Abstract
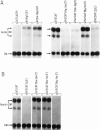
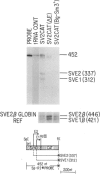
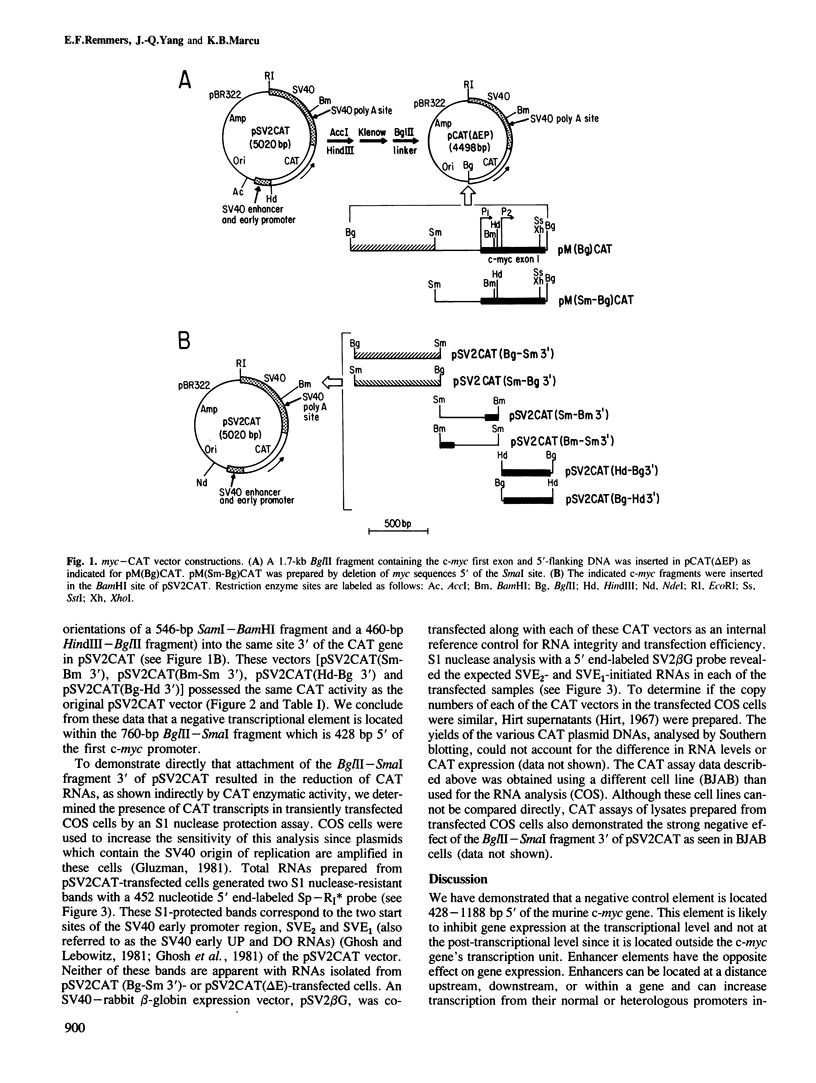
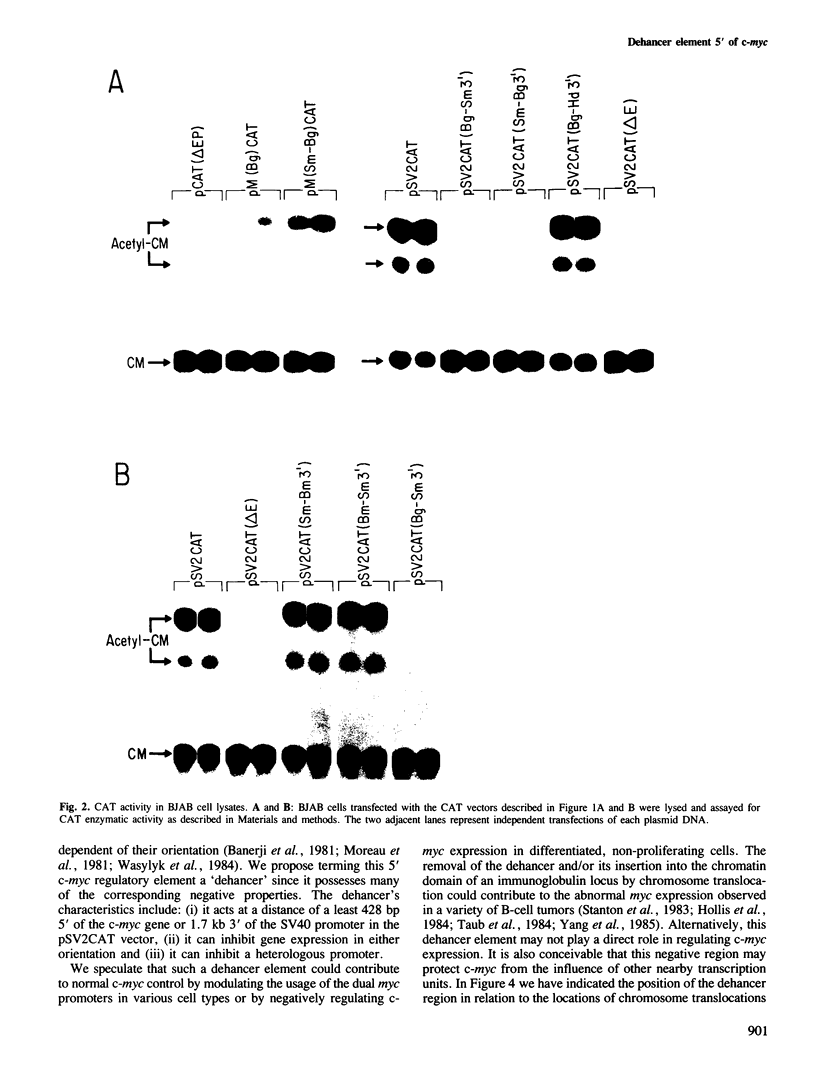
We have investigated the nature of regulatory sequences within the vicinity of the murine c-myc locus by analyzing the expression of myc-chloramphenicol acetyl transferase (CAT) vectors transfected into a human lymphoblastoid cell line (BJAB) and a monkey fibroblast line (COS). CAT enzymatic assays and S1 nuclease protection experiments reveal that a negative element resides 428-1188 bp 5' of the first c-myc promoter, P1. This 760-bp segment of 5'-flanking c-myc DNA dramatically inhibits CAT gene expression in the pSV2CAT vector when placed in either orientation approximately 1.7 kb 3' (and approximately 3.2 kb 5' on the circular plasmid) from the SV40 promoter region. By employing this strategy, we were unable to identify an analogous DNA segment that is closer to or within the first c-myc exon. We propose that this 5' c-myc region be termed a 'dehancer' since this negative element has the opposite properties of a transcriptional enhancer.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J., Nasmyth K. A., Strathern J. N., Klar A. J., Hicks J. B. Regulation of mating-type information in yeast. Negative control requiring sequences both 5' and 3' to the regulated region. J Mol Biol. 1984 Jul 5;176(3):307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bienz-Tadmor B., Zakut-Houri R., Libresco S., Givol D., Oren M. The 5' region of the p53 gene: evolutionary conservation and evidence for a negative regulatory element. EMBO J. 1985 Dec 1;4(12):3209–3213. doi: 10.1002/j.1460-2075.1985.tb04067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K. Characterization of a "silencer" in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985 May;41(1):41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. SV40 DNA transfection of cells in suspension: analysis of efficiency of transcription and translation of T-antigen. Gene. 1981 Mar;13(2):197–202. doi: 10.1016/0378-1119(81)90008-1. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Cory S., Adams J. M. Transposition of the immunoglobulin heavy chain enhancer to the myc oncogene in a murine plasmacytoma. Cell. 1985 Jan;40(1):71–79. doi: 10.1016/0092-8674(85)90310-1. [DOI] [PubMed] [Google Scholar]
- Fahrlander P. D., Piechaczyk M., Marcu K. B. Chromatin structure of the murine c-myc locus: implications for the regulation of normal and chromosomally translocated genes. EMBO J. 1985 Dec 1;4(12):3195–3202. doi: 10.1002/j.1460-2075.1985.tb04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P. Simian virus 40 early mRNA's contain multiple 5' termini upstream and downstream from a Hogness-Goldberg sequence; a shift in 5' termini during the lytic cycle is mediated by large T antigen. J Virol. 1981 Oct;40(1):224–240. doi: 10.1128/jvi.40.1.224-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hennighausen L., Siebenlist U., Danner D., Leder P., Rawlins D., Rosenfeld P., Kelly T., Jr High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985 Mar 21;314(6008):289–292. doi: 10.1038/314289a0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hollis G. F., Mitchell K. F., Battey J., Potter H., Taub R., Lenoir G. M., Leder P. A variant translocation places the lambda immunoglobulin genes 3' to the c-myc oncogene in Burkitt's lymphoma. Nature. 1984 Feb 23;307(5953):752–755. doi: 10.1038/307752a0. [DOI] [PubMed] [Google Scholar]
- Klein G. Specific chromosomal translocations and the genesis of B-cell-derived tumors in mice and men. Cell. 1983 Feb;32(2):311–315. doi: 10.1016/0092-8674(83)90449-x. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Li Y., Holland C. A., Hartley J. W., Hopkins N. Viral integration near c-myc in 10-20% of mcf 247-induced AKR lymphomas. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6808–6811. doi: 10.1073/pnas.81.21.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosthaf L., Pawlita M., Gruss P. A viral enhancer element specifically active in human haematopoietic cells. Nature. 1985 Jun 13;315(6020):597–600. doi: 10.1038/315597a0. [DOI] [PubMed] [Google Scholar]
- O'Donnell P. V., Fleissner E., Lonial H., Koehne C. F., Reicin A. Early clonality and high-frequency proviral integration into the c-myc locus in AKR leukemias. J Virol. 1985 Aug;55(2):500–503. doi: 10.1128/jvi.55.2.500-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Haseltine W. A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985 Jul;41(3):813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Chambon P. Short and long range activation by the SV40 enhancer. Nucleic Acids Res. 1984 Jul 25;12(14):5589–5608. doi: 10.1093/nar/12.14.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Q., Bauer S. R., Mushinski J. F., Marcu K. B. Chromosome translocations clustered 5' of the murine c-myc gene qualitatively affect promoter usage: implications for the site of normal c-myc regulation. EMBO J. 1985 Jun;4(6):1441–1447. doi: 10.1002/j.1460-2075.1985.tb03800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Q., Mushinski J. F., Stanton L. W., Fahrlander P. D., Tesser P. C., Marcu K. B. Modes of c-myc oncogene activation in murine plasmacytomas. Curr Top Microbiol Immunol. 1984;113:146–153. doi: 10.1007/978-3-642-69860-6_25. [DOI] [PubMed] [Google Scholar]
- Zech L., Haglund U., Nilsson K., Klein G. Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int J Cancer. 1976 Jan 15;17(1):47–56. doi: 10.1002/ijc.2910170108. [DOI] [PubMed] [Google Scholar]