Abstract
Purpose
We developed an artificial intelligence (AI) model to detect immunotherapy -related adverse events (irAEs) from clinical narratives of electronic health records (EHRs) at the patient level.
Patients and Methods
Training data, used for internal validation of the AI model, comprised 1230 clinical notes from 30 patients at The Ohio State University James Cancer Hospital−20 patients who experienced irAEs and ten who did not. 3256 clinical notes of 50 patients were utilized for external validation of the AI model.
Results
Use of a leave-one-out cross-validation technique for internal validation among those 30 patients yielded accurate identification of 19 of 20 with irAEs (positive patients; 95% sensitivity) and correct dissociation of eight of ten without (negative patients; 80% specificity). External validation on 3256 clinical notes of 50 patients yielded high sensitivity (95%) but moderate specificity (64%). If we improve the model’s specificity to 100%, it could eliminate the need to manually review 2500 of those 3256 clinical notes (77%).
Conclusion
Combined use of this AI model with the manual review of clinical notes will improve both sensitivity and specificity in the detection of irAEs, decreasing workload and costs and facilitating the development of improved immunotherapies.
Keywords: immunotherapy, cancer, adverse events, machine learning, natural language processing, artificial intelligence, electronic health records
Introduction
Adverse drug events (ADEs) are harmful side effects of drug use that can prolong hospitalization, heighten healthcare costs, and significantly increase the number of morbidity and mortality cases.1–4 Notwithstanding, ADEs are frequently preventable,5,6 and their early identification and prevention are vital to ensure patient safety, mitigate healthcare costs, and improve therapies.7,8 Though the field of immunotherapy has revolutionized cancer treatment, enhancing survival rates across various types of cancer,9,10 therapeutic advancements have been associated with unfavorable outcomes,11 referred to as immunotherapy -related adverse events (irAEs). IrAEs can affect any organ system and commonly target the gastrointestinal tract, endocrine glands, skin, and liver, resulting in such conditions as colitis (eight to ten percent in some studies), diarrhea (27 to 54%), pneumonitis, thyroid abnormalities, rash/dermatitis (34 to 45%), hepatitis (five to ten percent), myalgia, and cardiotoxicity−side effects that can lead to severe and even life-threatening conditions.12–14 Throughout this manuscript, we will discuss ADEs in the context of immunotherapy and use the terms ADE and irAE interchangeably.
Electronic health records (EHRs) can contain longitudinal patient information, including details related to ADEs, that can be used as source data for the surveillance of post-marketing drug safety.14 However, our recently published study on irAEs, including colitis, hepatitis, and pneumonitis, revealed that the electronic health records of 46% of patients with irAEs that were noted during manual record review registered no ICD code evidence of these events.15 This illustrates the urgency for building an automated system capable of processing clinical texts and extracting pertinent information that avoids the time-consuming and labor-intensive task of manually extracting irAE data from clinical narratives.
Natural language processing (NLP), a branch of artificial intelligence (AI), has become popular in clinical research. NLP can automatically process text and extract valuable information. In both biomedical literature and EHRs, NLP has been extensively used for information extraction, such as named-entity recognition (NER) and relation extraction (RE).16,17 Early NLP favored rule-based methods, leading to the development of tools like KnowledgeMap,18 MedEx,19 and MedXN,20 but the preference has shifted to machine learning (ML) and deep learning (DL) methods for the extraction of information from clinical texts.16,17,21,22
ML and DL techniques, in particular, have transformed the detection of ADEs in EHRs, enabling the analysis and extraction of ADE data from vast amounts of unstructured data in clinical notes.21,22 As a result, by automating the detection process, AI technologies can reduce the workload for labor-intensive manual chart review, significantly improving efficiency and minimizing human error. Notwithstanding the substantial progress, particularly in ADE detection challenges, such as the 2018 National NLP Clinical Challenges (N2c2)23 and 2019 Medication and Adverse Drug Events from Electronic Health Records Challenge (MADE 1.0),24 most models primarily focus on ADE detection at the event rather than patient level, thereby limiting comprehensive ADE detection and failing to account for the longitudinal nature of ADEs.
The challenge of developing AI models lies in achieving high sensitivity at the patient level. For patients receiving immunotherapy, missing irAEs could lead to undetected and, therefore, unmanaged severe outcomes that compromise patient safety.13,25,26 However, efforts to identify irAEs at the individual patient level have inadequately emphasized the necessary high sensitivity.27
Our study aims to bridge these gaps by leveraging NLP and ML technologies to develop a high-sensitivity model capable of detecting patient irAEs from clinical notes in EHRs, and we validate the performance of our AI model through manual review. We also investigate how NLP and ML can reduce the workload for manual review to achieve high specificity in detecting irAEs.
Material and Methods
Data Source
As part of our collection of data for training our model, we obtained the clinical notes of 30 patients who had received immune checkpoint inhibitors (ICIs) at The Ohio State University James Cancer Hospital between 2011 and 2021. The patients were selected randomly, and for each patient, we collected the clinical notes from the first twelve months following the date when the first ICI dose was administered. The ICI dose date was obtained from structured data. We collected 1230 clinical notes from the 30 selected patients, which we used to train our model, and we collected 3256 clinical notes from 50 additional patients as validation data. The institutional review board of The Ohio State University approved this study (#2020C0145).
Data Annotation
The drug-ADE relationship is annotated in the training data at the sentence level in each note, after which the patient-level drug-ADE relationship is annotated. In the validation data, only the patient-level drug-ADE relationship is annotated. Therefore, the cost and labor of training data annotation are much more expensive than validation annotation. This is the primary reason the number of patient samples in validation is larger than the training sample. The annotation process involved examining each patient’s medical history and identifying relevant drug-ADE relationships. Two independent annotators, each with distinct expertise, performed the manual review. The first annotator was a graduate student with four years of experience developing corpora using EHRs, and the second was a clinical researcher with hands-on experience working with EHRs.
Guideline for Manual Annotation of Drug-ADE Relations
Annotation of a positive drug-ADE relation required clear evidence in the note that the adverse event was induced by the drug. Cases in which an ADE was related to multiple, or a combination of drugs required separate annotation of each drug to the ADE to draw a drug-ADE relationship.
Sentence-Level Annotation
The goal of sentence-level annotation, to annotate drug-ADE relations within a sentence, required annotators to examine only the relations in which the drug and adverse event appeared in the same sentence. Relations between drugs and adverse events across sentences were outside the scope of this study. After the first round of annotation, the two annotators resolved any disagreements in their assessments through discussion.
Patient-Level Annotation
We established a clear criterion to determine whether a patient was positive or negative for a drug-ADE relation. Patients were classified as positive if their clinical notes identified at least one positive drug-ADE relation and negative if the notes showed no indication of a positive drug-ADE relation. This manuscript will consistently apply this definition of positive and negative patients. After a thorough manual review of the clinical notes for all 30 patients that were included in the training data, 20 patients were identified as positive and ten as negative. Complete agreement of the two independent annotators in their assessments at the patient-level annotation based on the clinical notes created a reliable gold standard dataset. We developed an NLP pipeline for further analysis and detail its components in the following sections.
Natural Language Processing Pipeline
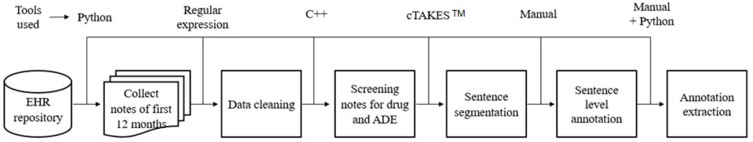
Our machine-learning model was fed clinical notes that underwent several processing steps through our NLP pipeline (Figure 1).
Figure 1.
Natural language processing (NLP) pipeline involving a combination of manual and automated methods. Specifically, sentence-level annotation was carried out manually, and all other steps were automated using various programming languages (such as Python and C++), tools, and techniques. Adverse Drug Events (ADE), adverse drug event; Electronic Health Records (EHR), health record.
Note Collection
We randomly selected 30 patients and collected 1230 clinical notes from the first twelve months after the first ICI dose was noted in structured data. We automated the collection process employing Python programming and maintained the order of the notes based on their respective dates.
Data Processing
The primary objective of the data processing procedure was to standardize the clinical notes, ensuring their consistency and suitability for further analysis. This included but was not limited to normalization of drug names and abbreviations and removal of uneven spaces to normalize space. We used regular-expressions28 techniques to perform the data processing task.
Screening Drug Names and Adverse Drug Events
The automated screening was performed to separate clinical notes containing the drug name and adverse drug event within the same note and make the dataset more relevant to our study. During the screening process, we included all possible mentions of the immune checkpoint inhibitor drugs, including both generic and brand names (Table 1). Our primary source of information for drug references was DrugBank,29 and we used the Common Terminology Criteria for Adverse Events (CTCAE)30 as our reference guide for collecting all potential references of the ADEs (Table 1). Thus, we enriched the list of drugs and ADEs. A physician further verified and updated the drug and ADE list to guarantee accuracy and reliability. We then utilized the finalized list to automate the screening process by implementing C++ programming.
Table 1.
Names of Immune Checkpoint Inhibitor (ICI) Drugs and Adverse Drug Events (ADEs) Used in This Study
| Drug | Adverse Drug Event |
|---|---|
| Atezolizumab Avelumab Cemiplimab Durvalumab Ipilimumab Nivolumab Pembrolizumab Tremelimumab |
Cardiotoxicity Cardiovascular disorders Colitis Hepatitis Myalgia/arthralgia Pneumonitis Thyroid abnormalities Rash/dermatitis Hepatobiliary disorders |
Sentence Segmentation
The Apache™ clinical Text Analysis and Knowledge Extraction System (cTAKES™)31 is an open-source tool that can perform several NLP tasks along with sentence segmentation. We used the system for sentence segmentation. Clinical notes contain many abbreviations, punctuation, and unexpected line breakers that make sentence segmentation very challenging. To address the challenges we encountered, we employed encryption of clinical notes. Specifically, we replaced characters that caused unexpected line breaks before performing sentence segmentation, thereby improving the performance of cTAKES™. After the segmentation, we decrypted the notes to preserve their original format.
Sentence-Level Annotation
Based on the sentence segmentation, we performed a sentence-level annotation.
Definition of Positive and Negative Drug-ADE Relationships and Annotation Extraction
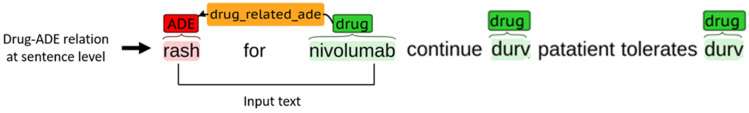
We considered all possible combinations between drugs and ADEs within a sentence to build the gold standard dataset of positive and negative relations. For instance, Figure 2 contains a sentence that mentions three drugs and one ADE. From this sentence, we can derive three potential drug-ADE pairs, with one pair indicating a positive relation and the remaining two suggesting negative relations. The annotators directly labeled the positive drug-ADE relation from the text through manual annotation. However, the negative drug-ADE associations were determined indirectly. Computationally, we generated all possible drug-ADE combinations and removed those positive relations manually identified by the annotators, and this left us with the negative drug-ADE relations. From every positive patient, we extracted both positive and negative drug-ADE relations. Negative patients had no positive drug-ADE relations. The two annotators disagreed regarding 23 drug-ADE relations after the first round of annotation and resolved their disagreements after discussion. Finally, 175 positive and 745 negative drug-ADE relations were extracted from the clinical notes of the 30 patients in the training data and served as the gold-standard positive and negative drug-ADE relations dataset. To every drug-ADE relation, we further assigned a patient identifier labeling the patient having the relation. This helped us separate the relations extracted from a particular patient.
Figure 2.
Example of sentence-level drug-adverse drug event (ADE) relation along with model input.
Data Processing for the Validation Set
We used the same NLP pipeline (Figure 1) to extract drug-ADE relation samples within the same sentences for the 50 validation patients. We considered all possible drug-ADE pairs within a sentence, similar to the training data while extracting those samples in our validation set. We did not conduct manual annotation at the sentence level for our validation set. Consequently, we obtained drug-ADE relationships with undefined labels, which we used to test the performance of our machine learning model.
Machine Learning Model
We built support vector machine (SVM), a supervised machine learning model to perform the analysis of our patient-level ADE detection in the training data.
Training and Test Data
We applied the leave-one-out cross-validation (LOOCV)32 method to detect drug-induced adverse events at the individual-patient level. To ascertain whether a patient had experienced drug-induced adverse events, the test dataset included all drug-ADE relations gathered from that particular patient. The training data comprised all other drug-ADE relations extracted from the remaining patient population. We then applied our SVM model on the training and testing set, performing this process separately for each patient.
Drug-ADE Prediction at the Patient Level
Our study categorized patients as positive if their clinical notes included at least one drug-ADE relationship and negative if none was found. We assessed the performance of our machine learning model based on its correct identification of these positive and negative patients.
Input of the Support Vector Machine Model and Feature Selection
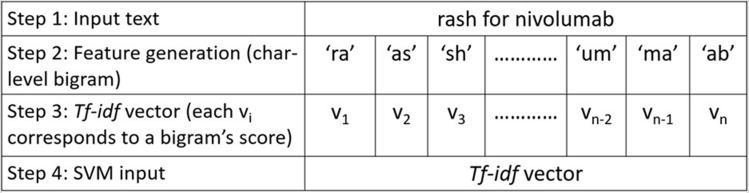
In SVM model development, the model input was the clinical text between a drug-ADE relation, including the drug and ADE names (Figure 2). We used character n-gram features to leverage the contextual information of a drug-ADE relation. All possible character n-grams were generated from the beginning to the end of our input text with a range of values for n, and those n-grams were then converted into term frequency-inverse document frequency (TF-IDF)33 vectors. We applied a grid search34 technique to obtain the best hyperparameter set from a range of values for the parameters c (rages from 0.001 to 1000) and gamma (ranges from 0.0001–100), applied different kernels, and eventually selected the radial basis function (RBF) kernel35 because it performed the best. Figure 3 illustrates the different steps in our SVM model with examples.
Figure 3.
Implementation of our Support vector machine (SVM) model with an example of a bigram. Here, we generated character-level bigrams of the input text, performed term frequency-inverse document frequency (TF-IDF) vectorization (v1, v2, v3 …. vn-1, vn), and then applied the SVM model and obtained the output indicating the class label (positive/negative).
Results
Evaluation of Prediction Performance on Internal Validation
Table 2 shows the SVM model’s performance for predicting negative patients. Because negative patients had no positive drug-ADE relation, the sensitivity value was 1.0 by default.36 Here, the specificity score indicates the model’s probability of identifying true-negative relations. As examples, the specificity score of 1 for Patient 1 indicates that the model accurately predicted both true-negative drug-ADE relations as negative, whereas the specificity of 0.92 for Patient 7 reveals that the model mistakenly predicted two of 25 true-negative drug-ADE relations as positive.
Table 2.
Results of Our Support Vector Machine (SVM) Model for Negative Patients. The Right-Most Column Indicates Whether the SVM Model Was Able to Correctly Identify a Negative Patient. The Same Model Was Run Separately for Each Negative Patient. We Can See That the SVM Model Was Able to Identify All the Negative Patients Except Those with Identifiers 7 and 10
| Patient ID | Number of Manually Labeled Drug-ADE Relations | Predicted Drug-ADE Relations | Specificity | Sensitivity | Patient-Level Model Prediction | ||
|---|---|---|---|---|---|---|---|
| + | - | Correctly Predicted +/ Predicted + | Correctly Predicted -/ Predicted - | ||||
| 1 | 0 | 2 | 0/0 | 2/2 | 1 | 1 | 0 |
| 2 | 0 | 7 | 0/0 | 7/7 | 1 | 1 | 0 |
| 3 | 0 | 1 | 0/0 | 1/1 | 1 | 1 | 0 |
| 4 | 0 | 23 | 0/0 | 23/23 | 1 | 1 | 0 |
| 5 | 0 | 1 | 0/0 | 1/1 | 1 | 1 | 0 |
| 6 | 0 | 5 | 0/0 | 5/5 | 1 | 1 | 0 |
| 7 | 0 | 25 | 0/2 | 23/23 | 0.92 | 1 | 1 |
| 8 | 0 | 17 | 0/0 | 17/17 | 1 | 1 | 0 |
| 9 | 0 | 17 | 0/0 | 17/17 | 1 | 1 | 0 |
| 10 | 0 | 29 | 0/4 | 25/25 | 0.86 | 1 | 1 |
To make patient-level prediction, we predict the patient as positive when the model predicts one positive drug-ADE relation for the patient. Table 2 highlights that eight of ten negative patients were predicted negative, and two were predicted positive.
Table 3 shows the model predictions for positive patients. Patient 11, for example, had five positive drug-ADE relations and 18 negative relations. The model predicted three drug-ADE relations that were true positive, and the model predicted 20 negative relations, 18 of which were true negative. Thus, the model demonstrated 60% sensitivity and 100% specificity in predicting drug-ADE relations for this patient. Because Patient 11 had three predicted drug-ADE relations, the patient was classified as positive. Patient 18 was predicted negative because the model predicted no drug-ADE, missing three true-positive relations and reflecting the model’s 100% specificity and 0% sensitivity. Interestingly, Patient 27 had one true-positive and 18 true-negative drug-ADE relations, but the model made six positive predictions, none of which was correct. Its 67% specificity and 0% sensitivity indicated very poor performance. However, because the model made a positive prediction, Patient 27 was correctly predicted as positive for the wrong reason.
Table 3.
Results of Our Support Vector Machine (SVM) Model for Positive Patients. The Same Model Was Run Separately for Each Positive Patient. Here We Can See That the SVM Model Was Able to Identify All the Positive Patients Except the Patient Identified as 18
| Patient Identifier | Number of Manual Labeled drug-ADE Relations | Predicted Drug-ADE Relations | Specificity | Sensitivity | Patient-Level Model Prediction | ||
|---|---|---|---|---|---|---|---|
| + | - | Correctly Predicted +/ Predicted + | Correctly Predicted -/ Predicted - | ||||
| 11 | 5 | 18 | 3/3 | 18/20 | 1.00 | 0.60 | 1 |
| 12 | 7 | 92 | 6/6 | 92/93 | 1.00 | 0.86 | 1 |
| 13 | 13 | 2 | 7/7 | 2/8 | 1.00 | 0.54 | 1 |
| 14 | 3 | 12 | 3/6 | 9/9 | 0.75 | 1.00 | 1 |
| 15 | 1 | 2 | 1/1 | 2/2 | 1.00 | 1.00 | 1 |
| 16 | 9 | 15 | 6/6 | 15/18 | 1.00 | 0.67 | 1 |
| 17 | 6 | 26 | 1/2 | 25/30 | 0.96 | 0.17 | 1 |
| 18 | 3 | 26 | 0/0 | 26/29 | 1.00 | 0.0 | 0 |
| 19 | 4 | 4 | 1/1 | 4/7 | 1.00 | 0.25 | 1 |
| 20 | 5 | 8 | 2/3 | 7/10 | 0.88 | 0.40 | 1 |
| 21 | 4 | 29 | 3/3 | 29/30 | 1.00 | 0.75 | 1 |
| 22 | 1 | 10 | 1/2 | 9/9 | 0.90 | 1.00 | 1 |
| 23 | 11 | 130 | 2/2 | 130/139 | 1.00 | 0.18 | 1 |
| 24 | 15 | 34 | 9/16 | 27/33 | 0.79 | 0.60 | 1 |
| 25 | 8 | 3 | 6/6 | 3/5 | 1.00 | 0.75 | 1 |
| 26 | 2 | 5 | 2/2 | 5/5 | 1.00 | 1.00 | 1 |
| 27 | 1 | 18 | 0/6 | 12/13 | 0.67 | 0.00 | 1 |
| 28 | 8 | 30 | 4/5 | 29/33 | 0.97 | 0.50 | 1 |
| 29 | 41 | 150 | 10/10 | 150/181 | 1.00 | 0.24 | 1 |
| 30 | 28 | 4 | 23/23 | 4/9 | 1.00 | 0.82 | 1 |
Overall, at the patient level, the model correctly predicted eight of ten negative patients (80% specificity) (Table 2) and 19 of 20 positive patients (95% sensitivity) (Table 3). Table 4 details our model’s performance metrics for the identification of drug-ADE relations. Achieving only 51% sensitivity and 97% specificity in predicting drug-ADE relations, the model had a much greater sensitivity in predicting drug-ADE at the patient level. However, the model demonstrated higher specificity at the individual drug-ADE-relation level than the patient level.
Table 4.
Performance Metrics for Our Model’s Identification of Drug-Adverse Drug Event Relations for the 30 Patients in the Internal Validation Set, Showcasing the Model’s Performance in Various Measures and Criteria
| Total Number of Relations | Manual Positive Label | Manual Negative Label | Correctly Predicted +/Predicted + | Correctly Predicted -/Predicted - | Specificity | Sensitivity |
|---|---|---|---|---|---|---|
| 920 | 175 | 745 | 90/116 | 719/804 | 0.97 | 0.51 |
Model Performance on the External Validation Set
We performed an external validation of our model using the data of a cohort of 50 new patients from the same data source employed to generate the data of the 30 patients used for the internal validation. A patient was labeled positive who demonstrated at least one true drug-ADE relation and otherwise labeled negative.
The SVM model was trained using the data of 30 patients, which contained 175 positive and 745 negative drug-ADE relations, and we then evaluated the model’s performance on the data of each of the 50 new patients. Model prediction at the individual-patient level involved predicting the number of positive and negative drug-ADE relations from all the notes of a single patient, with the presence of at least one predicted positive drug-ADE relation in the record predicting the patient as positive. Table 5 details our model’s performance metrics for those 50.
Table 5.
Performance Metrics of Our Model for the 50 Patients in the External Validation Set, Showcasing the Model’s Performance in Various Measures and Criteria
| Number of Notes for the 50 Patients | Manual Positive Label | Manual Negative Label | Predicted Positive Label | Predicted Negative Label | Notes in Predicted Positive Label | Specificity | Sensitivity |
|---|---|---|---|---|---|---|---|
| 3256 | 22 | 28 | 31 | 19 | 756 | 0.64 | 0.95 |
The SVM model predicted 31 patients as positive and 19 as negative. A physician who independently examined the clinical notes and identified the positive and negative patients then evaluated the model’s performance, comparing these results with the manual review findings. Manual review of clinical notes identified 22 true-positive and 28 true-negative patients. Twenty-one of the 31 patients predicted positive were true positive; and 18 of the 19 patients predicted negative were true negative. In the end, in the validation cohort, sensitivity was 95% and specificity, 64%. We think the external validation study confirms the high sensitivity, 95%, in the internal validation cohort. The 64% specificity in the external validation cohort was moderately lower than that in the internal validation group, 80%.
The SVM Model Can Significantly Reduce the Workload of Manual Review
The SVM model predicted 31 patients as positive based on positive drug-ADE relations predicted from 756 clinical notes from these patients. These notes were those remaining after our AI model filtered out 2500 of the initial 3256 notes through NLP steps to detect irAEs. Consequently, manual review of these residual 756 notes will allow us to detect ten false-positive patients of 31 patients predicted positive. This process allows us to maintain the 95% sensitivity while achieving 100% specificity.
Discussion
Erroneous predictions arose from the model’s inability to manage negation among causative terms and ambiguity surrounding the use of ADE terms in contexts unrelated to adverse events that complicated the classification process. In the first case, for instance, the causal term “related” in the relation “‘rash is unrelated to the treatment, and no difference related to durvalumab has been noticed’ was misinterpreted as a causal connection between the drug and the adverse event. In the second case, only 43 of 120 total occurrences of the term ‘cough’ in our internal validation data represented an ADE.
Furthermore, we believe broader context would provide our model with increased flexibility for accurate classification with greater number of positive and negative relations. Our internal validation set identified a positive patient with only one positive drug-ADE relation, resulting in a model misclassification. However, incorporation of drug-ADE relations across sentences would likely uncover additional positive drug-ADE relations for that patient. We plan to integrate drug-ADE relations across sentences in future work to avoid such issues and thereby enhance model performance.
Our study demonstrated the effectiveness of the traditional NLP method SVM in clinical text analysis, particularly as part of a screening tool to reduce the manual review workload. TF-IDF features with an SVM were chosen for their computational efficiency, suitability for the relatively small dataset used in this study, and ability to provide interpretable and reliable outputs. Additionally, our model can be easily integrated into clinical workflows without requiring extensive computational resources. Our model had a high sensitivity in the internal validation, correctly classifying 19 of 20 positive patients. However, in seven patients, sensitivity scores below 50% indicated the need for improved identification of positive relations; the model misclassified one of these patients. In our external validation, the model’s high sensitivity score (95%) indicated its ability to identify most positive cases correctly, but its 64% specificity recommends the need for improvement to reduce false-positive predictions. Pretraining machine learning models on large clinical or biomedical domain knowledge data sets could significantly enhance the identification of positive relations and reduce false positives, and we can better understand the contextual nuances surrounding relationships to improve our model’s accuracy by leveraging domain-specific insights. In future endeavors, we plan to utilize corpora trained in biomedical and clinical texts, such as BioBERT37 and ClinicalBERT,38 to incorporate domain knowledge.
Conclusion
This study effectively demonstrates the development and application of an AI model adept at detecting irAEs from clinical narratives in EHRs. Use of this model substantially streamlined the process of identifying patient-level irAEs, showing particularly strong sensitivity, a pivotal factor for effective detection. This impressive performance underpins its potential applicability in real-world settings to aid clinicians in the accurate identification of irAEs from patients’ EHRs. The model also drastically reduced the workload associated with the manual review of clinical notes, efficiently sifting out notes lacking related irAE information and achieving 95% sensitivity in the external validation set.
Acknowledgments
This study is a part of the PhD thesis4 of Md Muntasir Zitu, the lead author of this manuscript.
Funding Statement
Our work is not funded.
Data Sharing Statement
The datasets featured in this article are currently unavailable for public access, as they include clinical notes from Electronic Health Records (EHRs). Consequently, these datasets are not published at this time. For inquiries regarding access to the datasets, please contact LL, Lang.Li@osumc.edu.
Ethical Statement and Institutional Review Board
The study received IRB (#2020C0145) approval for secondary data analysis of existing data, which contains identifiable patient data. As the data were accessed and analyzed in a secure environment in the College of Medicine at the Ohio State University, the loss of confidentiality risk was minimized. Because this study analyzes a limited set of variables and patients from electronic medical records, according to IRB approval, it does not need informed consent from patients. Our study complies with the Declaration of Helsinki.
Disclosure
The author(s) report no conflicts of interest in this work.
References
- 1.Zitu MM, Zhang S, Owen DH, Chiang C, Li L. Generalizability of machine learning methods in detecting adverse drug events from clinical narratives in electronic medical records. Front Pharmacol. 2023;14:1218679. doi: 10.3389/fphar.2023.1218679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poudel DR, Acharya P, Ghimire S, Dhital R, Bharati R. Burden of hospitalizations related to adverse drug events in the USA: a retrospective analysis from a large inpatient database. Pharmacoepidemiol Drug Saf. 2017;26(6):635–641. doi: 10.1002/pds.4184 [DOI] [PubMed] [Google Scholar]
- 3.Binkheder S, Wu HY, Quinney SK, et al. PhenoDEF: a corpus for annotating sentences with information of phenotype definitions in biomedical literature. J Biomed Semantics. 2022;13(1):17. doi: 10.1186/s13326-022-00272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zitu MM Adverse Drug Event Detection from Clinical Narratives of Electronic Medical Records Using Artificial Intelligence. The Ohio State University; Accessed Dec 17, 2024. Available from: http://rave.ohiolink.edu/etdc/view?acc_num=osu168976942934742. [Google Scholar]
- 5.Watanabe JH, McInnis T, Hirsch JD. Cost of prescription drug-related morbidity and mortality. Ann Pharmacother. 2018;52(9):829–837. doi: 10.1177/1060028018765159 [DOI] [PubMed] [Google Scholar]
- 6.Rommers MK, Teepe-Twiss IM, Guchelaar HJ. Preventing adverse drug events in hospital practice: an overview. Pharmacoepidemiol Drug Saf. 2007;16(10):1129–1135. doi: 10.1002/pds.1440 [DOI] [PubMed] [Google Scholar]
- 7.Handler SM, Altman RL, Perera S, et al. A systematic review of the performance characteristics of clinical event monitor signals used to detect adverse drug events in the hospital setting. J Am Med Inform Assoc. 2007;14(4):451–458. doi: 10.1197/jamia.M2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushal R, Jha AK, Franz C, et al. Return on investment for a computerized physician order entry system. J Am Med Inform Assoc. 2006;13(3):261–266. doi: 10.1197/jamia.M1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauken KE, Dougan M, Rose NR, Lichtman AH, Sharpe AH. Adverse events following cancer immunotherapy: obstacles and opportunities. Trends Immunol. 2019;40(6):511–523. doi: 10.1016/j.it.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med. 2018;378(2):158–168. doi: 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- 13.Haanen J, Carbonnel F, Robert C, et al. Management of toxicities from immunotherapy: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(suppl 4):iv119–iv142. doi: 10.1093/annonc/mdx225 [DOI] [PubMed] [Google Scholar]
- 14.Puzanov I, Diab A, Abdallah K, et al. Managing toxicities associated with immune checkpoint inhibitors: consensus recommendations from the Society for Immunotherapy of Cancer (SITC) Toxicity Management Working Group. J Immunother Cancer. 2017;5(1):95. doi: 10.1186/s40425-017-0300-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nashed A, Zhang S, Chiang C-W, et al. Comparative assessment of manual chart review and ICD claims data in evaluating immunotherapy-related adverse events. Cancer Immunol Immunother. 2021;2021:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman AB, Peterson KS, Alba PR, DuVall SL, Patterson OV. Detecting adverse drug events with rapidly trained classification models. Drug Saf. 2019;42(1):147–156. doi: 10.1007/s40264-018-0763-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Q, Ji Z, Li Z, et al. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J Am Med Inform Assoc. 2020;27(1):13–21. doi: 10.1093/jamia/ocz063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denny JC, Irani PR, Wehbe FH, Smithers JD, Spickard III A. The KnowledgeMap project: development of a concept-based medical school curriculum database. AMIA Annu Symp Proc. 2003;2003:195–199. [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19–24. doi: 10.1197/jamia.M3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc. 2014;21(5):858–865. doi: 10.1136/amiajnl-2013-002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dandala B, Joopudi V, Devarakonda M. Adverse drug events detection in clinical notes by jointly modeling entities and relations using neural networks. Drug Saf. 2019;42(1):135–146. doi: 10.1007/s40264-018-0764-x [DOI] [PubMed] [Google Scholar]
- 22.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med. 2018;1(1):18. doi: 10.1038/s41746-018-0029-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inform Assoc. 2020;27(1):3–12. doi: 10.1093/jamia/ocz166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagannatha A, Liu F, Liu W, Yu H. Overview of the First Natural Language Processing Challenge for Extracting Medication, Indication, and Adverse Drug Events from Electronic Health Record Notes (MADE 1.0). Drug Saf. 2019;42(1):99–111. doi: 10.1007/s40264-018-0762-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santini FC, Rizvi H, Plodkowski AJ, et al. Safety and efficacy of re-treating with immunotherapy after immune-related adverse events in patients with NSCLC. Cancer Immunol Res. 2018;6(9):1093–1099. doi: 10.1158/2326-6066.CIR-17-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamieson L, Forster MD, Zaki K, et al. Immunotherapy and associated immune-related adverse events at a large UK centre: a mixed methods study. BMC Cancer. 2020;20(1):743. doi: 10.1186/s12885-020-07215-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta S, Belouali A, Shah NJ, Atkins MB, Madhavan S. Automated identification of patients with immune-related adverse events from clinical notes using word embedding and machine learning. JCO Clin Cancer Inform. 2021;5(5):541–549. doi: 10.1200/CCI.20.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bui DD, Zeng-Treitler Q. Learning regular expressions for clinical text classification. J Am Med Inform Assoc. 2014;21(5):850–857. doi: 10.1136/amiajnl-2013-002411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–906. doi: 10.1093/nar/gkm958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freites-Martinez A, Santana N, Arias-Santiago S, Viera A. Using the Common Terminology Criteria for Adverse Events (CTCAE - Version 5.0) to evaluate the severity of adverse events of anticancer therapies. Actas Dermosifiliogr. 2021;112(1):90–92. doi: 10.1016/j.ad.2019.05.009 [DOI] [PubMed] [Google Scholar]
- 31.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts DR, Bahn V, Ciuti S, et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography. 2017;40(8):913–929. doi: 10.1111/ecog.02881 [DOI] [Google Scholar]
- 33.Qaiser S, Ali R. Text mining: use of TF-IDF to examine the relevance of words to documents. Int J Comput Appl. 2018;181:25–29. doi: 10.5120/ijca2018917395 [DOI] [Google Scholar]
- 34.Radzi SFM, Karim MKA, Saripan MI, Rahman MAA, Isa INC, Ibahim MJ. Hyperparameter tuning and pipeline optimization via grid search method and tree-based AutoML in breast cancer prediction. J Pers Med. 2021;11(10):978. doi: 10.3390/jpm11100978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung KM, Kao WC, Sun CL, Wang LL, Lin CJ. Radius margin bounds for support vector machines with the RBF kernel. Neural Comput. 2003;15(11):2643–2681. doi: 10.1162/089976603322385108 [DOI] [PubMed] [Google Scholar]
- 36.Hand DJ, Christen P, Kirielle NF. F*: an interpretable transformation of the F-measure. Mach Learn. 2021;110(3):451–456. doi: 10.1007/s10994-021-05964-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alsentzer E, Murphy JR, Boag W, et al. Publicly Available Clinical BERT Embeddings. arXiv. 2019. arXiv:1904.03323. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets featured in this article are currently unavailable for public access, as they include clinical notes from Electronic Health Records (EHRs). Consequently, these datasets are not published at this time. For inquiries regarding access to the datasets, please contact LL, Lang.Li@osumc.edu.