Abstract
Recent genetic studies have linked numerous loci to psychiatric disorders. However, the biological pathways that connect these genetic associations to psychiatric disorders’ specific pathophysiological processes are largely unclear. Endophenotypes, first defined over five decades ago, are heritable traits, independent of disease state that are associated with a disease, encompassing a broad range of neurophysiological, biochemical, endocrinological, neuroanatomical, cognitive, and neuropsychological characteristics. Considering the advancements in genetics and genomics over recent decades, we propose a revised definition of endophenotypes as ‘genetically influenced phenotypes linked to disease or treatment characteristics and their related events.’ We also updated endophenotype criteria to include (1) reliable measurement, (2) association with the disease or its related events, and (3) genetic mediation. ‘Genetic mediation’ is introduced to differentiate between causality and pleiotropic effects and allows non-linear relationships. Furthermore, this updated Endophenotype 2.0 framework expands to encompass genetically regulated responses to disease-related factors, including environmental risks, illness progression, treatment responses, and resilience phenotypes, which may be state-dependent. This broadened definition paves the way for developing new endophenotypes crucial for genetic analyses in psychiatric disorders. Integrating genetics, genomics, and diverse endophenotypes into multi-dimensional mechanistic models is vital for advancing our understanding of psychiatric disorders. Crucially, elucidating the biological underpinnings of endophenotypes will enhance our grasp of psychiatric genetics, thereby improving disease risk prediction and treatment approaches.
Subject terms: Medical genetics, Psychiatric disorders
Endophenotype: classical definition, criteria, and successes
Gottesman and Shields [1] (1973) introduced endophenotypes as measurable phenotypes between illness and genotypes. Gershon, Rieder, and Goldin [2, 3] (1978, 1986) and Gottesman & Gould [4] (2003) specified the classical criteria of endophenotypes for psychiatric genetics listed in Table 1. Others further contributed to the discussion regarding its applications in psychiatric genetics [3, 5–7]. Neurophysiological, biochemical, endocrinological, neuroanatomical, cognitive, and other neuropsychological measures (including self-report questionnaire data) are commonly used endophenotypes. The commonly used endophenotypes for psychosis include measurements derived from electroencephalography (EEG), cognition, eye movements, and brain imaging. EEG responses to repeated external events (Event Related Responses; ERPs), such as auditory stimuli, have long been known as abnormalities in psychosis disorders and have been proposed as critical biological components of psychosis [8]. Impaired cognition is an important feature of schizophrenia (SCZ) [9, 10]. ERP and cognition abnormalities are present before the onset of psychosis symptoms [11–13].
Table 1.
Endophenotype 2.0 revisions: definition and criteria.
| Endophenotype 1.0 | Endophenotype 2.0 | Major changes from 1.0 to 2.0 | |
|---|---|---|---|
| Definition |
Genotype: Not defined. Originally referred to concordance in relatives. Endophenotype: A phenotype that is heritable, co-segregates with a psychiatric illness, yet is present even when the disease is not (i.e. state independent), and is found in non-affected family members at a higher rate than in the population. (per Gottesman & Gould 2003 [4], Rieder, Goldin and Gershon [2, 3]) A heritable intermediate phenotype associated with a disease. [Intermediate phenotype is a mechanism-related manifestation of a complex phenotype.] (per Goldman & Ducci 2007 update [6]). |
Genotype: state of variations in DNA sequence, including single nucleotide polymorphisms, copy number variants, tandem repeats, de novo mutations, and structural variants. Endophenotype: A genetically influenced phenotype between genotype and disease diagnosis. It is linked to disease or treatment characteristics and related events, which include disease symptoms, risk or resilience factors, and responses to treatment, with evidence from family or population data. |
• Expanded the definition to include illness-, risk-, resilience-, and treatment response related features. • Emphasized genetic mediation. • Added molecular and cellular endophenotypes, and many other new phenotypes • Population evidence is emphasized |
| Criteria |
1. The endophenotype is associated with illness in the population. 2. The endophenotype is heritable. 3. The endophenotype is primarily state independent (manifests in an individual whether or not illness is active). 4. Within families, endophenotype and illness co-segregate. 5. The endophenotype found in affected family members is found in non-affected family members at a higher rate than in the general population. (Per Rieder 1978 [3]) |
1. Reliable measurement in individuals 2. Associated with disease or treatment characteristics, and related events. 3. Genetically mediated |
• Removed the requirement of state independence; • Removed the requirement of evidence from family data; • Added illness-related features, which include risk, resilience, and treatment response; • Response to risk environments, such as stress and inflammation, is specifically added; • Genetic mediation can be in many forms, including non-linear relationships; • Causation is emphasized |
| Examples of major types | Neurophysiological, biochemical, endocrinological, neuroanatomical, cognitive, or neuropsychological (including configured self-report data) measures |
• Molecular endophenotypes: genomic and metabolic measurements (including expression, splicing, developmental, and age-related changes in these measurements) and findings derived or imputed from DNA, RNA, protein, or other molecular variations. • Cellular endophenotypes • Treatment- and course-related endophenotypes: response to drugs and response to non-drug treatment. • Response to risk-environment endophenotypes: psychological stress, toxin, insufficient nutrition, sleep condition, and inflammation. |
Historically, the meaning of “genotype” has evolved from measuring similarity in relatives to DNA sequence information, including base-pair level variations (SNPs), insertions, deletions, and structural variants. “Phenotype” refers to any assessable characteristic of an individual beyond genotype. Phenotypes are the products of genotypes, environmental factors, and their interactions throughout the lifespan, ranging from molecular and cellular measurements to high-order behavioral traits.
Genetic study of endophenotypes has been pursued actively in the past decades and has generated some successful examples in studies of attention-deficit/hyperactivity disorder (ADHD), substance use disorders (SUD), and major depression (MDD), as reviewed by Sanchez–Roige and Palmer [14]. Behavioral traits of cognition (including intelligence), eye movement control, emotion, mood, executive function, social function, and personality are impaired in multiple psychiatric disorders. Cognitive deficits are present in children who later develop SCZ [11, 12]. Several notable examples of genetic analysis of behavioral endophenotypes across psychiatric disorders have been reported: shared genetics between education attainment and SCZ [15], neuroticism has been positively correlated with psychotic experiences and with the risk of SCZ [16–18], and depression [19]. Intelligence [20] and “Big Five” personality measures (openness [21, 22], conscientiousness [22], extraversion [23], and neuroticism [24]) have GWAS loci overlapping with loci associated with various psychiatric disorders. The N-back working memory test [25] is one example of a standardized behavioral measure. GWAS of N-back test in 2298 participants estimated SNP-based heritability ranging between 31 and 41%, but no genome-wide significant SNP was reported. Another endophenotype commonly linked to mood disorders is facial expression recognition [26]. A GWAS of facial emotion recognition in 4097 subjects failed to detect genome-wide significant SNP association [27]. Seven genome-wide significant genetic loci have been reported for endophenotypes of spatial processing, abstraction and mental flexibility, antisaccade task, sensorimotor dexterity, face memory, and continuous performance test by the Consortium on the Genetics of Schizophrenia (COGS) Study (N = 1533) [28]. COGA published GWAS of EEG on 8810 individuals and reported one locus on 18q23 associated with low theta EEG coherence [29]. In a GWAS of diffusion tensor imaging (DTI) model-derived parameters along 21 cerebral white matter tracts in nearly 44,000 individuals, 151 SNPs associated with white matter microstructure were discovered [30]. The Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study (N = 849) identified three genome-wide significant associations for eye movement measures [31]. A GWAS of Prepulse inhibition (PPI) (N = 1493) identified two significant loci [32]. However, their meta-analysis (N = 2660) failed to capture any genome-wide significant loci for PPI [33].
Glahn et al. [7]. brought quantitative trait loci of gene expression (eQTL) or proteins (pQTL) into the scope of endophenotypes when there were very limited eQTL data in 2014. They discussed concerns about the classical assumptions regarding the genetics of endophenotypes as compared with the genetics of disease and proposed to study endophenotypes that are closer to gene action, like gene expression.
Consistent with Glahn et al., we propose that the genetic influence on endophenotypes will be stronger for traits closer to DNA than those more distal. Molecular QTLs exhibit significantly higher heritability than higher-order phenotypes, such as brain imaging, electrophysiology, or behavioral traits. The genetic contribution from individual SNPs tends to diminish as information progresses through the Central Dogma of molecular biology [34, 35]. Generally, as phenotypes become more distant from DNA sequences, more regulatory factors (including environmental factors) are involved, decreasing SNP heritability. Therefore, most endophenotypes are likely to exhibit genetic complexity. SNP heritability of these endophenotypes falls in the range between the two extremes represented by molecular traits and disorder diagnoses. Consequently, the sample size required to achieve sufficient statistical power may range from dozens to tens of thousands, even millions, depending on the specific characteristics of the endophenotypes, including their distance to DNA and the number of regulators involved. These values should be determined through population-based studies or large cohorts. Molecular endophenotypes such as gene expression and DNA methylation require much fewer samples for genome-wide study than are required for genome-wide associations with genotypes (GWAS) [36] (Table 2). Significant associations have been reported for eQTL with less than 100 samples, while disease GWAS typically requires tens of thousands.
Table 2.
Commonly used and emerging endophenotypes for psychiatric disorders.
| Biological levels | Endophenotypes | Major existing data | Sample size range in existing data (N) |
|---|---|---|---|
| Molecular | Brain gene expression | 26 studies including PsychENCODE, GTEx, ROSMAP | 44–1866 |
| Brain DNA methylation | 11 studies | 125–494 | |
| Brain protein abundance | Three studies, including PsychENCODE/BrainGVEX, ROSMAP | 133–835 | |
| Brain structure and function | Structural and functional MRI (neurological, structure of the nervous system, structure of brain)a | 373 studies including ENIGMA, BSNP | 327–26,577 |
| EEGb | 5 studies | 191–6644 | |
| Eye movementb | 3 studies | 60–6246 | |
| Cognition (cognitive)a | 78 studies | 441–386,276 | |
| Individual differences in psychological characteristics | Personality (psychiatric, mental function, temperament, and personality)a | 99 studies including UK Biobank | 5228–1,067,913 |
| Sleep (psychiatric, mental function, sleep functions)a | 55 studies | 721–449,732 | |
| Social interaction (social interactions)a | 20 studies | 86,941–452,302 |
Some endophenotype data have been collected along with genotype data in large-scale population-based studies, including the UK Biobank [37, 38], the Adolescent Brain Cognitive Development (ABCD) study [39], and All of Us [40]. These data are primarily questionnaires and non-endophenotype diagnoses, but there is some inclusion of brain imaging, cognition tests, and medical history or records. UK Biobank has phenotypes of health-related outcomes, clinical coding, questionnaires, physical measures, imaging data, Big Five Personality Traits, subjective well-being, anxiety, loneliness, depressive symptoms, intelligence, and education attainment. ABCD collects brain imaging data and neurocognitive measures, including crystallized and fluid reasoning, reading ability, working memory, behavioral inhibition, verbal learning and memory, sequencing skills, processing speed, attention, and visuospatial reasoning. Environmental factors are also emphasized in ABCD, particularly those related to substance use. All of Us collects data from electronic health records and digital health records, substance use, surveys of social behavior, and optional social network data [40].
The UK Biobank has been one of the most productive and used data sources. In addition to brain imaging studies [41–43], GWASs of UK Biobank data have yielded significant genetic associations for traits of cognitive functions and educational attainment, which is pertinent to Schizophrenia and neurodegenerative diseases [44] and many other mental and physical health-related traits [45]. Interestingly, the SNP heritability of intelligence-related traits is particularly strong in socioeconomically deprived people [46]. Other large biobanks are emerging, such as Vanderbilt Biobank (BioVU), the eMERGE (Electronic Medical Records and Genomics) Network, Million Veteran Program (MVP), China Kadoorie Biobank, FinnGen, and Danish National Biobanks. Global Biobank Meta-analysis Initiative (GBMI) is a collaborative network of 23 biobanks representing more than 2.2 million consented individuals with genetic data linked to electronic health records. Although psychiatry is not the major focus of these large biobanks, these biobanks have some endophenotypes, like brain imaging and cognition measures. However, these existing biobanks were designed with the influence of endophenotype 1.0. Endophenotype 2.0 data are mostly not part of these existing biobanks.
Limitations of earlier endophenotype definition and the need for endophenotype 2.0 with new definition and criteria
The classic endophenotype definition and criteria were created before the era of genome-wide association study (GWAS) and genomics. New understandings about disorders, including mental illnesses, their phenotypes, and underlying genetics, have emerged. Here, we term the classic endophenotype definitions Endophenotype 1.0 and call attention to the following four major limitations:
The state independent criterion. The original criteria for endophenotypes required state independence, which excludes many important traits, such as early development stage-specific or age-dependent traits that may have permanent effects but are only detectable in a certain period of life. Temporally dynamic phenotypes (such as prenatal neural differentiation, growth, and migration) may prove critical for understanding disease susceptibility. The developmental hypothesis of psychiatric disorders has been proposed since the 1820s [47] and gradually matured [48–50]. Thousands of genes have very different expression profiles in prenatal development stages than in adults [51, 52].
-
The illness-association criterion. The original criterion is to co-segregate with illnesses. But resilience, response to treatment or risk environment, and course of illness are important research areas for psychiatric genetics. Treatment may or may not be directly associated with disease risk. However, even the side effects of treatment are translationally relevant. Genetic studies of the drug responses have developed into pharmacogenetics. Responses to non-medication treatments, even placebo effects, may prove equally interesting. Moreover, the genetics of these factors may or may not overlap with the genetics of illness.
Human responses to environmental changes may be regulated by genetic factors. Cannabis use, environmental stress during pregnancy (including famine and certain viral infections), childhood, and adulthood, history of obstetric complications, and low serum folate level are recognized environmental risk factors for SCZ [53, 54]. The responses to these risk factors are known to vary among individuals, which implies possible genetic mediation [55].
Statistical associations of endophenotypes with the risk of illness would include both positive and negative associations. Negative associations with risk would be related to resilience endophenotypes. By resilience, we refer to processes that reduce the risk of illness in persons with other predisposing risk genes or environmental factors. Resilience to schizophrenia has been reported to have associated loci [56], but resilience-related endophenotypes are less studied than risk-related phenotypes. Much of resilience might simply be the alternate alleles to genetic risk factors or lack of exposure, but there might also be specific genetic factors for protective effects, such as genes that can drive behaviors related to active stress-coping [57, 58], cognitive abilities [59], and flexibility [60–63], which can be related to most psychiatric disorders. Genetics of resilience related to stress response have also been explored using GWAS [64, 65].
Study in families or populations. The original criterion is about family study for historical reasons. However, many endophenotype studies are more frequently performed in populations than families because of more accessible access to large samples. This is particularly important for the weak genetic effects of endophenotypes. Requiring the phenotypes to co-segregate within families and be more common in well-family members than in the population has been largely replaced by case-control comparisons or analysis of quantitative traits in population samples.
Measurable distinctions between pleiotropy and causation were poorly developed when Endophenotype 1.0 was created. An endophenotype may be associated with illness, and genetic variants may be associated with both the endophenotype and the illness, but the variants do not have a causative effect on illness; such associations are called pleiotropy. Genetic mediation or causal relationship must be differentiated from association to make endophenotype useful for understanding disease mechanisms.
Genetic mediation refers to the phenomenon of shared genetic factors connecting an endophenotype to the development or causation of a disorder. It highlights the genetic influences that operate through the endophenotype, contributing to the manifestation of the disorder. Genetic factors of an endophenotype per se may not be directly observed to be associated with illness, as reviewed by Thomas [66]. When an SNP (or gene) is associated with both endophenotype and illness, the presence of causal relationships can be evaluated by statistical analyses such as Mendelian Randomization (MR) [67] or mediation analyses of multi-dimensional data. Moreover, genetic mediation can take the form of non-linear effects, which could fail correlation tests, like in the study of treatment changes [68]. Li et al. examined the non-linear association between sleep duration, cognition, and mental health, and uncovered underlying brain structure and genetic mechanisms missed by linear approaches [69]. A guideline for high-dimensional mediation analysis on high-throughput genomics studies is available [70].
One may ask, what are the consequences of using an outdated definition? Why must we update the definition when most of the proposed Endophenotype 2.0 is expensive to study? Using outdated definitions in research leads to inefficient use of resources, diminished statistical power, poor reproducibility, and, more importantly, missed opportunities to uncover critical links that could reveal true disease mechanisms. By incorporating advancements in omics, cellular biology, neuroscience, and computational modeling, updated definitions like Endophenotype 2.0 can provide deeper mechanistic insights and enhanced predictive accuracy. Although studying these phenotypes can be expensive at present, history shows that bold conceptual shifts often drive the development of cost-effective approaches, ultimately making advanced methods economically viable and more accessible over time.
The updated concept of Endophenotype 2.0 eliminates the restrictions and limitations of the original definition and criteria, allowing novel and broader phenotypes to be included within the unified framework of the endophenotype concept. When a phenotype is classified as an endophenotype, established methods for studying endophenotypes become applicable, fostering new opportunities for interdisciplinary applications. For example, QTL (quantitative trait loci) analysis demonstrates how methods developed for eQTL research have been successfully extended to other endophenotypes, including techniques for genetic mapping, fine-mapping, causal analysis, and multi-dimensional omics integration. The imputation of unmeasured endophenotypes in large datasets leveraging a genetic study of a small, deeply-phenotyped cohort, using methods like PrediXcan [71], UTMOST [72], BrainXcan [73], and others [74], has shown to be highly effective. Additionally, endophenotypes enable the exploration of their interconnected relationships through networked analyses. Comparative and integrative analyses across multi-dimensional endophenotypes are now feasible because they share similarly defined structural frameworks. This adaptability facilitates the introduction of new analytical methods that emphasize causal inference and non-linear genetic mediation. New mechanisms could also be revealed by including previously excluded phenotypes, particularly those associated with state-dependent traits. This adaptability underscores the value of updating the definition.
Endophenotype 2.0 fills in mechanism gaps between genetic variants and psychiatric disorders, capturing missing biology
Psychiatric genetics has achieved significant progress in the past fifteen years. Hundreds of common SNPs and dozens of rare copy number variants (CNVs) have been associated with major psychiatric disorders. However, a widely recognized gap exists between genetic associations of common diseases and biology [75–78]. The biological processes related to genotypic associations have not been unraveled, referred as “missing biology”. Disease-associated genetic variants frequently do not pinpoint specific genes, as most of the associated SNPs are non-coding and regulatory elements can be far away from the gene body. Even for associations mapped to genes through genetic, genomic, and epigenomic studies, their relationships to high-order phenotypes and functions at the levels of cells, circuits, brain, and behavior are mostly unknown. Functional annotation of GWAS signals and assigning genetic associations to specific mechanisms are the most important follow-ups of GWAS.
Linking genetic variants to disorder through endophenotypes contributes to the mechanistic interpretation of disease. Meyer-Lindenberg and Weinberger demonstrated how imaging genetics help relate genes to the mechanism, brain circuitry, emotion, and risk of schizophrenia and depression [79]. However, it should be noted that we expect pleiotropy to be common. Most regulations and actions are in networked systems and, therefore, multi-factorial and complex, involving many endophenotypes in between.
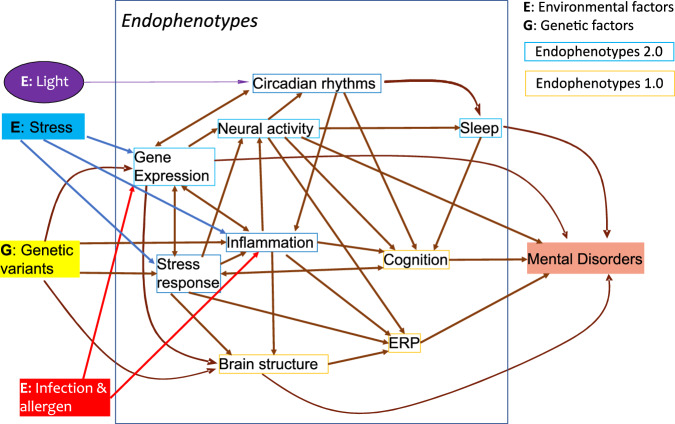
Different endophenotypes may interact to contribute to disease pathology, as illustrated in Fig. 1. At the same time, clinically distinct psychiatric disorders may share underlying mechanisms. The relationships among the endophenotypes will inform the mechanistic networks, pathways, and circuitries underlying diseases. Genomic Structural Equation Modelling (Genomic SEM) is a valuable tool that can be used to model associations among endophenotypes and to identify genetic variants that can affect cross-disorder liability [80].
Fig. 1. Environmental and Genetic Interactions with Endophenotypes 1.0 and 2.0.
The Hypothesized path analysis may resolve the mechanistic causal relationships from genetic variations (G) and environmental factors (E) to major endophenotypes (both 1.0 and 2.0) and, ultimately, disorders. The effect of each edge could be quantified in future studies.
Endophenotype 2.0 and the missing heritability gap
More than a decade ago, a gap in heritability estimates was termed “the missing heritability” in common diseases [78], where SNP-based heritability was found to be considerably smaller than heritability based on concordance of illness between relatives. For SCZ, twins and other concordances among relatives generate a heritability of 0.67 [81], but SNP-based heritability is 0.26 [82]. Now, heritability is empirically defined as the proportion of additive genetic variance in phenotype by case-control difference. The challenge of “missing heritability” can be reframed as a large part of case-control variance not being accounted for by additive genetic variance in the polygenic model. The additive genetic variance calculation is a “black box,” where effect sizes on illness and allele frequencies of all common SNPs enter the box, and the weighted summation of their effects exits the box.
One proposed explanation for missing heritability is that uncommon and rare variants contribute to familial concordance but only common variants are included in SNP-based heritability [83]. Another explanation is that SNP-based heritability only measures an event that occurs at conception (genotype generation), whereas illness concordance between relatives as adults is based on observed case-control status, which reflects the effects of common SNPs plus all the developmental and environmental exposure events that have occurred since conception. These events include gene-gene and gene-environment interactions, and their endophenotype products [84]. The additive effects model does not represent the indirect effects of genotypes interacting with other risk contributors.
Response to exposures, including protective environments and medication, is one major new category for Endophenotype 2.0. This new category allows us to explore mechanisms of a presumably major component of psychiatric genetics, genetic contributors that interact with the environment. Such endophenotypes may help uncover gene-gene interaction and gene-environment interaction. These non-additive interaction effects are not included in disease GWAS [85]. With the help of endophenotypes, some of the missed heritability accounted for by the non-additive effects may be recovered [84].
It is worth emphasizing that genes with strong effects on the endophenotype but weak effects on illness may nonetheless prove clinically useful, such as drug targets that can modify the endophenotype. A successful example is the drugs that modify a synthetic enzyme of serum cholesterol (HMG-CoA reductase, HMGCR) to prevent coronary artery disease (CAD). HMGCR is highly associated with LDL levels as identified by MR but only has a small effect on the disease CAD [86, 87].
Within the framework of endophenotype 2.0, the study of gene-environment interactions (GxE) opens new avenues for research. For example, genetic variants that regulate response to psychological stress may contribute to the risk of disorders. When these variants are mapped through endophenotypes, disease GxE analysis can be focused and powerful as the number of tests may be dramatically reduced. For example, an exposome score (ES), which estimates the sum of environmental risks [88], was tested for interaction with polygenic risk score (PRS) and confirmed GxE effects on psychosis [89]. In a similar vein, stress-related exposures were tested for interaction with PRS of depression and anxiety and confirmed the GxE effects on depression and anxiety [90]. While PRS and ES or equivalent summary scores are powerful measures for demonstrating the GxE effects on disease risk, they are blind to the granular mechanism of individual genes and pathways. There is still a significant opportunity for research into how particular genes interact with specific environmental risk factors.
Endophenotype 2.0 distinguishes causation from association
Association is not causation. Two parallel events stemming from a common cause can show an association without directly influencing each other. This non-causal relationship (horizontal pleiotropy) can complicate the interpretation of data and provide limited insight into the mechanisms underlying a disease. Understanding this distinction is crucial for accurate analysis and interpretation in scientific research. Estimating true causal effects can be achieved through vertical pleiotropy, where an SNP affects one endophenotype and, as a result, another. Conversely, genetic factors may directly influence the illness, bypassing the endophenotype in horizontal pleiotropy. These different genetic pathways can be analyzed using Mendelian Randomization (MR) methods [66, 67], enabling the assessment of pleiotropic and causal relationships.
MR provides tests of causality and can be applied to multi-dimensional data. Functional connections between genotypes and (epi-)genomic profiles have been studied in autopsy and fetal brains. Brain anatomic structures and physiology have also been associated with psychosis disorders. Nonetheless, most of these associations have yet to be tested for causation. An SNP associated with both illness and endophenotype can be used as an “instrument” to determine whether the association with illness is an indirect effect mediated by the endophenotype exposure or a direct effect on illness. The ratio of the regression coefficient for illness to the coefficient for the endophenotype is the MR measure of the SNP’s direct causality [66].
Genetic variants presumably exert their effects on illness through various endophenotypes. Thus, the challenging problem of determining the causality of an SNP on disease could be converted into two sub-problems (1) SNP to endophenotype and (2) endophenotype to disease causal relationships. MR can use inputs from either a single sample (“one-sample MR”) or two samples (“two-sample MR”) depending on the sources of the SNPs, exposure, and outcome data [91]. Two-sample MR is particularly useful because the endophenotype (exposure) data are often collected in a cohort different from the case-control (outcome) cohort. Applying two-sample MR to brain imaging and disease GWAS revealed significant causal relationships of structural features of the superior longitudinal fasciculus with anorexia nervosa (AN) and of the corpus callosum with Alzheimer’s disease [92].
Univariable MR studies the effect of a single exposure on an outcome such as disease diagnosis using methods such as Standard MR (Wald ratio) [93], inverse-variance weighted (IVW) [94], the weighted median [95], mode-based estimate (MBE) [96], contamination mixture [97], and the MR-Egger regression [95, 98]. IVW and MR-Egger use all available SNPs as instruments, assuming they are valid. Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) [99], MR-Lasso [100], and MR-robust [100] select only a subset of SNPs that are likely to be valid instruments. The IVW method is sensitive but assumes no horizontal pleiotropy. MR-Egger can detect and adjust for horizontal pleiotropy but is less sensitive than IVW. It should be noted that the quality of MR relies on the instrument strength, measured by F-statistics, which indicates the association between the SNPs and the exposure of interest. F-statistic >10 is a general guideline. Sensitivity analyses should be performed using methods like MR-Egger regression, weighted median, or heterogeneity tests to evaluate the robustness of the results based on different assumptions about pleiotropy.
Multivariable MR (MVMR) [101] allows for the simultaneous analysis of multiple exposures (endophenotypes) to estimate their direct and indirect effects on an outcome. Multivariate generalized linear models [102] and multivariate functional linear models [103] can test for genetic effects on multiple traits. Pleiotropy-robust methods for MVMR [101] and Pleiotropy Estimation and Test Bootstrap (PET-B) [104] are designed to estimate and test the extent of pleiotropy between a variant and multiple phenotypes. All these methods aim to improve the detection and characterization of pleiotropy compared to univariate approaches. MVMR revealed complex pleiotropic causal relationships among intelligence, education, bipolar disorder (BD), and SCZ [105]. MVMR also discovered that the genetic liability of smoking had a deleterious association with longevity, while genetic liabilities for major psychiatric disorders had no independent associations [106].
Each method has its pros and cons. Combining multivariate and univariate GWAS models could be useful for identifying and narrowing down candidate loci with potential pleiotropic effects for downstream experiments [107]. Guidelines for performing MR are available and continuously updated [108].
Emerging endophenotypes of psychiatric disorders
Endophenotypes can be classified by biological levels from molecular to whole organism. Genetics have effects on structure, function, and physiology at each of the biological levels. Behavioral and neurophysiological phenotypes are high-end endophenotypes. Some endophenotypes are well-accepted, such as those in Endophenotype 1.0 for psychosis disorders. Most emerging endophenotypes have only fragmentary evidence suggesting they may meet the three criteria of Endophenotype 2.0. Population GWASs from the UK Biobank and others have uncovered genetic associations with many traits. Some of them are also associated with psychiatric disorders, such as educational attainment [15] and sleep chronotype [109].
Examples of commonly used and emerging endophenotypes are shown in Table 2, along with some existing data collections/studies in the public domain. We compiled data from the GWAS Catalog [110] and GWAS Atlas [111]. Molecular trait QTLs were collected directly from the literature, as they were not in the typical collections of the GWAS databases.
Since more than 80% of the genetic variants associated with psychiatric disorders are non-coding, dysregulated gene expression is currently the major suspect for the underlying molecular changes, and such dysregulation is defined here as a molecular endophenotype. Multiple postmortem brain studies support this expression dysregulation model. Differentially expressed genes and splicing isoforms have been reported in SCZ, BD, and ASD postmortem brains [112, 113]. SNPs that regulate expression (eQTLs), RNA splicing (sQTLs), and DNA methylation (mQTLs) are enriched in major psychiatric disorders [114–124].
Specific subtypes of brain cells have been implicated in psychiatric disorders. Neuropharmacological studies have implicated dopaminergic, glutamatergic, and GABAergic neurons in SCZ [125]. Analyses of coexpression modules in postmortem brains of psychiatric patients suggest that neurons, oligodendrocytes, astrocytes, and microglia are involved in SCZ, BD, and ASD [113]. With the technological advances of single-cell sequencing and algorithms for deconvolution [126] of cell type in genomic analyses, deeper understandings are expected for the roles of each subtype of cells in specific brain regions.
New endophenotypes are expected or under development
-
Molecular phenotype data has accumulated quickly in the past ten years. Molecular phenotypes such as gene expression in eQTLs have been proven effective in capturing genetic regulators by their associated SNPs. Gene expression, splicing, gene networks, protein abundance, and chromatin accessibility have associations with disease and are thus molecular endophenotypes. Quantitative trait loci (QTL) for eQTLs, sQTLs, and mQTLs are commonly used to capture genetic regulators for thousands of genes and to explain GWAS results.
We expect molecular endophenotypes to snowball in the coming years with the fast-evolving sequencing technologies. These would include more molecular trait (-omics) QTLs mapped to early brain development and cell subtypes. eQTLs from postmortem tissues only explain a small portion of GWAS signals [127]. Dynamic (conditional or context-dependent) eQTLs that may capture regulators of gene expression under specific developmental stages or environments are expected to recover more functional annotation of GWAS. Recently, 672 fetal brain samples have been used to generate early developmental eQTL, sQTL, and isoform QTL data [128]. Single-cell RNA sequencing (scRNA-seq) profiled expression across three differentiation stages on 215 induced pluripotent stem cell (iPSC) lines that were differentiated toward a midbrain neural state and generated cell-type-specific eQTLs for early developmental stages [129].
-
Cellular phenotypes are not yet practical measurements but are conceivable future developments. Cellular function studies in cultured cells have been performed on some candidate genes, including POU3F [130, 131], DGCR5 [132], TRIM8 [131], DISC1 [133, 134], NRXN1 [135], SETD1A [136], TCF4 [137], ZNF804A [138], PCCB [139], GLT8D1, and CSNK2B [140], and others [141]. Expression dysregulation of these genes was found to affect cell differentiation [131, 135, 140], proliferation [131, 137, 140], electrophysiology [131, 140], and expression of other risk genes [132, 136–139]. More associated genes need to be studied for cellular phenotypes in both early developmental and matured brain subtype cells, which will be more challenging. Development and neural circuitry are also important for the etiology of psychiatric disorders. Fetal brain eQTLs showed unique contributions to the risks of SCZ and autistic spectrum disorder (ASD) [128, 142, 143], further emphasizing the importance of developmental biology.
iPSC-derived neuronal, glial cells and various organoids are used for modeling phenotypes of early-developing brains and have the advantage of preserving each donor’s genetic background, diagnoses, and possibly endophenotypes. Most current cellular models target individual candidate genes with a small number of cell lines, which can only assess the functionality of a few SNPs or genes. Improvements in scalability are on the horizon. A large-scale massively parallel reporter assay (MPRA) in iPSC-derived neurons was reported to study 240 GWAS loci simultaneously [144]. Hundreds of iPSC lines from patients and controls may soon be studied for genetic effects on cell proliferation, differentiation, electrophysiology, pharmacologic response, and calcium imaging to produce corresponding endophenotypes. iPSCs in mental disorders may become an area with heavy research investment in the coming years.
Brain imaging and EEG-based measures are the major 1.0 type endophenotypes in psychiatry today, as represented by the ENIGMA [145] and B-SNIP [146]. Various structural, connectivity, and activity metrics of specific brain regions have been implicated in distinct disorders [147–149]. Current methods can only be used for costly deep phenotyping. New portable instruments and mobile devices may increase the throughput and reduce costs in the future. Data quality and standardization across instruments and sites have been challenging, but early tests have delivered promising results [150, 151].
Treatment response is a category of Endophenotype 2.0, and individual variation in treatment response is well recognized. Several GWASs of treatment response have been published with dozens of loci that can survive multiple-testing corrections [152–154]. Various scales for symptoms [155–157] are commonly used in clinical trials to assess treatment effects. Although symptomatic improvement is the desired result, physiologic measurements and other changes in endophenotypes are related to the biological processes underlying treatment and have led to more effective predictors [8, 146].
Inflammation-related phenotypes are another Endophenotype 2.0 category related to the risk environment. They can reflect individual variation in response to inflammatory environmental insults, which include infections, allergens, vaccines, psychological stress, or wounds. Immune changes have been detected in peripheral blood and postmortem brains of patients with major psychiatric disorders [158–160]. For example, IL6, TNFα, VEGF, and CRP were found to be significantly higher in the plasma of psychosis probands than in controls [161]. Inflammatory factors were associated with blood-brain barrier pathology [162], neuroimaging, cognitive impairment, and symptom measures [161, 163]. Some immune changes are common across disorders, while others are disorder-specific [158, 164]. A critical question is how much individual variation in inflammation responses is regulated by genetics and further contributes to psychiatric risk. Using immunophenotyping flow cytometry in a genetic study of innate and adaptive immune traits with 497 adult female twins, 76% of 23,394 immune phenotypes showed a predominantly heritable influence [165]. Leukocyte subsets, CD39 on regulatory T cells, and CD32 on myeloid dendritic cell (DC) populations are examples of highly heritable traits detected in this study. Some of the inflammation-related factors were stable over time in healthy people [166–168]. 3′ UTR Alternative polyadenylation (APA) QTL (3′aQTL) was mapped in 18 human immune-related blood cell types and eight immune stimulation conditions [169]. Neural cell types have not yet been studied for genetic regulation of immune responses.
-
Psychological stress-related traits are another group of risk environment-related endophenotypes. Stress contributes to the risk of depression, anxiety, panic disorder, and post-traumatic stress disorder (PTSD) [170, 171]. SCZ patients report greater exposure to stressful life events than controls [172, 173]. Recent stressful life events were associated with the onset and progression of psychosis in some patients [174]. Similarly, psychological stress was linked to the onset, progress, and severity of BD, with particularly strong evidence for early-life stress [175]. The stress-regulated hypothalamic-pituitary-adrenal (HPA) axis system is associated with MDD through the classic dexamethasone suppression test [176], although the association is much more complex than initially proposed [177, 178]. A candidate gene study found that variation in the glucocorticoid receptor was associated with cortisol levels [179]. Defining and quantifying stress response for genetic research presents its complexities and challenges.
Questionnaire-based methods like the Childhood Trauma Questionnaire (CTQ) [180–183] and structured (or semi-structured) interviews [184–188] are commonly used to collect stress-related data. It should be noted that data could be biased by the people who administrate data collection. Objective measures that are free of influence from the data collector are desirable. Cortisol levels in the blood, saliva, or hair, EEG measures, blood pressure, heart rate, and skin conductance would be objective measures of stress response at the time of measurement. Chronic stress is more difficult to measure objectively than acute stress. There is ample literature studying the mental health and even epigenetic changes associated with psychological stressors. One study showed that the heritability of response to repeated stress could be high (h2 > 0.97) [189] in a twin study of the Trier Social Stress Test (TSST), which is a controlled acute stress experiment. Hair cortisol concentration was found to be highly heritable (h2 = 0.72) in a twin study [190]. Genetic studies of stress responses were reviewed in 2006 [191]. Progress has been made since. Meta-analysis of genome-wide interaction study (GWIS) of stress-sensitivity by modeling the interaction between SNPs and MDD status on neuroticism score in data of UK Biobank and Generation Scotland: Scottish Family Health Study identified one significant associated gene ZNF366 in gene-based tests [192]. ZNF366 was reported to regulate glucocorticoid receptor function [193]. Moreover, 3,820 glucocorticoid receptor-response-modulating cis-eQTLs were reported in the peripheral blood of 160 male individuals [194]. Out of these SNPs, MPRA validated 547 in glucocorticoid-responsive regulatory elements, with 233 showing allele-dependent activity [195].
Responses to environmental changes are essential for human survival and contribute to psychiatric disorders. These responses include responses to in-utero events in SCZ (infection and severe food restriction). The possibility that there are genetic components to these obstetric event responses was raised by Mittal et al. in 2008 [55], but to our knowledge, no molecular investigation has been conducted on this hypothesis. Chronotype, defined as an individual’s natural sleep timing preference, is a potential endophenotype. In addition to clock genes regulating circadian rhythms, GWAS of chronotype identified 351 significant loci. Moreover, schizophrenia, depressive symptoms, MDD, and intelligence were all negatively correlated with the morning chronotype [109]. However, genetic variants influencing sensitivity to varying durations of light exposure have not yet been investigated.
Two complementary approaches to the collection of endophenotype data
Deep and shallow phenotyping are two alternative approaches for collecting endophenotype data. They each have their own pros and cons as shown in Table 3.
Table 3.
Deep and shallow phenotyping.
| Deep | Shallow | |
|---|---|---|
| Design | Precise and comprehensive collection of data on multiple phenotypes in one cohort. | Minimal phenotyping, large samples. |
| Methods | Advanced instruments and standardization. | Minimal standardization. May rely on self-reports or reported diagnoses, mobile devices, medical records, etc. |
| Examples | Brain imaging, eye tracking, EEG, molecular and cellular traits, cognition | Diagnosis, self-reported behavioral and online questionnaire, digital phenotypes |
| Procedural costs in time and money | Expensive and lengthy phenotyping of each participant. | High throughput, low cost. |
| Pros | Quality (accuracy and precision) of measures can be better monitored | Can generate large datasets in multiple populations. |
| Cons | Small sample size: generalizability may be a concern. | Concerns about validity/quality of phenotypes. |
| Hybrid approaches | Electronic Health Records contain both standardized measurements and descriptive information. | |
Deep phenotyping can be defined as a precise and comprehensive collection of data on multiple phenotypes in one cohort. Such phenotyping will be time-consuming and require advanced instruments and standardization. Several major projects in psychiatry, including B-SNIP [146], ENIGMA [145], and ABCD [196], are collecting deep phenotyping data, including brain imaging, eye tracking, EEG, and cognitive function.
Shallow phenotyping collects phenotype data in large numbers of subjects but with less content related to illness and less emphasis on standardization. When it was realized that tens of thousands of subjects would be needed to establish associations of common SNPs with polygenic diseases, a minimal phenotyping strategy was adopted. Self-reported or medical record diagnosis, survey at a distance (online), or passive data recorded by a mobile device was used, but concerns remain on the validity or accuracy of the phenotypes [197]. In a study of “chronotype”, which aimed at studying persons with a variant sleep cycle, validation of sleep episode timing was obtained from activity meters in a subset of the UK Biobank samples. However, the relationship of chronotype to specific sleep laboratory measurements (such as sleep phase or REM onset) could not be determined [109].
Shallow phenotyping strategies without any validation of the measurements have been used for most genetic studies of psychiatric disorders in the past decade, with success in detecting disease-associated SNPs, but without demonstrated successes from large-scale importation of other medical record data. 23andMe and the UK Biobank collect phenotype data mostly through shallow phenotyping approaches without external validation. Correlation between endophenotypes can help validate some endophenotypes collected from shallow phenotyping approaches and even help to impute missing data. The shallow phenotyping likely boosted sample size with the cost of adding noises. The success of genetic mapping depends on the fast-growing sample size over the noises.
Electronic Health Records (EHR) represent a hybrid approach that encompasses both deep and shallow phenotypes. Typically, EHRs include rigorously standardized lab tests and physiological results, complemented by less validated data like natural language descriptions. However, the data quality in EHRs can vary significantly across different fields.
To facilitate the development of endophenotype 2.0, we created An Endophenotype Quality Index (EQI) with 12 scoring criteria (Supplementary Material). This index differs from Glahn et al.’s Endophenotype Ranking Value [198], which emphasizes the heritability of endophenotypes and illness and their correlations. Our EQI criteria add several practical measures, including stability, throughput, and costs. We give extra weight to objective and quantitative measures. We also consider generalizability related to age, population, sex, and clinical state. A good endophenotype should be suitable for affordable, high-throughput, large-sample studies with a clearly defined target population.
Uses of the genetics of endophenotypes
Genetic analysis of endophenotypes may assist functional annotation of GWAS signals in disorders and illustrate the pathways, biological processes, and mechanisms involved. We expect the integrative analysis of the genetics of endophenotypes with disease diagnosis will facilitate the discovery of novel risk genes and fine-mapping of causal variants and may also improve the accuracy of disease risk prediction.
Colocalization can be used for fine-mapping disease-associated SNPs. When a disease shares association signals with an endophenotype, the posterior probability of colocalization at a single causal variant can be estimated by a colocalization method like HyPrColoc [199] taking Linkage Disequilibrium (LD) into account.
An endophenotype can also serve as a functional annotation of an SNP when the SNP and its LD region are associated with a disorder. However, this assumption could be wrong because of horizontal pleiotropy. A causal analysis, such as MR, better supports this type of connection.
Risk prediction is an important goal of genetic study, and endophenotypes can play a role
PRS is the major measure for risk prediction today, although there are large overlaps between case and control groups. In a recent evaluation of risk prediction methods for PRS in SCZ [200], the best PRS predictor accounted for 9.9% of risk variance on a liability scale in populations of European descent (EUR). EUR-PRS-based prediction accuracy is much lower when applied to non-EUR because of population genetic structure. Another study on PRS prediction of psychosis in high-risk populations came to similar findings [201]. The current use of PRS in predicting psychiatric outcomes has been critically reviewed [202, 203]. Methods are actively developed to incorporate endophenotypes to improve PRS prediction on diagnosis [204].
Coronary Artery Disease (CAD) offers an interesting example of endophenotype-related risk prediction for SCZ and related psychosis disorders. CAD is a polygenic disorder with a set of risk factors first quantified in the Framingham Heart Study [205]. The risk factors include the components of serum cholesterol, blood pressure, cigarette smoking, sex, and age. Serum triglycerides, previously thought to be a CAD risk factor, failed the MR test [206]. The c-statistic, which is equivalent to AUC, is used to quantify risk prediction in CAD [207]. In a 1998 CAD risk study, the c-statistic was 0.74 for women and 0.77 for men. Using a PRS for CAD in 2020, Damask et al. found that the use of both high baseline LDL-C and high PRS yielded improved risk estimations in the placebo control group as compared with the use of either factor alone [208].
Psychosis disorders and other polygenic disorders may also benefit from the study of endophenotypes for treatment and prevention, as well as for investigation of the biology of illness [8]. Multivariate analysis of commonly used endophenotypes, including fitted regression models and machine learning [209] in the same individuals enables endophenotype intercorrelations to be used in risk prediction [210]. Multivariate regression can improve prediction results over models restricted to genotypes on the risk of psychosis and other disorders [210, 211]. When there are large numbers of phenotypes, analysis faces challenges of multiple-testing burdens and correlated phenotypes [212, 213]. Significance thresholds must be adjusted for correlations among the phenotypes, sometimes requiring simulation or permutation in addition to theoretical distribution analyses [214].
Multi-dimensional endophenotype data for subtyping diseases and resolving disease mechanisms
Multi-dimensional data of endophenotypes may be used to create subtypes of disease if one disorder can be partitioned into subgroups with specific risk factors and genetics and subsequently with optimal treatment choices. The B-SNIP project has developed biotypes of psychosis as physiologically coherent subgroups [162], as an important pilot effort that may reshape future diagnosis systems. High and low-inflammation subgroups of SCZ and BD were also proposed [215, 216]. Fast-evolving machine-learning methods will particularly benefit from structured multi-dimensional data in developing new disease classification systems.
With large and multi-dimensional data of endophenotypes and diseases from diverse sources, path analyses can be performed as imagined in Fig. 1. Path analysis utilizes multiple regression to evaluate causal relationships among parts of a system. Every path, or edge in a networked system, can be assessed for effect size and probability of mediation effect. The networked paths link genetic variants to many endophenotypes, ultimately to a disorder. These networks may be investigated as hypothesized biological processes underlying the disorder.
In summary, to address the limitations of the current endophenotype formulations dating back over a half-century, we propose Endophenotype 2.0 with an updated definition and criteria. Building larger cohorts of patients and controls with deep and shallow phenotyping of endophenotypes and studying their intertwined genetics and genomics may be a fruitful scientific endeavor for the coming years. Multi-dimensional genetic/genomic studies of endophenotypes will hopefully provide biological insights, establish mechanisms for psychiatric disorders, uncover more risk genes, and inform translational use of genetic findings. Improving existing endophenotypes and developing new ones that meet quality criteria are current challenges for the field. In the end, endophenotypes at different levels can be expected to work together to solve the mechanistic steps between genetics and disease.
Supplementary information
Quality score of endophenotypes for psychiatric genetics
Acknowledgements
SUNY Empire Innovation Program, NIH grants U01 MH122591, U01MH116489, R01MH110920, U01MH103340 (to CL) R21AG045789, R01MH103368-01A1 (to ESG) supported studies of molecular traits in postmortem brains and deep phenotyping in psychosis disorders and healthy controls. The Shaanxi Normal University and Central South University funds (to CL) contributed to studies of novel endophenotypes on stress responses and inflammation. All these projects inspired the formation of the concepts and ideas presented in this paper. We thank Dr. Richard Kopp for his close reading of an earlier draft. We also utilized Grammarly and ChatGPT to correct grammatical errors and suggest alternative word choices in English. Both authors have no conflict of interest to declare.
Author contributions
CL conceived the study and drafted the manuscript. ESG contributed additional ideas and assisted in revising and refining the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chunyu Liu, Email: liuch@upstate.edu.
Elliot S. Gershon, Email: egershon@bsd.uchicago.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-024-03195-1.
References
- 1.Gottesman II, Shields J. Genetic theorizing and schizophrenia. Br J Psychiatry J Ment Sci. 1973;122:15–30. [DOI] [PubMed] [Google Scholar]
- 2.Gershon ES, Goldin LR. Clinical methods in psychiatric genetics. I. Robustness of genetic marker investigative strategies. Acta Psychiatr Scand. 1986;74:113–8. [DOI] [PubMed] [Google Scholar]
- 3.Rieder RO. Gershon ES. Genetic strategies in biological psychiatry. Arch Gen Psychiatry. 1978;35:866–73. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J psychiatry. 2003;160:636–45. [DOI] [PubMed] [Google Scholar]
- 5.Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37:163–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman D, Ducci F. Deconstruction of vulnerability to complex diseases: enhanced effect sizes and power of intermediate phenotypes. ScientificWorldJournal. 2007;7:124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventos H, Blangero J, et al. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Javitt DC, Sweet RA. Auditory dysfunction in schizophrenia: integrating clinical and basic features. Nat Rev Neurosci. 2015;16:535–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheffield JM, Karcher NR, Barch DM. Cognitive deficits in psychotic disorders: a lifespan perspective. Neuropsychol Rev. 2018;28:509–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodberry KA, Giuliano AJ, Seidman LJ. Premorbid IQ in schizophrenia: a meta-analytic review. Am J psychiatry. 2008;165:579–87. [DOI] [PubMed] [Google Scholar]
- 11.Cannon M, Jones P, Huttunen MO, Tanskanen A, Huttunen T, Rabe-Hesketh S, et al. School performance in Finnish children and later development of schizophrenia: a population-based longitudinal study. Arch Gen Psychiatry. 1999;56:457–63. [DOI] [PubMed] [Google Scholar]
- 12.Cannon TD, Bearden CE, Hollister JM, Rosso IM, Sanchez LE, Hadley T. Childhood cognitive functioning in schizophrenia patients and their unaffected siblings: a prospective cohort study. Schizophr Bull. 2000;26:379–93. [DOI] [PubMed] [Google Scholar]
- 13.Friedman D, Vaughan HG Jr, Erlenmeyer-Kimling L. Cognitive brain potentials in children at risk for schizophrenia: preliminary findings. Schizophr Bull. 1982;8:514–31. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Roige S, Palmer AA. Emerging phenotyping strategies will advance our understanding of psychiatric genetics. Nat Neurosci. 2020;23:475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Hellard S, Wang Y, Witoelar A, Zuber V, Bettella F, Hugdahl K, et al. Identification of gene loci that overlap between schizophrenia and educational attainment. Schizophr Bull. 2017;43:654–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lysaker PH, Wilt MA, Plascak-Hallberg CD, Brenner CA, Clements CA. Personality dimensions in schizophrenia: associations with symptoms and coping. J Nerv Ment Dis. 2003;191:80–86. [DOI] [PubMed] [Google Scholar]
- 17.Shi J, Yao Y, Zhan C, Mao Z, Yin F, Zhao X. The relationship between big five personality traits and psychotic experience in a large non-clinical youth sample: the mediating role of emotion regulation. Front Psychiatry. 2018;9:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lahey BB. Public health significance of neuroticism. Am Psychol. 2009;64:241–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navrady LB, Ritchie SJ, Chan SWY, Kerr DM, Adams MJ, Hawkins EH, et al. Intelligence and neuroticism in relation to depression and psychological distress: Evidence from two large population cohorts. Eur Psychiatry. 2017;43:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smeland OB, Wang Y, Lo MT, Li W, Frei O, Witoelar A, et al. Identification of genetic loci shared between schizophrenia and the Big Five personality traits. Sci Rep. 2017;7:2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo MT, Hinds DA, Tung JY, Franz C, Fan CC, Wang Y, et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet. 2017;49:152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Middeldorp CM, de Moor MH, McGrath LM, Gordon SD, Blackwood DH, Costa PT, et al. The genetic association between personality and major depression or bipolar disorder. A polygenic score analysis using genome-wide association data. Transl Psychiatry. 2011;1:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DJ, Escott-Price V, Davies G, Bailey ME, Colodro-Conde L, Ward J, et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatry. 2016;21:749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meule A. Reporting and Interpreting Working Memory Performance in n-back Tasks. Front Psychol. 2017;8:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gur RC, Erwin RJ, Gur RE, Zwil AS, Heimberg C, Kraemer HC. Facial emotion discrimination: II. Behavioral findings in depression. Psychiatry Res. 1992;42:241–51. [DOI] [PubMed] [Google Scholar]
- 27.Coleman JRI, Lester KJ, Keers R, Munafo MR, Breen G, Eley TC. Genome-wide association study of facial emotion recognition in children and association with polygenic risk for mental health disorders. Am J Med Genet B Neuropsychiatr Genet. 2017;174:701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwood TA, Lazzeroni LC, Maihofer AX, Swerdlow NR, Calkins ME, Freedman R, et al. Genome-wide association of endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia (COGS) study. JAMA Psychiatry. 2019;76:1274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyers JL, Zhang J, Chorlian DB, Pandey AK, Kamarajan C, Wang JC, et al. A genome-wide association study of interhemispheric theta EEG coherence: implications for neural connectivity and alcohol use behavior. Mol Psychiatry. 2021;26:5040–52. [DOI] [PMC free article] [PubMed]
- 30.Zhao B, Li T, Yang Y, Wang X, Luo T, Shan Y, et al. Common genetic variation influencing human white matter microstructure. Science. 2021;372:eabf3736. [DOI] [PMC free article] [PubMed]
- 31.Lencer R, Mills LJ, Alliey-Rodriguez N, Shafee R, Lee AM, Reilly JL, et al. Genome-wide association studies of smooth pursuit and antisaccade eye movements in psychotic disorders: findings from the B-SNIP study. Transl Psychiatry. 2017;7:e1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roussos P, Giakoumaki SG, Zouraraki C, Fullard JF, Karagiorga VE, Tsapakis EM, et al. The relationship of common risk variants and polygenic risk for schizophrenia to sensorimotor gating. Biol Psychiatry. 2016;79:988–96. [DOI] [PubMed] [Google Scholar]
- 33.Quednow BB, Ejebe K, Wagner M, Giakoumaki SG, Bitsios P, Kumari V, et al. Meta-analysis on the association between genetic polymorphisms and prepulse inhibition of the acoustic startle response. Schizophr Res. 2018;198:52–59. [DOI] [PubMed] [Google Scholar]
- 34.Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Q, Jiang Y, Shieh AW, Zhou D, Chen R, Wang F, et al. The impact of common variants on gene expression in the human brain: from RNA to protein to schizophrenia risk. bioRxiv [Preprint]. 2023. 10.1101/2023.06.04.543603.
- 36.Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, Kelsoe J, et al. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J psychiatry. 2009;166:540–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutt RK, Hannon K, Easley TO, Griffis JC, Zhang W, Bijsterbosch JD. Mental health in the UK Biobank: a roadmap to self-report measures and neuroimaging correlates. Hum Brain Mapp. 2022;43:816–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jamshidi J, Schofield PR, Gatt JM, Fullerton JM. Phenotypic and genetic analysis of a wellbeing factor score in the UK Biobank and the impact of childhood maltreatment and psychiatric illness. Transl Psychiatry. 2022;12:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karcher NR, Barch DM. The ABCD study: understanding the development of risk for mental and physical health outcomes. Neuropsychopharmacology. 2021;46:131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coleman JRI. The validity of brief phenotyping in population biobanks for psychiatric genome-wide association studies on the biobank scale. Complex Psychiatry. 2021;7:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith SM, Douaud G, Chen W, Hanayik T, Alfaro-Almagro F, Sharp K, et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci. 2021;24:737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C. Multi-trait genome-wide analyses of the brain imaging phenotypes in UK Biobank. Genetics. 2020;215:947–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Mol Psychiatry. 2016;21:758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DC, Ritchie SJ, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry. 2016;21:1624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rask-Andersen M, Karlsson T, Ek WE, Johansson A. Modification of heritability for educational attainment and fluid intelligence by socioeconomic deprivation in the UK Biobank. Am J psychiatry. 2021;178:625–34. [DOI] [PubMed] [Google Scholar]
- 47.Morris-Rosendahl DJ, Crocq MA. Neurodevelopmental disorders-the history and future of a diagnostic concept. Dialogues Clin Neurosci. 2020;22:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinberger DR. The neurodevelopmental origins of schizophrenia in the penumbra of genomic medicine. World Psychiatry. 2017;16:225–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inui T, Kumagaya S, Myowa-Yamakoshi M. Neurodevelopmental hypothesis about the etiology of autism spectrum disorders. Front Hum Neurosci. 2017;11:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scandurra V, Emberti Gialloreti L, Barbanera F, Scordo MR, Pierini A, Canitano R. Neurodevelopmental disorders and adaptive functions: a study of children with autism spectrum disorders (ASD) and/or attention deficit and hyperactivity disorder (ADHD). Front Psychiatry. 2019;10:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:eaat7615. [DOI] [PMC free article] [PubMed]
- 53.Belbasis L, Köhler CA, Stefanis N, Stubbs B, van Os J, Vieta E, et al. Risk factors and peripheral biomarkers for schizophrenia spectrum disorders: an umbrella review of meta-analyses. Acta Psychiatr Scand. 2018;137:88–97. [DOI] [PubMed] [Google Scholar]
- 54.Lumey LH, Stein AD, Susser E. Prenatal famine and adult health. Annu Rev Public Health. 2011;32:237–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittal VA, Ellman LM, Cannon TD. Gene-Environment Interaction and Covariation in Schizophrenia: The Role of Obstetric Complications. Schizophr Bull. 2008;34:1083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess JL, Tylee DS, Mattheisen M, Børglum AD, Als TD, Grove J, et al. A polygenic resilience score moderates the genetic risk for schizophrenia. Mol Psychiatry. 2021;26:800–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saxon L, Makhashvili N, Chikovani I, Seguin M, McKee M, Patel V, et al. Coping strategies and mental health outcomes of conflict-affected persons in the Republic of Georgia. Epidemiol Psychiatr Sci. 2017;26:276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q, Zhou Y, Ho SMY. Active and avoidant coping profiles in children and their relationship with anxiety and depression during the COVID-19 pandemic. Sci Rep. 2022;12:13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Procopio F, Zhou Q, Wang Z, Gidziela A, Rimfeld K, Malanchini M, et al. The genetics of specific cognitive abilities. Intelligence. 2022;95:101689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gabrys RL, Tabri N, Anisman H, Matheson K. Cognitive control and flexibility in the context of stress and depressive symptoms: the cognitive control and flexibility questionnaire. Front Psychol. 2018;9:2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaghi MM, Vertes PE, Kitzbichler MG, Apergis-Schoute AM, van der Flier FE, Fineberg NA, et al. Specific frontostriatal circuits for impaired cognitive flexibility and goal-directed planning in obsessive-compulsive disorder: evidence from resting-state functional connectivity. Biol Psychiatry. 2017;81:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zong JG, Cao XY, Cao Y, Shi YF, Wang YN, Yan C, et al. Coping flexibility in college students with depressive symptoms. Health Qual Life Outcomes. 2010;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shrivastava A, Desousa A. Resilience: a psychobiological construct for psychiatric disorders. Indian J Psychiatry. 2016;58:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cusack SE, Aliev F, Bustamante D, Dick DM, Amstadter AB, Spit for Science Working G. A statistical genetic investigation of psychiatric resilience. Eur J Psychotraumatol. 2023;14:2178762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maul S, Giegling I, Fabbri C, Corponi F, Serretti A, Rujescu D. Genetics of resilience: implications from genome-wide association studies and candidate genes of the stress response system in posttraumatic stress disorder and depression. Am J Med Genet B Neuropsychiatr Genet. 2020;183:77–94. [DOI] [PubMed] [Google Scholar]
- 66.Thomas DC. Conti DV. Commentary: the concept of ‘Mendelian Randomization. Int J Epidemiol. 2004;33:21–25. [DOI] [PubMed] [Google Scholar]
- 67.Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45:1717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hofmann SG, Curtiss JE, Hayes SC. Beyond linear mediation: toward a dynamic network approach to study treatment processes. Clin Psychol Rev. 2020;76:101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Sahakian BJ, Kang J, Langley C, Zhang W, Xie C, et al. The brain structure and genetic mechanisms underlying the nonlinear association between sleep duration, cognition and mental health. Nat Aging. 2022;2:425–37. [DOI] [PubMed] [Google Scholar]
- 70.Zeng P, Shao Z, Zhou X. Statistical methods for mediation analysis in the era of high-throughput genomics: Current successes and future challenges. Comput Struct Biotechnol J. 2021;19:3209–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu Y, Li M, Lu Q, Weng H, Wang J, Zekavat SM, et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat Genet. 2019;51:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang Y, Melia O, Caroll TJ, Brettin T, Brown A, Im HK. BrainXcan identifies brain features associated with behavioral and psychiatric traits using large scale genetic and imaging data. medRxiv [Preprint]. 2022. 10.1101/2021.06.01.21258159.
- 74.Mai J, Lu M, Gao Q, Zeng J, Xiao J. Transcriptome-wide association studies: recent advances in methods, applications and available databases. Commun Biol. 2023;6:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, et al. Psychiatric genomics: an update and an agenda. Am J psychiatry. 2018;175:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. [DOI] [PubMed] [Google Scholar]
- 79.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–27. [DOI] [PubMed] [Google Scholar]
- 80.Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wray NR, Gottesman II. Using summary data from the danish national registers to estimate heritabilities for schizophrenia, bipolar disorder, and major depressive disorder. Front Genet. 2012;3:118–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baselmans BML, Yengo L, van Rheenen W, Wray NR. Risk in relatives, heritability, SNP-based heritability, and genetic correlations in psychiatric disorders: a review. Biol Psychiatry. 2021;89:11–19. [DOI] [PubMed] [Google Scholar]
- 83.Visscher PM, Yang J, Goddard ME. A commentary on ‘common SNPs explain a large proportion of the heritability for human height’ by Yang et al. (2010). Twin Res Hum Genet. 2010;13:517–24. [DOI] [PubMed] [Google Scholar]
- 84.Blanco-Gómez A, Castillo-Lluva S, Del Mar Sáez-Freire M, Hontecillas-Prieto L, Mao JH, Castellanos-Martín A, et al. Missing heritability of complex diseases: Enlightenment by genetic variants from intermediate phenotypes. Bioessays. 2016;38:664–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei WH, Hemani G, Haley CS. Detecting epistasis in human complex traits. Nat Rev Genet. 2014;15:722–33. [DOI] [PubMed] [Google Scholar]
- 86.Richardson TG, Leyden GM, Wang Q, Bell JA, Elsworth B, Davey Smith G, et al. Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol. 2022;20:e3001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koyama S, Ito K, Terao C, Akiyama M, Horikoshi M, Momozawa Y, et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet. 2020;52:1169–77. [DOI] [PubMed] [Google Scholar]
- 88.Guloksuz S, van Os J, Rutten BPF. The exposome paradigm and the complexities of environmental research in psychiatry. JAMA Psychiatry. 2018;75:985–6. [DOI] [PubMed] [Google Scholar]
- 89.Pries LK, Dal Ferro GA, van Os J, Delespaul P, Kenis G, Lin BD, et al. Examining the independent and joint effects of genomic and exposomic liabilities for schizophrenia across the psychosis spectrum. Epidemiol Psychiatr Sci. 2020;29:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang R, Lifelines CohortS, Hartman CA, Snieder H. Stress-related exposures amplify the effects of genetic susceptibility on depression and anxiety. Transl Psychiatry. 2023;13:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Consortium E-I. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song W, Qian W, Wang W, Yu S, Lin GN. Mendelian randomization studies of brain MRI yield insights into the pathogenesis of neuropsychiatric disorders. BMC Genom. 2021;22:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burgess S, Foley CN, Allara E, Staley JR, Howson JMM. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Verbanck M, Chen CY, Neale B, Do R. Publisher Correction: detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:1196. [DOI] [PubMed] [Google Scholar]
- 100.Rees JMB, Wood AM, Dudbridge F, Burgess S. Robust methods in Mendelian randomization via penalization of heterogeneous causal estimates. PLoS ONE. 2019;14:e0222362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grant AJ, Burgess S. Pleiotropy robust methods for multivariable Mendelian randomization. Stat Med. 2021;40:5813–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schaid DJ, Tong X, Batzler A, Sinnwell JP, Qing J, Biernacka JM. Multivariate generalized linear model for genetic pleiotropy. Biostatistics. 2019;20:111–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y, Liu A, Mills JL, Boehnke M, Wilson AF, Bailey-Wilson JE, et al. Pleiotropy analysis of quantitative traits at gene level by multivariate functional linear models. Genet Epidemiol. 2015;39:259–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lutz SM, Fingerlin TE, Hokanson JE, Lange C. A general approach to testing for pleiotropy with rare and common variants. Genet Epidemiol. 2017;41:163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Adams CD. A multivariable Mendelian randomization to appraise the pleiotropy between intelligence, education, and bipolar disorder in relation to schizophrenia. Sci Rep. 2020;10:6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosoff DB, Hamandi AM, Bell AS, Mavromatis LA, Park LM, Jung J, et al. Major psychiatric disorders, substance use behaviors, and longevity. JAMA Psychiatry. 2024;81:889–901. [DOI] [PMC free article] [PubMed]
- 107.Fernandes SB, Zhang KS, Jamann TM, Lipka AE. How well can multivariate and univariate GWAS distinguish between true and spurious pleiotropy? Front Genet. 2020;11:602526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 2019;4:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Watanabe K, Stringer S, Frei O, Mirkov MU, de Leeuw C, Polderman TJC, et al. Author Correction: a global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2020;52:353. [DOI] [PubMed] [Google Scholar]
- 112.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127. [DOI] [PMC free article] [PubMed]
- 114.Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dobbyn A, Huckins LM, Boocock J, Sloofman LG, Glicksberg BS, Giambartolomei C, et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am J Hum Genet. 2018;102:1169–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cai L, Huang T, Su J, Zhang X, Chen W, Zhang F, et al. Implications of newly identified brain eQTL genes and their interactors in schizophrenia. Mol Ther Nucleic Acids. 2018;12:433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bhalala OG, Nath AP, Consortium UKBE, Inouye M, Sibley CR. Identification of expression quantitative trait loci associated with schizophrenia and affective disorders in normal brain tissue. PLoS Genet. 2018;14:e1007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang W, Gamazon ER, Zhang X, Konkashbaev A, Liu C, Szilagyi KL, et al. SCAN database: facilitating integrative analyses of cytosine modification and expression QTL. Database. 2015;2015:bav025. [DOI] [PMC free article] [PubMed]
- 119.Davis LK, Gamazon ER, Kistner-Griffin E, Badner JA, Liu C, Cook EH, et al. Loci nominally associated with autism from genome-wide analysis show enrichment of brain expression quantitative trait loci but not lymphoblastoid cell line expression quantitative trait loci. Mol Autism. 2012;3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Davis LK, Yu D, Keenan CL, Gamazon ER, Konkashbaev AI, Derks EM, et al. Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genet. 2013;9:e1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18:340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scharf JM, Yu D, Mathews CA, Neale BM, Stewart SE, Fagerness JA, et al. Genome-wide association study of Tourette’s syndrome. Mol Psychiatry. 2013;18:721–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stewart SE, Yu D, Scharf JM, Neale BM, Fagerness JA, Mathews CA, et al. Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry. 2013;18:788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ji Y, Wei Q, Chen R, Wang Q, Tao R, Li B. Integration of multidimensional splicing data and GWAS summary statistics for risk gene discovery. PLoS Genet. 2022;18:e1009814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Javitt DC, Spencer KM, Thaker GK, Winterer G, Hajós M. Neurophysiological biomarkers for drug development in schizophrenia. Nat Rev Drug Discov. 2008;7:68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dai R, Chu T, Zhang M, Wang X, Jourdon A, Wu F, et al. Evaluating performance and applications of sample-wise cell deconvolution methods on human brain transcriptomic data. bioRxiv [Preprint]. 2023. 10.1101/2023.03.13.532468. [DOI] [PMC free article] [PubMed]
- 127.Umans BD, Battle A, Gilad Y. Where are the disease-associated eQTLs? Trends Genet. 2021;37:109–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wen C, Margolis M, Dai R, Zhang P, Przytycki PF, Vo DD, et al. Cross-ancestry atlas of gene, isoform, and splicing regulation in the developing human brain. Science. 2023;384:eadh0829. [DOI] [PubMed]
- 129.Jerber J, Seaton DD, Cuomo ASE, Kumasaka N, Haldane J, Steer J, et al. Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation. Nat Genet. 2021;53:304–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen C, Meng Q, Xia Y, Ding C, Wang L, Dai R, et al. The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci Transl Med. 2018;10:eaat8178. [DOI] [PMC free article] [PubMed]
- 131.Ding C, Zhang C, Kopp R, Kuney L, Meng Q, Wang L, et al. Transcription factor POU3F2 regulates TRIM8 expression contributing to cellular functions implicated in schizophrenia. Mol Psychiatry. 2021;26:3444–60. [DOI] [PMC free article] [PubMed]
- 132.Meng Q, Wang K, Brunetti T, Xia Y, Jiao C, Dai R, et al. The DGCR5 long noncoding RNA may regulate expression of several schizophrenia-related genes. Sci Transl Med. 2018;10:eaat6912. [DOI] [PMC free article] [PubMed]
- 133.Srikanth P, Han K, Callahan DG, Makovkina E, Muratore CR, Lalli MA, et al. Genomic DISC1 disruption in hiPSCs alters wnt signaling and neural cell fate. Cell Rep. 2015;12:1414–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zeng L, Zhang P, Shi L, Yamamoto V, Lu W, Wang K. Functional impacts of NRXN1 knockdown on neurodevelopment in stem cell models. PLoS ONE. 2013;8:e59685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cameron D, Blake DJ, Bray NJ, Hill MJ. Transcriptional changes following cellular knockdown of the schizophrenia risk gene SETD1A are enriched for common variant association with the disorder. Mol Neuropsychiatry. 2019;5:109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hill MJ, Killick R, Navarrete K, Maruszak A, McLaughlin GM, Williams BP, et al. Knockdown of the schizophrenia susceptibility gene TCF4 alters gene expression and proliferation of progenitor cells from the developing human neocortex. J Psychiatry Neurosci. 2017;42:181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hill MJ, Jeffries AR, Dobson RJ, Price J, Bray NJ. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum Mol Genet. 2012;21:1018–24. [DOI] [PubMed] [Google Scholar]
- 139.Zhang W, Zhang M, Xu Z, Yan H, Wang H, Jiang J, et al. Human forebrain organoid-based multi-omics analyses of PCCB as a schizophrenia associated gene linked to GABAergic pathways. Nat Commun. 2023;14:5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang CP, Li X, Wu Y, Shen Q, Zeng Y, Xiong Q, et al. Comprehensive integrative analyses identify GLT8D1 and CSNK2B as schizophrenia risk genes. Nat Commun. 2018;9:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hoffmann A, Ziller M, Spengler D. Focus on causality in ESC/iPSC-based modeling of psychiatric disorders. Cells. 2020;9:366. [DOI] [PMC free article] [PubMed]
- 142.O’Brien HE, Hannon E, Hill MJ, Toste CC, Robertson MJ, Morgan JE, et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Walker RL, Ramaswami G, Hartl C, Mancuso N, Gandal MJ, de la Torre-Ubieta L, et al. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell. 2020;181:745. [DOI] [PubMed] [Google Scholar]
- 144.Retallick-Townsley KG, Lee S, Cartwright S, Cohen S, Sen A, Jia M, et al. Dynamic stress- and inflammatory-based regulation of psychiatric risk loci in human neurons. bioRxiv [Preprint]. 2024. 10.1101/2024.07.09.602755.
- 145.Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tamminga CA, Pearlson G, Keshavan M, Sweeney J, Clementz B, Thaker G. Bipolar and schizophrenia network for intermediate phenotypes: outcomes across the psychosis continuum. Schizophr Bull. 2014;40:S131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karlsgodt KH, Sun D, Cannon TD. Structural and functional brain abnormalities in schizophrenia. Curr Dir Psychol Sci. 2010;19:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct. 2008;213:93–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sylvester CM, Yu Q, Srivastava AB, Marek S, Zheng A, Alexopoulos D, et al. Individual-specific functional connectivity of the amygdala: a substrate for precision psychiatry. Proc Natl Acad Sci USA. 2020;117:3808–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kam JWY, Griffin S, Shen A, Patel S, Hinrichs H, Heinze HJ, et al. Systematic comparison between a wireless EEG system with dry electrodes and a wired EEG system with wet electrodes. Neuroimage. 2019;184:119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hinrichs H, Scholz M, Baum AK, Kam JWY, Knight RT, Heinze H-J. Comparison between a wireless dry electrode EEG system with a conventional wired wet electrode EEG system for clinical applications. Sci Rep. 2020;10:5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wigmore EM, Hafferty JD, Hall LS, Howard DM, Clarke TK, Fabbri C, et al. Genome-wide association study of antidepressant treatment resistance in a population-based cohort using health service prescription data and meta-analysis with GENDEP. Pharmacogenomics J. 2020;20:329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li QS, Tian C, Hinds D. 23andMe. Genome-wide association studies of antidepressant class response and treatment-resistant depression. Transl Psychiatry. 2020;10:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu H, Yan H, Wang L, Li J, Tan L, Deng W, et al. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry. 2018;5:327–38. [DOI] [PubMed] [Google Scholar]
- 155.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13:261–76. [DOI] [PubMed] [Google Scholar]
- 157.Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: reliability, validity and sensitivity. Br J psychiatry J Ment Sci. 1978;133:429–35. [DOI] [PubMed] [Google Scholar]
- 158.Yuan N, Chen Y, Xia Y, Dai J, Liu C. Inflammation-related biomarkers in major psychiatric disorders: a cross-disorder assessment of reproducibility and specificity in 43 meta-analyses. Transl Psychiatry. 2019;9:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Drexhage RC, Weigelt K, van Beveren N, Cohen D, Versnel MA, Nolen WA, et al. Immune and neuroimmune alterations in mood disorders and schizophrenia. Int Rev Neurobiol. 2011;101:169–201. [DOI] [PubMed] [Google Scholar]
- 160.Giridharan VV, Sayana P, Pinjari OF, Ahmad N, da Rosa MI, Quevedo J, et al. Postmortem evidence of brain inflammatory markers in bipolar disorder: a systematic review. Mol Psychiatry. 2020;25:94–113. [DOI] [PubMed] [Google Scholar]
- 161.Lizano P, Lutz O, Xu Y, Rubin LH, Paskowitz L, Lee AM, et al. Multivariate relationships between peripheral inflammatory marker subtypes and cognitive and brain structural measures in psychosis. Mol Psychiatry. 2021;26:3430–43. [DOI] [PMC free article] [PubMed]
- 162.Tamminga CA, Clementz BA, Pearlson G, Keshavan M, Gershon ES, Ivleva EI, et al. Biotyping in psychosis: using multiple computational approaches with one data set. Neuropsychopharmacology. 2021;46:143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Misiak B, Stanczykiewicz B, Kotowicz K, Rybakowski JK, Samochowiec J, Frydecka D. Cytokines and C-reactive protein alterations with respect to cognitive impairment in schizophrenia and bipolar disorder: a systematic review. Schizophr Res. 2018;192:16–29. [DOI] [PubMed] [Google Scholar]
- 164.Chen Y, Dai J, Tang L, Mikhailova T, Liang Q, Li M et al. Neuroimmune transcriptome changes in patient brains of psychiatric and neurological disorders. Mol Psychiatry. 2023;28:710–21. [DOI] [PMC free article] [PubMed]
- 165.Mangino M, Roederer M, Beddall MH, Nestle FO, Spector TD. Innate and adaptive immune traits are differentially affected by genetic and environmental factors. Nat Commun. 2017;8:13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Epstein MM, Breen EC, Magpantay L, Detels R, Lepone L, Penugonda S, et al. Temporal stability of serum concentrations of cytokines and soluble receptors measured across two years in low-risk HIV-seronegative men. Cancer Epidemiol Biomark Prev. 2013;22:2009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ho GY, Xue XN, Burk RD, Kaplan RC, Cornell E, Cushman M. Variability of serum levels of tumor necrosis factor-alpha, interleukin 6, and soluble interleukin 6 receptor over 2 years in young women. Cytokine. 2005;30:1–6. [DOI] [PubMed] [Google Scholar]
- 168.Navarro SL, Brasky TM, Schwarz Y, Song X, Wang CY, Kristal AR, et al. Reliability of serum biomarkers of inflammation from repeated measures in healthy individuals. Cancer Epidemiol Biomark Prev. 2012;21:1167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li L, Ma X, Cui Y, Rotival M, Chen W, Zou X, et al. Immune-response 3’UTR alternative polyadenylation quantitative trait loci contribute to variation in human complex traits and diseases. Nat Commun. 2023;14:8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang L, Zhao Y, Wang Y, Liu L, Zhang X, Li B, et al. The effects of psychological stress on depression. Curr Neuropharmacol. 2015;13:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schneiderman N, Ironson G, Siegel SD. Stress and health: psychological, behavioral, and biological determinants. Annu Rev Clin Psychol. 2005;1:607–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brown GW, Birley JL. Crises and life changes and the onset of schizophrenia. J Health Soc Behav. 1968;9:203–14. [PubMed] [Google Scholar]
- 173.Norman RM, Malla AK. Stressful life events and schizophrenia. I: a review of the research. Br J Psychiatry J Ment Sci. 1993;162:161–6. [DOI] [PubMed] [Google Scholar]
- 174.Betz LT, Penzel N, Kambeitz-Ilankovic L, Rosen M, Chisholm K, Stainton A, et al. General psychopathology links burden of recent life events and psychotic symptoms in a network approach. NPJ Schizophr. 2020;6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Post RM, Leverich GS. The role of psychosocial stress in the onset and progression of bipolar disorder and its comorbidities: the need for earlier and alternative modes of therapeutic intervention. Dev Psychopathol. 2006;18:1181–211. [DOI] [PubMed] [Google Scholar]
- 176.Hayes PE, Ettigi P. Dexamethasone suppression test in diagnosis of depressive illness. Clin Pharmacol. 1983;2:538–45. [PubMed] [Google Scholar]
- 177.Fountoulakis KN, Gonda X, Rihmer Z, Fokas C, Iacovides A. Revisiting the Dexamethasone Suppression Test in unipolar major depression: an exploratory study. Ann Gen Psychiatry. 2008;7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nandam LS, Brazel M, Zhou M, Jhaveri DJ. Cortisol and major depressive disorder-translating findings from humans to animal models and back. Front Psychiatry. 2019;10:974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schatzberg AF, Keller J, Tennakoon L, Lembke A, Williams G, Kraemer FB, et al. HPA axis genetic variation, cortisol and psychosis in major depression. Mol Psychiatry. 2014;19:220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shields A, Wise L, Ruiz-Narvaez E, Seddighzadeh B, Byun H, Cozier Y, et al. Childhood abuse, promoter methylation of leukocyte NR3C1 and the potential modifying effect of emotional support. Epigenomics. 2016;8:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Farrell C, Doolin K, O’ Leary N, Jairaj C, Roddy D, Tozzi L, et al. DNA methylation differences at the glucocorticoid receptor gene in depression are related to functional alterations in hypothalamic-pituitary-adrenal axis activity and to early life emotional abuse. Psychiatry Res. 2018;265:341–8. [DOI] [PubMed] [Google Scholar]
- 182.Booij L, Szyf M, Carballedo A, Frey E, Morris D, Dymov S, et al. DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: a study in depressed patients and healthy controls. PloS ONE. 2015;10:e0119061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Alexander N, Kirschbaum C, Stalder T, Muehlhan M, Vogel S. No association between FKBP5 gene methylation and acute and long-term cortisol output. Transl Psychiatry. 2020;10:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Parent J, Parade S, Laumann L, Ridout K, Yang B, Marsit C, et al. Dynamic stress-related epigenetic regulation of the glucocorticoid receptor gene promoter during early development: The role of child maltreatment. Dev Psychopathol. 2017;29:1635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tyrka A, Ridout K, Parade S, Paquette A, Marsit C, Seifer R. Childhood maltreatment and methylation of FK506 binding protein 5 gene (FKBP5). Dev Psychopathol. 2015;27:1637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tyrka A, Parade S, Welch E, Ridout K, Price L, Marsit C, et al. Methylation of the leukocyte glucocorticoid receptor gene promoter in adults: associations with early adversity and depressive, anxiety and substance-use disorders. Transl Psychiatry. 2016;6:e848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Parade S, Novick A, Parent J, Seifer R, Klaver S, Marsit C, et al. Stress exposure and psychopathology alter methylation of the serotonin receptor 2A (HTR2A) gene in preschoolers. Dev Psychopathol. 2017;29:1619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bustamante A, Aiello A, Galea S, Ratanatharathorn A, Noronha C, Wildman D, et al. Glucocorticoid receptor DNA methylation, childhood maltreatment and major depression. J Affect Disord. 2016;206:181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Federenko IS, Nagamine M, Hellhammer DH, Wadhwa PD, Wust S. The heritability of hypothalamus pituitary adrenal axis responses to psychosocial stress is context dependent. J Clin Endocrinol Metab. 2004;89:6244–50. [DOI] [PubMed] [Google Scholar]
- 190.Rietschel L, Streit F, Zhu G, McAloney K, Frank J, Couvy-Duchesne B, et al. Hair cortisol in twins: heritability and genetic overlap with psychological variables and stress-system genes. Sci Rep. 2017;7:15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ising M, Holsboer F. Genetics of stress response and stress-related disorders. Dialogues Clin Neurosci. 2006;8:433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Arnau-Soler A, Adams MJ, Major Depressive Disorder Working Group of the Psychiatric Genomics GenerationS, Hayward C, Thomson PA. Genome-wide interaction study of a proxy for stress-sensitivity and its prediction of major depressive disorder. PLoS ONE. 2018;13:e0209160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hontelez S, Karthaus N, Looman MW, Ansems M, Adema GJ. DC-SCRIPT regulates glucocorticoid receptor function and expression of its target GILZ in dendritic cells. J Immunol. 2013;190:3172–9. [DOI] [PubMed] [Google Scholar]
- 194.Arloth J, Bogdan R, Weber P, Frishman G, Menke A, Wagner KV, et al. Genetic differences in the immediate transcriptome response to stress predict risk-related brain function and psychiatric disorders. Neuron. 2015;86:1189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Penner-Goeke S, Bothe M, Rek N, Kreitmaier P, Pohlchen D, Kuhnel A, et al. High-throughput screening of glucocorticoid-induced enhancer activity reveals mechanisms of stress-related psychiatric disorders. Proc Natl Acad Sci USA. 2023;120:e2305773120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Volkow ND, Koob GF, Croyle RT, Bianchi DW, Gordon JA, Koroshetz WJ, et al. The conception of the ABCD study: from substance use to a broad NIH collaboration. Dev Cogn Neurosci. 2018;32:4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cai N, Revez JA, Adams MJ, Andlauer TFM, Breen G, Byrne EM, et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat Genet. 2020;52:437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Glahn DC, Curran JE, Winkler AM, Carless MA, Kent JW Jr, Charlesworth JC, et al. High dimensional endophenotype ranking in the search for major depression risk genes. Biol Psychiatry. 2012;71:6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Foley CN, Staley JR, Breen PG, Sun BB, Kirk PDW, Burgess S, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021;12:764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ni G, Zeng J, Revez JA, Wang Y, T. G, Restaudi R, et al. A Comparison of Ten Polygenic Score Methods for Psychiatric Disorders Applied Across Multiple Cohorts. Biol Psychiatry. 2020;90:611–20. [DOI] [PMC free article] [PubMed]
- 201.Perkins DO, Olde Loohuis L, Barbee J, Ford J, Jeffries CD, Addington J, et al. Polygenic risk score contribution to psychosis prediction in a target population of persons at clinical high risk. Am J psychiatry. 2020;177:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Anderson JS, Shade J, DiBlasi E, Shabalin AA, Docherty AR. Polygenic risk scoring and prediction of mental health outcomes. Curr Opin Psychol. 2019;27:77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Murray GK, Lin T, Austin J, McGrath JJ, Hickie IB, Wray NR. Could polygenic risk scores be useful in psychiatry?: A review. JAMA Psychiatry. 2021;78:210–9. [DOI] [PubMed] [Google Scholar]
- 204.Kharitonova EV, Sun Q, Ockerman F, Chen B, Zhou LY, Cao H, et al. EndoPRS: incorporating endophenotype information to improve polygenic risk scores for clinical endpoints. medRxiv [Preprint]. 2024. 10.1101/2024.05.23.24307839.
- 205.Kannel WB, Dawber TR, Kagan A, Revotskie N, Stokes J 3rd. Factors of risk in the development of coronary heart disease-six year follow-up experience. the Framingham Study. Ann Intern Med. 1961;55:33–50. [DOI] [PubMed] [Google Scholar]
- 206.Tan YD, Xiao P, Guda C. In-depth Mendelian randomization analysis of causal factors for coronary artery disease. Sci Rep. 2020;10:9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–47. [DOI] [PubMed] [Google Scholar]
- 208.Damask A, Steg PG, Schwartz GG, Szarek M, Hagström E, Badimon L, et al. Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES trial. Circulation. 2020;141:624–36. [DOI] [PubMed] [Google Scholar]
- 209.Koppe G, Meyer-Lindenberg A, Durstewitz D. Deep learning for small and big data in psychiatry. Neuropsychopharmacology. 2021;46:176–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gershon ES, Lee SH, Zhou X, Sweeney JA, Tamminga CA, Pearlson GD, et al. An opportunity for primary prevention research in psychotic disorders. Schizophr Res. 2021;243:433–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lee SH, Clark S, Van der Werf J. Estimation of genomic prediction accuracy from reference populations with varying degrees of relationship. PLoS ONE. 2017;12:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dahl A, Iotchkova V, Baud A, Johansson A, Gyllensten U, Soranzo N, et al. A multiple-phenotype imputation method for genetic studies. Nat Genet. 2016;48:466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hormozdiari F, Kang EY, Bilow M, Ben-David E, Vulpe C, McLachlan S, et al. Imputing phenotypes for genome-wide association studies. Am J Hum Genet. 2016;99:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Asif H, Alliey-Rodriguez N, Keedy S, Tamminga CA, Sweeney JA, Pearlson G, et al. GWAS significance thresholds for deep phenotyping studies can depend upon minor allele frequencies and sample size. Mol Psychiatry. 2021;26:2048–55. [DOI] [PMC free article] [PubMed]
- 215.Zhu Y, Owens SJ, Murphy CE, Ajulu K, Rothmond D, Purves-Tyson T, et al. Inflammation-related transcripts define “high” and “low” subgroups of individuals with schizophrenia and bipolar disorder in the midbrain. Brain Behav Immun. 2022;105:149–59. [DOI] [PubMed] [Google Scholar]
- 216.Fillman SG, Weickert TW, Lenroot RK, Catts SV, Bruggemann JM, Catts VS, et al. Elevated peripheral cytokines characterize a subgroup of people with schizophrenia displaying poor verbal fluency and reduced Broca’s area volume. Mol Psychiatry. 2016;21:1090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality score of endophenotypes for psychiatric genetics