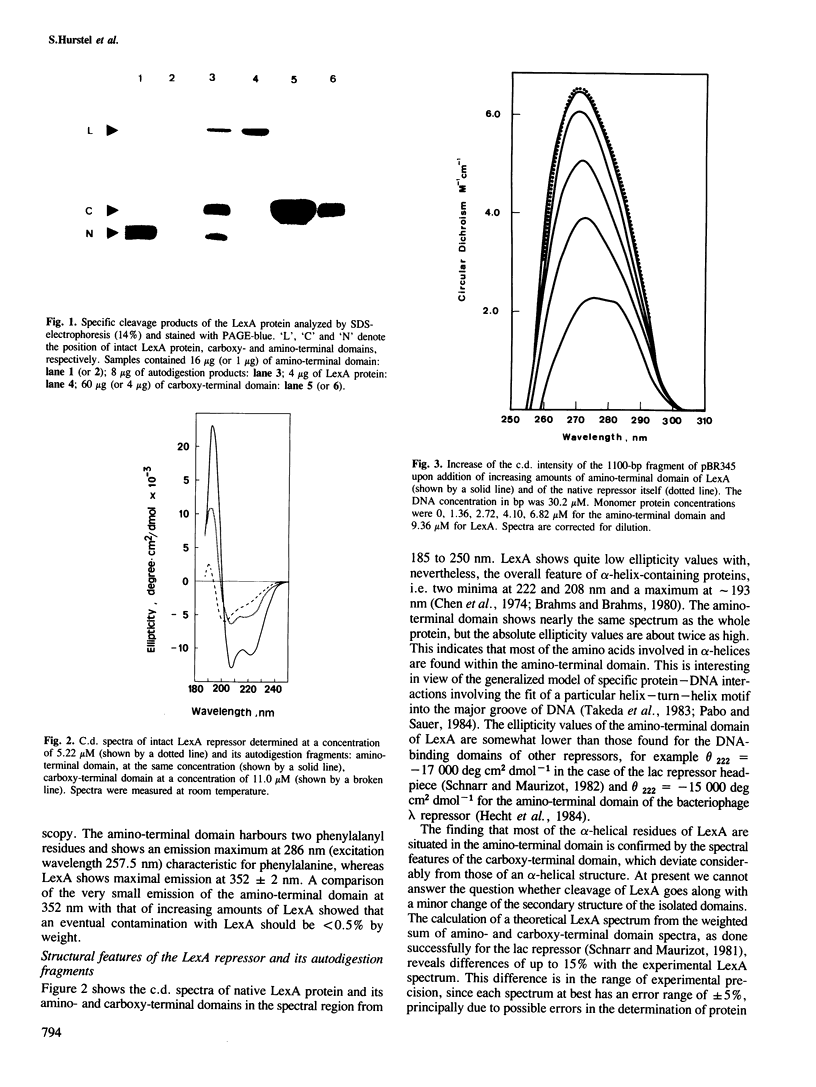
Abstract
Both the amino-terminal and the carboxy-terminal domain of the LexA repressor have been purified using the LexA protein autodigestion reaction at alkaline pH, which leads to the same specific products as the physiological RecA-catalyzed proteolysis of repressor. We show by circular dichroism (c.d) that, upon non-specific binding to DNA, the purified amino-terminal domain induces a very similar if not identical conformational change of the DNA as does the entire repressor. The positive c.d. signal increases approximately 3-fold if the DNA lattice is fully saturated with protein. Further, the amino-terminal domain of the LexA protein binds specifically to the operator of the recA gene, producing qualitatively the same effects on the methylation pattern of the guanine bases by dimethylsulfate as the entire repressor, consisting of a methylation inhibition effect at four distal operator guanines and a slight enhancement at the central bases. The spacing between these contacts suggests that LexA does not bind to the operator along the same face of the DNA helix. As shown by c.d. studies the amino-terminal domain harbours a substantial amount of residues in alpha-helical conformation, a prerequisite for DNA recognition via a helix--turn--helix structural motif as proposed for many other regulatory proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. A phage repressor-operator complex at 7 A resolution. Nature. 1985 Aug 15;316(6029):596–601. doi: 10.1038/316596a0. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Betlach M. C., Heyneker H. L., Shine J., Rodriguez R. L., Boyer H. W. Origin of replication of pBR345 plasmid DNA. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5265–5269. doi: 10.1073/pnas.74.12.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms S., Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J Mol Biol. 1980 Apr;138(2):149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A. P., Revzin A., von Hippel P. H. Molecular parameters characterizing the interaction of Escherichia coli lac repressor with non-operator DNA and inducer. Biochemistry. 1977 Nov 1;16(22):4757–4768. doi: 10.1021/bi00641a001. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T., Chau K. H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974 Jul 30;13(16):3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Eliason J. L., Weiss M. A., Ptashne M. NH2-terminal arm of phage lambda repressor contributes energy and specificity to repressor binding and determines the effects of operator mutations. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2339–2343. doi: 10.1073/pnas.82.8.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. H., Sturtevant J. M., Sauer R. T. Effect of single amino acid replacements on the thermal stability of the NH2-terminal domain of phage lambda repressor. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5685–5689. doi: 10.1073/pnas.81.18.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Nakatani T., Hase T., Matsubara H., Ogawa H. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell. 1981 Dec;27(3 Pt 2):515–522. doi: 10.1016/0092-8674(81)90393-7. [DOI] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Ogawa H. Nucleotide sequence of the lexA gene of E. coli. Cell. 1981 Mar;23(3):689–697. doi: 10.1016/0092-8674(81)90432-3. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger J. H., Elledge S. J., Walker G. C. Isolation and characterization of Tn5 insertion mutations in the lexA gene of Escherichia coli. J Bacteriol. 1983 Mar;153(3):1368–1378. doi: 10.1128/jb.153.3.1368-1378.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Edmiston S. H., Pacelli L. Z., Mount D. W. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Hill S. A. Deletions within a hinge region of a specific DNA-binding protein. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2301–2305. doi: 10.1073/pnas.82.8.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W., Yanisch-Perron C. R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W. The SOS regulatory system: control of its state by the level of RecA protease. J Mol Biol. 1983 Jul 15;167(4):791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- Markham B. E., Little J. W., Mount D. W. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981 Aug 25;9(16):4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizot J. C., Charlier M., Hélène C. Lac repressor binding to poly (d(A-T)). Conformational changes. Biochem Biophys Res Commun. 1974 Oct 8;60(3):951–957. doi: 10.1016/0006-291x(74)90406-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miki T., Ebina Y., Kishi F., Nakazawa A. Organization of the lexA gene of Escherichia coli and nucleotide sequence of the regulatory region. Nucleic Acids Res. 1981 Feb 11;9(3):529–543. doi: 10.1093/nar/9.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Matthews B. W. Many gene-regulatory proteins appear to have a similar alpha-helical fold that binds DNA and evolved from a common precursor. J Mol Evol. 1983;19(2):109–114. doi: 10.1007/BF02300748. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Krovatin W., Jeffrey A., Sauer R. T. The N-terminal arms of lambda repressor wrap around the operator DNA. Nature. 1982 Jul 29;298(5873):441–443. doi: 10.1038/298441a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Schlotmann M., Beyreuther K. Degradation of the DNA-binding domain of wild-type and i-d lac repressors in Escherichia coli. Eur J Biochem. 1979 Mar 15;95(1):39–49. doi: 10.1111/j.1432-1033.1979.tb12937.x. [DOI] [PubMed] [Google Scholar]
- Schnarr M., Daune M. Cooperative and salt-resistant binding of lexA protein to non-operator DNA. FEBS Lett. 1984 Jun 11;171(2):207–210. doi: 10.1016/0014-5793(84)80489-5. [DOI] [PubMed] [Google Scholar]
- Schnarr M., Durand M., Maurizot J. C. Nonspecific interaction of the lac repressor headpiece with deoxyribonucleic acid: fluorescence and circular dichroism studies. Biochemistry. 1983 Jul 19;22(15):3563–3570. doi: 10.1021/bi00284a005. [DOI] [PubMed] [Google Scholar]
- Schnarr M., Maurizot J. C. Secondary structure of the lac repressor headpiece. Possibilities and limitations of a joint infrared and circular dichroism study. Eur J Biochem. 1982 Nov 15;128(2-3):515–520. [PubMed] [Google Scholar]
- Schnarr M., Maurizot J. C. Unfolding of lac repressor and its proteolytic fragment by urea: headpieces stabilize the core within lac repressor. Biochemistry. 1981 Oct 13;20(21):6164–6169. doi: 10.1021/bi00524a039. [DOI] [PubMed] [Google Scholar]
- Schnarr M., Pouyet J., Granger-Schnarr M., Daune M. Large-scale purification, oligomerization equilibria, and specific interaction of the LexA repressor of Escherichia coli. Biochemistry. 1985 May 21;24(11):2812–2818. doi: 10.1021/bi00332a032. [DOI] [PubMed] [Google Scholar]
- Scopes R. K. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974 May;59(1):277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- Sprecher C. A., Baase W. A., Johnson W. C., Jr Conformation and circular dichroism of DNA. Biopolymers. 1979 Apr;18(4):1009–1019. doi: 10.1002/bip.1979.360180418. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Daune M., Schnarr M. Fluorescence study of the RecA-dependent proteolysis of LexA, the repressor of the SOS system in Escherichia coli. FEBS Lett. 1986 Feb 17;196(2):215–218. doi: 10.1016/0014-5793(86)80249-6. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Ohlendorf D. H., Anderson W. F., Matthews B. W. DNA-binding proteins. Science. 1983 Sep 9;221(4615):1020–1026. doi: 10.1126/science.6308768. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]