling and synaptic dysfunction in neurodegenerative diseases. Neural Regen. Res. 18, 237–243 10.4103/1673-5374.346540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klevanski, M., Saar, M., Baumkotter, F., Weyer, S.W., Kins, S. and Muller, U.C. (2014) Differential role of APP and APLPs for neuromuscular synaptic morphology and function. Mol. Cell. Neurosci. 61, 201–210 10.1016/j.mcn.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 83.Weyer, S.W., Klevanski, M., Delekate, A., Voikar, V., Aydin, D., Hick, M.et al. (2011) APP and APLP2 are essential at PNS and CNS synapses for transmission, spatial learning and LTP. EMBO J. 30, 2266–2280 10.1038/emboj.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klevanski, M., Herrmann, U., Weyer, S.W., Fol, R., Cartier, N., Wolfer, D.P.et al. (2015) The APP intracellular domain is required for normal synaptic morphology, synaptic plasticity, and hippocampus-dependent behavior. J. Neurosci. 35, 16018–16033 10.1523/JNEUROSCI.2009-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Strecker, P., Ludewig, S., Rust, M., Mundinger, T.A., Gorlich, A., Krachan, E.G.et al. (2016) FE65 and FE65L1 share common synaptic functions and genetically interact with the APP family in neuromuscular junction formation. Sci. Rep. 6, 25652 10.1038/srep25652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ashley, J., Packard, M., Ataman, B. and Budnik, V. (2005) Fasciclin II signals new synapse formation through amyloid precursor protein and the scaffolding protein dX11/Mint. J. Neurosci. 25, 5943–5955 10.1523/JNEUROSCI.1144-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang, Z., Wang, B., Yang, L., Guo, Q., Aithmitti, N., Songyang, Z.et al. (2009) Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J. Neurosci. 29, 10788–10801 10.1523/JNEUROSCI.2132-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Markovinovic, A., Greig, J., Martin-Guerrero, S.M., Salam, S. and Paillusson, S. (2022) Endoplasmic reticulum-mitochondria signaling in neurons and neurodegenerative diseases. J. Cell Sci. 135, jcs248534 10.1242/jcs.248534 [DOI] [PubMed] [Google Scholar]
- 89.Belyaev, N.D., Nalivaeva, N.N., Makova, N.Z. and Turner, A.J. (2009) Neprilysin gene expression requires binding of the amyloid precursor protein intracellular domain to its promoter: implications for Alzheimer disease. EMBO Rep. 10, 94–100 10.1038/embor.2008.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim, H.S., Kim, E.M., Lee, J.P., Park, C.H., Kim, S., Seo, J.H.et al. (2003) C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J. 17, 1951–1953 10.1096/fj.03-0106fje [DOI] [PubMed] [Google Scholar]
- 91.Leist, M. and Nicotera, P. (1998) Apoptosis, excitotoxicity, and neuropathology. Exp. Cell Res. 239, 183–201 10.1006/excr.1997.4026 [DOI] [PubMed] [Google Scholar]
- 92.Lau, A. and Tymianski, M. (2010) Glutamate receptors, neurotoxicity and neurodegeneration. Pflugers Arch. 460, 525–542 10.1007/s00424-010-0809-1 [DOI] [PubMed] [Google Scholar]
- 93.Nakayama, K., Ohkawara, T., Hiratochi, M., Koh, C.S. and Nagase, H. (2008) The intracellular domain of amyloid precursor protein induces neuron-specific apoptosis. Neurosci. Lett. 444, 127–131 10.1016/j.neulet.2008.08.034 [DOI] [PubMed] [Google Scholar]
- 94.Passer, B., Pellegrini, L., Russo, C., Siegel, R.M., Lenardo, M.J., Schettini, G.et al. (2000) Generation of an apoptotic intracellular peptide by gamma-secretase cleavage of Alzheimer's amyloid beta protein precursor. J. Alzheimers Dis. 2, 289–301 10.3233/JAD-2000-23-408 [DOI] [PubMed] [Google Scholar]
- 95.Alves da Costa, C., Sunyach, C., Pardossi-Piquard, R., Sevalle, J., Vincent, B., Boyer, N.et al. (2006) Presenilin-dependent gamma-secretase-mediated control of p53-associated cell death in Alzheimer's disease. J. Neurosci. 26, 6377–6385 10.1523/JNEUROSCI.0651-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li, M., Pehar, M., Liu, Y., Bhattacharyya, A., Zhang, S.C., O'Riordan, K.J.et al. (2015) The amyloid precursor protein (APP) intracellular domain regulates translation of p44, a short isoform of p53, through an IRES-dependent mechanism. Neurobiol. Aging 36, 2725–2736 10.1016/j.neurobiolaging.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Greenwood, E.K., Angelova, D.M., Buchner, H.M.I. and Brown, D.R. (2022) The AICD fragment of APP initiates a FoxO3a mediated response via FANCD2. Mol. Cell. Neurosci. 122, 103760 10.1016/j.mcn.2022.103760 [DOI] [PubMed] [Google Scholar]
- 98.Kogel, D., Concannon, C.G., Muller, T., Konig, H., Bonner, C., Poeschel, S.et al. (2012) The APP intracellular domain (AICD) potentiates ER stress-induced apoptosis. Neurobiol. Aging 33, 2200–2209 10.1016/j.neurobiolaging.2011.06.012 [DOI] [PubMed] [Google Scholar]
- 99.Nakaya, T., Kawai, T. and Suzuki, T. (2009) Metabolic stabilization of p53 by FE65 in the nuclear matrix of osmotically stressed cells. FEBS J. 276, 6364–6374 10.1111/j.1742-4658.2009.07349.x [DOI] [PubMed] [Google Scholar]
- 100.Zambrano, N., Minopoli, G., de Candia, P. and Russo, T. (1998) The Fe65 adaptor protein interacts through its PID1 domain with the transcription factor CP2/LSF/LBP1. J. Biol. Chem. 273, 20128–20133 10.1074/jbc.273.32.20128 [DOI] [PubMed] [Google Scholar]
- 101.Lucas, J.J., Hernandez, F., Gomez-Ramos, P., Moran, M.A., Hen, R. and Avila, J. (2001) Decreased nuclear beta-catenin, tau hyperphosphorylation and neurodegeneration in GSK-3beta conditional transgenic mice. EMBO J. 20, 27–39 10.1093/emboj/20.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.L'Episcopo, F., Drouin-Ouellet, J., Tirolo, C., Pulvirenti, A., Giugno, R., Testa, N.et al. (2016) GSK-3beta-induced Tau pathology drives hippocampal neuronal cell death in Huntington's disease: involvement of astrocyte-neuron interactions. Cell Death Dis. 7, e2206 10.1038/cddis.2016.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schrotter, A., Mastalski, T., Nensa, F.M., Neumann, M., Loosse, C., Pfeiffer, K.et al. (2013) FE65 regulates and interacts with the Bloom syndrome protein in dynamic nuclear spheres - potential relevance to Alzheimer's disease. J. Cell Sci. 126, 2480–2492 10.1242/jcs.121004 [DOI] [PubMed] [Google Scholar]
- 104.Folch, J., Junyent, F., Verdaguer, E., Auladell, C., Pizarro, J.G., Beas-Zarate, C.et al. (2012) Role of cell cycle re-entry in neurons: a common apoptotic mechanism of neuronal cell death. Neurotox. Res. 22, 195–207 10.1007/s12640-011-9277-4 [DOI] [PubMed] [Google Scholar]
- 105.Tokarz, P., Kaarniranta, K. and Blasiak, J. (2016) Role of the cell cycle re-initiation in DNA damage response of post-mitotic cells and its implication in the pathogenesis of neurodegenerative diseases. Rejuvenation Res. 19, 131–139 10.1089/rej.2015.1717 [DOI] [PubMed] [Google Scholar]
- 106.Kajiwara, Y., Akram, A., Katsel, P., Haroutunian, V., Schmeidler, J., Beecham, G.et al. (2009) FE65 binds Teashirt, inhibiting expression of the primate-specific caspase-4. PLoS One 4, e5071 10.1371/journal.pone.0005071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hitomi, J., Katayama, T., Eguchi, Y., Kudo, T., Taniguchi, M., Koyama, Y.et al. (2004) Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J. Cell Biol. 165, 347–356 10.1083/jcb.200310015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kajiwara, Y., McKenzie, A., Dorr, N., Gama Sosa, M.A., Elder, G., Schmeidler, J.et al. (2016) The human-specific CASP4 gene product contributes to Alzheimer-related synaptic and behavioural deficits. Hum. Mol. Genet. 25, 4315–4327 10.1093/hmg/ddw265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dong, Z., Zhou, L., Del Villar, K., Ghanevati, M., Tashjian, V. and Miller, C.A. (2005) JIP1 regulates neuronal apoptosis in response to stress. Brain Res. Mol. Brain Res. 134, 282–293 10.1016/j.molbrainres.2004.10.039 [DOI] [PubMed] [Google Scholar]
- 110.Haefliger, J.A., Tawadros, T., Meylan, L., Gurun, S.L., Roehrich, M.E., Martin, D.et al. (2003) The scaffold protein IB1/JIP-1 is a critical mediator of cytokine-induced apoptosis in pancreatic beta cells. J. Cell Sci. 116, 1463–1469 10.1242/jcs.00356 [DOI] [PubMed] [Google Scholar]
- 111.Scheinfeld, M.H., Ghersi, E., Davies, P. and D'Adamio, L. (2003) Amyloid beta protein precursor is phosphorylated by JNK-1 independent of, yet facilitated by, JNK-interacting protein (JIP)-1. J. Biol. Chem. 278, 42058–42063 10.1074/jbc.M304853200 [DOI] [PubMed] [Google Scholar]
- 112.Chang, K.A., Kim, H.S., Ha, T.Y., Ha, J.W., Shin, K.Y., Jeong, Y.H.et al. (2006) Phosphorylation of amyloid precursor protein (APP) at Thr668 regulates the nuclear translocation of the APP intracellular domain and induces neurodegeneration. Mol. Cell. Biol. 26, 4327–4338 10.1128/MCB.02393-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Triaca, V., Sposato, V., Bolasco, G., Ciotti, M.T., Pelicci, P., Bruni, A.C.et al. (2016) NGF controls APP cleavage by downregulating APP phosphorylation at Thr668: relevance for Alzheimer's disease. Aging Cell 15, 661–672 10.1111/acel.12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ahmad, M.A., Kareem, O., Khushtar, M., Akbar, M., Haque, M.R., Iqubal, A.et al. (2022) Neuroinflammation: a potential risk for dementia. Int. J. Mol. Sci. 23, 616 10.3390/ijms23020616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kouli, A., Spindler, L.R.B., Fryer, T.D., Hong, Y.T., Malpetti, M., Aigbirhio, F.I.et al. (2024) Neuroinflammation is linked to dementia risk in Parkinson's disease. Brain 147, 923–935 10.1093/brain/awad322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weisman, D., Hakimian, E. and Ho, G.J. (2006) Interleukins, inflammation, and mechanisms of Alzheimer's disease. Vitam. Horm. 74, 505–530 10.1016/S0083-6729(06)74020-1 [DOI] [PubMed] [Google Scholar]
- 117.Swardfager, W., Lanctot, K., Rothenburg, L., Wong, A., Cappell, J. and Herrmann, N. (2010) A meta-analysis of cytokines in Alzheimer's disease. Biol. Psychiatry 68, 930–941 10.1016/j.biopsych.2010.06.012 [DOI] [PubMed] [Google Scholar]
- 118.Wang, W.Y., Tan, M.S., Yu, J.T. and Tan, L. (2015) Role of pro-inflammatory cytokines released from microglia in Alzheimer's disease. Ann. Transl. Med. 3, 136 10.3978/j.issn.2305-5839.2015.03.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Haass, C., Hung, A.Y. and Selkoe, D.J. (1991) Processing of beta-amyloid precursor protein in microglia and astrocytes favors an internal localization over constitutive secretion. J. Neurosci. 11, 3783–3793 10.1523/JNEUROSCI.11-12-03783.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spitzer, P., Walter, M., Goth, C., Oberstein, T.J., Linning, P., Knolker, H.J.et al. (2020) Pharmacological inhibition of amyloidogenic APP processing and knock-down of APP in primary human macrophages impairs the secretion of cytokines. Front. Immunol. 11, 1967 10.3389/fimmu.2020.01967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Porrini, V., Lanzillotta, A., Branca, C., Benarese, M., Parrella, E., Lorenzini, L.et al. (2015) CHF5074 (CSP-1103) induces microglia alternative activation in plaque-free Tg2576 mice and primary glial cultures exposed to beta-amyloid. Neuroscience 302, 112–120 10.1016/j.neuroscience.2014.10.029 [DOI] [PubMed] [Google Scholar]
- 122.Imbimbo, B.P., Hutter-Paier, B., Villetti, G., Facchinetti, F., Cenacchi, V., Volta, R.et al. (2009) CHF5074, a novel gamma-secretase modulator, attenuates brain beta-amyloid pathology and learning deficit in a mouse model of Alzheimer's disease. Br. J. Pharmacol. 156, 982–993 10.1111/j.1476-5381.2008.00097.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chakrabarti, A., Chatterjee, A., Sengupta, M.B., Chattopadhyay, P. and Mukhopadhyay, D. (2014) Altered levels of amyloid precursor protein intracellular domain-interacting proteins in Alzheimer disease. Alzheimer Dis. Assoc. Disord. 28, 283–290 10.1097/WAD.0000000000000011 [DOI] [PubMed] [Google Scholar]
- 124.Dean, T., Mendiola, A.S., Yan, Z., Meza-Acevedo, R., Cabriga, B., Akassoglou, K.et al. (2024) Fibrin promotes oxidative stress and neuronal loss in traumatic brain injury via innate immune activation. J Neuroinflammation. 21, 94 10.1186/s12974-024-03092-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shichita, T., Hasegawa, E., Kimura, A., Morita, R., Sakaguchi, R., Takada, I.et al. (2012) Peroxiredoxin family proteins are key initiators of post-ischemic inflammation in the brain. Nat. Med. 18, 911–917 10.1038/nm.2749 [DOI] [PubMed] [Google Scholar]
- 126.Morizawa, Y.M., Hirayama, Y., Ohno, N., Shibata, S., Shigetomi, E., Sui, Y.et al. (2017) Reactive astrocytes function as phagocytes after brain ischemia via ABCA1-mediated pathway. Nat. Commun. 8, 28 10.1038/s41467-017-00037-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sollberger, G., Strittmatter, G.E., Kistowska, M., French, L.E. and Beer, H.D. (2012) Caspase-4 is required for activation of inflammasomes. J. Immunol. 188, 1992–2000 10.4049/jimmunol.1101620 [DOI] [PubMed] [Google Scholar]
- 128.Misrani, A., Tabassum, S. and Yang, L. (2021) Mitochondrial dysfunction and oxidative stress in Alzheimer's disease. Front. Aging Neurosci. 13, 617588 10.3389/fnagi.2021.617588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Venkataraman, A.V., Mansur, A., Rizzo, G., Bishop, C., Lewis, Y., Kocagoncu, E.et al. (2022) Widespread cell stress and mitochondrial dysfunction occur in patients with early Alzheimer's disease. Sci. Transl. Med. 14, eabk1051 10.1126/scitranslmed.abk1051 [DOI] [PubMed] [Google Scholar]
- 130.Bernardo, T.C., Marques-Aleixo, I., Beleza, J., Oliveira, P.J., Ascensao, A. and Magalhaes, J. (2016) Physical exercise and brain mitochondrial fitness: the possible role against Alzheimer's disease. Brain Pathol. 26, 648–663 10.1111/bpa.12403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ungvari, Z., Sonntag, W.E., de Cabo, R., Baur, J.A. and Csiszar, A. (2011) Mitochondrial protection by resveratrol. Exerc. Sport Sci. Rev. 39, 128–132 10.1097/JES.0b013e3182141f80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Han, Y., Chu, X., Cui, L., Fu, S., Gao, C., Li, Y.et al. (2020) Neuronal mitochondria-targeted therapy for Alzheimer's disease by systemic delivery of resveratrol using dual-modified novel biomimetic nanosystems. Drug Deliv. 27, 502–518 10.1080/10717544.2020.1745328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ward, M.W., Concannon, C.G., Whyte, J., Walsh, C.M., Corley, B. and Prehn, J.H. (2010) The amyloid precursor protein intracellular domain(AICD) disrupts actin dynamics and mitochondrial bioenergetics. J. Neurochem. 113, 275–284 10.1111/j.1471-4159.2010.06615.x [DOI] [PubMed] [Google Scholar]
- 134.Nensa, F.M., Neumann, M.H., Schrotter, A., Przyborski, A., Mastalski, T., Susdalzew, S.et al. (2014) Amyloid beta a4 precursor protein-binding family B member 1 (FE65) interactomics revealed synaptic vesicle glycoprotein 2A (SV2A) and sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2) as new binding proteins in the human brain. Mol. Cell. Proteomics 13, 475–488 10.1074/mcp.M113.029280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Green, K.N., Demuro, A., Akbari, Y., Hitt, B.D., Smith, I.F., Parker, I.et al. (2008) SERCA pump activity is physiologically regulated by presenilin and regulates amyloid beta production. J. Cell Biol. 181, 1107–1116 10.1083/jcb.200706171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leissring, M.A., Murphy, M.P., Mead, T.R., Akbari, Y., Sugarman, M.C., Jannatipour, M.et al. (2002) A physiologic signaling role for the gamma -secretase-derived intracellular fragment of APP. Proc. Natl Acad. Sci. U.S.A. 99, 4697–4702 10.1073/pnas.072033799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu, Z., Li, H., Su, J., Xu, S., Zhu, F., Ai, J.et al. (2019) Numb depletion promotes Drp1-mediated mitochondrial fission and exacerbates mitochondrial fragmentation and dysfunction in acute kidney injury. Antioxid. Redox Signal. 30, 1797–1816 10.1089/ars.2017.7432 [DOI] [PubMed] [Google Scholar]
- 138.Hu, Y.B., Ren, R.J., Zhang, Y.F., Huang, Y., Cui, H.L., Ma, C.et al. (2019) Rho-associated coiled-coil kinase 1 activation mediates amyloid precursor protein site-specific Ser655 phosphorylation and triggers amyloid pathology. Aging Cell 18, e13001 10.1111/acel.13001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lau, D.H.W., Paillusson, S., Hartopp, N., Rupawala, H., Morotz, G.M., Gomez-Suaga, P.et al. (2020) Disruption of endoplasmic reticulum-mitochondria tethering proteins in post-mortem Alzheimer's disease brain. Neurobiol. Dis. 143, 105020 10.1016/j.nbd.2020.105020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Montesinos, J., Pera, M., Larrea, D., Guardia-Laguarta, C., Agrawal, R.R., Velasco, K.R.et al. (2020) The Alzheimer's disease-associated C99 fragment of APP regulates cellular cholesterol trafficking. EMBO J. 39, e103791 10.15252/embj.2019103791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Morcillo, P., Kabra, K., Velasco, K., Cordero, H., Jennings, S., Yun, T.D.et al. (2024) Aberrant ER-mitochondria communication is a common pathomechanism in mitochondrial disease. Cell Death Dis. 15, 405 10.1038/s41419-024-06781-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marchi, S., Patergnani, S., Missiroli, S., Morciano, G., Rimessi, A., Wieckowski, M.R.et al. (2018) Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium 69, 62–72 10.1016/j.ceca.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 143.Sassano, M.L., Felipe-Abrio, B. and Agostinis, P. (2022) ER-mitochondria contact sites; a multifaceted factory for Ca(2+) signaling and lipid transport. Front. Cell Dev. Biol. 10, 988014 10.3389/fcell.2022.988014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carreras-Sureda, A., Kroemer, G., Cardenas, J.C. and Hetz, C. (2022) Balancing energy and protein homeostasis at ER-mitochondria contact sites. Sci. Signal. 15, eabm7524 10.1126/scisignal.abm7524 [DOI] [PubMed] [Google Scholar]
- 145.Pera, M., Larrea, D., Guardia-Laguarta, C., Montesinos, J., Velasco, K.R., Agrawal, R.R.et al. (2017) Increased localization of APP-C99 in mitochondria-associated ER membranes causes mitochondrial dysfunction in Alzheimer disease. EMBO J. 36, 3356–3371 10.15252/embj.201796797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu, P., Chang, J.C., Zhou, X., Wang, W., Bamkole, M., Wong, E.et al. (2021) GSAP regulates lipid homeostasis and mitochondrial function associated with Alzheimer's disease. J. Exp. Med. 218, e20202446 10.1084/jem.20202446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chau, D.D., Ng, L.L., Zhai, Y. and Lau, K.F. (2023) Amyloid precursor protein and its interacting proteins in neurodevelopment. Biochem. Soc. Trans. 51, 1647–1659 10.1042/BST20221527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zheng, H., Jiang, M., Trumbauer, M.E., Sirinathsinghji, D.J., Hopkins, R., Smith, D.W.et al. (1995) beta-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell 81, 525–531 10.1016/0092-8674(95)90073-X [DOI] [PubMed] [Google Scholar]
- 149.Heber, S., Herms, J., Gajic, V., Hainfellner, J., Aguzzi, A., Rulicke, T.et al. (2000) Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J. Neurosci. 20, 7951–7963 10.1523/JNEUROSCI.20-21-07951.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Walsh, D.M., Fadeeva, J.V., LaVoie, M.J., Paliga, K., Eggert, S., Kimberly, W.T.et al. (2003) gamma-Secretase cleavage and binding to FE65 regulate the nuclear translocation of the intracellular C-terminal domain (ICD) of the APP family of proteins. Biochemistry 42, 6664–6673 10.1021/bi027375c [DOI] [PubMed] [Google Scholar]
- 151.Gersbacher, M.T., Goodger, Z.V., Trutzel, A., Bundschuh, D., Nitsch, R.M. and Konietzko, U. (2013) Turnover of amyloid precursor protein family members determines their nuclear signaling capability. PLoS One 8, e69363 10.1371/journal.pone.0069363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Turk, M. and Baumeister, W. (2020) The promise and the challenges of cryo-electron tomography. FEBS Lett. 594, 3243–3261 10.1002/1873-3468.13948 [DOI] [PubMed] [Google Scholar]
- 153.Tamayev, R., Zhou, D. and D'Adamio, L. (2009) The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol. Neurodegener. 4, 28 10.1186/1750-1326-4-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou, D., Zambrano, N., Russo, T. and D'Adamio, L. (2009) Phosphorylation of a tyrosine in the amyloid-beta protein precursor intracellular domain inhibits Fe65 binding and signaling. J. Alzheimers Dis. 16, 301–307 10.3233/JAD-2009-0970 [DOI] [PubMed] [Google Scholar]
- 155.Das, S., Raychaudhuri, M., Sen, U. and Mukhopadhyay, D. (2011) Functional implications of the conformational switch in AICD peptide upon binding to Grb2-SH2 domain. J. Mol. Biol. 414, 217–230 10.1016/j.jmb.2011.09.046 [DOI] [PubMed] [Google Scholar]
- 156.Oishi, M., Nairn, A.C., Czernik, A.J., Lim, G.S., Isohara, T., Gandy, S.E.et al. (1997) The cytoplasmic domain of Alzheimer's amyloid precursor protein is phosphorylated at Thr654, Ser655, and Thr668 in adult rat brain and cultured cells. Mol. Med. 3, 111–123 10.1007/BF03401803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee, M.S., Kao, S.C., Lemere, C.A., Xia, W., Tseng, H.C., Zhou, Y.et al. (2003) APP processing is regulated by cytoplasmic phosphorylation. J. Cell Biol. 163, 83–95 10.1083/jcb.200301115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chow, W.N., Ngo, J.C., Li, W., Chen, Y.W., Tam, K.M., Chan, H.Y.et al. (2015) Phosphorylation of FE65 Ser610 by serum- and glucocorticoid-induced kinase 1 modulates Alzheimer's disease amyloid precursor protein processing. Biochem. J. 470, 303–317 10.1042/BJ20141485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jowsey, P.A. and Blain, P.G. (2015) Fe65 Ser228 is phosphorylated by ATM/ATR and inhibits Fe65-APP-mediated gene transcription. Biochem. J. 465, 413–421 10.1042/BJ20140656 [DOI] [PubMed] [Google Scholar]
- 160.Noble, W., Planel, E., Zehr, C., Olm, V., Meyerson, J., Suleman, F.et al. (2005) Inhibition of glycogen synthase kinase-3 by lithium correlates with reduced tauopathy and degeneration in vivo. Proc. Natl Acad. Sci. U.S.A. 102, 6990–6995 10.1073/pnas.0500466102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Phiel, C.J., Wilson, C.A., Lee, V.M. and Klein, P.S. (2003) GSK-3alpha regulates production of Alzheimer's disease amyloid-beta peptides. Nature 423, 435–439 10.1038/nature01640 [DOI] [PubMed] [Google Scholar]
- 162.Bartling, C.R.O., Jensen, T.M.T., Henry, S.M., Colliander, A.L., Sereikaite, V., Wenzler, M.et al. (2021) Targeting the APP-Mint2 protein-protein interaction with a peptide-based inhibitor reduces amyloid-beta formation. J. Am. Chem. Soc. 143, 891–901 10.1021/jacs.0c10696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Szogi, T., Schuster, I., Borbely, E., Gyebrovszki, A., Bozso, Z., Gera, J.et al. (2019) Effects of the pentapeptide P33 on memory and synaptic plasticity in APP/PS1 transgenic mice: a novel mechanism presenting the protein Fe65 as a target. Int. J. Mol. Sci. 20, 3050 10.3390/ijms20123050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Brandimarti, R., Irollo, E. and Meucci, O. (2023) The US9-derived protein gPTB9TM modulates APP processing without targeting secretase activities. Mol. Neurobiol. 60, 1811–1825 10.1007/s12035-022-03153-2 [DOI] [PMC free article] [PubMed] [Google Scholar]