Abstract
Background
Diarrhea caused by non-O1/O139-group V. cholerae (NOVC) tends to be mild and can be readily overlooked. In this report, a NOVC strain designated XXM was isolated from the blood of a 68-year-old male undergoing surgical treatment for a bile duct malignancy in October 2023.
Methods
XXM was identified through a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Virulence genes were detected using a V. cholerae ctxA/ctxB virulence gene dual real-time fluorescent PCR kit. AST-GN13 and AST-GN334 cards were used to test the resistance against 16 antibiotics with a Vitek2 compact system. The genomic and phylogenetic characteristics of XXM were established through whole genome sequencing (WGS).
Results
Serum agglutination tests revealed the isolate to be a non-O1/non-O139 strain. The strain was sensitive to all 16 tested antibiotics and did not carry the ctxA/ctxB gene. MLST analyses identified the XXM strain as ST1538. WGS analyses identified 8 classes of virulence genes with different functions. A total of 3.541 bacterial genes, including 3.482 from V. cholerae, were annoted using the Non-Redundant Protein Sequence (NR) database. Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses annotated 32 genes including 17 key proteins involved in the V. cholerae biofilm pathway. Comparative analyses using the Pathogen Host Interactions Database (PHI) identified the YbeY gene. Evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) was used to annotate 3280 genes in 21 categories. Phylogenetic analyses revealed that strain XXM was closely related to V. cholerae strain Man9.
Conclusion
The XXM carries multiple virulence genes, and this genomic analysis of the XXM in comparison with other NOVC strains provides important information for an improved understanding of the pathogenicity of NOVC in clinical samples.
Keywords: Vibrio cholerae, non-O1/O139-group, whole genome sequencing, bloodstream infection, molecular characterization
Introduction
Vibrio cholerae is the bacterial pathogen responsible for cholera. Of the over 200 V. cholerae serotypes recorded to date, only serogroups O1 and O139 caused global pandemics following their spread from the delta of the river Ganges.1 Approximately 2.9 million individuals throughout the globe suffer from cholera annually, resulting in 95,000 deaths, and the vast majority of which occur in developing countries.2
V. cholerae infections can cause fulminant diarrhea attributable to the virulence genes. The ctxA/ctxB encodes cholera enterotoxin, which causes the extensive loss of fluids and electrolytes. TCP gene cluster encodes a toxin-coregulated pilus, which is important for the intestinal colonization of these bacteria. Non-O1 and non-O139 V. cholerae (NOVC) strains, however, rarely produce cholera enterotoxin. While these NOVC strains cause watery diarrhea and severe illness in some cases, they tend to cause milder infections than the typical presentation of cholera,3 and generally consist of enteric infections, including sepsis.4 Individuals with hematological or hepatic diseases face greater risk of NOVC infection,4,5 exhibiting a mortality rate of 12.1–39%.5,6
In this study, a NOVC strain was isolated from a patient undergoing surgery to treat bile duct malignancy. A retrospective analysis of infection-related information was conducted for this patient, and whole genome sequencing (WGS) was employed for the molecular characterization of the isolated V. cholerae strain.
Materials and Methods
Strain Isolation and Antibiotic Susceptibility Testing
A 5–10 mL sample of peripheral blood was collected from the patient in a blood culture vial (bioMérieux, France). Culture positivity was recorded after 11 h 35 min, and the microbes were then further cultured on blood agar and MacConkey agar plates (Kangtai, Wenzhou), which were incubated for 24–48 h at 35°C in a 5% CO2 incubator. The isolate was identified as V. cholerae through matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (bioMérieux, France) testing. Further examination using O139 V. cholerae antiserum and Inaba and Ogawa mixed antiserum (Denka Seiken, Japan) was performed, identifying the strain as a NOVC strain (designed as XMM). Virulence genes encoded by this strain were detected with a V. cholerae ctxA/ctxB virulence gene dual real-time fluorescent PCR kit (Beijing Applied Biological Technologies, China). The resistance of this strain to 16 antibiotics (Amikacin, Amoxicillin-Clavulanic acid, Cefepime, Cefoperazone-Sulbactam, Cefoxitin, Ceftazidime, Cefuroxime, Levofloxacin, Piperacillin-Tazobactam, Tetracycline, Sulfamethoxazole-Rimethoprim, Ampicillin, Ampicillin-Sulbactam, Cefazolin, Imipenem, Gentamicin) was evaluated with the Vitek2 compact system with the AST-GN13 and AST-GN334 cards. The minimum inhibitory concentration (MIC) values for these antibiotics were obtained automatically from this system, and phenotypes were determined based on the Clinical and Laboratory Standards Institute guidelines (Performance Standards for Antimicrobial Susceptibility Testing, 33rd edition). Escherichia coli ATCC25922 served as the reference strain.
DNA Extraction and Genome Assembly
A HiPure Bacterial DNA Kit (Magentec, China) was used to extract genomic DNA. The TruSeq DNA Sample Preparation Kit (Illumina, USA) and the Template Prep Kit (Pacific Biosciences, USA) were used for library preparation. WGS was performed with the Nanopore PromrthION48 platform and the Illumina Novaseq platform (Personal Biotechnology Company, China). After removing adapter-containing sequences and filtering the reads using Adapter Removal7 and SOAPec,8 the filtered reads were assembled using SPAdes9 and A5-miseq10 for scaffold and contig construction. Data from sequencing with the Nanopore platform were assembled with Flye11 and the Unicycler software.12 All of the assembled results were then integrated to compile complete genomic sequences with the Pilon software.13
Whole Genome Sequencing Analyses
GeneMarkS v4.3214 was used for gene prediction, while tRNAs, rRNAs, and other ncRNAs were, respectively, identified with tRNAscan-SE,15 Barrnap (v 0.9) and Rfam.16 Genomic island predictions were made with IslandViewer4,17 while pathogenicity and antibiotic resistance-related genes were, respectively, retrieved with the VFDB (Virulence Factors of Pathogenic Bacteria) database18 and CARD (The Comprehensive Antibiotic Resistance) database.19 The virulence genes of XXM were compared with those of the reference strain RFB16 negative for ctxA/ctxB virulence genes (accession number: GCA_008369605.1) and two NOVC strains isolated from blood samples in China (VCHL017, accession number: GCA_022758165.1; 2352495169, accession number: GCA_029906585.1). Functional annotation was achieved via BLAST searching against the NR (Non-Redundant Protein Database),20 KEGG (Kyoto Encyclopedia of Gene and Genomes),21 COG (Cluster of Orthologous Groups of proteins),22 Swissprot, Pathogen–Host Interactions Database (PHI), and CGview23 databases. PubMLST (https://pubmlst.org/) was used to establish the sequence type for the isolate. Genomic sequencing data were inputted into the UBGC v3.0 software (https://www.ezbiocloud.net/tools/ubcg), leading to the identification of 92 core genes that were used to construct a phylogenetic tree with the maximum likelihood method (Supplementary Table 1). The sequences of XXM and other strains were uploaded to the EzBioCloud (https://www.ezbiocloud.net/tools/ani) to compare the level of similarity. The arrangements of the genomes of the XXM, Man9 (accession number: GCA_030295265.1), and the reference strain RFB16 strains were analyzed using Mauve version 20150226 build 10 (c) package by dividing the genome into multiple locally collinear blocks (LCBs).24
Results
Case Presentation
In October 2023, a 68-year-old male patient developed a fever (maximum temperature: 39.0°C) accompanied by a headache that lasted for 10 minutes and resolved after nausea and two episodes of vomiting. This patient’s medical history indicated that they had undergone laparoscopic pancreaticoduodenectomy for lower choledochal tumors 7 months ago. Laboratory findings showed elevated procalcitonin (4.59 ng/mL), neutrophil percentage (92.2%), glutamic oxaloacetic transaminase level (112 U/L), and glutamic pyruvic transaminase level (57 U/L). However, his hypersensitive C-reactive protein levels and white blood cell counts were in the normal range. Cefoperazone-sulbactam (2.00 g) intravenous infusion was administered every 8 hours on the first day. On the third day after admission, gram-negative bacilli were detected in laboratory blood cultures and identified as V. cholerae (designated as strain XXM), with a Non-O1/O139 serotype; no ctxA/ctxB virulence genes were detected. This strain was sensitive to 16 tested antibiotic agents (Table 1). Therefore, the patient’s treatment regimen was modified on day 4 to ceftriaxone (2.00 g) intravenous infusion every 24 hours. After 4 days, his temperature had returned to the normal range, his headache symptoms had abated, and his procalcitonin levels had decreased.
Table 1.
Antimicrobial Susceptibility Testing for V. cholerae Strain XXM
| Antimicrobial agents | MIC(μg/mL) |
|---|---|
| Amikacin | ≤ 2 |
| Amoxicillin-Clavulanic acid | 4 |
| Cefepime | ≤ 0.12 |
| Cefoperazone-Sulbactam | ≤ 8 |
| Cefoxitin | ≤ 4 |
| Ceftazidime | ≤ 0.12 |
| Cefuroxime | ≤ 1 |
| Levofloxacin | ≤ 0.12 |
| Piperacillin-Tazobactam | ≤ 4 |
| Tetracycline | ≤ 0.5 |
| Sulfamethoxazole-Rimethoprim | ≤ 20 |
| Ampicillin | ≤ 2 |
| Ampicillin-Sulbactam | ≤ 2 |
| Cefazolin | ≤ 4 |
| Imipenem | 4 |
| Gentamicin | ≤ 1 |
Genome Assembly and Gene Predictions
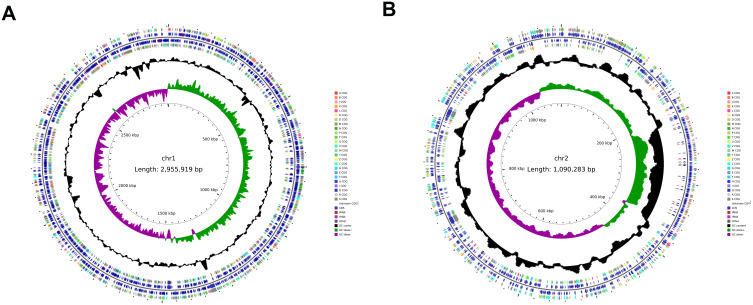
The genome of this V. cholerae isolate consisted of two chromosomes, respectively, measuring 2,955,919 bp and 1,090,283 bp, with respective GC content of 47.90% and 46.85%. These two chromosomes, respectively, harbored 2575 and 1012 open reading frames. In total, 104 tRNAs, 31 rRNAs, and 47 other ncRNAs were identified across the genome. The final assembled genome was submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/) with the accession number: CP166972-CP166973.
Virulence and Antibiotic-Resistance Genes in the Genome
Using a threshold 90–100% sequence identity, 8 classes of virulence genes with different functions were identified. Comparison with the reference strain RFB16 with negative ctxA/ctxB virulence genes and two NOVC strains isolated from blood samples in China, showed an absence of the gbpA and rtxA virulence genes in the isolate (Table 2). When comparative analyses were performed at a 80–100% identity level, 6 resistance genes were detected, 4 of which were distributed across 3 gene islands (Table 3).
Table 2.
Distribution of Virulence Genes in Different Strains
| VF Function | Common VF genes | Additional Genes in XXM | Additional Genes in RFB16 | Additional Genes in VCHL017 | Additional Genes in 2352495169 |
|---|---|---|---|---|---|
| Effector delivery system | clpB/vasG epsC epsD epsE epsF epsG epsH epsI epsJ epsK epsL epsM epsN icmF/vasK vasA vasB vasC vasD vasE vasF vasH vasI vasJ vasL VC0395_RS15455 VC0395_RS15460 VCA0109 vgrG-2 vipA/mglA vipB/mglB | hcp-1 hcp-2 vgrG-3 | hcp-2 VCA0122 vcrD2 vgrG-3 | hcp-1 | hcp-1 vgrG-3 |
| Motility | cheA cheB cheR cheV cheW cheY cheZ flaA flaB flaC flaD flaE flaG flaI fleN/flhG fleR/flrC fleS/flrB flgA flgB flgC flgD flgE flgF flgG flgH flgI flgJ flgK flgL flgM flgN flgO flgP flgT flhA flhB flhF fliA fliD fliE fliF fliG fliH fliI fliJ fliK fliL fliM fliN fliO fliP fliQ fliR fliS flrA motA motB motX motY | ||||
| Adherence | mshB mshC mshD mshE mshF mshG mshH mshI mshJ mshK mshL mshM mshN pilB pilC pilD/vcpD mshB | gbpA | gbpA | gbpA | |
| biofilm | cqsA luxS VC_RS04610 VC_RS04620 VC_RS09110 vpsH vpsI vpsj vpsP vpsQ vpsU | vpsC vpsD vpsE vpsF vpsG | |||
| Exotoxin | rtxB rtxC rtxD hlyA tlh | rtxA | rtxA | rtxA | |
| Exoenzyme | hap/vvp | nanH | nanH | ||
| Immune modulation | cpsA cpsB cpsC cpsD cpsF | wbfY wbjD/wecB wecC | wbjD/wecB wecC | wbjD/wecB wecC | |
| Nutritional/Metabolic factor | hasR hutR irgA vctA vctC vctD vctG vctP vibA vibB vibC vibE vibF vibH viuA viuB viuC viuD viuG viuP | vibD | vibD | hutA vibD | hutA |
Table 3.
Prediction of Antibiotic-Resistance Genes Encoded by V. cholerae Strain XXM
| Gene | Describe | Identity | Gene Island |
|---|---|---|---|
| EF-Tu | Elfamycin resistant | 100 | chr1 IS11 |
| StrA | Resistant to Streptomycin | 94.35 | chr1 IS10 |
| rpoB | Resistance to rifampicin | 85.32 | chr1 IS10 |
| CRP | Antibiotic efflux | 95.24 | / |
| parE | Resistance to fluoroquinolones | 80.73 | / |
| qnrVC4 | Resistance to quinolone | 100 | chr2 IS1 |
Protein Feature Analyses
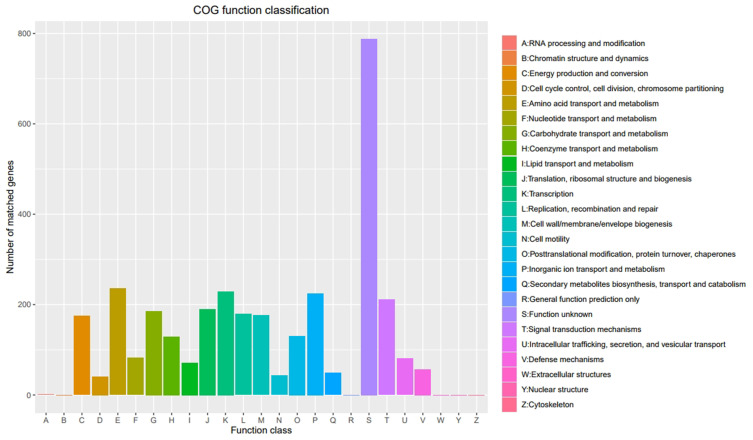
NR annotation was achieved for 3581 bacterial genes, including 3481 V. cholerae genes (Supplementary Table 2). KEGG analyses led to the annotation of 32 genes associated with the V. cholerae biofilm formation pathway (Supplementary Table 3). PHI comparisons at a 90–100% identity level revealed 31 reduced virulence genes, 2 unaffected virulence genes (hns CspV), and 1 instance of loss of pathogenicity for V. cholerae gene (YbeY) (Table 4). eggNOG annotation was achieved for 3280 genes in 21 categories (Figure 1), and the results were arranged into a genomospheric map (Figure 2).
Table 4.
Annotation of Strain XXM Genes Using a PHI Analysis
| Function class | Gene Name |
|---|---|
| Reduced virulence | tatA tatB tatC spoT AphA leuO relA VcpD VC2340 fadD LonA gntR vexD VC1348 VprA VC1295 relV aphB LuxO ToxR oadB vexF CsrA Hfq edd gntK gntU eda vexB hlyA VCA0895 VCA0931 ohrR |
| Unaffected pathogenicity | hns CspV |
| Loss pathogenicity | YbeY |
Figure 1.
Cluster of orthologous groups (COG) functional annotation of V. cholerae XXM genes. Annotated entries are shown on the abscissa with the number of matched genes on the ordinate. The color coding of COG functions is shown in the legend.
Figure 2.
Circular genome map of V. cholerae XXM. (A) Larger chromosome; (B) Smaller chromosomes. From inside to outside, the first circle represents the scale, the second circle indicates the GC Skew (green values indicate >0; purple values <0), the third ring represents the GC content (the outer black ring indicates greater than the average, while the inner black ring indicates less than the average), the fourth and seventh circles represent the COG to which each CDS belongs, and the fifth and sixth circles represent the positions of CDS (blue), tRNAs (red), and rRNAs (purple) in the genome.
MLST Analyses and Genomic Comparisons
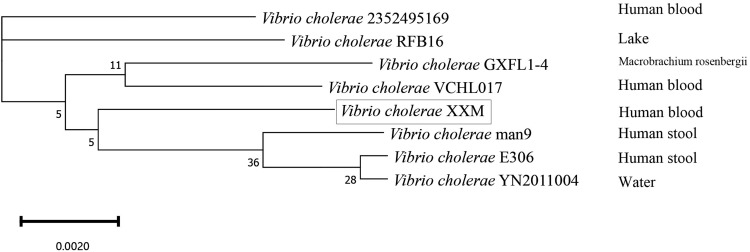
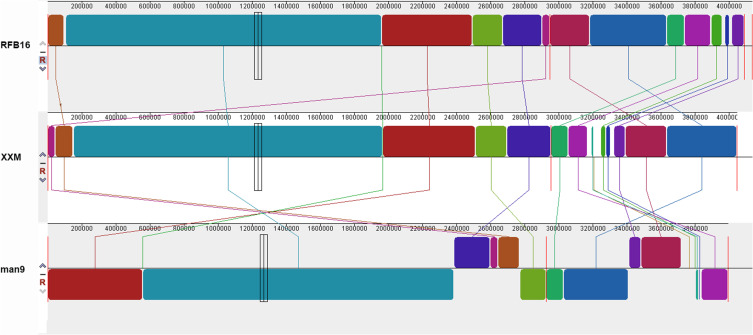
MLST analyses identified the XXM strain as ST1538, while the phylogenetic tree identified Man9 as the strain most closely related to XXM (Figure 3), with an Average Nucleotide Identity (ANI) of 98.26%. The XXM strain and RFB16 were largely identical (ANI 98.32%), with some blocks of rearrangement and insertion (chr2: 214,426–237,502, 250,579–292,117, 347,501–369,039), but no instances of inversion. Both rearrangements and inversions were evident in the Man9 strain relative to the RFB16 (ANI 98.27%) (Figure 4).
Figure 3.
Phylogenetic tree of 8 V.cholerae strains. The source from which the strains were isolated is indicated on the right, and rectangular one was for this study. The NCBI accession numbers of the V. cholera strains are provided in Supplementary Table 1.
Figure 4.
Mauve alignment of VCHL017 with five other V. cholerae genomes. The Mauve software divides the genome into several locally collinear blocks, with homologous regions of the genome colored identically and connected by lines.
Discussion
In summary, a NOVC strain was herein isolated from the blood of an older male with a history of malignancy. Hematologic and hepatic diseases have previously been identified as risk factors for NOVC infection.25 This strain exhibited good sensitivity to β-lactam and quinolone antibiotics during phenotypic testing, although WGS revealed the presence of the quinolone resistance genes parE and qnrVC4. This is consistent with results from a prior report in which an isolate exhibited phenotypic sensitivity to quinolones despite the detection of the qnrVC4 and parE genes.26 This may be attributable to the fact that certain parE mutations are not linked to quinolone resistance.27
The ctxA/ctxB genes and the TCP gene cluster are absent in strain XXM, consistent with the fact that the patient only experienced mild gastrointestinal symptoms. However, various virulence factors were detected in this strain. Effector delivery systems, including the type II and IV secretion systems, are key virulence factors that can support nutrient acquisition, the formation of biofilms, and the enhancement of strain pathogenicity.28,29 Motility can enhance virulence by improving the odds of V. cholerae binding to the intestinal mucosa, with nonmotile strains exhibiting impaired virulence linked to a reduced capacity for adsorption to the surfaces of mouse intestinal segments in prior reports.30 Other strains reported to cause bloodstream infections5,31,32 contained the cytotoxic rtxBCD and hlyA genes, all of which are RTX (Repeats in ToXin) toxins,33 that function by the penetration and permeabilization of host cell membranes, resulting in infection.34 The gbpA gene was not detected in this strain compared with the reference strain RFB16 and two blood isolates. Studies have shown that the gbpA gene encoding an important virulence factors linked to bacterial colonization identified pandemic isolates, is not ubiquitously present in V. cholerae strains.35 Besides, 33 genes associated with V. cholerae biofilm formation were identified in this strain. Bacterial accumulation in biofilms can support persistent infections and limit the efficacy of antimicrobial drugs.36,37 Managing such cases thus requires adequate doses of standard antibiotics together with the timely replacement of the needle to mitigate the potential for biofilm formation as a result of bacterial residues. PHI annotations revealed 31 genes with reduced virulence consistent with a reduction in their activity or consequent loss of function linked to an overall decrease in bacterial pathogenicity, the YbeY gene encodes an endoribonuclease involved in ribosome biosynthesis. Deleting this gene can have an adverse effect on the growth and replication of bacteria,38 and it may thus represent a promising new target for drug development efforts.
MLST identified the XXM strain as ST1538, which has previously been isolated in Thailand.39 The source of this strain may have been as a food or water contaminant, given that V. cholerae can be found in the freshwater rivers of Zhejiang Province.40 These environmentally derived strains generally exhibit an absence of any apparent selection pressure, with MLST typing revealing a diverse array of bacterial community characteristics, and no prevalent types.40 ST167 and ST173 were previously identified as the major epidemic types of non-pandemic O1 V. cholerae in Zhejiang Province, China.39 Phylogenetic tree analyses identified the Man9 strain first isolated in Sasebo City, Nagasaki Prefecture in 1946 from an individual who had returned from northeastern China as the closest relative to the XXM strain. China suffered severe outbreaks of cholera in 1940 and 1946, and it is possible that the Man9 lineage may have been the causative agent.41
Limitations
The patient was hospitalized without a stool culture test to determine whether Vibrio cholerae invaded the bloodstream from the intestine or other ways of invading the bloodstream.
Conclusion
The V. cholerae XXM strain was isolated from blood samples collected from patients after tumor resection surgery. While the strain was observed to carry a variety of virulence factors, it did not contain the ctxA/ctxB genes. Phylogenetic analysis showed that the strain was most closely related to a cholera strain prevalent in China in the 1940s. Although NOVC is easy to ignore in clinical practice, it is important to monitor NOVC strains isolated from sites other than the digestive tract, as these strains have the potential to exert their pathogenic effects through multiple virulence factors. The whole genome analysis of NOVC strains will help to understand the virulence characteristics and evolution of strains, and also provide help for clinical treatment.
Ethics Approval
Informed consent to participate in this case study and to publish the findings was obtained from the patient. This study and disclosure of patient clinical data were approved by Dongyang People’s Hospital Ethics Committee (No. 2024-YX-231). The analysis used routinely collected anonymized programmatic data.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Clemens JD, Nair GB, Ahmed T, Qadri F, Holmgren J. Cholera. Lancet. 2017;390(10101):1539–1549. doi: 10.1016/S0140-6736(17)30559-7 [DOI] [PubMed] [Google Scholar]
- 2.Uwishema O, Okereke M, Onyeaka H, et al. Threats and outbreaks of cholera in Africa amidst COVID-19 pandemic: a double burden on Africa’s health systems. Trop Med Health. 2021;49(1):93. doi: 10.1186/s41182-021-00376-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutta D, Chowdhury G, Pazhani GP, et al. Vibrio cholerae non-O1, non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg Infect Dis. 2013;19(3):464–467. doi: 10.3201/eid1903.121156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez JY, Duarte C, Rodriguez GJ, et al. Bacteremia by non-O1/non-O139 Vibrio cholerae: case description and literature review. Biomedica. 2023;43(3):323–329. doi: 10.7705/biomedica.6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Wu Y, Sun X, et al. Non-O1/non-O139 Vibrio cholerae bacteraemia in mainland China from 2005 to 2019: clinical, epidemiological and genetic characteristics. Epidemiol Infect. 2020;148:e186. doi: 10.1017/S0950268820001545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen YT, Tang HJ, Chao CM, Lai CC, Zhou D. Clinical manifestations of non-O1 Vibrio cholerae infections. PLoS One. 2015;10(1):e0116904. doi: 10.1371/journal.pone.0116904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31(4):587–589. doi: 10.1093/bioinformatics/btu661 [DOI] [PubMed] [Google Scholar]
- 11.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–546. doi: 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- 12.Wick RR, Judd LM, Gorrie CL, Holt KE, Phillippy AM. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–2618. doi: 10.1093/nar/29.12.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalvari I, Argasinska J, Quinones-Olvera N, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46(D1):D335–D342. doi: 10.1093/nar/gkx1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertelli C, Laird MR, Williams KP, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–W35. doi: 10.1093/nar/gkx343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016;44(D1):D694–697. doi: 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McArthur AG, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348–3357. doi: 10.1128/AAC.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 21.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–185. doi: 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–269. doi: 10.1093/nar/gku1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21(4):537–539. doi: 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- 24.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Lu Y, Qian H, et al. Non-O1, Non-O139 Vibrio cholerae (NOVC) Bacteremia: case Report and Literature Review, 2015-2019. Infect Drug Resist. 2020;13:1009–1016. doi: 10.2147/IDR.S245806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Qian J, Ke Q, Wang Y, Liu Y, Bao D. Bacteremia Caused by a Serotype Ob5 Vibrio cholerae Strain in a Cirrhotic Patient in China. Microbiol Spectr. 2023;11(4):e0205423. doi: 10.1128/spectrum.02054-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Yin L, Lv C, et al. Parallel Evolution to Elucidate the Contributions of PA0625 and parE to Ciprofloxacin Sensitivity in Pseudomonas aeruginosa. Microorganisms. 2022;11(1). doi: 10.3390/microorganisms11010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulthurst S. The Type VI secretion system: a versatile bacterial weapon. Microbiology. 2019;165(5):503–515. doi: 10.1099/mic.0.000789 [DOI] [PubMed] [Google Scholar]
- 29.Naskar S, Hohl M, Tassinari M, Low HH. The structure and mechanism of the bacterial type II secretion system. Mol Microbiol. 2021;115(3):412–424. doi: 10.1111/mmi.14664 [DOI] [PubMed] [Google Scholar]
- 30.Guentzel MN, Berry LJ. Motility as a virulence factor for Vibrio cholerae. Infect Immun. 1975;11(5):890–897. doi: 10.1128/iai.11.5.890-897.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Hao J, Zeng Z, Jinbo L. Identification and genomic analysis of a Vibrio cholerae strain isolated from a patient with bloodstream infection. Heliyon. 2022;8(11):e11572. doi: 10.1016/j.heliyon.2022.e11572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang J, Li S, Zhang M, Li F, Tang Y, Yang F. Whole Genome Analysis of a Non-O1, Non-O139 Vibrio cholerae Detected from Human Blood in China. Infect Drug Resist. 2023;16:5453–5461. doi: 10.2147/IDR.S420095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benz R. RTX-Toxins. Toxins (Basel). 2020;12(6). doi: 10.3390/toxins12060359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filipi K, Rahman WU, Osickova A, Osicka R. Kingella kingae RtxA Cytotoxin in the Context of Other RTX Toxins. Microorganisms. 2022;10(3). doi: 10.3390/microorganisms10030518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fennell TG, Blackwell GA, Thomson NR, Dorman MJ. gbpA and chiA genes are not uniformly distributed amongst diverse Vibrio cholerae. Microb Genom. 2021;7(6). doi: 10.1099/mgen.0.000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart PS. Biophysics of biofilm infection. Pathog Dis. 2014;70(3):212–218. doi: 10.1111/2049-632X.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkatesan N, Perumal G, Doble M. Bacterial resistance in biofilm-associated bacteria. Future Microbiol. 2015;10(11):1743–1750. doi: 10.2217/fmb.15.69 [DOI] [PubMed] [Google Scholar]
- 38.Leskinen K, Varjosalo M, Skurnik M. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology. 2015;161(Pt 2):285–299. doi: 10.1099/mic.0.083097-0 [DOI] [PubMed] [Google Scholar]
- 39.Luo Y, Payne M, Kaur S, Octavia S, Jiang J, Lan R. Emergence and genomic insights of non-pandemic O1 Vibrio cholerae in Zhejiang, China. Microbiol Spectr. 2023;11(6):e0261523. doi: 10.1128/spectrum.02615-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y, Wang H, Liang J, et al. Population Structure and Multidrug Resistance of Non-O1/Non-O139 Vibrio cholerae in Freshwater Rivers in Zhejiang, China. Microb Ecol. 2021;82(2):319–333. doi: 10.1007/s00248-020-01645-z [DOI] [PubMed] [Google Scholar]
- 41.Kuninobu KI, Takemura T, Takizawa Y, Hasebe F, Yamashiro T. Whole-genome analysis of a Vibrio cholerae O1 biotype classical strain isolated in 1946 in Sasebo city, Nagasaki prefecture, from a returnee from the northeast part of China. Trop Med Health. 2023;51(1):5. doi: 10.1186/s41182-023-00500-4 [DOI] [PMC free article] [PubMed] [Google Scholar]