Abstract
Sodium butyrate (NaBu), well-known as a histone deacetylase inhibitor and for its capacity to impede cell growth, can enhance the production of a specific protein, such as an antibody, in recombinant Chinese hamster ovary (CHO) cell cultures. In this study, two CHO cell lines, namely K1 and DG44, along with their corresponding mAb-producing lines, K1-Pr and DG44-Pr, were cultivated with or without NaBu. A SWATH-based profiling method was employed to analyze the proteome. Cells cultured in the presence of NaBu exhibited a reduction in mitosis and gene expression, supported by their culture data demonstrating growth inhibition. The presence of NaBu corresponded to upregulation of intracellular trafficking and secretion pathways, aligned with an observed increase in mAb production, and was associated with an elevated glycosylation pathway and a slight alteration in the glycosylation profile of the mAbs. Increased fatty acid oxidation, redox interactions, and lipid biosynthesis were also observed and are likely attributable to the metabolism of NaBu. A comprehensive understanding of the systemic effects of NaBu will facilitate the discovery of strategies to enhance or prolong the productivity of CHO cells.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13568-024-01807-z.
Keywords: Butyric acid, CHO cells, SWATH-MS, Redox, mAbs
Introduction
Biologics are rapidly growing in market share with the sales of biologics expected to be dominant (Senior 2023). To improve the development and production processes, a lot of efforts are dedicated to the implementation of quality by design approach, leading to safer and more effective products of higher quality (Newcombe 2014; Ortiz-Enriquez et al. 2016; Rizzo et al. 2022). Recombinant monoclonal antibodies (mAbs) covered 36% of the recombinant biopharmaceutical proteins in 2021 and over 70% of those are produced in CHO cells (Lalonde and Durocher 2017). CHO cells produce high yields of recombinant proteins necessary for large-scale biotherapeutics manufacturing and are able to mimic human glycans in the production of glycoproteins (Lai et al. 2013).
Various research has been performed to increase the yield of CHO cells producing recombinant proteins, spanning from cell line engineering to treatment with specific compound (Lai et al. 2013). One of the molecules positively influencing CHO productivity is sodium butyrate (NaBu), the sodium salt of butyric acid. NaBu is known for its capacity to inhibit cell growth by arresting the cell cycle at the G0/G1 phase, thereby increasing the specific productivity of CHO cells which is reciprocal to cell growth and resulting in a rise of recombinant protein production. This is achieved by altering transcriptional regulation of certain genes through histone hyperacetylation (Davie 2003) as NaBu is a non-competitive inhibitor of class 1 and 2 histone deacetylases. However, NaBu also binds to pro-apoptotic proteins, leading to an activation of the apoptotic signals in CHO (Li et al. 2022) and multiple cancer cells (Hague et al. 1993; Jiang et al. 2012; Salimi et al. 2017). While a pro-autophagic effect was discovered, this was proposed as a pro-survival effect rather than a cell death mechanism of NaBu-treated cells to eliminate dysfunctional mitochondria (Lee and Lee 2012). A combination of NaBu treatment with apoptosis inhibitors as an attempt to mitigate the deleterious effects succeeded in promoting higher transgene expression compared to NaBu treatment alone (Li et al. 2022).
In an effort to improve the development and production pipeline in bioprocessing, we believe that a comprehensive omics-based systemic analysis of the effects of NaBu on CHO cells may facilitate greater understanding of its mechanism of action and open the path to further improvement in CHO production yield. Untargeted proteomics performed with sequential window acquisition of all theoretical mass spectra by mass spectrometry (SWATH-MS)– a data-independent acquisition (DIA) method– offers the chance to see thousands of proteins contemporaneously, allowing us to take a panoramic picture of the biology of the cells (Schubert et al. 2015; Ludwig et al. 2018). Subsequent pathway analyzes can identify essential leads towards higher NaBu-induced productivity, and reduce research and development time (Walker et al. 2017; Park et al. 2021). DIA-MS analysis is limited by the availability of a good quality spectral library, which is dependent on comprehensive data dependent acquisition and laborious curation. To support the application of DIA-MS for future research into recombinant biotherapeutic proteins production, our group has previously prepared extensive spectral libraries for CHO proteome and harvested cell culture fluid (Sim et al. 2020). In recent years, DIA-MS has been increasingly applied in bioprocess research to facilitate more efficient processes and production of safer biotherapeutics (Walker et al. 2017; Orellana et al. 2017; López-Pedrouso et al. 2021). In this paper DIA was used to understand the double effect of NaBu on mAb-producing CHO cells, on one side it promotes an increased yield of production and on the other it contributes to an accumulation of oxidative stress, leading to apoptosis.
Material and methods
Reagents and material
Parental cell line CHO-K1 and CHO-DG44 were purchased from Merck (SAFC, Hub Carlsbad CA) and Thermo Fisher Scientific (Grand Island, NY) respectively. Sequencing-grade porcine trypsin was purchased from Promega Corporation (Madison, WI), and Rapid PNGase F from New England Biolabs (Ipswich, MA). Halt™ protease inhibitor cocktail and Pierce™ BCA Protein Assay Kit were purchased from Thermo Fisher Scientific (Waltham, MA); Glycoworks RapiFluor-MS N-glycan Kit, RFMS-labeled dextran and 50 mM ammonium formate solution were from Waters Corporation (Milford, MA); S-trap™ mini spin columns from ProtiFi™ (Huntington, NY). Acetonitrile (ACN) was from Merck (Burlington, MA), and all other reagents were purchased from Sigma-Aldrich (St Louis, MO, US) unless stated otherwise.
Cell Culture
All cell lines were cultured in a fully defined protein-free culture medium in 125 mL disposable Erlenmeyer shake flasks (Corning, Acton, MA) and incubated on a shaker platform at 110 rpm in a humidified 37 °C/8% CO2 incubator. Viable cell density and viability were measured by Vi-CELL™ XR cell viability analyzer (Beckman Coulter, Brea, CA) as per manufacturer’s instructions. The trastuzumab-producing CHO-K1 and CHO-DG44 cells, which were in-house generated stable cell lines by Dr. Yang Yuan Sheng (Cell Line Development group), were treated with or without 1 mM NaBu for 48 h and subjected to SWATH-MS analysis.
Protein extraction and digestion
Cells were pelleted at 1,500 × g, washed thrice with ice-cold phosphate-buffered saline (PBS) and resuspended in cell lysis buffer (5% sodium dodecyl sulfate (SDS), 50 mM triethylammonium bicarbonate buffer (TEAB), pH 8.5, and Halt™ protease inhibitor cocktail). Cell lysates were further disrupted using an ultrasonic processor (Model: UP50H, Hielscher Inc., Teltow, DE) at 100% amplitude with 0.5 s pulse on and 0.5 s pulse off for 20 times on ice and clarified by high-speed centrifugation at 20,000 × g for 10 min. Protein concentration was determined using the Pierce™ BCA Protein Assay Kit according to manufacturer’s instructions. Subsequently, 200 µg of protein from each sample was digested with 1:25 trypsin to protein ratio on S-trap™ mini spin columns (ProtiFi™, Huntington, NY) according to manufacturer’s protocol, and peptide eluates were dried down using SpeedVac (Savant Instruments, Holbrook, NY).
SWATH-MS data-independent acquisition
SWATH-MS acquisition was performed on nanoAcquity UPLC system (Waters) with ACQUITY UPLC M-class BEH C18 peptide column (75 µm internal diameter (i.d.) × 200 mm length, 1.7 µm particle size, 130 Å pore size) coupled to a TripleTOF 6600 mass spectrometer (SCIEX, Framingham, MA). Indexed retention time (iRT) peptides from Biognosys AG (Schlieren, CH) were added to all samples for retention time normalisation following manufacturer’s instructions. The LC system was operated with buffer A (0.1% FA/water) and B (0.1% FA/ACN) at a flow rate of 300 nL/min. A two-step gradient (5–30/30–40% B) was performed over 106 min. The acquisition was performed with a 100-variable window scheme generated with the SWATH Variable Window Calculator 1.0 (SCIEX). An overlap of 1 m/z was set at the lower limit of each window. The MS1 scan range covered 400–1,250 m/z for 250 ms and MS2 spectra were acquired in high-sensitivity mode from 50 to 2,000 m/z for 30 ms, resulting in a total cycle time of 3.3 s. To maintain the similarity between data-dependent acquisition (DDA) and DIA, the collision energy (CE) equation was conserved (0.0625 × m/z–10.5) (Sim et al. 2020). The CE of a doubly charged precursor centered in the middle of the isolation window was applied to all the charge states, with a collision energy spread (CES) of 5 eV.
In-gel trypsin digest and glycan release
In-gel trypsin digestion of excised SDS-PAGE separated gel bands containing mAb protein was performed as previously described (Chng et al. 2015) and extracted peptides were evaporated to dryness in a SpeedVac. Glycans were released and labeled using GlycoWorks RapiFluor-MS N-glycan Kit according to manufacturer’s instructions. Peptides were re-solubilized in water and GlycoWorks Rapid Buffer and incubation with 1.2 μL Rapid PNGase F at 50 °C for 10 min. RapiFluor-MS (RFMS) labeling solution was added to cooled samples and left at room temperature for 5 min. Samples were cleaned using a GlycoWorks HILIC μElution Plate and reconstituted in 25% dimethylformamide (DMF)/52.5% ACN (v/v) in water.
HILIC-UPLC-FLR with ESI–MS
RFMS-labeled glycans were analyzed by HILIC-UPLC-FLR ESI–MS using a Waters ACQUITY UPLC H-Class coupled online to a Waters Xevo G2-S Q-TOF controlled by UNIFI Scientific Information System. RFMS-labeled glycans were injected onto an ACQUITY UPLC® Glycan BEH Amide Column (130 Å, 1.7 μm, 2.1 mm × 150 mm) at a flow rate of 0.4 mL/min and column temperature of 60 °C. Buffer A was 50 mM ammonium formate (pH 4.4) and Buffer B was 100% ACN. After an initial column equilibration of 2 min, glycans were separated using a linear gradient of 20%-49% of Buffer A for 40 min, increasing to 100% over 1.5 min, followed by 3 min washing. Fluorescence detection was used for glycan quantitation (Ex 265 nm/ Em 425 nm). For MS detection, samples were analyzed in sensitivity mode and spectra were acquired in positive ion mode with a full MS scan over a range of 400–2000 m/z and accumulation time of 1 s. The instrument conditions were as follows: 2.75 kV electrospray ionisation capillary voltage, 15 V sample cone voltage, 120 °C ion source temperature, 300 °C desolvation temperature and 800 L/h desolvation gas flow.
Construction of CHO global spectral library
Previously published DDA data (Sim et al. 2020) were submitted for database searches via Spectronaut v17 (Biognosys). The spectra were searched against Cricetulus griseus proteome from UniProt (UP000001075_10029, 1 sequence per protein (1SPP), updated at 28–11-2022) containing 23,883 entries. 60 DDA WIFF files from wild-type CHO cells were searched to create an ion library using the Pulsar engine. Peptide, protein, and peptide-spectrum match (PSM) false discovery rates (FDR) were set at 1%. Fragment ions m/z range was set between 100 and 1800, precursors were selected with a minimum of 3 and a maximum of 6 fragments per peptide. A maximum of 2 missed cleavages was permitted.
Data analysis
SWATH analysis was accomplished using Spectronaut 17 (Biognosys). The DIA spectra data were searched against the CHO global spectral library followed by C. griseus database (UniProt UP000001075_10029 1SPP). For identification, the precursor q-value cutoff was set at 0.01 and posterior error probability (PEP) was set to 0.2. The protein q-value cutoff at the experiment level was set at 0.01 and at the run level at 0.05. The run-wise PEP was set at 0.75. For quantitation, only precursors identified in at least 20% of the runs were retained with background signal selected as the imputation strategy for missing values. Differentially abundant candidates were tested with an unpaired t-test assuming equal variance, and a group-wise multiple testing correction was applied. The candidates were filtered for Absolute AVG Log2Ratio > 0.58, q-value < 0.05 and Unique Peptides > 2. Subsequent pathway analysis was performed using Cytoscape and its collection of applications: stringApp, EnrichmentMap and AutoAnnotate (Shannon et al., 2003).
The upregulated and downregulated candidates were analyzed separately for each condition of interest. The list of the differentially regulated proteins was queried in String with 0.4 confidence score cut-off against the C. griseus organism database. A network of enriched proteins was retrieved from String and a subnetwork was created with the core enriched proteins. Log2FC and q-values were imported in the node tables to visualize up- and downregulated proteins in the pathways. A String Enrichment analysis was performed on the subnetwork. The enrichment results were filtered for Gene Ontology (GO) processes and redundant terms were removed. The enrichment maps were calculated based on String Enrichment analysis, with similarity score cutoff set at 0.4 for downregulation and at 0.3 for upregulation. AvgLog2ratio was imported in the enrichment map. The AutoAnnotation app was used on the enrichment map to highlight the groups. The groups of interest were isolated for further functional GO processes terms enrichment analysis.
Data availability
The data that support the findings of this study are available on PRIDE with the following identifier PXD048575.
Results
NaBu's effect on cell viability, growth, and mAb production
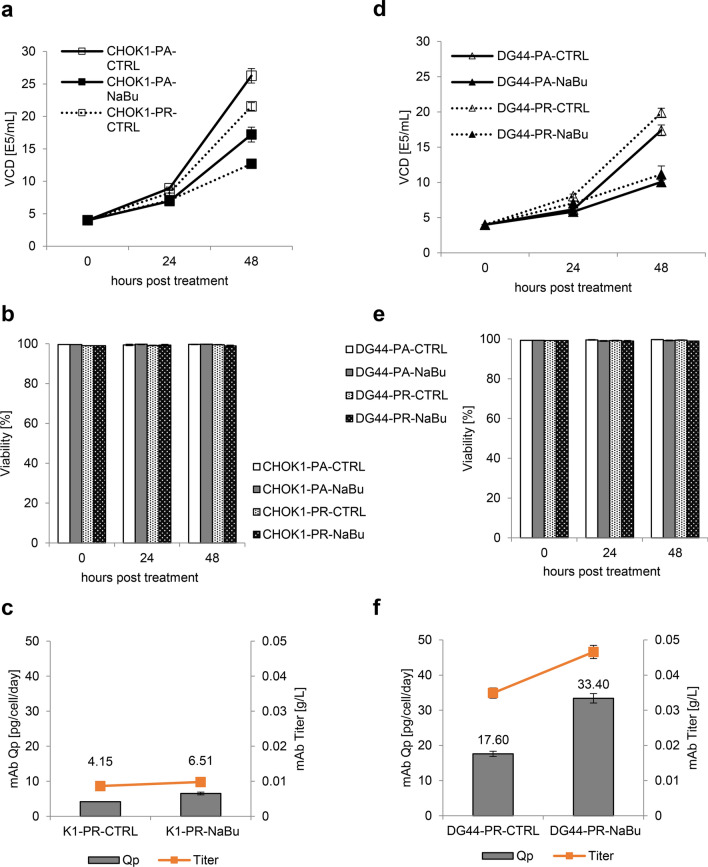
The effects of NaBu on CHO cells at a systemic level were studied by SWATH-MS-based untargeted analysis of the proteomes of two CHO cell lines, K1 and DG44: both untransfected parental (Pa) and mAb-producing (Pr) cells were cultured with or without NaBu supplementation. The viability of all cell lines assayed at 0, 24 and 48 h in culture was similar (Fig. 1B, E), but NaBu-treated cells were increasing at a slower rate at 24 h and 48 h compared to untreated control cells (Fig. 1A, D). These observations corroborated NaBu’s known function as a growth inhibitor (Davie 2003). NaBu is also known to enhance the synthesis of recombinant proteins (Han et al. 2005; Lee and Lee 2012; Hong et al. 2014), and this was reflected in NaBu-treated cells having augmented yield and titer compared to the control cells (Fig. 1C, F). A more marked effect upon NaBu treatment was observed in DG44-Pr compared to K1-Pr, with ~ 90% increase in specific productivity in DG44-Pr and ~ 57% in K1-Pr. The ability of NaBu to inhibit DNA replication and cell growth in CHO cells, and thereby increasing cellular resources for the synthesis of recombinant proteins was also shown by other groups (Hua et al. 2022). Furthermore, other means of arresting CHO cells in G0/G1 phases were also demonstrated to influence protein synthesis and increase recombinant protein production in CHO cells. Lu et al. (2023) showed the efficacy of apilimod in blocking the cell cycle in G0/G1 to favour protein production, while Lataster et al. (2023) promoted protein production by blocking the cells in G0/G1 using an optogenetic approach to influence p21, a cell cycle inhibitor. An additional confirmation of the improved productivity of CHO cells in G0/G1 phase was given by the work of Dutton et al. (2006). The positive link between G0/G1 cell cycle phase and productivity may also explain in part the larger increase in specific productivity observed in DG44-Pr compared to K1-Pr cells upon NaBu treatment, as we have previously detected lower expression of cell-cycle progression genes and proteins (and hence greater sensitivity to the disruptive effect of NaBu on cell replication) in DG44 cell line (Lakshmanan et al. 2019).
Fig. 1.
Cell culture viability and yield. The effects of sodium butyrate (NaBu) on A,D viable cell density, B,E viability and C,F mAb production yield in CHO-K1 and CHO-DG44 parental ad production cells. Viable cell density and viability were analyzed by trypan blue exclusion and measured at day 0, 1 and 2 after inoculation. The titer (g/L) and specific productivity (pg/cell/day) were determined by ELISA assay. All error bars showed the standard deviation of three biological replicates
PCA and analysis workflow
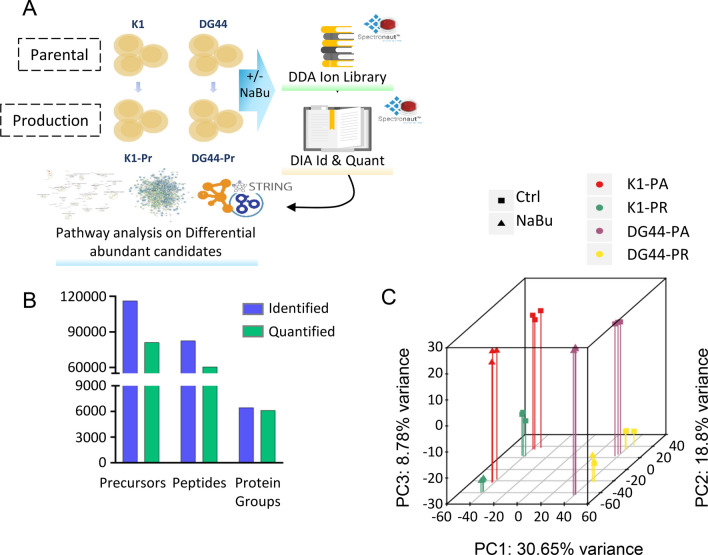
SWATH data analysis was performed using Spectronaut 17 and a CHO ion library of 9,367 proteins generated by reprocessing data previously acquired at our laboratory by Sim et al. (2020) (Fig. 2A). Analysis of all samples yielded 6,145 (95.1%) of quantifiable proteins at a q-value cutoff of 0.01 (Fig. 2B). Principal component analysis (PCA) in Fig. 2C showed distinct clustering for each sample, with cell lineage primarily contributing to variation in PC1 (30.65%), NaBu treatment in PC2 (18.80%) and mAb production in PC3 (8.78%), and a total variance of 58.23% explained by the three PCs. Differentially expressed proteins between untreated control and NaBu-treated cells were subsequently subjected to pathway analysis using Cytoscape (Fig. 2A). Similar pathways were being affected following NaBu treatment in the parental (Suppl. Table S1 and S2) and their corresponding mAb-producing cells (Figs. 3 and 4), with the latter showing more marked changes and thus focused upon to illustrate the effects of NaBu treatment in K1 and DG44 cell lines.
Fig. 2.
CHO Treatment and SWATH analysis. A Parental (K1-Pa and DG44-Pa) and production (K1-Pr and DG44-Pr) CHO cell lines untreated or treated with NaBu. 60 CHO global library DDA runs were reprocessed with Spectronaut 17 to create the ion library. CHO SWATH runs were analyzed in Spectronaut 17 and differentially abundant candidates were selected. A pathways regulation analysis was performed using String and Gene ontology through Cytoscape. B Precursors, peptides and proteins identified at 0.01 q-value (Blue) and quantified (Green). C: 3D principal component analysis (PCA) of the runs calculated by PCAGO (https://github.com/rumangerst/pcago-unified) (Holzer, 2019). Component 1 covers 30.65% of variance, component 2 the 18.8% and component 3 the 8.78%, explaining the 58.23% of the total variance
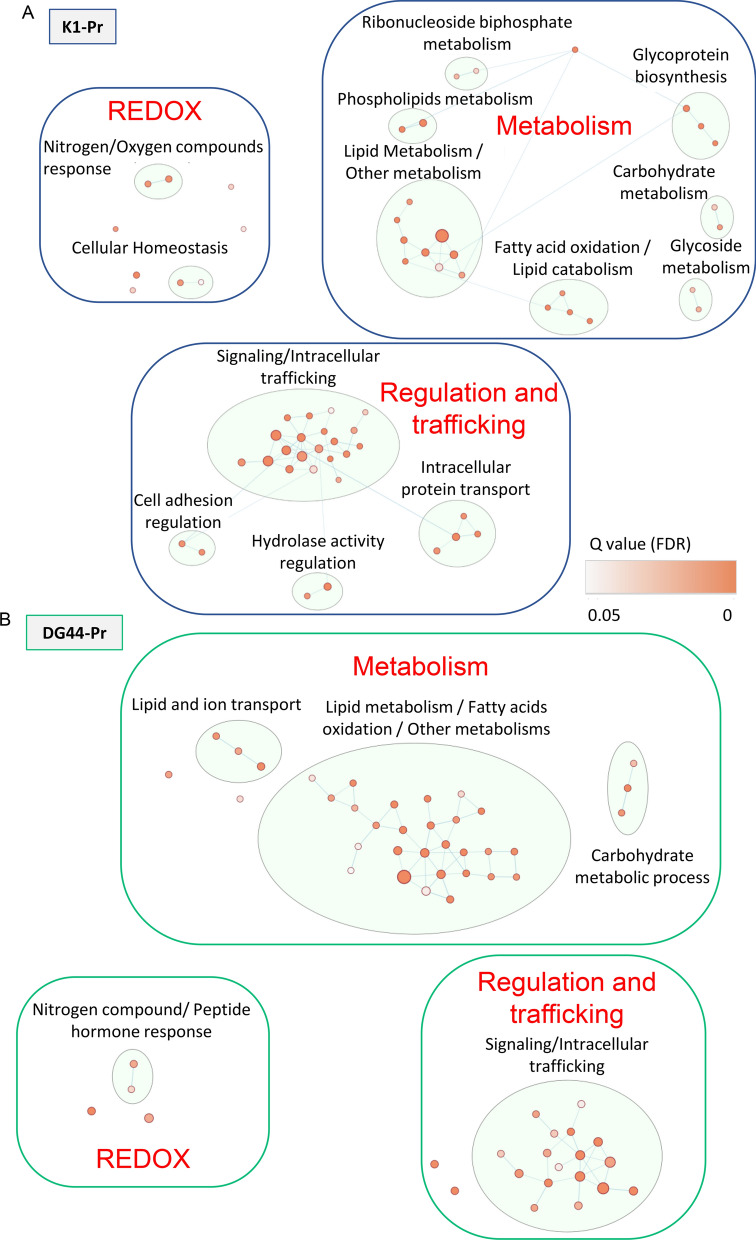
Fig. 3.
NaBu upregulated Lipid metabolism, REDOX exchanges and Signalling/Intracellular trafficking in CHO cells. Differentially upregulated abundant proteins were imported into Cytoscape. A network was retrieved through the StringApp and a String functional enrichment was performed. An enrichment map was created using the Enrichment Map app on the enriched GO biological processes. Clusters of interest are reported for A K1-Pr and for B DG44-Pr. The dimension of the nodes is determined by the number of the proteins representing the process and the color represents the q-value (FDR). The node cutoff was set at q-value 0.05 and the similarity cutoff (Edge cutoff) was set at 0.3. Nodes details are reported in Table 1 and 2
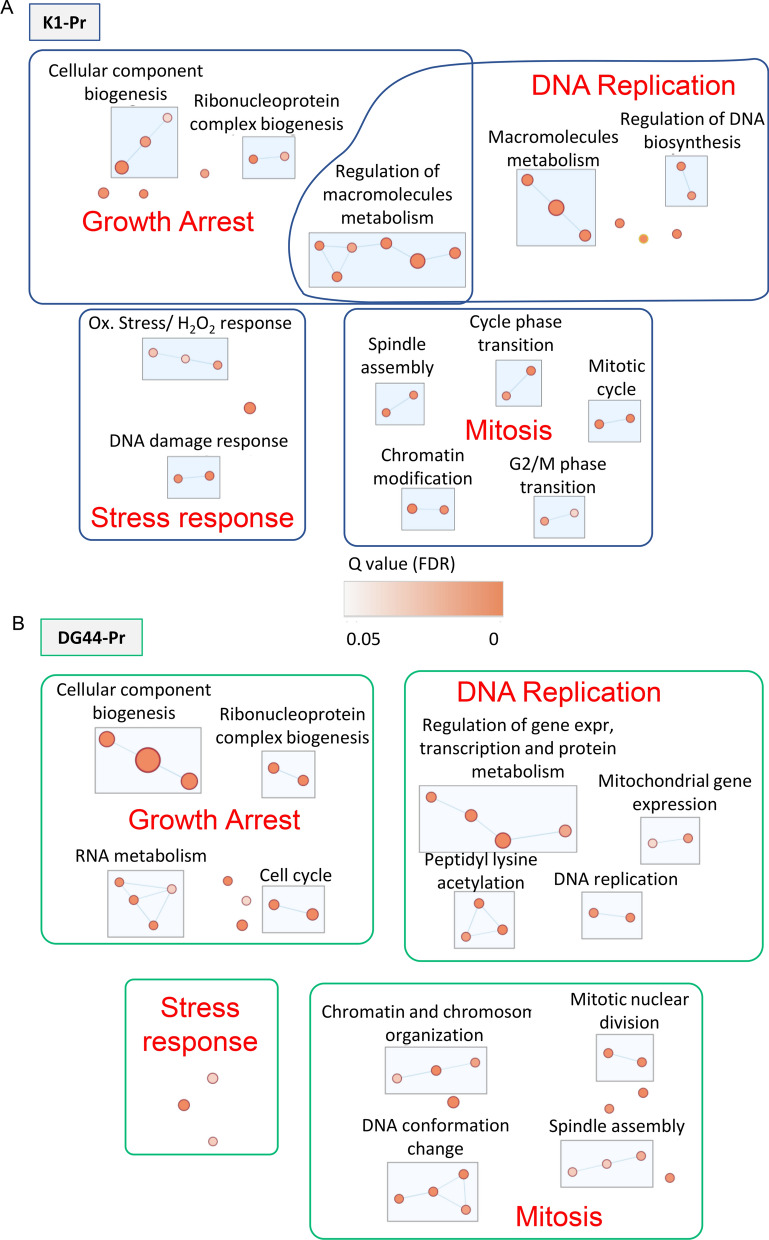
Fig. 4.
NaBu induced inhibition of gene expression, cell cycle, mitosis and gene expression-related stress response in CHO cells. Differentially downregulated proteins were imported into Cytoscape. A network was retrieved through the StringApp and a String functional enrichment was performed. An enrichment map was created on the enriched GO biological processes. Clusters of interest are reported for A K1-Pr and for B DG44-Pr. The dimension of the nodes is determined by the number of the proteins representing the process and the color represents the q-value (FDR). The node cutoff was set at q-value 0.05 and the similarity cutoff (Edge cutoff) was set at 0.4. Nodes details are reported in Table 3 and 4
NaBu upregulated pathways and areas
NaBu treatment in mAb-producing cells upregulated pathways which converged in three macro-areas: metabolism, intracellular trafficking, and oxidation–reduction (redox) (Table 1, Fig. 3). Within metabolism, the more pronounced changes were observed in lipid metabolism and fatty acids oxidation for both the producing cell lines. DG44-Pr hits a higher number of GO processes and proteins compared to K1-Pr. The lipid and ion transport cluster suggests increased lipid movements in or across the cells under the influence of NaBu. Other metabolic pathways were also upregulated in DG44-Pr under NaBu. K1-Pr shows a distinct cluster of upregulation of glycoprotein biosynthesis, in accordance with increased mAb production shown in Fig. 1C. The positively regulated N-linked glycosylation pathway of K1-Pr was shown in Suppl. Fig. S1A. The same pathway was statistically filtered out in DG44-Pr, but the most upregulated protein (phosphomannomutase—G3IJJ7) was upregulated in both the cell lines. Several studies have shown that the addition of NaBu in cell culture resulted in the alteration of glycosylation on therapeutic proteins (Rodriguez et al. 2005; Cabrera et al. 2005; Hong et al. 2014). A typical N-glycan profile was shown here with a high percentage of bi-antennary glycans and core fucosylation in the mAbs produced from K1-Pr and DG44-Pr cells (Suppl. Fig. S1B, C). Upon NaBu treatment, the N-glycan profile of mAbs showed slightly higher galactosylation and lower fucosylation compared to the products from untreated cultures (Suppl. Fig. S1D, E). Nevertheless, the overall pattern of glycosylation remained similar in the presence or absence of NaBu. Finally, glycoside metabolism and ribonucleoside bisphosphate metabolism clusters were upregulated in K1-Pr, with the main process of the latter upregulated in DG44-Pr too.
Table 1.
Upregulated pathways in NaBu-treated CHO K1-Pr cells
| K1-Pr | |||||
|---|---|---|---|---|---|
| Primary area | Cluster | GO process | Description | FDR q-value | Protein number |
| Metabolism | Lipid Metabolism / Other metabolism | GO:0044281 | Small molecule metabolic process | 1.9E-15 | 121 |
| GO:0008152 | Metabolic process | 9.8E-12 | 365 | ||
| GO:0006629 | Lipid metabolic process | 1.1E-07 | 79 | ||
| GO:0009058 | Biosynthetic process | 6.3E-07 | 131 | ||
| GO:0032787 | Monocarboxylic acid metabolic process | 6.7E-05 | 39 | ||
| GO:0008610 | Lipid biosynthetic process | 7.2E-04 | 34 | ||
| GO:0044283 | Small molecule biosynthetic process | 7.3E-03 | 33 | ||
| GO:1,901,362 | Organic cyclic compound biosynthetic process | 1.1E-02 | 51 | ||
| GO:1,901,360 | Organic cyclic compound metabolic process | 3.9E-02 | 121 | ||
| Fatty acid oxidation / Lipid catabolism | GO:0044242 | Cellular lipid catabolic process | 1.3E-04 | 21 | |
| GO:0006520 | Cellular amino acid metabolic process | 5.3E-04 | 24 | ||
| GO:0046395 | Carboxylic acid catabolic process | 5.3E-04 | 22 | ||
| GO:0019395 | Fatty acid oxidation | 5.4E-03 | 11 | ||
| Glycoprotein Biosynthesis | GO:1,901,135 | Carbohydrate derivative metabolic process | 1.5E-09 | 72 | |
| GO:0009101 | Glycoprotein biosynthetic process | 1.1E-05 | 30 | ||
| GO:0006487 | Protein n-linked glycosylation | 6.0E-04 | 12 | ||
| Phospholipids Metabolism | GO:0006793 | Phosphorus metabolic process | 1.1E-05 | 104 | |
| GO:0019637 | Organophosphate metabolic process | 1.9E-04 | 51 | ||
| Carbohydrate metabolism | GO:0019318 | Hexose metabolic process | 9.6E-03 | 14 | |
| GO:0005975 | Carbohydrate metabolic process | 2.9E-02 | 29 | ||
| Glycoside metabolism | GO:1,901,657 | Glycosyl compound metabolic process | 1.9E-02 | 12 | |
| GO:0016137 | Glycoside metabolic process | 2.6E-02 | 5 | ||
| Ribonucleoside biphosphate metabolism | GO:0015936 | Coenzyme a metabolic process | 2.0E-02 | 5 | |
| GO:0033875 | Ribonucleoside bisphosphate metabolic process | 3.2E-02 | 11 | ||
| Individual nodes of interest | GO:0055086 | Nucleobase-containing small molecule metabolic process | 3.6E-04 | 35 | |
| Regulation and trafficking | Signaling / Intracellular trafficking | GO:0051179 | Localization | 1.5E-09 | 242 |
| GO:0032879 | Regulation of localization | 8.0E-07 | 147 | ||
| GO:0065008 | Regulation of biological quality | 3.2E-06 | 185 | ||
| GO:0016043 | Cellular component organization | 4.4E-06 | 233 | ||
| GO:0060627 | Regulation of vesicle-mediated transport | 2.0E-04 | 42 | ||
| GO:0010941 | Regulation of cell death | 2.1E-04 | 89 | ||
| GO:0051128 | Regulation of cellular component organization | 5.3E-04 | 120 | ||
| GO:0032880 | Regulation of protein localization | 6.6E-04 | 58 | ||
| GO:0043065 | Positive regulation of apoptotic process | 8.6E-04 | 43 | ||
| GO:0060341 | Regulation of cellular localization | 9.5E-04 | 62 | ||
| GO:0051050 | Positive regulation of transport | 1.3E-03 | 57 | ||
| GO:0048585 | Negative regulation of response to stimulus | 2.3E-03 | 80 | ||
| GO:0022607 | Cellular component assembly | 4.2E-03 | 104 | ||
| GO:0048522 | Positive regulation of cellular process | 6.1E-03 | 226 | ||
| GO:0023051 | Regulation of signaling | 9.7E-03 | 140 | ||
| GO:0080134 | Regulation of response to stress | 1.2E-02 | 64 | ||
| GO:0030100 | Regulation of endocytosis | 1.5E-02 | 19 | ||
| GO:0050727 | Regulation of inflammatory response | 2.9E-02 | 23 | ||
| GO:0050793 | Regulation of developmental process | 3.6E-02 | 116 | ||
| GO:1,903,530 | Regulation of secretion by cell | 4.4E-02 | 37 | ||
| Intracellular protein transport | GO:0008104 | Protein localization | 4.5E-07 | 115 | |
| GO:0006886 | Intracellular protein transport | 7.4E-04 | 51 | ||
| GO:0033365 | Protein localization to organelle | 4.3E-03 | 41 | ||
| GO:0016192 | Vesicle-mediated transport | 4.7E-03 | 67 | ||
| Cell adhesion regulation | GO:0030155 | Regulation of cell adhesion | 3.4E-03 | 44 | |
| GO:0051270 | Regulation of cellular component movement | 4.2E-03 | 58 | ||
| Hydrolase activity regulation | GO:0050790 | Regulation of catalytic activity | 1.1E-04 | 114 | |
| GO:0051345 | Positive regulation of hydrolase activity | 6.2E-03 | 42 | ||
| Redox | Nitrogen/Oxygen compounds response | GO:1,901,700 | Response to oxygen-containing compound | 4.2E-03 | 75 |
| GO:1,901,698 | Response to nitrogen compound | 6.3E-03 | 55 | ||
| Cellular homeostasis | GO:0019725 | Cellular homeostasis | 7.0E-03 | 48 | |
| GO:0070838 | Divalent metal ion transport | 4.6E-02 | 21 | ||
| Individual nodes of interest | GO:0055114 | Oxidation–reduction process | 4.4E-06 | 67 | |
| GO:0036295 | Cellular response to increased oxygen levels | 8.3E-03 | 5 | ||
| GO:2,001,233 | Regulation of apoptotic signaling pathway | 3.1E-02 | 26 | ||
| GO:0010508 | Positive regulation of autophagy | 3.2E-02 | 13 | ||
| GO:0045454 | Cell redox homeostasis | 4.0E-02 | 7 | ||
The table describes in detail the areas influenced by NaBu in Fig. 3A. Clusters characterize the different areas, and each cluster is described by the GO processes supporting the upregulation. For each GO process the description, the number of the proteins and the FDR corrected q-value is reported
Intracellular trafficking is supported by many interconnected GO processes grouping for the common features of signaling and intracellular trafficking. This result agreed with Fig. 1C, highlighting increased secretory activity from augmented mAb production. At the same time, it revealed the possible effects of autophagy as well, evidenced by increased vacuole mobilization in the cells. Increased production of mAb and metabolic activities by the treatment promoted multiplication of redox exchanges inside the cells, especially those involving the endoplasmic reticulum (ER) and mitochondria (Ha et al. 2018). The redox cluster showed activation of the response mechanism to stresses in the cells for both nitrogenous compounds and hydrogen peroxide. The enrichment of the single GO processes with their FDR values for both DG44-Pr and K1-Pr is reported in Table 1.
NaBu downregulated pathways and areas
NaBu was not only inducing an upregulation of several pathways, but also a downregulation of other pathways, as shown in Fig. 4. The main downregulatory effects of NaBu were clustered into four areas of potential interest: DNA replication, mitosis, growth arrest and stress response. Both cell lines shared the ribonucleoprotein complex biogenesis cluster, highlighting a common propensity of NaBu treatment to reduce the biosynthesis of constituent macromolecules for other cellular activities. This was complemented by a decrease in proteins involved in cellular component biogenesis for K1-Pr and in proteins important for the cell cycle in DG44-Pr. Taken together, they indicated a growth arrest of the NaBu-treated cells. While the apparent effect of NaBu on treated CHO cells was an increase in mAb production, de facto, it was exerting its effect at the level of DNA replication (Lee and Lee 2012; Jiang et al. 2012; Li et al. 2022). In the analyzed cell lines, NaBu induced an inhibition of gene expression and mitotic mechanisms, hinting that the cells were stranded in the G1 phase (Fig. 4). Our pathway analysis confirms the known effects of NaBu on the cell cycle (Semaan et al. 2020; Avello et al. 2022). The arrest of the cell cycle is compatible with the effects seen on CHO cell viability and mAb production. High cell viability shown in Fig. 1 confirmed that the cells were viable at the moment of the experiment; the lower number of cells compared to the control was due to the growth arrest as seen in the pathway analysis. The augmented mAb production may be due to increased availability of resources with the blocking of the cell cycle and the consequent redirection of cellular machinery for protein production. A similar behaviour was observed when the cell cycle was arrested by thymidine or NaBu (Al-Rubeai et al. 1992; Avello et al. 2022). The downregulation of the DNA replication was supported by DG44-Pr data, where different processes clustered in the peptidyl lysine acetylation group. The downregulation of this GO process corresponded to greater histone acetylation due to the inhibition of histone deacetylases, and increased acetylation of histones is known to alter DNA replication (Kruh 1981; Davie 2003). Western blot of control and NaBu-treated K1 and DG44 cells showed a marked increase in the acetylation of Histone H4 upon NaBu treatment (Suppl. Fig. S2), thus confirming the known effect of NaBu on cells and supporting the inhibition of DNA replication (Tables 2, 3 and 4).
Table 2.
Upregulated pathways in NaBu-treated CHO DG44-Pr cells
| DG44-Pr | |||||
|---|---|---|---|---|---|
| Primary area | Cluster | GO process | Description | FDR q-value | Protein number |
| Metabolism | Lipid metabolism / Fatty acids oxidation / Other metabolisms | GO:0044281 | Small molecule metabolic process | 2.3E-15 | 98 |
| GO:0008152 | Metabolic process | 2.7E-10 | 273 | ||
| GO:0006629 | Lipid metabolic process | 8.7E-07 | 62 | ||
| GO:0009056 | Catabolic process | 1.3E-06 | 85 | ||
| GO:0044283 | Small molecule biosynthetic process | 1.7E-05 | 34 | ||
| GO:1,901,135 | Carbohydrate derivative metabolic process | 2.2E-05 | 48 | ||
| GO:0019752 | Carboxylic acid metabolic process | 2.8E-05 | 45 | ||
| GO:0044242 | Cellular lipid catabolic process | 1.6E-04 | 18 | ||
| GO:0009058 | Biosynthetic process | 2.2E-04 | 93 | ||
| GO:0019637 | Organophosphate metabolic process | 3.0E-04 | 41 | ||
| GO:0008610 | Lipid biosynthetic process | 3.2E-04 | 29 | ||
| GO:0044282 | Small molecule catabolic process | 3.2E-04 | 24 | ||
| GO:1,901,362 | Organic cyclic compound biosynthetic process | 6.4E-04 | 45 | ||
| GO:1,901,136 | Carbohydrate derivative catabolic process | 1.5E-03 | 14 | ||
| GO:1,901,615 | Organic hydroxy compound metabolic process | 2.0E-03 | 26 | ||
| GO:0006694 | Steroid biosynthetic process | 4.9E-03 | 11 | ||
| GO:0072521 | Purine-containing compound metabolic process | 6.0E-03 | 21 | ||
| GO:0006631 | Fatty acid metabolic process | 6.0E-03 | 21 | ||
| GO:0016125 | Sterol metabolic process | 6.0E-03 | 12 | ||
| GO:0046165 | Alcohol biosynthetic process | 7.1E-03 | 11 | ||
| GO:0046434 | Organophosphate catabolic process | 1.1E-02 | 12 | ||
| GO:0072523 | Purine-containing compound catabolic process | 1.9E-02 | 7 | ||
| GO:0019395 | Fatty acid oxidation | 3.5E-02 | 8 | ||
| GO:0046475 | Glycerophospholipid catabolic process | 4.0E-02 | 5 | ||
| GO:1,901,360 | Organic cyclic compound metabolic process | 4.3E-02 | 92 | ||
| GO:0033875 | Ribonucleoside bisphosphate metabolic process | 4.4E-02 | 9 | ||
| GO:0015936 | Coenzyme a metabolic process | 4.8E-02 | 4 | ||
| Carbohydrate metabolic process | GO:0005975 | Carbohydrate metabolic process | 3.8E-04 | 29 | |
| GO:0044262 | Cellular carbohydrate metabolic process | 5.7E-03 | 14 | ||
| GO:0019318 | Hexose metabolic process | 2.3E-02 | 11 | ||
| Lipid and ion transport | GO:0006811 | Ion transport | 1.5E-03 | 56 | |
| GO:0010876 | Lipid localization | 5.4E-03 | 22 | ||
| GO:0015711 | Organic anion transport | 1.3E-02 | 24 | ||
| Individual nodes of interest | GO:0016192 | Vesicle-mediated transport | 4.0E-04 | 57 | |
| GO:0019216 | Regulation of lipid metabolic process | 1.1E-02 | 21 | ||
| GO:0045923 | Positive regulation of fatty acid metabolic process | 3.7E-02 | 6 | ||
| Regulation and trafficking | Signaling / Intracellular trafficking | GO:0051179 | Localization | 5.1E-11 | 193 |
| GO:0032879 | Regulation of localization | 1.2E-06 | 115 | ||
| GO:0008104 | Protein localization | 5.8E-06 | 87 | ||
| GO:0051128 | Regulation of cellular component organization | 2.0E-05 | 100 | ||
| GO:2,000,145 | Regulation of cell motility | 1.2E-04 | 49 | ||
| GO:0065008 | Regulation of biological quality | 2.2E-04 | 135 | ||
| GO:0010941 | Regulation of cell death | 7.5E-04 | 68 | ||
| GO:0050790 | Regulation of catalytic activity | 6.0E-03 | 81 | ||
| GO:0016043 | Cellular component organization | 1.1E-02 | 159 | ||
| GO:0060627 | Regulation of vesicle-mediated transport | 1.1E-02 | 29 | ||
| GO:0043065 | Positive regulation of apoptotic process | 1.3E-02 | 31 | ||
| GO:0048585 | Negative regulation of response to stimulus | 1.4E-02 | 59 | ||
| GO:0042592 | Homeostatic process | 1.8E-02 | 60 | ||
| GO:0051050 | Positive regulation of transport | 2.9E-02 | 40 | ||
| GO:0051345 | Positive regulation of hydrolase activity | 2.9E-02 | 31 | ||
| GO:0022603 | Regulation of anatomical structure morphogenesis | 4.4E-02 | 43 | ||
| GO:0032880 | Regulation of protein localization | 4.4E-02 | 39 | ||
| Individual nodes of interest | GO:0006928 | Movement of cell or subcellular component | 1.2E-03 | 60 | |
| Redox | Nitrogen compound / peptide hormone response | GO:1,901,698 | Response to nitrogen compound | 1.4E-02 | 42 |
| GO:0043434 | Response to peptide hormone | 3.4E-02 | 19 | ||
| Individual nodes of interest | GO:0055114 | Oxidation–reduction process | 3.5E-10 | 65 | |
| GO:0006950 | Response to stress | 1.5E-02 | 103 | ||
The table describes in detail the areas influenced by NaBu in Fig. 3B. Clusters characterize the different areas, and each cluster is described by the GO processes supporting the upregulation. For each GO process the description, the number of the proteins and the FDR corrected q-value is reported
Table 3.
Downregulated pathways in NaBu-treated CHO K1-Pr cells
| K1-Pr | |||||
|---|---|---|---|---|---|
| Primary area | Cluster | GO process | Description | FDR q-value | Protein number |
| Growth arrest | Cellular component biogenesis | GO:0071840 | Cellular component organization or biogenesis | 9.4E-05 | 170 |
| GO:0044085 | Cellular component biogenesis | 8.6E-03 | 82 | ||
| GO:0034622 | Cellular protein-containing complex assembly | 3.4E-02 | 33 | ||
| Ribonucleoprotein complex biogenesis | GO:0034470 | ncRNA processing | 2.2E-04 | 26 | |
| GO:0022613 | Ribonucleoprotein complex biogenesis | 2.1E-02 | 24 | ||
| Regulation of macromolecules metabolism | GO:0060255 | Regulation of macromolecule metabolic process | 7.0E-10 | 213 | |
| GO:0051246 | Regulation of protein metabolic process | 1.3E-04 | 102 | ||
| GO:0010605 | Negative regulation of macromolecule metabolic process | 1.4E-04 | 100 | ||
| GO:0051338 | Regulation of transferase activity | 8.9E-04 | 44 | ||
| GO:0044093 | Positive regulation of molecular function | 2.0E-03 | 65 | ||
| GO:0031401 | Positive regulation of protein modification process | 1.2E-02 | 48 | ||
| Individual nodes of interest | GO:0051301 | Cell division | 1.9E-03 | 29 | |
| GO:0009058 | Biosynthetic process | 3.3E-03 | 83 | ||
| GO:0033044 | Regulation of chromosome organization | 1.1E-02 | 21 | ||
| DNA replication | Macromolecules metabolism | GO:0043170 | Macromolecule metabolic process | 2.2E-21 | 240 |
| GO:0090304 | Nucleic acid metabolic process | 6.3E-16 | 110 | ||
| GO:0006464 | Cellular protein modification process | 9.0E-08 | 112 | ||
| Regulation of DNA biosynthesis | GO:0051052 | Regulation of DNA metabolic process | 2.0E-03 | 23 | |
| GO:2,000,278 | Regulation of DNA biosynthetic process | 7.4E-03 | 11 | ||
| Individual nodes of interest | GO:0006397 | mRNA processing | 2.9E-06 | 33 | |
| GO:0006259 | DNA metabolic process | 2.2E-05 | 40 | ||
| GO:0009057 | Macromolecule catabolic process | 9.1E-04 | 42 | ||
| Mitosis | Chromatin modification | GO:0051276 | Chromosome organization | 1.9E-06 | |
| GO:0016569 | Covalent chromatin modification | 3.5E-03 | |||
| Spindle assembly | GO:1,902,850 | Microtubule cytoskeleton organization involved in mitosis | 1.3E-03 | ||
| GO:0051225 | Spindle assembly | 3.5E-03 | 56 | ||
| Mitotic cycle | GO:0000278 | Mitotic cell cycle | 4.8E-06 | 23 | |
| GO:0140014 | Mitotic nuclear division | 3.1E-05 | 12 | ||
| Cycle phase transition | GO:0010564 | Regulation of cell cycle process | 5.1E-05 | 10 | |
| GO:1,901,990 | Regulation of mitotic cell cycle phase transition | 8.3E-03 | 36 | ||
| G2/M phase transition | GO:1,902,749 | Regulation of cell cycle G2/M phase transition | 1.1E-02 | 17 | |
| GO:0072425 | Signal transduction involved in G2 DNA damage checkpoint | 3.3E-02 | 36 | ||
| Stress response | Ox. stress / H2O2 response | GO:0006979 | Response to oxidative stress | 1.1E-02 | 17 |
| GO:0010035 | Response to inorganic substance | 2.5E-02 | 10 | ||
| GO:0042542 | Response to hydrogen peroxide | 3.2E-02 | 4 | ||
| DNA damage response | GO:0080135 | Regulation of cellular response to stress | 6.5E-05 | ||
| GO:2,001,020 | Regulation of response to DNA damage stimulus | 3.3E-03 | |||
| Individual nodes of interest | GO:0006950 | Response to stress | 1.5E-05 | ||
The table describes in detail the areas influenced by NaBu in Fig. 4A. Clusters characterize the different areas, and each cluster is described by the GO processes supporting the downregulation. For each GO process the description, the number of the proteins and the FDR corrected q-value is reported
Table 4.
Downregulated pathways in NaBu-treated CHO DG44-Pr cells
| DG44-Pr | |||||
|---|---|---|---|---|---|
| Primary area | Cluster | GO process | Description | FDR q-value | Protein number |
| Growth arrest | Cellular component biogenesis | GO:0071840 | Cellular component organization or biogenesis | 2.7E-15 | 214 |
| GO:0044260 | Cellular macromolecule metabolic process | 5.6E-13 | 185 | ||
| GO:0009987 | Cellular process | 2.8E-10 | 438 | ||
| RNA metabolism | GO:0034661 | ncRNA catabolic process | 1.0E-03 | 7 | |
| GO:0090501 | RNA phosphodiester bond hydrolysis | 2.1E-03 | 14 | ||
| GO:0016073 | snRNA metabolic process | 2.9E-03 | 7 | ||
| GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 3.0E-02 | 6 | ||
| Ribonucleoprotein complex biogenesis | GO:0034660 | ncRNA metabolic process | 2.3E-19 | 56 | |
| GO:0022613 | Ribonucleoprotein complex biogenesis | 1.5E-14 | 49 | ||
| Cell cycle | GO:0007049 | Cell cycle | 1.5E-13 | 79 | |
| GO:0051301 | Cell division | 6.8E-08 | 40 | ||
| Individual nodes of interest | GO:0051726 | Regulation of cell cycle | 3.3E-06 | 55 | |
| GO:0042273 | Ribosomal large subunit biogenesis | 1.7E-04 | 13 | ||
| GO:1,902,750 | Negative regulation of cell cycle G2/m phase transition | 3.4E-02 | 7 | ||
| DNA replication | Regulation of gene expr, transcription and protein metabolism | GO:0010629 | Negative regulation of gene expression | 1.2E-08 | 91 |
| GO:0051171 | Regulation of nitrogen compound metabolic process | 2.8E-05 | 185 | ||
| GO:0000122 | Negative regulation of transcription by RNA polymerase II | 2.9E-03 | 41 | ||
| GO:0032268 | Regulation of cellular protein metabolic process | 1.4E-02 | 90 | ||
| Peptidyl lysine acetylation | GO:0018205 | Peptidyl-lysine modification | 3.1E-06 | 26 | |
| GO:0016569 | Covalent chromatin modification | 1.4E-05 | 29 | ||
| GO:0018394 | Peptidyl-lysine acetylation | 8.4E-03 | 12 | ||
| Mitochondrial gene expression | GO:0140053 | Mitochondrial gene expression | 1.0E-02 | 10 | |
| GO:0000963 | Mitochondrial RNA processing | 3.4E-02 | 4 | ||
| DNA replication | GO:0033260 | Nuclear DNA replication | 3.0E-03 | 6 | |
| GO:0006260 | DNA replication | 5.2E-03 | 14 | ||
| Mitosis | DNA conformation change | GO:0071103 | DNA conformation change | 3.9E-04 | 22 |
| GO:0006333 | Chromatin assembly or disassembly | 1.4E-03 | 16 | ||
| GO:0070828 | Heterochromatin organization | 2.0E-03 | 9 | ||
| GO:0065004 | protein-DNA complex assembly | 9.8E-03 | 14 | ||
| Chromatin and chromosome organization | GO:0033044 | Regulation of chromosome organization | 2.2E-06 | 30 | |
| GO:2,001,251 | Negative regulation of chromosome organization | 1.1E-02 | 12 | ||
| GO:1,902,275 | Regulation of chromatin organization | 2.7E-02 | 14 | ||
| Spindle assembly | GO:0032465 | Regulation of cytokinesis | 1.6E-02 | 9 | |
| GO:0051255 | Spindle midzone assembly | 2.9E-02 | 4 | ||
| GO:0000915 | Actomyosin contractile ring assembly | 2.9E-02 | 3 | ||
| Mitotic nuclear division | GO:0051983 | Regulation of chromosome segregation | 1.4E-03 | 12 | |
| GO:0007088 | Regulation of mitotic nuclear division | 3.2E-03 | 14 | ||
| Individual nodes of interest | GO:0051276 | Chromosome organization | 1.7E-15 | 77 | |
| GO:0140014 | Mitotic nuclear division | 7.6E-08 | 21 | ||
| GO:1,902,850 | Microtubule cytoskeleton organization involved in mitosis | 5.3E-03 | 11 | ||
| GO:0000281 | Mitotic cytokinesis | 5.5E-03 | 9 | ||
| Stress response | Individual nodes of interest | GO:0033554 | Cellular response to stress | 2.6E-05 | 68 |
| GO:0000012 | Single strand break repair | 2.9E-02 | 4 | ||
| GO:0080134 | Regulation of response to stress | 3.1E-02 | 47 | ||
The table describes in detail the areas influenced by NaBu in Fig. 4B. Clusters characterize the different areas, and each cluster is described by the GO processes supporting the downregulation. For each GO process the description, the number of the proteins and the FDR corrected q-value is reported
From the results in both K1-Pr and DG44-Pr, a clear inhibition of mitotic activities in these CHO cells as a consequence of NaBu treatment was evidenced by multiple clusters of downregulated pathways (Fig. 4). Despite slight differences in the processes, there were 5 pathway clusters displaying a common behaviour. Pathways associated with mitotic nuclear division and chromatin and chromosome organisation/modification were inhibited in both cell lines upon NaBu treatment. Furthermore, diminished spindle assembly activity was reported for K1-Pr that could be coupled with actomyosin contractile ring inhibition. DG44-Pr exhibited diminished spindle assembly and heterochromatin formation, which is important for sister chromatid cohesion.
Stress response was the last downregulated macro area observed in NaBu-treated cells (Fig. 4). Two defined clusters denoting a minor response to DNA damage and a decreased response to oxidative stress/H2O2 were observed in K1-Pr cells. No cluster was observed in DG44-Pr, but the trend of decreased stress response was maintained by the single processes.
Discussion
This work aimed to decipher the systemic effects of NaBu on CHO cells. Understanding the problems arising from NaBu treatment can pave the road to new solutions, improving its beneficial effects or mitigating adverse outcomes from its administration to mAb-producing cells. Global proteome changes profiled by SWATH-MS, together with cell culture and product glycosylation data, were analyzed to obtain a systemic view of the pathways altered by NaBu in the cells. The spectral ion library was recompiled from previously acquired DDA runs (Sim et al. 2020) using the latest version of Spectronaut Pulsar engine with enhanced capabilities to improve the library without lowering the quality of the data. While this global library, built from existing DDA data of fractionated wild type CHO cell samples, provided an extensive coverage of the CHO proteome and allowed for a fast analysis of the experiment, constructing a project specific library from samples including all the experimental conditions will likely have a higher statistical power and may lead to better results (Ludwig et al. 2018).
The known effects of NaBu, inhibition of cell proliferation and influence on histone modification, shown in this DIA-based proteomics study corroborated with the transcriptomic analysis by Schulze et al. (2022), the gene expression analysis by Wippermann et al. (2017) and the DDA proteomics analysis by Müller et al. (2017). The nature of NaBu, being a short-chained fatty acid (SCFA), has to be taken into consideration in the interpretation of the metabolic upregulation observed. A disproportion between fatty acid uptake and oxidation can lead to the accumulation of fatty acids and lipids in cytosol (van Eunen et al. 2013). An abundance of long-chained fatty acids is known to induce a lipid overload effect (Nguyen et al. 2008; Alsabeeh et al. 2018). Fatty acids are processed through the beta-oxidation cycle, and they can further undergo a de novo lipid biogenesis phase leading to the accumulation of lipid droplets or secretion of lipoproteins (Menendez and Lupu 2007; Walther and Farese 2012; van Eunen et al. 2013). These biological activities are implicated with an upregulation of mitochondrial activities and oxidative processes in the cells. The increased redox exchanges, another cluster that we found to be upregulated, commonly provoke the accumulation of oxidative stress and development of mitochondrial dysfunction (Nakamura et al. 2012; Wang et al. 2013; Ezquerro et al. 2020; Rios-Morales et al. 2022). The machineries connected with FA (FA oxidation and lipids synthesis) are upregulated in the analysis. As seen, it resulted in a natural increase in redox exchanges and oxidative stress accumulation. At the same time, the hyperactive protein synthesis machinery of the ER contributed to a further accumulation of oxidative stress, which is another source of future ER stress (Ozcan et al. 2004; Ha et al. 2018; Ezquerro et al. 2020; Gast et al. 2021). Cellular stress response machinery such as the production of glutathione are used to manage redox stress and working to keep the situation under tolerable limits.
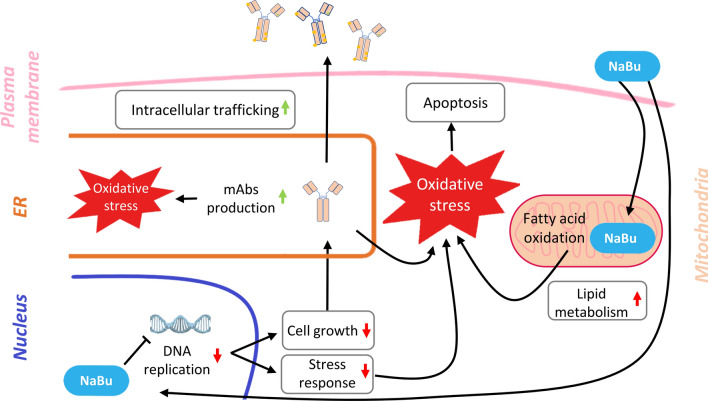
The increased production of the recombinant protein seen in NaBu cells leads to increased secretion, resulting in additional stress for the secretory machinery (Lim et al. 2010). NaBu treatment is also known to induce autophagy activation in CHO cells (Lee and Lee 2012). The major metabolic activities, together with the boosted production of mAb, the response to oxidative stress and the activation of autophagy are all mechanisms behind the upregulation of the intracellular signaling and trafficking, the third positively modulated area highlighted during the analysis. The downside of NaBu treatment was that its inhibitory effects seemed to impair the ability of the cells to activate an important response mechanism such as the redox stress one. On the contrary, it has been reported that NaBu activates Nrf2 (Dong et al. 2017), a transcription factor activated by the increase of oxidative stress in the cytoplasm leading to antioxidant response (Bryan et al. 2013). The activation was seen in mice experiments and not in cell culturing experiments as in our case, where we found a downregulation of proteins connected with the antioxidant response. This downregulation is a major problem, considering NaBu itself is expected to be an indirect source of oxidative stress. Based on previous studies and our results, we propose a working model for the effects of NaBu on mAb-producing cells and potential mitigations that can be taken to prolong production time and yield (Fig. 5). The partial response of cells to redox stress impairs their ability to control oxidative stress accumulation, promoted by increased burden on mitochondrial respiratory machinery and fatty acids beta-oxidation. The ER, which is one of the responsible for buffering oxidative stress, may not be available to manage the escalation in redox activity with it being consigned for mAb synthesis. This may intensify ER stress and further oxidative stress accumulation (Ha et al. 2018). In this situation, the antioxidant response should be activated at capacity, but its partial reaction due to NaBu treatment hampers the ability of the cells to control the oxidative stress levels. These complicated circumstances likely push the cells to activate their apoptotic and autophagic mechanisms, starting the processes that will eventually lead to cell death (Fig. 5). Hypothetically, an attempt to control NaBu-induced oxidative stress accumulation in these cells may prevent the onset of early apoptosis and allow longer production times. Our conclusion would explain the optimal results obtained on CHO culturing in bioreactors by Han et al. (2005). With the addition of the antioxidant N-acetylcysteine, they managed to prolong the culture for almost 200 h, doubling the production yield compared to cells solely treated with NaBu.
Fig. 5.
Model of butyric acid effect on CHO cells mAbs production. NaBu downregulates the DNA replication affecting the cell growth, which indirectly promotes mAbs production. With a slower growth, cells dedicate resources and machineries to the production and secretion of mAbs, increasing the intracellular trafficking and the oxidative stress deriving from the protein synthesis in the ER. However, the supplemented NaBu is also a fatty acid that has to be oxidized in the mitochondria, resulting in an upregulation of the lipid metabolism of the cells and of the oxidative stress deriving from the mitochondrial activity. The combination of the increased oxidative stress from different sources with the downregulation of the antioxidant response results in an uncontrolled accumulation of oxidative stress in CHO cells, leading to an early apoptosis
Supplementary Information
Acknowledgements
We thank all the members of our laboratory, particularly Wang Loo Chien, for the constructive criticisms and the useful suggestions.
Author contributions
K.S. and C.L. designed and executed the experiments, M.G. performed the proteomics data analysis and drafted the manuscript, Y.K. K.W-K. T.N-K. S.T. X.B. corrected the manuscript, K.W-K. T.N-K. performed the glycan analysis, X.B. conceived and supervised the study and S.T. guided the study. All authors read and approved the final version.
Funding
The study was financially supported by AB SCIEX Pte Ltd. and Industry Alignment Fund (IAF) from Biomedical Research Council (BMRC) of A*STAR (Agency for Science, Technology and Research), Singapore (IAF111189).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Al-Rubeai M, Emery AN, Chalder S, Jan DC (1992) Specific monoclonal antibody productivity and the cell cycle-comparisons of batch, continuous and perfusion cultures. Cytotechnology 9:85–97. 10.1007/BF02521735 [DOI] [PubMed] [Google Scholar]
- Alsabeeh N, Chausse B, Kakimoto PA, Kowaltowski AJ, Shirihai O (2018) Cell culture models of fatty acid overload: Problems and solutions. Biochim Biophys Acta Mol Cell Biol Lipids 1863:143–151. 10.1016/j.bbalip.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avello V, Torres M, Vergara M, Berrios J, Valdez-Cruz NA, Acevedo C, Molina Sampayo M, Dickson AJ, Altamirano C (2022) Enhanced recombinant protein production in CHO cell continuous cultures under growth-inhibiting conditions is associated with an arrested cell cycle in G1/G0 phase. PLoS ONE 17:e0277620. 10.1371/journal.pone.0277620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan HK, Olayanju A, Goldring CE, Park BK (2013) The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85:705–717. 10.1016/j.bcp.2012.11.016 [DOI] [PubMed] [Google Scholar]
- Cabrera G, Cremata JA, Valdés R, García R, González Y, Montesino R, Gómez H, González M (2005) Influence of culture conditions on the N-glycosylation of a monoclonal antibody specific for recombinant hepatitis B surface antigen. Biotechnol Appl Biochem 41:67–76. 10.1042/BA20040032 [DOI] [PubMed] [Google Scholar]
- Chng J, Wang T, Nian R, Lau A, Hoi KM, Ho SCL, Gagnon P, Bi X, Yang Y (2015) Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. Mabs 7:403–412. 10.1080/19420862.2015.1008351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie JR (2003) Inhibition of histone deacetylase activity by butyrate. J Nutr 133:2485S-2493S. 10.1093/jn/133.7.2485S [DOI] [PubMed] [Google Scholar]
- Dong W, Jia Y, Liu X, Zhang H, Li T, Huang W, Chen X, Wang F, Sun W, Wu H (2017) Sodium butyrate activates NRF2 to ameliorate diabetic nephropathy possibly via inhibition of HDAC. J Endocrinol 232:71–83. 10.1530/JOE-16-0322 [DOI] [PubMed] [Google Scholar]
- Dutton RL, Scharer J, Moo-Young M (2006) Cell cycle phase dependent productivity of a recombinant Chinese hamster ovary cell line. Cytotechnology 52:55–69. 10.1007/s10616-006-9041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezquerro S, Becerril S, Tuero C, Méndez-Giménez L, Mocha F, Moncada R, Valentí V, Cienfuegos JA, Catalán V, Gómez-Ambrosi J, Piper Hanley K, Frühbeck G, Rodríguez A (2020) Role of ghrelin isoforms in the mitigation of hepatic inflammation, mitochondrial dysfunction, and endoplasmic reticulum stress after bariatric surgery in rats. Int J Obes (Lond) 44:475–487. 10.1038/s41366-019-0420-2 [DOI] [PubMed] [Google Scholar]
- Gast V, Campbell K, Picazo C, Engqvist M, Siewers V, Molin M (2021) The Yeast eIF2 Kinase Gcn2 Facilitates H2O2-Mediated Feedback Inhibition of Both Protein Synthesis and Endoplasmic Reticulum Oxidative Folding during Recombinant Protein Production. Appl Environ Microbiol 87:1–16. 10.1128/AEM.00301-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha TK, Hansen AH, Kol S, Kildegaard HF, Lee GM (2018) Baicalein reduces oxidative stress in CHO cell cultures and improves recombinant antibody productivity. Biotechnol J. 10.1002/BIOT.201700425 [DOI] [PubMed] [Google Scholar]
- Hague A, Manning AM, Hanlon KA, Hart D, Paraskeva C, Huschtscha LI (1993) Sodium butyrate induces apoptosis in human colonic tumour cell lines in a p53-independent pathway: implications for the possible role of dietary fibre in the prevention of large-bowel cancer. Int J Cancer 55:498–505. 10.1002/IJC.2910550329 [DOI] [PubMed] [Google Scholar]
- Han KO, Moon KS, Yang J, Ho CY, Ji SA, Jong ML, Ji TK, Ji UY, Tae HB (2005) Effect of N-Acetylcystein on butyrate-treated Chinese hamster ovary cells to improve the production of recombinant human interferon-beta-1a. Biotechnol Prog 21:1154–1164. 10.1021/BP050057V [DOI] [PubMed] [Google Scholar]
- Hong JK, Lee SM, Kim KY, Lee GM (2014) Effect of sodium butyrate on the assembly, charge variants, and galactosylation of antibody produced in recombinant Chinese hamster ovary cells. Appl Microbiol Biotechnol 98:5417–5425. 10.1007/S00253-014-5596-8 [DOI] [PubMed] [Google Scholar]
- Hua J, Xu H, Zhang Y, Ge J, Liu M, Wang Y, Wei Y, Shi Y, Hou LL, Jiang H (2022) Enhancement of recombinant human IL-24 (rhIL-24) protein production from site-specific integrated engineered CHO cells by sodium butyrate treatment. Bioprocess Biosyst Eng 45:1979–1991. 10.1007/S00449-022-02801-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Guo Q, Wu J, Guo B, Wang Y, Zhao S, Lou H, Yu X, Mei X, Wu C, Qiao S, Wu Y (2012) Dual effects of sodium butyrate on hepatocellular carcinoma cells. Mol Biol Rep 39:6235–6242. 10.1007/S11033-011-1443-5 [DOI] [PubMed] [Google Scholar]
- Kruh J (1981) Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol Cell Biochem 42:65–82. 10.1007/BF00222695/METRICS [DOI] [PubMed] [Google Scholar]
- Lai T, Yang Y, Ng SK (2013) Advances in mammalian cell line development technologies for recombinant protein production. Pharm 6:579–603. 10.3390/PH6050579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan M, Kok YJ, Lee AP, Kyriakopoulos S, Lim HL, Teo G, Poh SL, Tang WQ, Hong J, Tan AH-M, Bi X, Ho YS, Zhang P, Ng SK, Lee D-Y (2019) Multi-omics profiling of CHO parental hosts reveals cell line-specific variations in bioprocessing traits. Biotechnol Bioeng. 10.1002/bit.27014 [DOI] [PubMed] [Google Scholar]
- Lalonde ME, Durocher Y (2017) Therapeutic glycoprotein production in mammalian cells. J Biotechnol 251:128–140. 10.1016/J.JBIOTEC.2017.04.028 [DOI] [PubMed] [Google Scholar]
- Lataster L, Huber HM, Böttcher C, Föller S, Takors R, Radziwill G (2023) Cell cycle control by optogenetically regulated cell cycle inhibitor protein p21. Biology (Basel) 12:1194. 10.3390/BIOLOGY12091194/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Lee GM (2012) Effect of sodium butyrate on autophagy and apoptosis in Chinese hamster ovary cells. Biotechnol Prog 28:349–357. 10.1002/btpr.1512 [DOI] [PubMed] [Google Scholar]
- Li W-F, Fan Z-L, Wang X-Y, Lin Y, Wang T-Y (2022) Combination of sodium butyrate and decitabine promotes transgene expression in CHO cells via apoptosis inhibition. N Biotechnol 69:8–17. 10.1016/j.nbt.2022.02.004 [DOI] [PubMed] [Google Scholar]
- Lim Y, Wong NSC, Lee YY, Ku SCY, Wong DCF, Yap MGS (2010) Engineering mammalian cells in bioprocessing– current achievements and future perspectives. Biotechnol Appl Biochem 55:175–189. 10.1042/BA20090363 [DOI] [PubMed] [Google Scholar]
- López-Pedrouso M, Borrajo P, Amarowicz R, Lorenzo JM, Franco D (2021) Peptidomic analysis of antioxidant peptides from porcine liver hydrolysates using SWATH-MS. J Proteomics 232:104037. 10.1016/J.JPROT.2020.104037 [DOI] [PubMed] [Google Scholar]
- Lu JT, Xiao MK, Feng YY, Wang XY, Le QL, Chai YR, Wang TY, Jia YL (2023) Apilimod enhances specific productivity in recombinant CHO cells through cell cycle arrest and mediation of autophagy. Biotechnol J 18:2200147. 10.1002/BIOT.202200147 [DOI] [PubMed] [Google Scholar]
- Ludwig C, Gillet L, Rosenberger G, Amon S, Collins BC, Aebersold R (2018) Data-independent acquisition-based SWATH - MS for quantitative proteomics: a tutorial. Mol Syst Biol 14:1–23. 10.15252/msb.20178126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez JA, Lupu R (2007) Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7:763–777. 10.1038/nrc2222 [DOI] [PubMed] [Google Scholar]
- Müller B, Heinrich C, Jabs W, Kaspar-Schönefeld S, Schmidt A, Rodrigues de Carvalho N, Albaum SP, Baessmann C, Noll T, Hoffrogge R (2017) Label-free protein quantification of sodium butyrate treated CHO cells by ESI-UHR-TOF-MS. J Biotechnol 257:87–98. 10.1016/J.JBIOTEC.2017.03.032 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Matoba S, Iwai-Kanai E, Kimata M, Hoshino A, Nakaoka M, Katamura M, Okawa Y, Ariyoshi M, Mita Y, Ikeda K, Okigaki M, Adachi S, Tanaka H, Takamatsu T, Matsubara H (2012) P53 promotes cardiac dysfunction in diabetic mellitus caused by excessive mitochondrial respiration-mediated reactive oxygen species generation and lipid accumulationlipid accumulation. Circ Hear Fail 5:106–115. 10.1161/CIRCHEARTFAILURE.111.961565 [DOI] [PubMed] [Google Scholar]
- Newcombe AR (2014) The Evolution of quality by design (QbD) for biologics. PDA J Pharm Sci Technol 68:320–322. 10.5731/PDAJPST.2014.00989 [DOI] [PubMed] [Google Scholar]
- Nguyen P, Leray V, Diez M, Serisier S, Bloc H, Le J, Siliart B, Dumon H (2008) Liver lipid metabolism. J Anim Physiol Anim Nutr (Berl) 92:272–283. 10.1111/j.1439-0396.2007.00752.x [DOI] [PubMed] [Google Scholar]
- Orellana CA, Marcellin E, Gray PP, Nielsen LK (2017) Overexpression of the regulatory subunit of glutamate-cysteine ligase enhances monoclonal antibody production in CHO cells. Biotechnol Bioeng 114:1825–1836. 10.1002/BIT.26316 [DOI] [PubMed] [Google Scholar]
- Ortiz-Enriquez C, de Romero-Díaz AJ, Hernández-Moreno AV, Cueto-Rojas HF, Miranda-Hernández MP, López-Morales CA, Pérez NO, Salazar-Ceballos R, Cruz-García N, Flores-Ortiz LF, Medina-Rivero E (2016) Optimization of a recombinant human growth hormone purification process using quality by design. Prep Biochem Biotechnol 46:815–821. 10.1080/10826068.2015.1135467 [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee A-H, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS (2004) Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306:457–461. 10.1126/science.1103160 [DOI] [PubMed] [Google Scholar]
- Park SY, Egan S, Cura AJ, Aron KL, Xu X, Zheng M, Borys M, Ghose S, Li Z, Lee K (2021) Untargeted proteomics reveals upregulation of stress response pathways during CHO-based monoclonal antibody manufacturing process leading to disulfide bond reduction. Mabs. 10.1080/19420862.2021.1963094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Morales M, Vieira-Lara MA, Homan E, Langelaar-Makkinje M, Gerding A, Li Z, Huijkman N, Rensen PCN, Wolters JC, Reijngoud DJ, Bakker BM (2022) Butyrate oxidation attenuates the butyrate-induced improvement of insulin sensitivity in myotubes. Biochim Biophys Acta - Mol Basis Dis 1868:166476. 10.1016/J.BBADIS.2022.166476 [DOI] [PubMed] [Google Scholar]
- Rizzo D, Cerofolini L, Giuntini S, Iozzino L, Pergola C, Sacco F, Palmese A, Ravera E, Luchinat C, Baroni F, Fragai M (2022) Epitope mapping and binding assessment by solid-state nmr provide a way for the development of biologics under the quality by design paradigm. J Am Chem Soc 144:10006–10016. 10.1021/JACS.2C03232/SUPPL_FILE/JA2C03232_SI_001.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Spearman M, Huzel N, Butler M (2005) Enhanced production of monomeric interferon-beta by CHO cells through the control of culture conditions. Biotechnol Prog 21:22–30. 10.1021/BP049807B [DOI] [PubMed] [Google Scholar]
- Salimi V, Shahsavari Z, Safizadeh B, Hosseini A, Khademian N, Tavakoli-Yaraki M (2017) Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis 16:1–11. 10.1186/S12944-017-0593-4/FIGURES/6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert OT, Gillet LC, Collins BC, Navarro P, Rosenberger G, Wolski WE, Lam H, Amodei D, Mallick P, Maclean B, Aebersold R (2015) Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc 10:426–441. 10.1038/nprot.2015.015 [DOI] [PubMed] [Google Scholar]
- Schulze M, Kumar Y, Rattay M, Niemann J, Wijffels RH, Martens DE (2022) Transcriptomic analysis reveals mode of action of butyric acid supplementation in an intensified CHO cell fed-batch process. Biotechnol Bioeng 119:2359–2373. 10.1002/BIT.28150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaan J, El-Hakim S, Ibrahim JN, Safi R, Elnar AA, El Boustany C (2020) Comparative effect of sodium butyrate and sodium propionate on proliferation, cell cycle and apoptosis in human breast cancer cells MCF-7. Breast Cancer 27:696–705. 10.1007/S12282-020-01063-6 [DOI] [PubMed] [Google Scholar]
- Senior M (2023) Fresh from the biotech pipeline: fewer approvals, but biologics gain share. Nat Biotechnol 41:174–182. 10.1038/s41587-022-01630-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim KH, Liu LCY, Tan HT, Tan K, Ng D, Zhang W, Yang Y, Tate S, Bi X (2020) A comprehensive CHO SWATH-MS spectral library for robust quantitative profiling of 10,000 proteins. Sci Data. 10.1038/S41597-020-00594-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eunen K, Simons SMJ, Gerding A, Bleeker A, den Besten G, Touw CML, Houten SM, Groen BK, Krab K, Reijngoud DJ, Bakker BM (2013) Biochemical competition makes fatty-acid β-oxidation vulnerable to substrate overload. PLoS Comput Biol. 10.1371/journal.pcbi.1003186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DE, Yang F, Carver J, Joe K, Michels DA, Yu XC (2017) A modular and adaptive mass spectrometry-based platform for support of bioprocess development toward optimal host cell protein clearance. Mabs 9:654. 10.1080/19420862.2017.1303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV (2012) Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81:687. 10.1146/ANNUREV-BIOCHEM-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Wang CC, Huang HC, Wei YH (2013) Mitochondrial dysfunction leads to impairment of insulin sensitivity and adiponectin secretion in adipocytes. FEBS J 280:1039–1050. 10.1111/febs.12096 [DOI] [PubMed] [Google Scholar]
- Wippermann A, Rupp O, Brinkrolf K, Hoffrogge R, Noll T (2017) Integrative analysis of DNA methylation and gene expression in butyrate-treated CHO cells. J Biotechnol 257:150–161. 10.1016/J.JBIOTEC.2016.11.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on PRIDE with the following identifier PXD048575.