Abstract
Study of mechanisms by which antibodies recognize different viral strains is necessary for the development of new drugs and vaccines to treat COVID-19 and other infections. Here, we report 2.5 Å cryo-EM structure of the SARS-CoV-2 Delta trimeric S-protein in complex with Fab of the recombinant analog of REGN10987 neutralizing antibody. S-protein adopts “two RBD-down and one RBD-up” conformation. Fab interacts with RBDs in both conformations, blocking the recognition of angiotensin converting enzyme-2. Three-dimensional variability analysis reveals high mobility of the RBD/Fab regions. Interaction of REGN10987 with Wuhan, Delta, Omicron BA.1, and mutated variants of RBDs is analyzed by microscale thermophoresis, molecular dynamics simulations, and ΔG calculations with umbrella sampling and one-dimensional potential of mean force. Variability in molecular dynamics trajectories results in a large scatter of calculated ΔG values, but Boltzmann weighting provides an acceptable correlation with experiment. REGN10987 evasion of the Omicron variant is found to be due to the additive effect of the N440K and G446S mutations located at the RBD/Fab binding interface with a small effect of Q498R mutation. Our study explains the influence of known-to-date SARS-CoV-2 RBD mutations on REGN10987 recognition and highlights the importance of dynamics data beyond the static structure of the RBD/Fab complex.
Subject terms: Cryoelectron microscopy, Molecular modelling
Cryo-EM structure of the SARS-CoV-2 Delta Sprotein in complex with the Fab of neutralizing REGN10987, MD simulations of Omicron and other RBD variants, and ΔG calculations explain the role of individual RBD mutations in REGN10987 evasion.
Introduction
The recent combat with COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has given rise to new strategies for design of robust neutralizing antibodies (nAbs)1–4. In particular, the studies of the relationship between viral genetic changes, transmissivity, and immune evasion5–7 have equipped the researchers with escape mutation maps8,9, allowing the development of prognostic tools to assess the danger of emerging SARS-CoV-2 variants and the efficacy of therapeutic antibodies against these variants10,11. All these tools are based on structural studies of complexes of the viral spike-protein (S-protein) with its receptor, angiotensin converting enzyme 2 (ACE2), and nAbs.
The S-protein is responsible for a membrane fusion of the virus with host cells. It is produced as a single-chain precursor that trimerizes and is subsequently cleaved by a furin-like protease into the receptor-binding subunit S1 and the fusion subunit S212,13. S1 folds into four domains—the N-terminal domain (NTD), the receptor-binding domain (RBD), and two C-terminal subdomains (SD1 and SD2), wrapped around S2 in a prefusion conformation14,15. Each RBD can adopt two distinct conformations: the ‘up’’ state, which is accessible to ACE2, and the ‘down’’ state, which is receptor-inaccessible14,16,17. The ‘down’’ state of the RBD could protect the viral receptor-binding site from host immune responses13,14. After the RBD binding to ACE2 on the host cell surface and the second proteolytic cleavage within S2 (S2′ site)18, S1 dissociates and S2 experiences extensive conformational changes and forms a post-fusion structure19,20 promoting the fusion of the viral particle with the host cell and infection.
To date, nAbs isolated from convalescent donors are considered emergency therapeutic agents or preventing agents for immunodeficient patients21,22. Most nAbs bind to either NTD or RBD22. For nAbs targeting RBD, the epitopes tend to overlap with the receptor-binding motif (RBM) preventing viral binding to ACE223. One of the well-known RBD-targeting nAbs is REGN10987 (imdevimab)24. In the reported 3.9 Å cryo-EM structure in the complex with the wild-type (WT) Wuhan RBD (PDB 6XDG)24, REGN10987 is located at the edge of the RBM and has a small overlap with the ACE2 binding site. According to the classification proposed by Barnes et al. 21, REGN10987 belongs to the nAbs from Class 3, which bind outside the ACE2 binding site and recognize both the ‘up’’ and ‘down’’ RBDs, while according to the Dejnirattisai et al. classification25, REGN10987 binds to the ‘right shoulder’’ of the RBD. REGN10987 exhibits IC50 of ~40 pM in neutralizing assays with the WT S-protein26. Treatment with a single antibody elicits selective pressure and can lead to the development of viral resistance, whereas antibody cocktails rarely promote the nAb-escaping virus variants26. So, a cocktail of REGN10987 and REGN10933 (casirivimab, an nAb targeting another epitope) is used in several countries for the emergency treatment of patients with COVID-19, including that caused by the Delta variant27,28.
The Delta variant (B.1.617.2), first reported in October 2020, shows enhanced transmissibility compared with the previously described virus variants and reduced sensitivity to therapeutic nAbs and host immunity elicited by the first-generation vaccines29,30. The enhanced interaction with ACE2, more efficient attachment to target cells, and faster fusion kinetics of the Delta variant29,31 are likely due to two RBD mutations (L452R and T478K)31,32. Neither of these mutations is located at the putative REGN10987 binding epitope, and the retention of REGN10987 activity against the Delta variant was confirmed by neutralization assays30.
The Omicron BA.1 variant (B.1.1.529), first reported in November 2021, has 15 mutations in RBD and shares only one of them (T478K) with the Delta variant. Omicron mutations can be categorized into neutral, affinity-diminishing, and compensatory ones, resulting in a negligible change in the ACE2 binding affinity33. ACE2 binding assays in vitro showed comparable Kd values in the low nanomolar range for the Omicron and Delta S-proteins, which were both lower than those for the Wuhan S-protein33. The Omicron mutations span the RBM and allow different virus variants, including BA.1, to escape REGN10987 and most other nAbs34–41.
Here we report the cryo-EM structure of the full-length Delta SARS-CoV-2 S-protein in the complex with the Fab of the recombinant analog of REGN10987. This fragment, hereafter referred to as REGN10987-Fab, differs from the original antibody in the light chain constant region. We describe the dynamics of the S-protein/REGN10987-Fab complex and investigate the role of individual RBD mutations in the REGN10987 evasion by the Omicron BA.1 and other SARS-CoV-2 variants. The data obtained will be useful for development of new therapeutic antibodies and prognostic tools to combat emerging variants of SARS-CoV-2 and other viruses.
Results
Cryo-EM structure of the trimeric Delta SARS-Cov-2 S-protein in the complex with REGN10987-Fab
The structure of the full-length trimeric Delta S-protein with the stabilizing mutations K986P and V987P14 and the furin cleavage site 682RRAR685 mutated to GSAG14 was determined in the complex with REGN10987-Fab (Supplementary Table 1) to an overall resolution of 2.53 Å (Fig. 1b, c, Table 1, Supplementary Figs. 1 and 2). The resolution was not homogeneous in different parts of the structure. The core of the S2 subunit was resolved significantly better than the NTDs and RBDs of the S1 subunit, Fabs, and N-linked glycan moieties (Supplementary Fig. 2). Similarly to the previous report14, the loop containing the S1/S2 junction and furin cleavage site (residues 677-688, Fig. 1a) was unresolved in the EM-maps in the all three S protomers. We used the full-length variant of the Delta S-protein solubilized in LMNG detergent, however, no density corresponding to the heptad repeat 2 (HR2) and trans-membrane (TM) regions were observed (Fig. 1 and Supplementary Fig. 2).
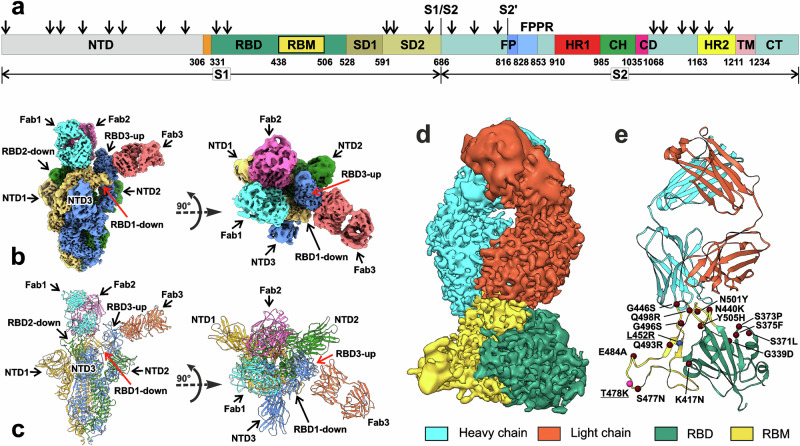
Fig. 1. Cryo-EM structure of the full-length Delta SARS-CoV-2 S-protein in the complex with REGN10987-Fab.
a Schematic representation of the sequence of full-length SARS-CoV-2 S-protein. Segments of S1 and S2 subunits include: NTD N-terminal domain, RBD receptor-binding domain, RBM receptor-binding module, SD1 and SD2, C-terminal subdomains 1 and 2; S1/S2, S1/S2 cleavage site; S2′, S2′ cleavage site; FP fusion peptide, FPPR fusion peptide proximal region, HR1 heptad repeat 1, CH central helix region, CD connector domain, HR2 heptad repeat 2, TM transmembrane helix, CT cytoplasmic tail; and arrow symbols for glycans. b, c Cryo-EM map of the Delta S-protein/Fab complex refined to 2.5 Å resolution and pseudoatomic model of the complex after local refinement of RBD/Fab regions. Three protomers (1, 2, and 3) of S-protein and the attached Fabs are color coded. RBD1/Fab1 and RBD2/Fab2 complexes are in ‘down’’ conformation, while RBD3/Fab3—in ‘up’’. d, e Cryo-EM map and modeled structure of the RBD1/Fab1 complex after focused refinement to 3.2 Å. The sites of Delta (blue and magenta spheres, underlined) and Omicron BA.1 (magenta and brown spheres) mutations are shown.
Table 1.
Cryo-EM data collection, refinement, and validation statistics
| Delta S protein/REGN10987-Fab (EMDB-14750) (PDB 7ZJL) | |
|---|---|
| Data collection and processing | |
| Magnification | 130,000 |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 50 |
| Defocus range (μm) | −0.8 to −1.6 |
| Pixel size (Å) | 0.929 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 147,977 |
| Final particle images (no.) | 44,323 |
| Map resolution (Å) | 2.53 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 2.0–9.0 |
| Refinement | |
| Initial model used (PDB code) | 7SBL, 6XDG |
| Model resolution (Å) | 2.60 |
| FSC threshold | 0.143 |
| Model resolution range (Å) | 2.0–9.0 |
| Map sharpening B factor (Å2) | −36.6 |
| Model composition | |
| Non-hydrogen atoms | 35,140 |
| Protein residues | 4551 |
| Ligands | 0 |
| B factors (Å2) | |
| Protein | 74.33 |
| Ligand | |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.847 |
| Validation | |
| MolProbity score | 0.81 |
| Clashscore | 0.07 |
| Poor rotamers (%) | 0.48 |
| Ramachandran plot | |
| Favored (%) | 95.95 |
| Allowed (%) | 4.01 |
| Disallowed (%) | 0.04 |
In our structure, S-protein adopted the one-RBD-up S-open conformation. Two RBDs (RBD1 and RBD2) were in the ‘down’’ state and the RBD3 was in the ‘up’’ state. Two Fabs bound to the RBD-down were located at the top of the S1 subunit, while the third Fab bound to the RBD3-up approached the S-protein complex laterally (Fig. 1b, c). A conformational distribution with ~70% of one-RBD-up and ~30% of three-RBD-down states has been observed previously for the apo Delta S-protein29,31. Moreover, the single-molecule Förster resonance energy transfer (smFRET) imaging showed that the apo-form of the WT Wuhan S-protein exhibits dynamic interconversions between the three-RBD-down S-closed (60%) and one-RBD-up S-open (40%) states42,43, while the D614G mutation (which is present in the Delta variant) increases the population of the S-open state to 60%43. Thus, the REGN10987-Fab binding likely captures the Delta S-protein in the most populated conformation.
The “630 loop” of the S1 subunit (residues 624-639 of SD2) and the fusion-peptide proximal region of the S2 subunit (FPPR, residues 829–853, the segment immediately downstream the fusion-peptide, Fig. 1a) have been proposed to control the structural rearrangement (opening/closing) of the S1 subunit20,44. These elements are located on the both sides of the RBD-down. They are structured in the three-RBD-down S-closed conformation and partially disordered in the one-RBD-up conformation, where the 630/FPPR pair surrounding the RBD1-down is ordered29,44. In our structure, densities corresponding to all three pairs of the 630/FPPR elements were not observed, reflecting increased dynamics within the S-protein/Fab complex.
Focused refinement of the RBD/Fab complexes
In the obtained EM-map, the RBD/Fab regions demonstrated low resolution ~9 Å. To increase resolution, we used a focused refinement with masks on the individual RBD/Fab complexes, resulting in resolution of 3.15 Å for RBD1/Fab1, 3.32 Å for RBD2/Fab2, and 3.38 Å for RBD3/Fab3 complexes (Fig. 1d and Supplementary Figs. 3–5). The pseudoatomic model of the S-protein/REGN10987-Fab complex was built using the combined EM-map (Fig. 1c).
Resolution achieved was sufficient for unambiguous modeling of the amino acid side chains and several H-bonds at the RBD-Fab interaction interfaces (Fig. 2), which were absent in the previous 3.9 Å structure of WT-RBD in the complex with REGN10933 and REGN10987 Fabs (PDB 6XDG)24. The differences in the interaction interfaces of REGN10987-Fab with RBD in the ‘up’’ and ‘down’’ states were insignificant (Supplementary Fig. 6a, Supplementary Data 1-1), with RMSD values for CA atoms in the RBD loops N437-N450 and Q498-Y508 RBD (responsible for the Fab binding) being ~0.38 Å. Comparison with the 6XDG structure24 revealed the RMSD of ~0.7 Å (Supplementary Fig. 6b). Analysis of the MD trajectories of the RBD/Fab complex (see below) revealed a significantly lower RMSD value in this region between the MD frames and our structure (1.15 ± 0.27 Å, mean ± SD, minimal value ~0.57 Å) than with the 6XDG structure (1.44 ± 0.26 Å, minimal value ~0.79 Å, Supplementary Fig. 6c, d). Thus, the observed structural differences can be attributed to the lower resolution of the previous structure24 rather than the dynamics of the RBD/Fab binding interface.
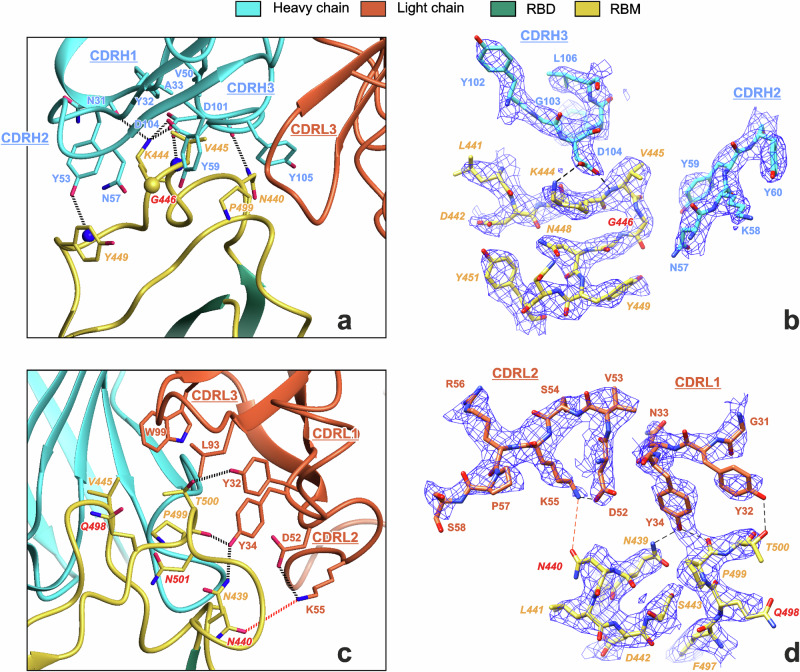
Fig. 2. Cryo-EM data reveal an interaction interface between REGN10987-Fab and Delta-RBD.
Interaction of the heavy (a, b) and light (c, d) chains of REGN10987-Fab with the Delta-RBD. Residues of RBD are in italic font. Residues mutated in the Omicron BA.1 variant are shown in red font. Hydrogen bonds are shown by black dotted/dashed lines. The red dotted/dashed lines illustrate proximity of N440 and K55L sidechains (distance 4.6 Å). CA atom of Gly446, backbone NH atoms, and backbone CO-groups are shown as yellow sphere, blue spheres, and sticks, respectively. Complementarity-determining regions (CDRs) of heavy and light chains are labeled.
Adjacent RBD-up and RBD-down in the S-open structures of the WT45, Beta46, Kappa46, and Omicron47 variants form intermolecular contacts that may promote the RBD erection, thereby facilitating its interaction with ACE2. In our combined model, hydrogen bonds were observed between the sidechains and the mainchain of the RBM residues A475, S477, and K478 of RBD1-down and Y369 and N370 of RBD3-up, as well as hydrophobic contact between F486 (RBD1-down) and F377 (RBD3-up) (Supplementary Fig. 7a). However, these contacts were not confirmed by direct EM-map analysis due to low resolution (5–7 Å) of the A475-N487 (RBD1-down) and Y369-F377 (RBD3-up) loops.
In the S-closed state of the WT S-protein, the neighboring RBDs-down interact with each other by hydrogen bonds between R403, Q493, and Y505 of one RBD and S373, S371, and Y369 of the other one48. In addition, the inter-RBD contacts were observed between N-linked N343 glycan and the Y489, F490, and F456 sidechains, as well as the D364-R457 salt bridge49. Here, the focused refinement of the (RBD1-down/Fab+RBD2-down/Fab) region with the joint mask (resolution ~3.8 Å) did not reveal any density corresponding to the RBD1/RBD2 interaction (Supplementary Fig. 7b). Moreover, the distance between CA atoms of D364 and R457 in the combined model was ~21 Å, pointing on the absence of the RBD1/RBD2 contacts. In addition, our structure did not reveal densities corresponding to free fatty acid molecules, which in the WT S-protein are suggested to lock pairs of the adjacent RBDs in the ‘down’’ state50. This agrees with the loose packing of ‘down’’ RBD1 and RBD2 in the S-protein/REGN10987-Fab complex.
Previous cryo-EM study of the apo Delta S-protein revealed two distinct one-RBD-up conformations that differ mainly in the spatial positioning of NTD1, NTD2, and RBD3-up (PDB 7SBL and 7SBO)29. Fab binding did not significantly alter the overall structure of the Delta S-protein, although minor changes were observed in the position and conformation of the NTD, RBD, and SD1 domains (Supplementary Fig. 8). Thus, the RBD3-up conformation in our structure was similar to that in the 7SBL structure, whereas the RBD1-down conformation was similar to the 7SBO structure. The largest differences from both apo structures were observed for RBD2-down, SD1 of the second subunit (SD1-2), and the spatially proximate NTD1 (Supplementary Fig. 8).
Conformational dynamics of the trimeric Delta S-protein in the complex with REGN10987-Fab
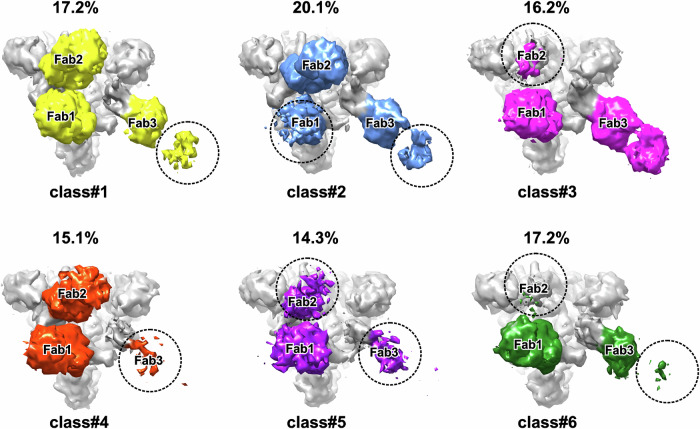
3D classification of the complete set of collected particles resulted in six classes with almost equal populations. In each class, three Fabs were bound to RBDs (Fig. 3). In the most classes, the densities of Fabs bound to ‘down’’ RBD1 and RBD2 were more intense than the density of Fab bound to RBD3-up (Fig. 3). Full Fab1 density was present in all classes except class #2, where only N-terminal domain density was observed. Full density for Fab2 was observed in three classes (#1, #2, #4), whereas partial density was presented in the other cases. Full Fab3 density was observed only in class #3, while partial density was found in the remaining classes (Fig. 3). The molar ratio (S-protein subunit): Fab in our sample was 1:1. Thus, the observed partial Fab density in some 3D classes indicates a large amplitude of the antibody domain motions rather than partial binding of the Fabs to RBDs. The mobility of the RBD/Fab regions decreased in the order RBD3 > RBD2 > RBD1.
Fig. 3. Overview of 3D classes derived from the Delta S-protein/REGN10987-Fab dataset.
Top views of the S-protein/Fab complexes are shown. Dashed circles indicate missing regions of the EM density map.
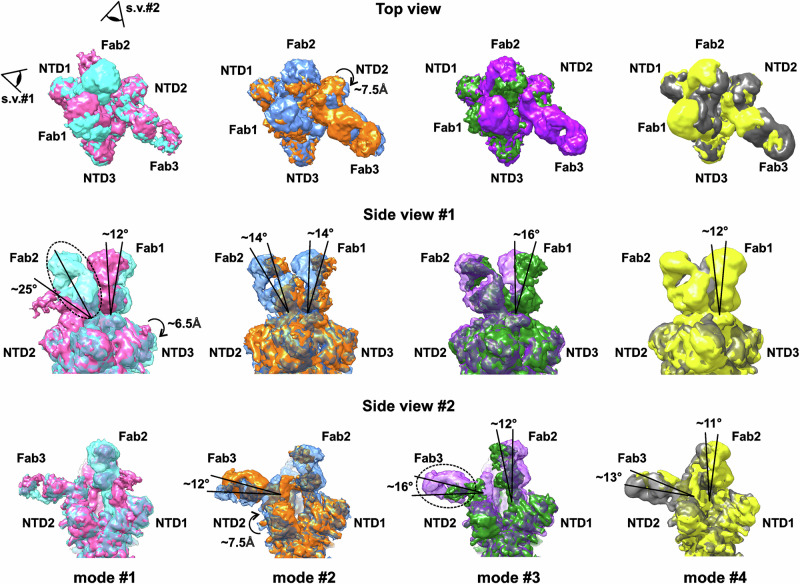
3D variability analysis (3DVA) of the 9 Å low-pass filtered structures in cryoSPARC51 revealed four modes of high-amplitude motions (Fig. 4 and Supplementary Movies 1–4). Mode #1 displayed high-amplitude motion of the RBD2/Fab2 fragment (inclination within at least 25°, precise measurement is impossible due to the vanishing density of Fab2). This motion induced the concerted tilt of the RBD1/Fab1 fragment (~12° towards RBD2/Fab2) and a ~6.5 Å displacement of NTD3 adjacent to RBD1/Fab1. A concerted low-amplitude motion of the RBD3/Fab3 fragment was also observed. The observed high-amplitude deviation of Fab2 from the vertical axis may be associated with dynamic rearrangements of RBD2, namely, with the movements from the ‘down’’ state towards the ‘up’’ state.
Fig. 4. Four major modes of motion in the Delta S-protein/REGN10987-Fab complex detected by 3D variability analysis (3DVA).
Modes of motion are arranged by columns. For each mode, the two extreme structures are shown in different colors. Dashed ellipses indicate missing regions of the EM density map. The tilting motions of the RBD/Fab complexes and displacements of NTD are shown.
In contrast to mode #1, modes #2, #3, and #4 described flapping motions of the RBD3/Fab3 complex with moderate amplitude (inclination of 12–16°). This motion led to the disappearance of Fab3 density in mode #3. The fluctuations in the position of RBD3/Fab3 were associated with the concerted tilting motions of the RBD1/Fab1 and RBD2/Fab2 fragments. The tilt amplitude of the RBD1/Fab1 and RBD2/Fab2 was similar (11–16°) in different modes, while the direction was different. In mode #2, Fab1 and Fab2 moved synchronously in the same plane, while in modes #3 and #4, Fabs moved in perpendicular planes. In addition, in mode #3, RBD1/Fab1 and RBD2/Fab2 swung parallel to each over in opposite directions, while in mode #4, they synchronously approached and moved away from each over. For NTDs, the largest amplitude of motions with a shift of ~7.5 Å was observed for NTD2, spatially close to the RBD3/Fab3 complex (mode #2). The obtained dynamics data confirmed the high mobility of the RBD2/Fab2 and RBD3/Fab3 complexes.
Delta-RBD/REGN10987-Fab interface
REGN10987-Fab interacted with Delta-RBD on one side of the RBM, forming multiple H-bonds, ionic bridges, and hydrophobic contacts with two RBM loops (N439-Y449 and Q498-N501, Fig. 2a, c, Supplementary Data 1-1). The first loop primarily contacted the Fab heavy chain, while the second loop almost exclusively interacted with the Fab light chain. The contact area of the RBD with the heavy chain of the antibody (~500 Å2) was more than twice larger than with the light chain (~210 Å2). The Fab heavy and light chains interacted with the RBD with all three complementarity-determining regions (CDRs, Fig. 2a, c).
The interaction of the Fab heavy chain (indicated by superscript H) with the RBD was stabilized by a set of ionic interactions and H-bonds centered around the positively charged K444 residue of the RBD. The K444 sidechain formed ionic bridges with the acidic groups of Fab residues D101H and D104H and H-bond with the N31H carbonyl. In addition, the negatively charged D104H sidechain formed the H-bond with the HN group of the RBD residue V445 (Fig. 2a, b), while the antibody Y53H sidechain formed the H-bond with the HN group of RBD Y449. Several sidechain-sidechain hydrophobic contacts, where the C-C distances were less than 5 Å, (V445-A33H, V445-V50H, V445-Y59H, Y449-Y53H, and P499-Y105H), as well as strong packing (G446-N57H), or possible π-cation interactions (K444-Y53H and K444-Y32H) between the RBD and the Fab heavy chain were also observed (Fig. 2a and Supplementary Data 1-1).
Interaction of the antibody’s light chain (indicated by superscript L) with the RBD was stabilized by H-bonds between the hydroxy groups of residues Y32L and Y34L and the sidechains of residues T500 and N439 and the carbonyl of P499 of the RBD (Fig. 2c, d). In addition, several packing/hydrophobic interactions (T500-Y32L, P499-Y34L, P499-L93L, and V445-W99L) were possible. The participation of RBD T500 in hydrogen bonding network or hydrophobic interactions depends on the orientation of its sidechain, which was not unambiguously determined by the EM density map (Fig. 2d).
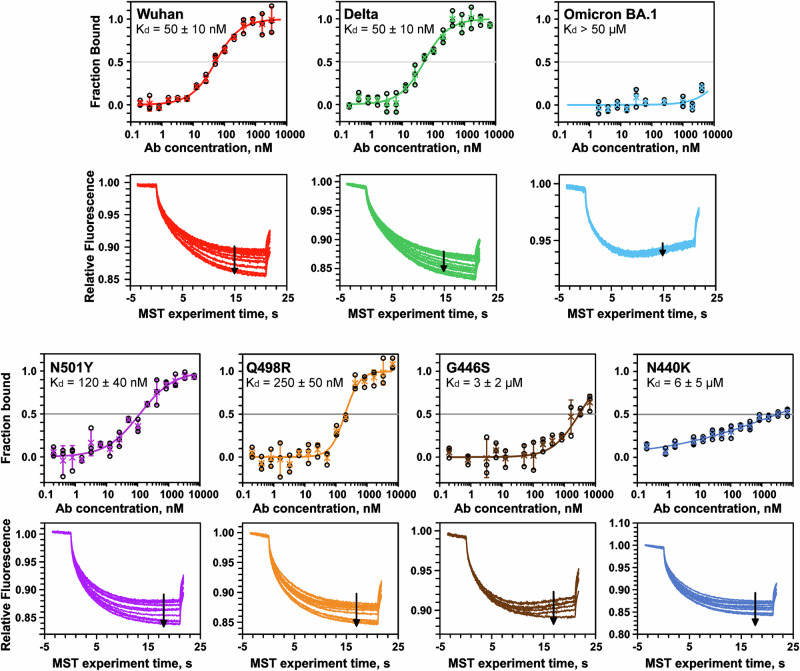
REGN10987 affinity to the mutant RBDs studied by microscale thermophoresis (MST)
Omicron BA.1 SARS-CoV-2 variant escapes neutralization by REGN1098735–40. From 15 mutations presented in the BA.1 RBD compared with the WT Wuhan variant (Fig. 1e, red/brown spheres) only four (N440K, G446S, Q498R, and N501Y) are located at the RBD/REGN10987-Fab interface and are absent in the Delta variant (Fig. 1e). Analysis of the Delta-RBD/REGN10987-Fab complex revealed that the G446S Omicron mutation can cause steric clash with the N57H sidechain35,41 (Fig. 2a, b), while the N440K mutation can result in electrostatic repulsion with K55L (ref. 39, Fig. 2c, d). Thus, at least two Omicron mutations can destabilize the S-protein/REGN10987 complex.
To investigate the effect of the Omicron RBD mutations on REGN10987 evasion, we produced recombinant RBDs of the Wuhan, Delta, and Omicron variants, as well as four Delta RBD variants containing point mutations from Omicron: Delta/N440K, Delta/G446S, Delta/Q498R, and Delta/N501Y. For the Delta RBD containing the other 10 mutations from Omicron, except N440, G446, Q498, and N501 (named Delta/‘‘Others’’ variant), we were unable to achieve sufficient expression level. The RBDs interaction with the recombinant REGN10987 analog was characterized by MST (Fig. 5). We found similar affinity of the antibody to Delta and Wuhan RBDs (Kd = 50 ± 10 nM) and no affinity to Omicron RBD (Kd > 50 μM). Obtained results agree with Kd of the REGN10987 binding to the WT-RBD containing a hexahistidine tag (45 nM)24 and are consistent with >10,000-fold increase in Kd for the Omicron BA.1 variant compared to the WT variant observed in the binding assay8 and with ~8000-fold increase in IC50 for virus neutralization52. A significant decrease in affinity to the REGN10987 analog was observed for mutants Q498R and N501Y (Kd ~250 nM and 120 nM, respectively) and weak binding was observed for mutants N440K and G446S (Kd ~6 μM and 3 μM, respectively).
Fig. 5. Microscale thermophoresis (MST) study of the REGN10987 affinity to the mutant RBDs.
Top panels: binding curves, datapoints are mean ± SD (n = 3 independent experiments). Bottom panels: MST traces. Arrows indicate increase in antibody concentrations. Obtained Kd values (±standard error of fit) are shown.
Molecular dynamics of REGN10987-Fab complexes with Wuhan, Omicron, and mutant RBDs
To analyze how the Omicron mutations influence the stability of the RBD/REGN10987-Fab complex, we examined the complexes of the Wuhan, Omicron BA.1, Delta/N440K, Delta/G446S, Delta/Q498R, Delta/N501Y, and Delta/‘‘Others’’ RBD variants using MD simulations. To obtain the characteristics of the complexes in equilibrium, three 1 µs MD replicas were calculated for each complex, and 500–1000 ns fragments of the trajectories were analyzed (Supplementary Figs. 9 and 10). Snapshots from the MD trajectories are shown in Fig. 6 and Supplementary Fig. 11, while lifetimes of the observed RBD/Fab intermolecular contacts are collected in Supplementary Data 1-2, 1-3, 1-4, 1-5, 1-6, 1-7 and 1-8. All RBD/Fab complexes were stable during 1 µs MD in all 7 × 3 = 21 replicas and demonstrated the contact areas in the range of 450–900 Å2 (Supplementary Fig. 9, averaged over 50 ns window). At each time point, the complexes were stabilized by at least three ‘classical’’ noncovalent interactions (ionic, H-bond, π-cation, or stacking) and not less than four sidechain-sidechain hydrophobic contacts.
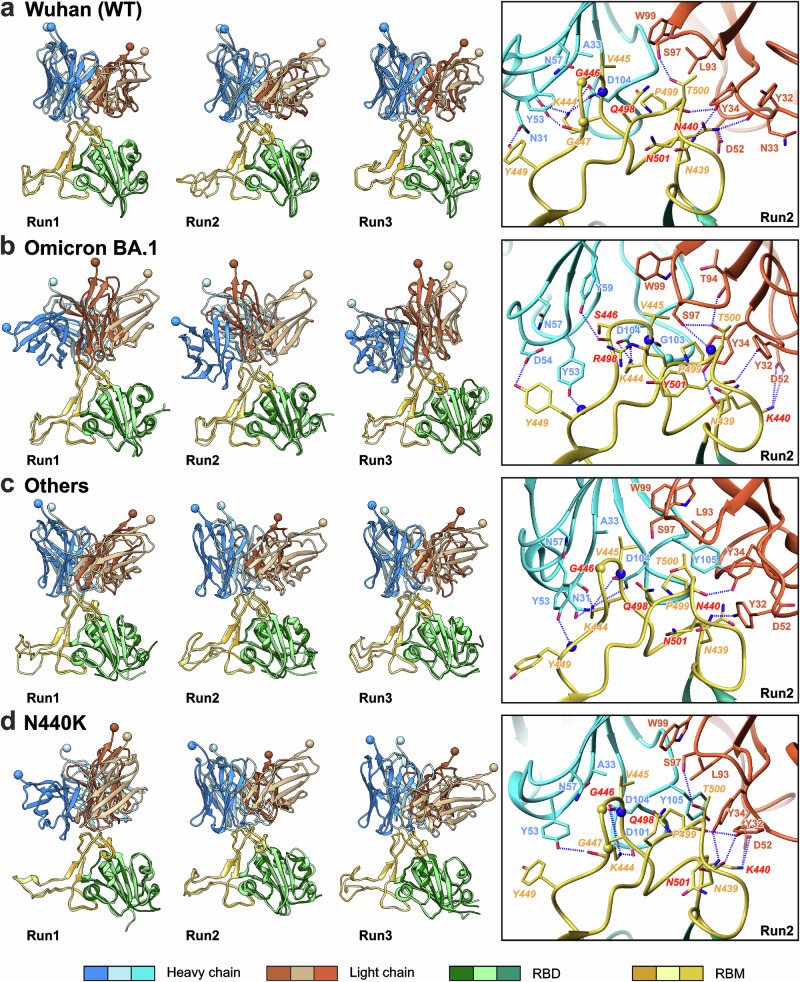
Fig. 6. Extreme (left panels) and mean (right panels, boxed) structures of the first eigenmode of motion from the 500–1000 ns part of the RBD/REGN10987-Fab MD trajectories.
RBD complexes with the Fab’s N-terminal domain are shown for three MD replicas (Run1 to Run3) for the RBD variants Wuhan (a), Omicron BA.1 (b), Delta/‘‘Others’’ (c), and Delta/N440K (d). Residues mutated in the Omicron BA.1 variant are shown in red font. CA atoms of the Gly RBD residues, backbone NH atoms, and backbone CO-groups are shown as yellow spheres, blue spheres, and sticks, respectively. To illustrate the amplitude of motions, the C′ atoms of residues A121H and G111L of the Fab’s N-terminal domain are shown as spheres.
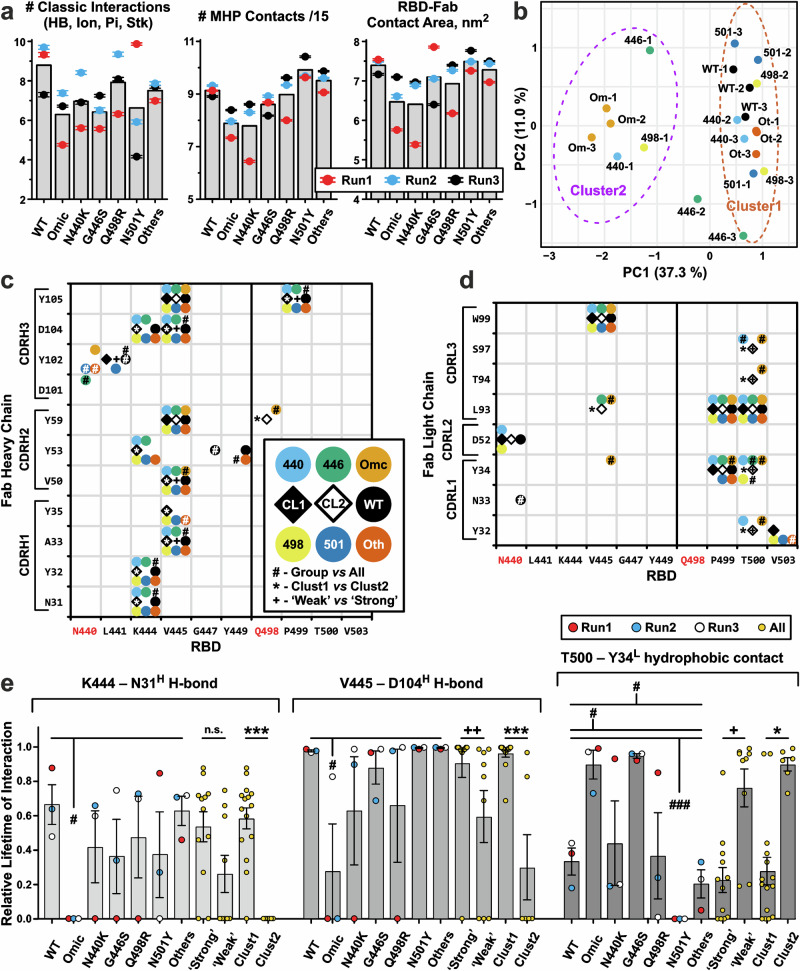
Comparison of replicas (Fig. 7a) revealed that the Omicron, N440K, G446S, and N501Y RBD variants formed fewer ‘classical’’ interactions with Fab, than the Wuhan, Q498R, and ‘Others’’ variants (6.6 vs 8.1 interactions, respectively). This was partly due to the lower number of contacts in some replicas. A similar situation was observed for molecular hydrophobicity potential (MHP) contacts53, which include hydrophobic-hydrophobic and polar-polar intermolecular interactions, and for the contact area in the complex (Fig. 7a). Thus, the Omicron and N440K RBD variants formed weaker complexes (average number of MHP contacts 120 vs 140 for the other RBD variants; average contact area 650 Å2 vs 730 Å2).
Fig. 7. Intermolecular interactions in the 500–1000 ns parts of the RBD/REGN10987-Fab MD trajectories.
a Average characteristics of MD trajectories (three replicas for each RBD variant, Run1 to Run3). The total number of ‘classic’’ RBD-Fab interactions (ionic, H-bond, π-cation, and stacking) and molecular hydrophobicity potential (MHP) contacts divided by 15, as well as the RBD-Fab contact area in the complex are shown. The scaling factor ‘15’’ was used to approximately equalize the number of ‘classic’’ and MHP contacts. MHP contacts include hydrophobic-hydrophobic and polar-polar intermolecular interactions. Data are mean ± S.E.M. (n = 500, 1 point per 1 ns of MD simulation). Bars denote means calculated over the three replicas. b Clustering of the MD replicas by K-means algorithm. Ellipses on a PCA plot show the 95% confidence interval of the clusters. Each MD trajectory was represented by a vector containing the lifetimes of all observed RBD-FAB contacts (Supplementary Data 1-9). Maps of the intermolecular contacts of the heavy (c) and light (d) Fab chains with RBDs. Interactions with an average (over the three replicas) lifetime ≥35% are shown as circles of different colors for different RBD variants. The 35% threshold was chosen to exclude situations where an interaction is presented in only one replica. Open and filled diamonds indicate interactions observed in 35% of trajectories from Cluster2 (trajectories Omic-1, Omic-2, Omic-3, 440-1, 446-1, and 498-1) and Cluster1 (all others trajectories), respectively. e Lifetimes (mean ± S.E.M., n = 3 independent MD replicas) of some intermolecular interactions normalized to the MD replica length. (c–e) #, *, + (p < 0.05), ++ (p < 0.01), and ###, ***(p < 0.001) indicate significant difference in the interaction lifetimes in the following comparisons: (#) the replicas of a particular RBD variant vs all other replicas (cumulatively); (*) Cluster1 vs Cluster2 replicas; and (+) MD trajectories of ‘Weak’’ vs ‘Strong’’ binders, respectively, according to the Mann–Whitney test. No correction for multiple comparisons was used. Group of ‘Strong’’ binders includes WT, ‘Others’’, Q498R, and N501Y RBDs. Group of ‘Weak’’ binders includes Omicron, N440K, and G446S RBDs. In addition, it was required that the difference in average interaction lifetime between the compared groups was >25%.
All RBD variants (including Omicron) in each MD replica retained the general structure of the complex with Fab, but the detailed set of stabilizing contacts varied between the replicas (Supplementary Data 1-9). Clustering of the replicas on the basis of observed intermolecular contacts using Principal Component Analysis (PCA) and the K-means method (Fig. 7b) showed that the trajectories can be divided into two clusters: Cluster2, including all three Omicron replicas and the first replicas of the N440K, G446S, and Q498R variants, and Cluster1, including all other trajectories. The resulting clustering also illustrates the replicas convergence. The replicas for the Wuhan, Omicron, and ‘Others’’ RBD variants were densely distributed in the PCA plot and hence had good convergence, while the N440K, G446S, Q498R, and N501Y replicas showed wide distributions, indicating low convergence. Additional analysis of the replica convergence is presented in the Supplementary Information and Supplementary Data 1-10.
Analysis of the first eigenmode of motions in the calculated MD traces revealed wobbling movements of Fabs in the RBD binding sites (Fig. 6 and Supplementary Fig. 11, left panels). Comparison of the extreme structures revealed a relatively small amplitude of the motions in the case of the Wuhan, G446S, Q498R, N501Y, and ‘Others’’ RBD variants and a larger amplitude for the Omicron and N440K variants (Fig. 6, left panels). However, root-mean-square fluctuation (RMSF) analysis revealed the highest mobility for the N440K and G446S RBD variants in the complex with Fab, both across the entire RBD and the antibody binding site (Supplementary Figs. 12 and 13). In contrast, the Omicron RBD demonstrated the lowest conformational fluctuations, inferior to those of the Wuhan RBD.
Thus, the dynamics of the Fab complexes with the N440K and Omicron RBD variants demonstrating the lowest affinity for the REGN109876 analog (Fig. 5) differed dramatically. N440K-RBD/Fab exhibited significant variability in the RBD structure and overall complex topology, whereas the Omicron-RBD/Fab complex demonstrated a dynamically confined RBD structure but large variations in the complex topology. The common features of both complexes were a small number of stabilizing intermolecular contacts and a small contact area in the complex compared with other RBD variants.
Intermolecular interactions in the RBD/REGN10987-Fab complexes
The structural similarity of the Fab complexes with the different RBD variants was confirmed by analyzing intermolecular interactions in the MD trajectories (Fig. 7c, d, and Supplementary Fig. 14). Several hydrophobic contacts with significant lifetime (≥35%) were observed for the all RBD variants: V445-V50H, V445-Y59H, V445-Y105H, V445-W99L, P499-L93L, and T500-L93L (RBD/Fab). Additionally, several polar interactions with the Fab heavy chain were conserved for the all RBD variants except Omicron: K444-N31H, K444-Y32H, K444-Y53H, K444-D104H, V445-A33H, V445-D104H. On the other hand, some interactions allowed us to distinguish the specific RBD variants (except Q498R) from the rest of the variants (Fig. 7c, d, # signs). The MD replicas of Cluster1 and Cluster2 were distinguished by 14 intermolecular interactions (Fig. 7c, d, * signs), whereas only nine interactions distinguished the MD trajectories of the RBD variants that, according to the MST data, bind Fab weakly (‘‘Weak’’: Omicron, N440K, and G446S) and strongly (‘‘Strong’’: Wuhan, Q498R, N501Y, and ‘Others’’, Fig. 7c, d, + signs). Lifetime of some of these interactions is shown in Fig. 7e. MD trajectories of the Omicron variant differed from those of all other RBD variants by lifetime and presence of K444-N31H and V445-D104H H-bonds. The Cluster2 trajectories also demonstrated significantly lower lifetime of these H-bonds. On the other hand, lifetime of the hydrophobic contact T500-Y34L was significantly different in the following comparisons: Omicron vs All, G446S vs All, N501Y vs All, Cluster1 vs Cluster2, and ‘Weak’’ vs ‘Strong’’ group (Fig. 7e, right). This contact was observed throughout almost the entire trajectories of the Omicron and G446S variants, 20–80% of the trajectories of the Wuhan, N440K, Q498R, and ‘Others’’ variants, and was not observed for the N501Y variant.
In the case of the Omicron RBD, the mutated residues formed multiple intermolecular contacts with the both Fab chains. Long-lived (lifetime ≥35%) π-cation interactions: K440/Y102H and R498/Y59H were observed (Fig. 7c, d), as well as several transient (lifetime <35%) interactions: ionic bridge K440/D52L, H-bonds between S446 and N57H and D104H, and hydrophobic contact Y501/I96L. However, the intermolecular contacts between Fab and the Delta-RBDs with Omicron point mutations did not always correspond to those in the Omicron/Fab complex. Thus, in contrast to the Omicron variant, the residue K440 in the N440K variant formed the long-lived ionic bridge with D52L. Residue S446 in the G446S variant formed the transient H-bond not with D104H but with Y59H; R498 in the Q498R variant formed the transient ionic bridge with D28L; and Y501 in the N501Y variant did not participate in significant intermolecular interactions with lifetime >10% (Supplementary Data 1-3, 1-4, 1-5, 1-6, 1-7). From the mutated residues, only K440 and R498 were capable to form long-lived intermolecular contacts (Fig. 7c, d). Residue N440 in the several RBD variants, including Wuhan, formed contacts with the same Fab residues (Y102H and D52L) as the mutated residue K440 in the Omicron and N440K variants. This means that the structure of the RBD-Fab complex does not change significantly upon the N440K mutation.
Influence of different RBD mutations on free energy of dissociation of REGN10987-Fab complexes
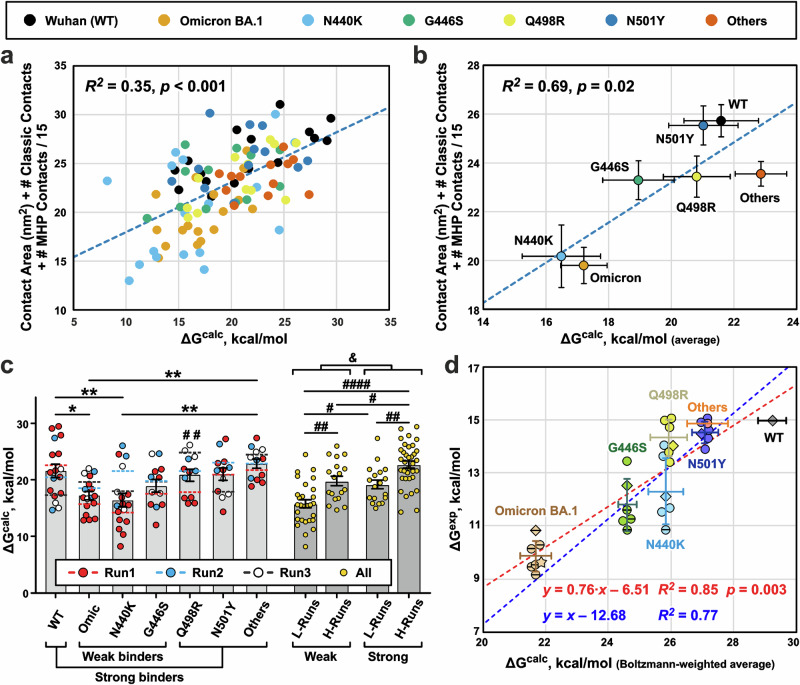
Umbrella sampling simulations and weighted histogram analysis method (WHAM)54,55 were used to determine the free energy changes (ΔG) during dissociation of the complexes of the N-terminal REGN10987-Fab domain with the different RBD variants. The reaction coordinate ξ corresponded to the distance between the centers of mass of the RBD and Fab, projected onto the principal axis Z of the inertia tensor of the complex. (The direction of the vector connecting the centers of mass approximately coincided with the direction of the Z-axis.) Using 43 sampling windows along the Z-axis, one-dimensional potential of mean force (PMF) curve was obtained, from which the free energy of dissociation (ΔGcalc) was calculated (Supplementary Fig. 15). To estimate the uncertainty of the ΔGcalc values, the calculations were repeated using different initial structures taken from the 500–1000 ns region of the MD replicas. For each RBD variant, from 12 to 17 calculations were performed (99 calculations in total, Supplementary Data 1-11).
The ΔGcalc values obtained for the different RBDs showed large scatters, often exceeding 10 kcal/mol (Fig. 8a, c). One of possible reasons for this scatter was the difference in the initial conformations used for the ΔG calculations56. Indeed, the ΔGcalc values demonstrated the significant moderate positive correlation with the structural characteristics of the initial complex (Fig. 8a, b). The squared correlation coefficient R2 between the ΔGcalc values and the sum of the number of ‘classical’’ intermolecular interactions, MHP contacts, and contact area in the complex was 0.35 (n = 99, p < 0.001, Fig. 8a and Supplementary Data 1-12), while R2 for the ΔGcalc values averaged over the RBD variants (arithmetic mean) was 0.69 (n = 7, p = 0.02, Fig. 8b). The lowest ΔGcalc values were obtained for replica 440-1 (Fig. 8c), which was characterized by the smallest RBD/Fab contact area (Fig. 7a). Moreover, the variability in the ΔGcalc values between the replicas exceeded the differences associated with the RBD variants. Dividing the ΔGcalc dataset into 2 × 2 categories (‘‘Weak’’/‘‘Strong’’) binders (RBD factor) and ‘H-Runs’’/‘‘L-Runs’’ (replica factor, MD replicas with high and low average ΔGcalc values) and analyzing with two-way ANOVA revealed the greater variation due to the replica factor (16.0%, p = 3.6 × 10−6) than due to the RBD factor (11.3%, p = 7.9 × 10−5, Fig. 8c).
Fig. 8. Calculation of free energy changes (ΔGcalc) upon dissociation of RBD/REGN10987-Fab complexes.
a ΔGcalc values calculated for the different RBD variants using different initial conformations (n = 99, in total) positively correlate with the structural characteristics of these conformations (Supplementary Data 1-11 and 1-12). b Same as (a), but averaged for each RBD variant (arithmetic mean ± S.E.M., n = 17 for Wuhan, Omicron, N440K RBD variants, and n = 12 for other RBD variants). c Calculated ΔGcalc values vs RBD variant/MD replica (Run1 to Run3). Bars are the averages (arithmetic mean ± S.E.M., the n numbers are the same as in (b)) for the RBD variants, and dashed lines are means for the replicas. *(p < 0.05) and **(p < 0.01) indicate significant differences between the RBD variants according to one-way ANOVA/Tukey test. #(p < 0.05), ##(p < 0.01), and ####(p < 0.0001) indicate significant differences between the four groups of MD replicas according to two-way ANOVA/Tukey test. Groups: Weak+L-Runs (Omic-1, 440-1, and 446-1), Weak+H-Runs (Omic-2, Omic-3, 440-2, 440-3, 446-2, and 446-3), Strong+L-Runs (WT-3, 498-1, 501-3, and Oth-1), Strong+H-Runs (WT-1, WT-2, 498-2, 498-3, 501-1, 501-2, Oth-2, and Oth-3). &(p = 1.7 × 10−6, n = 99 independent ΔG calculations, two-tailed t-test) indicates significant difference between ‘Weak’’ and ‘Strong’’ binders. d Boltzmann-weighted averaged ΔGcalc values (abscissa, Boltzmann-weighted mean ± Boltzmann-weighted SD, N = 12–17, Supplementary Data 1-13) for different RBD variants show a strong positive correlation with the averaged experimental ΔGexp values (ordinate, arithmetic mean ± SD, N = 6, Supplementary Data 1-14). ΔGexp values recalculated from the MST data (Fig. 5) are shown as diamonds. ΔGexp values recalculated from EC50 and Kd values reported in the literature8,35,38,39,57 are shown as circles. Datapoints for the N440K, G446S, and Omicron BA.1 variants corresponding to lower estimates of EC50 or Kd values8,35,38,39,57 are shown by horizontally crossed circles and diamond. ΔGexp value for the Omicron BA.1 variant recalculated from the measured EC50 value52 is shown as a star. Only one experimental data point is present for the ‘Others’’ RBD variant. The different ΔGexp data sets were aligned assuming that the dissociation free energy for WT RBD is 15 kcal/mol. Therefore, only one data point is shown for the WT variant. Blue and red dashed lines represent regressions to linear models with unitary and variable slope, respectively.
Assuming that the ΔGcalc values are functions of the initial conformation and represent the energy of the complex dissociation in that conformation, we can eliminate the influence of ‘bad’’ replicas or ‘weak’’ complexes sampled during MD by using Boltzmann-weighted averaging. In this case, the low ΔGcalc values do not contribute much and the average ΔG value will be closer to the maximal ΔGcalc observed in the dataset rather than their arithmetic mean. The resulting Boltzmann-weighted average ΔGcalc values (Fig. 8d and Supplementary Data 1-13) were in qualitative agreement with the RBD/REGN10987 affinities measured by MST (Fig. 5) and with the literature data7,8 (Supplementary Data 1-14). The Omicron BA.1 and Wuhan variants showed the weakest and strongest affinity, respectively, while the ΔGcalc values for the other RBD variants increased in the following order: Omicron << G446S < N440K ≤ Q498R < N501Y ≤ “Others” < Wuhan.
To compare the ΔGcalc values with experimental ones, we converted the Kd values determined by MST (Fig. 5), as well as the EC50 and Kd values for the REGN10987 interaction with different SARS-CoV-2 or RBD variants reported in the literature7,8,35,38,39,57, to the energies of dissociation (ΔGexp, Fig. 8d). To reduce the variability between different experimental datasets, the ΔGexp values were adjusted so that the value for the WT variant was 15 kcal/mol (this corresponds to EC50 of REGN10987 of 2 ng/ml—the value commonly observed in pseudovirus neutralization experiments). Some of the reported EC50 and Kd values for the N440K and G446S mutants and the Omicron variant were estimates of the values from below (corresponding ΔGexp values are shown by horizontally crossed symbols in Fig. 8d). The additional ΔGexp value for the Omicron variant (Fig. 8d, star) was taken from the work52, where EC50 of neutralization was measured for REGN10987. Boltzmann-averaged ΔGcalc values showed a significant strong positive correlation with the compiled ΔGexp dataset (Fig. 8d, red dashed line, R2 = 0.85, n = 7, p = 0.003). The slope of this linear correlation was less than unity (0.76 ± 0.14, fitted value ± SE), but the difference from unity was not significant (95% CI 0.40–1.13). Fitting the data using a linear model with unit slope (ΔGexp = ΔGcalc + C0, where C0 is a constant) also provided a strong positive correlation (Fig. 8d, blue dashed line, R2 = 0.77). According to the F-test, the use of a more complex model with a variable slope was not statistically justified (p = 0.16). The reason for the systematic shift of the calculated ΔGcalc values compared to the experimental ΔGexp values (non-zero C0 value) remains unclear.
Discussion
Structural data on the interaction of nAbs with different variants of the SARS-CoV-2 S-protein are required to predict the activity of developed therapeutic antibodies and to design new ones against emerging variants of SARS-CoV-2 and other viruses. To our knowledge, the structure of the REGN10987 complex with any variant of the trimeric S-protein has not been reported previously. The only available structure was the low resolution cryo-EM structure of the REGN10987-Fab complex with the WT-RBD24. However, information about the binding epitopes may be lost when considering the isolated RBDs, which adopt ‘down’’ or ‘up’’ conformations in the full-length S-protein trimer with hidden or exposed ACE2 binding sites, respectively17. It has been proposed that REGN10987 recognizes the WT-RBD in the both conformations58, but little was known about its interaction with the RBD of other SARS-CoV-2 variants. The data obtained here show that REGN10987-Fab effectively interacts with the both ‘up’’ and ‘down’’ RBD states of the Delta variant (Fig. 1b, c) and argue in favor of weak binding to the Omicron BA.1 RBD (Fig. 6b).
Comparison with the apo form of the Delta S-protein29,31 revealed that the REGN10987-Fab binding increases the Delta S-protein dynamics. Indeed, the 3DVA analysis showed translational movements of the NTDs and dynamic tilts of the RBD/Fab fragments (Fig. 4). In some 3D classes, the high-amplitude motions resulted in a weakening of the density for the individual RBD/Fab fragments (Fig. 3). The highest mobility was observed for the RBD3-up/Fab3 fragment, possibly reflecting the greater mobility inherent to the RBD in the ‘up’’ state compared to the ‘down’’ state, which is required for better recognition and binding to ACE2.
The next most mobile domain was the RBD2-down (Fig. 3). High-amplitude (~25°) motions of the RBD2/Fab2 fragment, in which the Fab2 tilts to the horizontal axis, were observed by 3DVA (Fig. 4, mode #1). Probably, these motions were related to the attempts of the RBD2 to adopt the ‘up’’ state. Another reason for the increased dynamics of the RBD2/Fab2 fragment may be the spatial proximity of the NTD2 to the mobile RBD3-up. At the same time, the RBD1-down has direct contacts with the RBD3-up (Supplementary Fig. 7a), so one would expect that the RBD1 responds to the all RBD3 movements, but this was not the case. The density corresponding to the RBD1/Fab1 complex was clearly visible in five from six 3D classes, and the 3DVA analysis revealed only the small movements of this complex (within 12–16°, Fig. 4).
Some RBD regions had low resolution even after focused refinement, for example, the A475-C488 loop from the RBM of the RBD1-down interacting with the Y369-F377 region from the neighboring RBD3-up (Supplementary Fig. 7a). Interestingly, these two regions demonstrated the highest mobility in the MD trajectories of the different RBD variants in the complex with the REGN10987-Fab (Supplementary Fig. 12). Thus, the local dynamics of the isolated RBD-Fab complexes reflects the dynamics of the RBD-Fab fragments within the trimeric S-protein.
Obtained structural and dynamic data suggest two independent mechanisms of the virus neutralization by REGN10987. First, the antibody binding site partially overlaps with the ACE2 binding interface on the Delta-RBD (Supplementary Fig. 16). Thus, the REGN10987 binding directly prevents the S-protein interaction with the target receptor. A second mechanism can be proposed considering dynamics of the S-protein/Fab complex. In contrast to the apo S-proteins containing the D614G mutation (D614G, Beta, Delta, Kappa, and Omicron BA.1 variants), which typically adopt multiple conformations, including the S-closed state16,29,31,44,46,59, we observed the Delta S-protein/Fab complex in only one conformation (one-RBD-up S-open, Fig. 1). Probably, the REGN10987-Fab binding shifts the S-protein equilibrium towards open states, as supported by the fluctuations of the ‘down’’ RBD2/Fab2 complex towards the ‘up’’ state (Fig. 4, mode #1). The lack of the contacts between the RBD1-down and RBD2-down (Supplementary Fig. 7b) also means that the REGN10987-Fab binding loosens the S-protein packing, which, together with the S1-subunit transition to the more open conformation, may lead to its shedding and S2 rearrangement to the postfusion structure20. Thus, the REGN10987-Fab binding could convert the S-protein into the inactive form prior to interaction with ACE2. Although, in our study, this scenario could not be realized because we used the special S-protein mutant with non-cleavable linkage between the S1 and S2 subunits14.
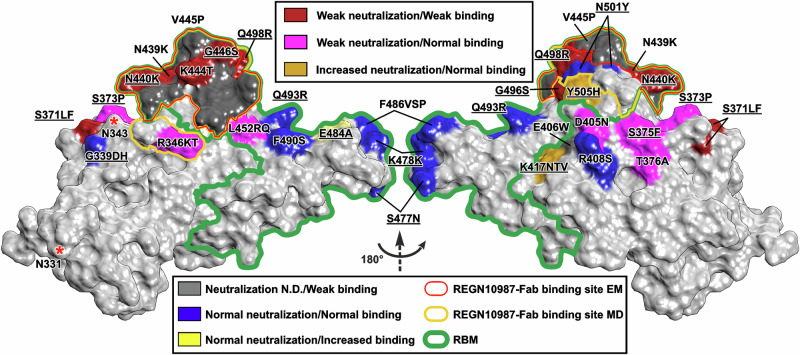
In our cryo-EM structure, limited set of the RBM residues (N439-L441, S443-N450, P499, and T500) form contacts with REGN10987-Fab (Fig. 9, red contour). The ‘contact spot’’ observed by MD is larger and additionally includes T345, R346, Q498, N501-V503, and Q506 (Fig. 9, yellow contour). This ‘contact spot’’ includes four mutations found in the Omicron BA.1 variant (N440K, G446S, Q498R, and N501Y) and four mutations found in other SARS-CoV-2 strains (R346K/T52,60, N439K61, K444T7, V445P7), which impair the REGN10987 binding to the RBD34,62 (Fig. 9, red and gray) and neutralization efficiency7,62 (Fig. 9, red).
Fig. 9. Map of the RBD mutations found in different Omicron variants.
RBM, the site where ACE2 binds, as well as REGN10987-Fab binding sites from cryo-EM structure and MD simulations are shown by green, red, and yellow contours, respectively. Mutations are colored according to their effect on virus neutralization by REGN10987, and according to their effect on the antibody binding. The coloring scheme is shown in the legend. Neutralization data were taken from7,52,60,61. The binding data were taken from8,34,61,62. Mutations found in the Omicron BA.1 variant are underlined. Glycosylation sites are marked with asterisks.
The REGN10987/RBD interaction was most strongly affected by the G446S and N440K substitutions (60- and 120-fold increase in the Kd value, respectively, Fig. 5), which together led to almost complete antibody evasion of the Omicron BA.1 variant (Figs. 5, 7a, and 8d). As it was proposed previously35,39,41, replacement of G446 to the larger serine residue results in the steric clash with the N57H sidechain, whereas N440K substitution results in repulsion from the proximal K55L residue (Fig. 2d). At the same time, this repulsion is compensated by the formation of the new ionic interaction with D52L (Fig. 6b, d). Comparison of the dissociation energies of the Fab complexes with the N440K, G446S, and Omicron RBD variants relative to the WT-RBD (, 3.4 ± 0.7, 4.6 ± 0.5, and 7.6 ± 0.7 kcal/mol, respectively, weighted mean ± weighted SD) revealed an approximate additivity of the values for the N440K and G446S mutations. This indicates that there is no strong epistatic interaction between these mutations, which is consistent with the results of the recent mutagenesis study8.
Previously, no inhibition by the REGN10987 antibody of the Omicron variants simultaneously harboring the N440K and G446S mutations (BA.1, BA.1.1, BA.2.75, BA.3, XBB, and BQ1.1) was detected34,35,38–40,52,60,63, whereas the Omicron variants with only N440K of these mutations (BA.2, BA.2.12.1, BA.2.38, BA.2.76, BA.4/5) were weakly inhibited38,52,60,63. In many cases, the weak BA.1/REGN10987 interaction (if any) was not detected in the neutralization or binding assays due to working within a limited range of the antibody concentrations (Fig. 8d, horizontally crossed symbols). Only in two works the weak BA.1/REGN10987 interaction was detected in the binding41 and neutralization52 experiments, although the binding parameters or complete inhibition curve were not obtained. The reported ~8000-fold increase in the IC50 value for the BA.1 variant relative to the Wuhan/D614G variant52 (Fig. 8d, star) is consistent with estimates of the IC50 increase obtained in other works (3000- to 16,000-fold)7. This, together with the results of our MD simulations and binding energy calculations (Figs. 6b, 7a and 8d), suggests the possibility of the formation of the weak BA.1/REGN10987 complex.
Mutations K444T (BQ.1.1) and V445P (XBB) found in other Omicron variants probably also result in the REGN10987 inactivation7,34,62. These two residues lie at the center of the REGN10987 epitope next to the critical G446 residue (Fig. 9) and form multiple polar and hydrophobic interactions with the antibody heavy chain (Fig. 7c). Interestingly, the XBB Omicron variant simultaneously contains two mutations that ensure the complete REGN10987 evasion: V445P and the N440K/G446S combination. In contrast, the N439K mutation observed in the non-Omicron virus variants (B.1.141, B.1.258, and AV.1) only partially attenuates the neutralizing activity and REGN10987 binding to the RBD61. Residue N439 is adjacent to another critical residue of the REGN10987 binding site—N440, and in the most of the MD replicas of the all RBD variants (except G446S) forms the H-bond with the Y34L sidechain (average lifetime 17–34%, Supplementary Data 1-2, 1-3, 1-4, 1-5, 1-6, 1-7 and 1-8). Thus, this hydrogen bonding is important, but not strictly required for maintaining of the complex structure.
The Q498R Omicron BA.1 mutation ( = 3.3 ± 0.7 kcal/mol) probably also contributes to the REGN10987 recognition. However, intermolecular long-lived interactions involving the R498 residue were observed in the MD trajectories of the Delta/Q498R and Omicron variants (Supplementary Data 1-3 and 1-6). Thus, the Q498R substitution is compensated by the new favorable contacts, which leads to a relatively small increase in the Kd value (5-fold, Fig. 5).
All other Omicron BA.1 mutations, including N501Y, probably have a little effect on the interaction with REGN10987 ( = 2.2 ± 0.6 and 2.1 ± 0.8 kcal/mol for the Delta/N501Y and ‘Others’’ RBD variants, respectively). The N501 residue is located close to the REGN10987 binding site, but it does not form direct contacts with Fab in the cryo-EM structure (Fig. 2c). In some MD trajectories, it forms only transient intermolecular H-bonds or hydrophobic contacts (Supplementary Data 1-2, 1-3, 1-4, 1-5, 1-6, 1-7 and 1-8). According to the literature7 and our MST data (Fig. 5), this mutation does not significantly affect the pseudovirus neutralization and antibody affinity. Interestingly, the N501Y substitution has been described as a compensatory mutation restoring the RBD affinity to ACE2, which was impaired by other mutations7–9. The N501Y mutation appeared already in the SARS-CoV-2 Alpha strain, for which a significant enhancement in affinity for ACE2 as well as increased infection and transmission have been reported64.
Consistent with our ΔG calculations, other RBD mutations described for the SARS-CoV-2 variants that lie outside the REGN10987 binding interface do not affect the antibody’s affinity in the most cases8,34,62 (Fig. 9, except the mutations S371L/F, G496S, E484A, and E406W (the E406W mutation is absent in known virus variants)). However, there is a ring of nearby residues whose substitution increases or decreases the REGN10987 neutralizing activity7,52,60,61 without changing its affinity8,34,61,62 (Fig. 9, magenta and gold). For example, the residue at the position 346 is likely responsible for the weak neutralizing activity of REGN10987 against the Omicron variant BA.1 (R346) but not the Omicron BA.1.1 (K346)52, and for the stronger neutralization of Omicron BA.2 (R346) compared to Omicron BA.2.76 (T346)60. R346 did not interact with REGN10987-Fab in our cryo-EM structure, but the long-lived intermolecular ionic interaction R346-D101H was observed in some MD trajectories for the Wuhan and G446S variants (Supplementary Data 1-2 and 1-5). Probably, other residues from the ‘magenta/gold ring’’ (Fig. 9) can also form the similar dynamic contacts with the antibody, but these were not sampled in our MD simulations. Such dynamic ‘substructural’’ interactions with the antibody could shift the conformational equilibrium of the trimeric S-protein toward a weaker (or stronger) interaction with ACE2 or toward a more open conformation, resulting in idle shedding of the S1 subunits, which could affect the virus neutralization without significant influence on the antibody affinity measured on the isolated RBD or trimeric S-protein.
The S371 residue is located outside the RBM (Fig. 9), and its mutations affect the activity of many nAbs targeting the RBD35. It was suggested that the S371L substitution interferes with N343 glycan35, whose conformation is important for interactions between the adjacent RBDs in the ‘down’’ state and regulates the RBD transition to the ‘up’’ state49. Comparison of the average structures from the MD simulations of the Wuhan and Omicron variants (Supplementary Fig. 17b) suggested that the mutations G339D, S371L, S373P, and S375F affect the conformation of the S366-S375 loop of the RBD and can cause a displacement of the N343 residue (and probably of the attached glycan) by 2 Å toward the REGN10987-Fab binding site. This may create a steric hindrance to the Omicron-RBD/REGN10987 interaction. The Omicron BA.2 variant compared to BA.1 contains two substitutions in this RBD region: S371F and T376A38,57 (Fig. 9). We suggest that these substitutions may also affect the ‘down’’-‘‘up’’ transition of the RBD and the RBD/REGN10987 interaction, and thus can be partially responsible for the antibody evasion.
One of the most surprising findings of the present study is that the ΔG values for dissociation of the RBD/REGN10987-Fab complexes calculated using the classical umbrella sampling approach are varied depending on the initial complex conformation. This scatter (amplitude ~10 kcal/mol) completely obscures the variation in the dissociation free energy due to the RBD mutations (2–7 kcal/mol, Supplementary Data 1-11). Notably, the classical protocol for the ΔG calculation usually involves a single calculation without variation in the initial conformation and the interaction pathway of the molecules. However, both these factors can significantly influence the resulting ΔG value56,65. To address these problems, it was proposed to use a multiple pathway sampling with adaptive-biasing approaches instead of umbrella sampling65.
Here we showed that a simple Boltzmann-weighted averaging of a relatively large set (12–17 values) of the ΔG values calculated using conventional umbrella sampling from different initial conformations taken from MD trajectories yields the free energy of dissociation of the RBD/Fab complex with the estimated accuracy of ~0.5 kcal/mol (Boltzmann-weighted SD, Supplementary Data 1-13). Moreover, the obtained Boltzmann-averaged ΔG values were in a good agreement (R2 ~ 0.8, Fig. 8d). with the compiled dataset of the experimental values that included results of the neutralization and binding studies of the REGN10987 antibody8,35,38,39,52,57 and our binding data (Fig. 5).
The main limitation of the proposed approach is the large systematic shift (amplitude ~13 kcal/mol) between the calculated and experimental ΔG values (Fig. 8d). This can be due to the tendency of Boltzmann-weighting to select the high-energy values. As a result, the calculated averages may depend on the outliers where the ΔG calculation was influenced by steric hindrances to molecule dissociation, resulting in an artificially large change in the PMF profile. However, as shown in Fig. 8c, the calculated ΔG values are fairly uniformly distributed up to the maximum value, and there are no individual outliers towards the large ΔG values. Other possible sources of an error in the ΔG calculations are underestimation of the entropic contributions associated with the rotational degrees of freedom of the interacting molecules66, incomplete accounting of the long-range electrostatic interactions, which could not decay to zero at a maximal separation of the molecules67, and insufficient sampling of the possible dissociation pathways of the molecules65. In any case, we cannot neglect these factors, and the proposed ΔG calculation scheme requires ‘calibration’’ using some known ΔG values to determine this ΔG shift.
In summary, we provided the high-resolution structure of the full-length Delta SARS-CoV-2 S-protein in the complex with REGN10987-Fab. We analyzed the antibody interaction with the Wuhan, Delta, Omicron BA.1, and mutated RBD variants by MST and MD simulations and explained the influence of the known-to-date SARS-CoV-2 RBD mutations on the REGN10987 antibody recognition. We proposed the mechanism of the REGN10987 action. Only three Omicron BA.1 mutations (G446S, N440K, and Q498R) reduced the REGN10987 affinity, whereas N501Y and other mutations found in BA.1 had a very little impact. Our work emphasizes that the static structure of the RBD/antibody complex is not sufficient to understand the mechanisms of an antibody action and evasion. Data obtained will help in a design of new therapeutic antibodies.
Methods
Recombinant proteins
SARS CoV-2 full-length spike-protein B.1.617.2 (Delta) variant in 0.001% LMNG was purchased from Cube Biotech (Germany). In addition to the Delta mutations (T19R, del 157-158, L452R, T478K, D614G, P681R, and D950N), the S-protein contained following modifications: furin cleavage site 682RRAR685 mutated to GSAG, two stabilizing mutations K986P and V987P, C-terminal Rho1D4 tag fused with spacer GSSG to the protein sequence.
To obtain recombinant analog of REGN10987 antibody, variable regions of the REGN10987 antibody’s gene were assembled from synthetic oligonucleotides (Supplementary Table 2, Lytech Co. Ltd, Moscow, Russia) and incorporated into the pVITRO1-dV-IgG1/κ plasmid (Addgene, USA), which encoded light and heavy chains of IgG1 with signal peptides, according to the published protocol68. As a result, the variable regions of IgG1 were replaced by variable regions of the REGN10987 antibody (Supplementary Table 1). The resulting plasmid was transfected into Expi293F cells using the Expi293 Expression System Kit (ThermoFisher Scientific, USA). After transfection, the cells were cultivated for 72 h in 5% CO2 at 37 °C upon stirring at 125 rpm. Next, the cells were collected by centrifugation and the protein was isolated from culture medium using Protein G Sepharose 4 Fast Flow (Cytiva, USA). The Fab fragment was obtained by papain hydrolysis of the antibody and purified using A-Sepharose (Roche, Switzerland) according the manufacturer’s protocol. Papain was obtained from Sigma-Aldrich (MilliporeSigma, USA).
To obtain the recombinant RBD of the Wuhan SARS-CoV-2 variant, the original sequence was amplified from the pCAGGS-SpikeRBD-His6 plasmid kindly provided by Florian Krammer69. DNA sequences encoding the RBDs of the Omicron, Delta, and Delta mutant variants were synthesized using PCR-based site-directed mutagenesis (the list of oligonucleotide sequences is provided in Supplementary Data 1-15, oligonucleotides were synthesized by Lytech Co. Ltd) and incorporated into the pcDNA-3.4 plasmid (ThermoFisher Scientific, USA) with an additional multiple cloning site digested by XbaI/XhoI. The resulting plasmids, encoding the RBD with a signal peptide and a C-terminal 6His-Tag, was transfected into Expi293F cells, and the cells were cultivated and collected as described above. The proteins were isolated from culture medium by metal chelate chromatography using a Ni2+-Sepharose High Performance resin (GE Healthcare, USA).
Cryo-EM sample preparation, data collection, and processing
The Delta S-protein in 20 mM HEPES, 150 mM NaCl, pH 7.5, 0.001% LMNG at concentration 1.3 mg/ml (~9.2 µM) was mixed with REGN10987-Fab from the stock in 20 mM HEPES, 150 mM NaCl, pH 7.5, 0.001% LMNG with concentration 6 mg/ml (~130 µM) to molar ratio 1:1. The mixture was incubated 30 min at room temperature, and 3.5 µl of the S-protein/Fab complex was applied to Quantifoil R1.2/1.3 200-mesh grids (Quantifoil Micro Tools GmbH, Germany), which were glow discharged for 30 s at 20 mA using GloQube Glow Discharge system (Quorum Technologies, UK). The grids were blotted for 6 s using blot force 0 and then plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 100% humidity and 4 °C.
Data were collected on Thermo Scientific Krios™G4 Cryo-TEM equipped with an E-CFEG, a Thermo Scientific Selectris™ X Energy Filter, and a Falcon 4 Detector operated in Electron-Event Representation (EER) mode. A total of 1434 movies were recorded using EPU software (Thermo Fisher Scientific) with 6 µm aberration-free image shift (AFIS) at nominal magnification of ×130,000 and a pixel size of 0.93 Å/pixel, with a defocus range between 0.8 and 1.6 µm at a total dose of 50 e−/Å2 per movie. The slit width of the energy filter was set to 10 eV.
All movies were imported into Relion 3.170 for preprocessing (Supplementary Fig. 1). Motion correction was performed using Relion’s implementation of the MC2 algorithm71, then CTF was estimated in CTFFIND72 using the sum of power spectrums from the movie frames. Next, crYOLO 1.7.673 was used for particle picking, resulting in 147977 particles which were extracted at 3.716 Å pixel size for 3D classification using global search against EM map of SARS-CoV-2 Delta variant spike protein (EMD-24982)29 as an initial model. 44323 particles were selected and re-extracted with a pixel size of 1.18 Å, followed by 3D refinement without symmetry and with C3 imposed, resulting in a 2.78 Å structure. “C3” map was used to consequentially refine defocus, anisotropic magnification, and high-order aberrations followed by reconstruction with already found angles and offsets, which lead to the resolution of 2.76 Å. This symmetry-imposed map was then used for particle polishing using a mask covering only well resolved areas of the S-protein. Polished particles were used for another round of 3D refinement and aberration refinement, archiving 2.36 Å resolution. Particles were imported into cryoSPARC 3.351, aberration values were re-estimated and Non-uniform refinement74 was performed both with and without symmetry, resulting in the 2.26 Å (C3) and 2.53 Å (C1) structures. Particle poses from C1 refinement and the corresponding structure were then used for signal subtraction and local refinement for each RBD/Fab region of the structure, providing 3.15–3.38 Å resolution. Finally, to address significant flexibility of the complex, 3D classification was performed without alignment (PCA initialization) with six classes and 3D Variability analysis75 with four modes and а low pass of 9 Å. Local sharpening was applied to all structures for further analysis.
All structures without a symmetry (S-protein with RBDs masked out, and 3 refined RBD/Fab regions after local refinement) were added to the same grid in Chimera v1.1576 prior to modeling. For an initial model, the structure of the Delta variant S-protein of SARS-CoV-2 with a similar conformation of RBDs was used (PDB: 7SBL29). As the previous structure of REGN10987-Fab (PDB: 6XDG24) was built using a significantly lower resolution map, AlphaFold277 was used to predict a structure of the Fab. Further modeling was performed in ISOLDE v1.378, Phenix79, and Coot v9.680. Visualization and analysis of the structures was performed using Chimera v1.1576, ChimeraX v1.381, and MolMol82.
Molecular dynamics of the RBD/REGN10987-Fab complexes
Starting structures for MD simulations of the different RBD variants in complex with REGN10987-Fab were calculated by introducing mutations in the cryo-EM structure of the Delta RBD/REGN10987-Fab complex using the PyMOL program (Version 2.5 Schrödinger, LLC). MD simulations were carried out in Gromacs version 2020.6 software83 compiled with Nvidia GPU acceleration support. All-atom Amber99sb-ildn84 forcefield was used for proteins, tip3p model was used for water.
Ionization states of amino acid residues were selected according to pH 7.5 (i.e., charged N- and C-termini, Arg, Asp, Glu, and Lys residues). Since pKa values of all His residues, calculated using DelphiPka software85, were in range 6.2–6.8, we selected uncharged states for all His residues. Choosing between Nδ1-H and Nε2-H tautomers of His was done manually by checking H-bonds: His192 and His201 of the Fab light chain are Nδ1-H and other His are Nε2-H.
The RBDs without glycan moieties were used for MD calculations. As it was shown recently, the absence of glycan moieties does not strongly affect the conformation of the Delta and Omicron RBDs86. The starting structures were placed in the center of the dodecahedron box at a distance of at least 1.2 nm from the edge of the box and solvated with water. Na+ and Cl− ions were added to reach up to 150 mM salt concentration and then Cl− ions were added to neutralize the system charge.
All simulations were performed with periodic boundary conditions, Verlet cutoff scheme, plain cutoff of 1.2 nm for van-der-Waals and particle-mesh Ewald method with potential-shift-verlet-modifier for Coulomb interactions, and long-range dispersion corrections for energy and pressure. Simulation included four stages: energy minimization in double precision, heating from 5 to 298 K during 1 ns with 1 fs timestep using V-rescale thermostat and fixed positions of protein heavy atoms (except atoms of mutated residues), equilibration during 40 ns with timestep 2 fs using V-rescale thermostat (298 K) and Parrinello-Rahman isotropic barostat (1 bar), three production runs (replicas) 1000 ns each with the same parameters and different initial velocities. Lincs algorithm was employed to constrain the bonds with H-atoms in MD simulations except for the heating phase.
PCA analysis of the MD replicas on the basis of intermolecular RBD-FAB contacts (Supplementary Data 1-9) was performed using the web-based version of the MetaboAnalyst software87.
Calculation of dissociation free energies (ΔGcalc)
To reduce the system size, ΔGcalc values were calculated for the different RBD variants (residues 333–526) and the N-terminal domain of Fab (light chain: residues 3–111; heavy chain: residues 1–121). Structures of the complexes were extracted from the second part (500–1000 ns) of MD trajectories. All complexes were oriented along the principal axes of inertia tensor so that the largest size was along the Z axis. Complexes were centered in identical rectangular boxes satisfying the following criteria: distance between the complex and box edge is at least 1.2 nm; boxes have identical sizes along the X and Y axes. Then the simulation boxes were extended in the Z direction by 8 nm and finally had size 9.34 × 9.34 × 18.71 nm. The complexes were solvated, ions were added, energy minimization and heating were performed using the same protocol as in MD simulation of the RBD/Fab complexes. Backbone atoms of 5-stranded β-sheet of the RBD (residues 354–358, 375–380, 393–403, 431–437, and 507–516) were fixed and distance along the Z axis between the centers of mass of the RBD and the Fab’s N-terminal domain was considered as a ‘reaction coordinate’’ ξ for the potential of mean force (PMF) method54,55. Initial configurations for umbrella sampling were generated by running MD simulation with timestep of 2 fs during 3 ns with applying ‘constant velocity’’ 2 nm/ns to the ‘reaction coordinate’’ distance and saving conformation each 1 ps. ‘Reaction coordinate’’ distances were calculated for all saved conformations, and 43 conformations for each system were selected as starting structures for umbrella sampling run. The selection procedure was as follows: minimum distance was rounded to the nearest multiple of 0.01 nm, desired distances were 20 distances from the rounded minimum with step of 0.05 nm, then 10 distances with step of 0.1 nm, and 13 distances with step of 0.2 nm. Conformations with the distances nearest to desired one were selected. In some cases, two or three additional conformations were selected in regions with lack of sampling.
Umbrella sampling simulations were performed for all selected starting structures during 5 ns, using Berendsen thermostat (298 K) and isotropic barostat (1 bar), saving the coordinates and forces every 10 ps. The ‘reaction coordinate’’ ξ was restrained at a starting position with harmonic potential with force constant 1000 kJ/mol/nm2. All other MD parameters were the same as in MD simulation of the RBD/Fab complexes. Trajectories from 1 ns to 5 ns were analyzed using the WHAM to calculate PMF profile along reaction coordinate55. Errors in PMF profiles were estimated by Bayesian bootstrapping of complete histograms with 200 starts. Final ΔGcalc values were calculated by averaging of the PMF values at the region, where the ‘reaction coordinate’’ distance was >7 nm. ΔGcalc errors were estimated as the sum in quadrature of average PMF error and averaging error. The typical error in the energy calculation was about 1 kcal/mol. At the same time, the errors introduced by variations in the initial conformations used to calculate ΔG were higher and reached a level of ~4 kcal/mol (unweighted SD) or ~0.5 kcal/mol (Boltzmann-weighted SD) only after averaging 12–17 values calculated with different initial structures (Supplementary Data 1-11 and 1-13). Therefore, averaging errors were used to represent the uncertainty in the ΔGcalc values.
Note that the WHAM, implemented in the Gromacs software, adds a kB·T·ln(4π·ξ2) term to the PMF to eliminate the entropic decrease in the PMF due to the increase in the number of configurations on a sphere of radius ξ. So, the umbrella sampling algorithm used is slightly different from the classical one54. Fab orientation and conformation were not constrained during sampling, whereas in RBD the position of the secondary structure elements was fixed by fixing their backbone atoms (see above). Thus, the calculated PMF profiles contained all ΔG terms except the free energy changes due to the change in RBD conformation upon Fab dissociation.
Microscale thermophoresis
MST assay was performed to verify the RBD interactions with the REGN10987 antibody. Recombinant His-tagged RBD of Wuhan, Delta, Delta/N440K, Delta/G446S, Delta/Q498R, Delta/N501Y, and Omicron SARS-CoV-2 variants were labeled using Red-NHS 2nd Generation kit (Nanotemper, Germany) according to the manufacturer’s protocols. Labeled RBDs were mixed with the antibody in the working buffer (10 mM sodium phosphate, pH 7.4, 140 mM NaCl, 3 mM KCl, 0.05% Tween-20) to a final RBD concentration of 40 nM and a final antibody concentration of 0–6.6 µM. The mixtures were stored at room temperature for 15 min prior to MST measurements. MST curves were registered at 25 °C using Monolith NT.115 (NanoTemper, Germany) equipped with a RED/GREEN detector with MST monitoring in RED mode. The dependence of the normalized fluorescence on the antibody concentration was analyzed using the MO. Affinity Analysis software (NanoTemper, Germany), and Kd values were obtained by fitting the experimental data to equation:
where ∆F is antibody-induced change in relative fluorescence, [RBD] is the total RBD concentration (40 nM), and [Ab] is the total antibody concentration.
The Kd values obtained by MST and EC50 and Kd values reported in the literature8,35,38,39,52,57 (EC50 data is reviewed in ref. 7) were converted to the free energy of the RBD/Fab complex dissociation (ΔGexp) using following equations:
where and —dissociation constants of the REGN10987 complexes with the mutant and WT RBDs reported in an original publication; and —the 50% inhibition concentrations of pseudoviruses containing mutant and WT RBDs by REGN10987 reported in an original publication; —the free energy of dissociation of the WT-RBD/REGN10987-Fab complex, taken to be 15 kcal/mol. The value 15 kcal/mol corresponds to EC50 of neutralization of 2 ng/ml or Kd of the RBD-REGN10987 complex dissociation of ~14 pM. The Wuhan or Wuhan/D614G SARS-CoV-2 variants were considered as wildtype variants (WT variants).
Statistics and reproducibility
Sample numbers (n), the type of averaging used, and the type of error bars are indicated in the figure legends. No exclusion criteria were applied for experimental data. The normality of the data was assessed using Shapiro–Wilk test. The data having normal distribution were analyzed using the one- or two-way ANOVA with appropriate multiple comparisons post-hoc test, or two-tailed t-test. The datasets where half or more sub-groups did not pass the normality test (e.g., interaction lifetimes) were analyzed using the two-tailed Mann-Whitney test. Differences in the data were considered statistically significant at p < 0.05. Analysis was performed using the GraphPad Prism 9.0 software (San-Diego, USA).
To ensure that the results were reproducible, three independent MST measurements were performed for each RBD variant using independently prepared solutions, but using RBD samples prepared during the same recombinant production. All MD simulations were also performed in three independent replicates. The ΔG calculations for each RBD variant were independently repeated at least 12 times. Structure determination by cryo-EM was considered reproducible.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
The work was supported by the Shenzhen High-level University Construction Funding. E.N.L. and M.P.K. are part of an innovative drug development team based on structural biology and bioinformatics at MSU-BIT University in Shenzhen (#2022KCXTD034). We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Lopukhin Federal Research, and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency for RBD expression and purification.
Author contributions
E.N.L., A.M.V., and Z.O.S. designed the study. E.B.P., D.E.N., M.V.K., V.A.M., D.A.S., D.D.K., E.N.G., J.I.S., V.N.L., and A.M.V. conducted experiments and analyzed the data. E.N.L., E.B.P., D.E.N., A.M.V., and Z.O.S. wrote the manuscript. E.N.L., Z.O.S., and M.P.K. acquired funding and supervised the project. All authors read and approved the final manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Janesh Kumar and Laura Rodríguez Pérez. A peer review file is available.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Source data for Figs. 5, 7 and 8 are provided in Supplementary Data 2. The obtained cryo-EM data were deposited in Electron Microscopy Data Bank (EMDB) under the accession number EMD-14750. Atomic coordinates for the S-protein-Delta/REGN10987-Fab complex have been deposited in RCSB Protein Data Bank (PDB) under the accession code 7ZJL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ekaterina N. Lyukmanova, Evgeny B. Pichkur, Dmitry E. Nolde.
Contributor Information
Ekaterina N. Lyukmanova, Email: lyukmanova_ekaterina@smbu.edu.cn
Zakhar O. Shenkarev, Email: zakhar-shenkarev@yandex.ru
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07422-9.
References
- 1.Focosi, D. et al. Monoclonal antibody therapies against SARS-CoV-2. Lancet Infect. Dis.22, e311–e326 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo, M. et al. Multi-omics for COVID-19: driving development of therapeutics and vaccines. Natl. Sci. Rev.10, nwad161 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padhi, A. K., Kalita, P., Maurya, S., Poluri, K. M. & Tripathi, T. From de novo design to redesign: harnessing computational protein design for understanding SARS-CoV-2 molecular mechanisms and developing therapeutics. J. Phys. Chem. B127, 8717–8735 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Liang, L., Wang, B., Zhang, Q., Zhang, S. & Zhang, S. Antibody drugs targeting SARS-CoV-2: time for a rethink? Biomed. Pharmacother.176, 116900 (2024). [DOI] [PubMed] [Google Scholar]
- 5.Carabelli, A. M. et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol.21, 162–177 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol.21, 361–379 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Xue, S., Han, Y., Wu, F. & Wang, Q. Mutations in the SARS-CoV-2 spike receptor binding domain and their delicate balance between ACE2 affinity and antibody evasion. Protein Cell15, 403–418 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moulana, A. et al. The landscape of antibody binding affinity in SARS-CoV-2 Omicron BA.1 evolution. eLife12, e83442 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao, Z., Zhang, L., Duan, Y., Tang, X. & Lu, J. Molecular insights into the adaptive evolution of SARS-CoV-2 spike protein. J. Infect.88, 106121 (2024). [DOI] [PubMed] [Google Scholar]
- 10.Wang, D., Huot, M., Mohanty, V. & Shakhnovich, E. I. Biophysical principles predict fitness of SARS-CoV-2 variants. Proc. Natl. Acad. Sci. USA121, e2314518121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shanker, V. R., Bruun, T. U. J., Hie, B. L. & Kim, P. S. Unsupervised evolution of protein and antibody complexes with a structure-informed language model. Science385, 46–53 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosch, B. J., van der Zee, R., de Haan, C. A. M. & Rottier, P. J. M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol.77, 8801–8811 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang, J. et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA.117, 11727–11734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benton, D. J. et al. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. USA118, e2022586118 (2021). [DOI] [PMC free article] [PubMed]
- 17.Yan, R. et al. Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2. Cell Res.31, 717–719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tortorici, M. A. & Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res.105, 93–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai, Y. et al. Distinct conformational states of SARS-CoV-2 spike protein. Science369, 1586–1592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, Y. et al. Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat. Rev. Immunol.23, 189–199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavor, E., Choong, Y. K., Er, S. Y., Sivaraman, H. & Sivaraman, J. Structural basis of SARS-CoV-2 and SARS-CoV antibody interactions. Trends Immunol.41, 1006–1022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, J. et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science369, 1010–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dejnirattisai, W. et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell184, 2183–2200.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baum, A. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science369, 1014–1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Brien, M. P. et al. Subcutaneous REGEN-COV antibody combination to prevent Covid-19. N. Engl. J. Med.385, 1184–1195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyashita, N. et al. Clinical efficacy of casirivimab-imdevimab antibody combination treatment in patients with COVID-19 Delta variant. J. Infect. Chemother.28, 1344–1346 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, J. et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science374, 1353–1360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Planas, D. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Wang, Y. et al. Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat. Commun.13, 871 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saville, J. W. et al. Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants. Nat. Commun.13, 742 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannar, D. et al. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science375, 760–764 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao, Y. et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature602, 657–663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, L. et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature602, 676–681 (2022). [DOI] [PubMed] [Google Scholar]
- 36.VanBlargan, L. A. et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 10.1038/s41591-021-01678-y (2022). [DOI] [PMC free article] [PubMed]
- 37.Dejnirattisai, W. et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell185, 467–484.e15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ai, J. et al. Antibody evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell Host Microbe30, 1077–1083.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tada, T. et al. Increased resistance of SARS-CoV-2 Omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. EBioMedicine78, 103944 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, T. et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science376, eabn8897 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCallum, M. et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science375, 864–868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, M. et al. Real-time conformational dynamics of SARS-CoV-2 spikes on virus particles. Cell Host Microbe28, 880–891 (2020). [DOI] [PMC free article] [PubMed]
- 43.Díaz-Salinas, M. et al. Conformational dynamics and allosteric modulation of the SARS-CoV-2 spike. eLife11, e75433 (2022). [DOI] [PMC free article] [PubMed]
- 44.Zhang, J. et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science372, 525–530 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, C. et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv.7, eabe5575 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Y. et al. Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM. Nat. Commun.12, 7345 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin, W. et al. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science375, 1048–1053 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wrobel, A. G. et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol.27, 763–767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sztain, T. et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem.13, 963–968 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toelzer, C. et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science370, 725–730 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Wang, Q. et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature608, 603–608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efremov, R. G. et al. Molecular lipophilicity in protein modeling and drug design. Curr. Med. Chem.14, 393–415 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Woo, H.-J. & Roux, B. Calculation of absolute protein-ligand binding free energy from computer simulations. Proc. Natl. Acad. Sci. USA102, 6825–6830 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hub, J. S., de Groot, B. L. & van der Spoel, D. g_wham—a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput.6, 3713–3720 (2010). [Google Scholar]
- 56.You, W., Tang, Z. & Chang, C. A. Potential mean force from umbrella sampling simulations: what can we learn and what is missed? J. Chem. Theory Comput.15, 2433–2443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Y. et al. Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies. Cell Discov.8, 104 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du, S. et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell183, 1013–1023.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gobeil, S. M.-C. et al. Structural diversity of the SARS-CoV-2 Omicron spike. Mol. Cell82, 2050–2068.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao, Y. et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe30, 1527–1539.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomson, E. C. et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell184, 1171–1187.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starr, T. N. et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science371, 850–854 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iketani, S. et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature604, 553–556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu, Y. et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature602, 294–299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aho, N., Groenhof, G. & Buslaev, P. Do all paths lead to Rome? How reliable is umbrella sampling along a single path? J. Chem. Theory Comput.20, 6674–6686 (2024). [DOI] [PubMed] [Google Scholar]
- 66.Gumbart, J. C., Roux, B. & Chipot, C. Efficient determination of protein-protein standard binding free energies from first principles. J. Chem. Theory Comput.9, 3789–3798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suh, D., Jo, S., Jiang, W., Chipot, C. & Roux, B. String method for protein-protein binding free-energy calculations. J. Chem. Theory Comput.15, 5829–5844 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Dodev, T. S. et al. A tool kit for rapid cloning and expression of recombinant antibodies. Sci. Rep.4, 5885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amanat, F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med.26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol.192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol.2, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Punjani, A. & Fleet, D. J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol.213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 76.Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. Struct. Biol.74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. Struct. Biol.75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr.66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci.30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph.14, 29–32 (1996). 51–55. [DOI] [PubMed] [Google Scholar]
- 83.Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX1–2, 19–25 (2015). [Google Scholar]
- 84.Lindorff-Larsen, K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins78, 1950–1958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang, L., Zhang, M. & Alexov, E. DelPhiPKa web server: predicting pKa of proteins, RNAs and DNAs. Bioinformatics32, 614–615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ives, C. M. et al. Role of N343 glycosylation on the SARS-CoV-2 S RBD structure and co-receptor binding across variants of concern. eLife13, RP95708 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pang, Z. et al. MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res.52, W398–W406 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Source data for Figs. 5, 7 and 8 are provided in Supplementary Data 2. The obtained cryo-EM data were deposited in Electron Microscopy Data Bank (EMDB) under the accession number EMD-14750. Atomic coordinates for the S-protein-Delta/REGN10987-Fab complex have been deposited in RCSB Protein Data Bank (PDB) under the accession code 7ZJL.