Abstract
Long non-coding RNAs (lncRNAs), which are RNA molecules longer than 200 nucleotides that do not encode proteins, are implicated in a variety of biological processes, including growth and development. Despite research into the role of lncRNAs in skeletal muscle development, the regulatory mechanisms governing ovine skeletal muscle development remain unclear. In this study, we analyzed the expression profiles of lncRNAs in skeletal muscle from 90-day-old embryos (F90), 1-month-old lambs (L30), and 3-year-old adult sheep (A3Y) using RNA sequencing. In total, 4 738 lncRNAs were identified, including 997 that were differentially expressed. Short-time series expression miner analysis identified eight significant expression profiles and a subset of lncRNAs potentially involved in muscle development. Kyoto Encyclopedia of Genes and Genomes enrichment analysis revealed that the predicted target genes of these lncRNAs were primarily enriched in pathways associated with muscle development, such as the cAMP and Wnt signaling pathways. Notably, the expression of lncRNA GTL2 was found to decrease during muscle development. Moreover, GTL2 was highly expressed during the differentiation of skeletal muscle satellite cells (SCs) and was shown to modulate ovine myogenesis by affecting the phosphorylation levels of PKA and CREB. Additionally, GTL2 was found to regulate both the proliferation and differentiation of SCs via the PKA-CREB signaling pathway. Overall, this study provides a valuable resource and offers novel insights into the functional roles and regulatory mechanisms of lncRNAs in ovine skeletal muscle growth and development.
Keywords: Skeletal muscle, LncRNA, Skeletal muscle satellite cells, Sheep, PKA-CREB pathway
INTRODUCTION
Mutton is a vital protein source for humans, favored for its high nutritional value, flavor, and low-calorie content. Skeletal muscle development is critical for determining both muscle mass and meat quality. Muscle growth is primarily shaped by the prenatal increase in fiber number and postnatal expansion in muscle volume ( Costa et al., 2021; Wei et al., 2014). Myogenesis, the process of muscle formation, is extremely complex and precisely regulated, involving protein-coding genes, transcription factors, epigenetic modifiers, and non-coding RNAs (ncRNAs) ( Brun et al., 2022; Cao et al., 2023; Chen et al., 2024; Wang et al., 2022b). Understanding the molecular mechanisms that regulate skeletal muscle development is essential for improving the production performance of livestock.
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts exceeding 200 bp in length, with the majority lacking protein-coding potential. However, recent studies have reported that certain lncRNAs can encode micropeptides ( Barczak et al., 2023; Papaioannou et al., 2019), many of which have been implicated in muscle development ( Lin et al., 2019; Perelló-Amorós et al., 2022). For instance, knockdown of LEMP (lncRNA encoded micropeptide), a 56 amino acid micropeptide encoded by MyolncR4, impairs the differentiation of C2C12 cells, while LEMP knockout in mice results in defective skeletal muscle development ( Wang et al., 2020b). Moreover, increasing evidence has shown that lncRNAs are involved in muscle growth and development ( Butchart et al., 2016), adipogenesis ( Huang et al., 2019; Ma et al., 2023; Raza et al., 2022; Wang et al., 2020a), osteogenic differentiation ( Sun et al., 2021; Wu et al., 2020b; Yu et al., 2020), and other biological processes ( Ning et al., 2024; Zhang et al., 2023). Recent research has highlighted the crucial roles of lncRNAs in the regulation of skeletal muscle growth and development in humans ( Miller et al., 2024; Simionescu-Bankston & Kumar, 2016), mice ( Butchart et al., 2016; Matsumoto et al., 2017; Wang et al., 2022c; Yue et al., 2023; Zhang et al., 2016; Zhou et al., 2017), zebrafish ( Zhou et al., 2024), and other animals. In mice, lncRNA 2 310 043L19Rik promotes the proliferation and inhibits the differentiation of myoblasts through silencing miR-125a-5p expression ( Li et al., 2020). Furthermore, Lnc021 enhances myoblast proliferation in vitro through DHX36 and EIF3B interaction ( Chen et al., 2022). In cattle, lnc23 ( Chen et al., 2021) and lncRNA H19 ( Xu et al., 2017) participate in the differentiation of bovine skeletal muscle satellite cells (SCs). In ovines, lnc-SEMT ( Wei et al., 2018), lncRNA CTTN-IT1 ( Wu et al., 2020a), and other lncRNAs participate in the regulation of muscle growth and development. LncRNAs can also affect myogenesis by binding with proteins, as seen with lncRNA IGF2 AS ( Song et al., 2020) and lnc403 ( Zhang et al., 2020b).
LncRNA GTL2, also known as Meg3, is a maternally expressed gene located on ovine chromosome 18 ( Charlier et al., 2001; Fleming-Waddell et al., 2009). Previous studies have shown the lncRNA GTL2 plays an important role in regulating myoblast differentiation and muscle regeneration ( Dill et al., 2021; Liu et al., 2023; Wang et al., 2021). For instance, the expression of lncRNA GTL2 in muscle tissue is regulated by DNA methylation ( Fan et al., 2022). Moreover, lncRNA GTL2 can act as a competitive endogenous RNA (ceRNA) by interacting with microRNAs (miRNAs) to regulate muscle growth and development. For example, lncRNA-GTL2 promotes bovine skeletal muscle differentiation by interacting with miRNA-135 ( Liu et al., 2019). These findings underscore the important effects of GTL2 in skeletal muscle development. However, the exact mechanism by which lncRNA GTL2 influences ovine myogenesis remains largely undetermined, necessitating further investigation into the functions and regulatory pathways of lncRNAs in ovine muscle development.
In the present study, the expression profiles of lncRNAs in sheep skeletal muscle at three different developmental stages (90-day-old embryos, 1-month-old lambs, and 3-year-old adult sheep) were constructed. Several key pathways and predicted lncRNAs related to ovine muscle growth and development were identified. Notably, lncRNA GTL2 was found to be highly expressed in the longissimus dorsi muscle of embryos. Our findings revealed that lncRNA GTL2 regulated the proliferation and differentiation of ovine skeletal muscle SCs via the PKA-CREB pathway. Overall, this study provides novel insights into the molecular mechanisms underlying ovine skeletal muscle development and myogenesis.
MATERIALS AND METHODS
Animals and sample preparation
Nine unrelated, healthy Duolang sheep, maintained under the same feeding conditions, were selected from the Changping Experimental Base of the Institute of Animal Science, Chinese Academy of Agricultural Sciences. For the collection of 90-day-old-embryos (F90), three fetuses were obtained from three pregnant ewes during induced abortion procedures, and longissimus dorsi muscle tissues were collected from the fetuses. For the 1-month-old lambs (L30) and 3-year-old adult sheep (A3Y), longissimus dorsi muscle samples were collected after humane euthanasia by carotid artery exsanguination, with three experimental replicates per group. All muscle samples were collected into liquid nitrogen and stored at –80℃ for RNA sequencing (RNA-seq). All animal experiments were conducted in accordance with the regulations and guidelines formulated by the Institute of Animal Science, Chinese Academy of Agricultural Sciences (IASCAAS-2 021-56).
RNA-seq library preparation and sequencing
Total RNA was extracted from the longissimus dorsi muscle tissue of nine individuals using TRIzol reagent (15596018, Invitrogen, USA). RNA quality and concentration were assessed using agarose gel electrophoresis, an Agilent Bioanalyzer 2100 (Agilent, USA), and a NanoDrop 2000 (Thermo Fisher, USA). Only RNA samples with a concentration≥100 ng/μL, total amount≥3 μg, and RNA integrity number (RIN)≥7.0 were retained for further analysis. The RNA-seq library was constructed using a Ribo-Zero rRNA Removal Kit (Illumina, USA). The remaining RNA was fragmented into smaller sequences using fragmentation buffers. These RNA fragments were then reverse-transcribed to generate cDNA, followed by second-strand synthesis using DNA Polymerase I (F530S, Thermo Fisher, USA), RNase H (EN0202, Thermo Fisher, USA), and dNTPs (R72501, Thermo Fisher, USA). The synthesized second-strand cDNA was amplified to construct the sequencing library. The U-labeled second-strand cDNA was degraded with the USER enzyme (M5505S, New England Biolabs, USA), and the resulting polymerase chain reaction (PCR) products were purified using AMPure XP beads (A63880, Beckman, USA). Finally, library quality was assessed using the Agilent Bioanalyzer 2100 system. Sequencing was performed on the Illumina HiSeq 2500 v.4 platform (Illumina, USA) in paired-end mode (125PE). After sequencing, 125 bp/150 bp paired-end reads were generated for all nine samples.
Identification of lncRNAs and mRNAs
First, the raw reads were filtered using NGSQC Toolkit (v.2.3.3) ( Patel & Jain, 2012) to remove reads with N>10% and low-quality sequences. High-quality clean reads were then obtained using FastQC (v.0.10.1) ( Brown et al., 2017). These clean reads were mapped to the ovine reference genome (Oar v.4.0) using TopHat tool (v.2.1.0) ( Trapnell et al., 2009). Subsequently, all transcript assemblies were merged into a reference transcriptome using Cufflinks (v.2.2.1) ( Trapnell et al., 2010). To identify candidate lncRNAs, transcripts shorter than 200 bp were removed, with only those containing more than two exons retained. Transcripts with fewer than three read counts were discarded. To filter out potential coding transcripts, the coding potential of the remaining transcripts was assessed using the Coding Potential Calculator (CPC) ( Kong et al., 2007), Coding-Non-Coding Index (CNCI) ( Sun et al., 2013), and Pfam Scan (Pfam) ( Finn et al., 2014). Only transcripts that lacked coding potential in all three tools (CPC, CNCI, and Pfam) were considered as non-coding RNAs.
In addition, clean reads were mapped to specific positions on the ovine reference genome (Oar v.4.0) using TopHat tool (v.2.1.0). The expression levels of all transcriptomes (28 094 mRNAs) from the nine samples were calculated using StringTie ( Pertea et al., 2015) and edgeR ( Robinson et al., 2010).
Differentially expressed (DE) lncRNAs
The expression levels of lncRNAs were quantified using the FPKM (Fragments Per Kilobase per Million) metric, calculated using StringTie and edgeR. The DE lncRNAs were identified using the R package edgeR in the comparison groups. The screening criteria for DE lncRNAs were set to |Fold Change|≥1, P<0.05, and false discovery rate (FDR)<0.01.
Functional enrichment analysis
The functional roles of lncRNAs were inferred based on their potential regulation of protein-coding genes through dominant cis and trans mechanisms ( Ferrer & Dimitrova, 2024). To explore the biological functions of the lncRNAs, their putative target genes were predicted using the LncTar tool (http://www.cuilab.cn/lnctar) ( Li et al., 2015). The predicted target genes of the DE lncRNAs were subjected to Gene Ontology (GO, https://biit.cs.ut.ee/gprofiler/gost) and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://kobas.cbi.pku.edu.cn/genelist/) functional enrichment analyses, with GO terms and KEGG pathways considered significant at P<0.05.
LncRNA-mRNA co-expression network analysis
To understand the co-expression relationships between lncRNAs and mRNAs ( Liao et al., 2011), a network of DE lncRNAs and their predicted target genes was visualized using Cytoscape (v.3.7.1) ( Shannon et al., 2003) to highlight key biological interactions involving hub genes. LncRNA-mRNA pairs were selected based on Pearson correlation coefficients (PCCs), with pairs showing |PCC≥0.99| used to construct the co-expression network.
Short time-series expression miner (STEM) clustering analysis
The DE lncRNAs were clustered using STEM ( Ernst & Bar-Joseph, 2006; Ernst et al., 2005) based on FPKM values, and lncRNAs with similar expression patterns were estimated using default parameters. The DE lncRNAs were clustered into distinct expression profiles according to their trends across time points. P-values were calculated by assessing the number of genes assigned in each profile using the true ordering of time points, with only colored profiles considered significant.
Sheep skeletal muscle SC isolation and culture
Sheep skeletal muscle SCs were isolated following previously described methods ( Wu et al., 2012; Zhao et al., 2018). In short, hindlimb muscle tissue from fetal sheep was minced into 1 mm 3 pieces and digested with 0.1% type I collagenase (218 021, Invitrogen, USA) for 1 h at 37℃, followed by digestion with 0.25% trypsin-EDTA (25 200 072, Invitrogen, USA) at 37℃ for 30 min. After digestion, the cell suspensions were filtered through a 70 μm filter and centrifuge 1000 r/min for 5 min to collect SCs at room temperature . The isolated SCs were cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12, 11320033, Invitrogen, USA) containing 10% horse serum (26050088, Invitrogen, USA), 20% fetal bovine serum (10099141, Invitrogen, USA), and 1% penicillin/streptomycin (15070063, Invitrogen, USA). When the SCs reached approximately 80% confluence, the growth medium was replaced with DMEM/F12 containing 2% horse serum and 1% penicillin/streptomycin to induce myoblast differentiation in vitro. The cells were cultured at 37℃ and 5% CO 2 in growth medium.
Quantitative real-time PCR (qPCR)
Total RNA from longissimus dorsi muscle tissue and SCs was extracted using TRIzol reagent according to the manufacturer’s instructions. Total RNA (2 μg) was reverse-transcribed to synthesize cDNA using HiScript III All-in-one RT SuperMix Perfect for qPCR (R333-01, Vazyme, China). Quantitative real-time PCR (qPCR) was performed using 10 μL of 2×Taq Pro Universal SYBR qPCR Master Mix (Q712-03, Vazyme, China), 0.8 μL of 20 μmol/L primer, 2 μL of cDNA, and 7.2 μL of RNase-free water. The qPCR steps were as follows: one cycle at 95℃ for 30 s, followed by 40 cycles at 95℃ for 10 s and 60℃ for 30 s. The relative expression levels of genes and lncRNAs were calculated using the 2 –∆∆CT method ( Bubner & Baldwin, 2004). Glyceraldehyde-3-phosphate dehydrogenase ( GAPDH) was used as the reference gene, and all RNA samples were analyzed in triplicate. The primers used for selected genes are listed in Supplementary Table S1.
Cell transfection
Small interfering RNAs (siRNAs) were transfected into SCs using Lipofectamine RNAiMAX (13778150, Invitrogen, USA) according to the manufacturer’s instructions. Both siRNAs and negative control siRNAs were designed and purchased from Generay (China). The following siRNA sequences were obtained: sense 5’-GAAAUGUUGUAGAUAUAAATT-3’ and antisense 5’-UUUAUAUCUACAACAUUUCTT-3’ of TCONS_00 544 451.
Plasmids were transfected into SCs or 293T cells using Lipofectamine 3000 reagent (L3000015, Invitrogen, USA) following the manufacturer’s instructions. The TCONS_00 544 451 sequence was amplified in vitro, then cloned into a pcDNA3.1(+) vector (Generay, China) to generate recombinant plasmids, including negative control, over-expression TCONS_00 544 451, pcDNA3.1(+)-GFP, pcDNA3.1(+)-GFPmut (start codon ATGGTG mutated to ATTGTT), 5’ UTR-ORF-GFP, and 5’ UTR-ORFmut-GFP (start codon ATG mutated to ATT).
CCK-8 assay
A cell proliferation assay was conducted using the Cell Counting Kit-8 (CK04, Dojindo, Japan) following the manufacturer’s protocols. Briefly, SCs were seeded into 96-well plates. After 24 h, 48 h, 72 h, and 96 h, fresh medium consisting of 90 μL of growth medium and 10 μL of CCK-8 reagent was added to the wells, followed by incubation at 37℃ for 4 h in a 5% CO 2 atmosphere. After incubation, optical density (OD) was determined at 450 nm using a microplate reader.
5-Ethynyl-2’-deoxyuridine (EdU) assay
After 48 h of transfection, SCs were labeled using an EdU assay kit (C10310-1, RiboBio, China) following the manufacturer’s protocols. The SCs were incubated with EdU-containing medium for 2 h, then fixed by 4% paraformaldehyde (P1110-500, Solarbio, China) for 30 min. Subsequently, Apollo ® staining was performed at room temperature in the dark for 30 min to label synthetic DNA. Finally, the SCs were counterstained with 4’,6-diamidino-2-phenylindole (DAPI, C0065-50mL, Solarbio, China), and inverted fluorescence images were acquired using a confocal microscope.
Flow cytometry (FCM)
After 48 h of transfection, SCs were washed in cold phosphate-buffered saline (PBS) and harvested by digestion with 0.25% trypsin-EDTA solution. The cells were fixed overnight in 70% absolute ethanol pre-cooled at –20℃. The cell cycle was then assessed using a cell cycle staining kit (MultiSciences, China) and analyzed by flow cytometry (Beckman, USA).
Western blotting
Cell samples were lysed using RIPA buffer (R0010, Solarbio, China) and protein concentration was detected using a BCA Protein Assay Kit (P0012, Beyotime, China). Protein samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE, PG112, epizyme, China), transferred to polyvinylidene fluoride (PVDF) membranes (FFP39, Beyotime, China), and immunoblotted with various primary antibodies at 4℃ overnight, including GFP (1:1 000, 66002-1-Ig, Proteintech, China), CDK1 (1:1 000, 65 182-1-lg, Proteintech, China), CDK2 (1:1 000, 10122-1-AP, Proteintech, China), PKA (1:1 000, 4782S, Cell Signaling Technology, USA), p-PKA (1:1 000, 9621S, Cell Signaling Technology, USA), CREB (1:1 000, 9197S, Cell Signaling Technology, USA), p-CREB (1:1 000, 9198S, Cell Signaling Technology, USA), MyoD (1:1 000, 18943-1-AP, Proteintech, China), MyoG (1:1 000, ab1835, Abcam, UK), SRF (1:1 000, 5147S, Cell Signaling Technology, USA), GAPDH (1:4 000, HRP-60004, Proteintech, China), and β-actin (1:4 000, 66009-1-Ig, Proteintech, China). After washing with Tris Buffered Saline with Tween-20 (TBST, HX1893, Huaxingbio, China), the membranes were incubated with secondary antibodies (1:5 000, Solarbio, China) for 1 h at room temperature. Western blotting was performed using a High-Sig ECL Kit (180-501, TANON, China) for chemiluminescence.
Immunofluorescence
After 24 h of transfection, SCs were induced to fuse and differentiate with 2% horse serum DMEM/F12. After 3 days of differentiation, the cells were fixed with 4% paraformaldehyde for 40 min at room temperature, followed by washing with PBS. Cell membranes were then permeabilized with 0.5% Triton X-100 (C0065, Solarbio, China) in PBS for 10 min at 37℃. Blocking was performed with 3% goat serum (C01-03 001, Bioss, China) at 37℃ for 1 h. The cells were then incubated overnight with the primary antibody MyHC (1:200, sc-376157, Santa Cruz Biotechnology, USA) at 4℃. The next day, the primary antibody was removed, and the cells were washed with PBS three times (5 min each). The cells were then incubated with fluorescent secondary antibody (1:100, A11001, Invitrogen, USA) at 37℃ for 1 h. Nuclei were counterstained using DAPI for 5 min at room temperature. Finally, fluorescence images of SCs were acquired using a confocal microscope.
Statistical analysis
All graphs were generated using GraphPad Prism v.8.0. Experimental data are presented as the mean±standard error of the mean (SEM). Differences between the negative control and treatment groups were analyzed using a two-tailed Student t-test. Statistical significance was indicated as follows: *: P<0.05; **: P<0.01; ***: P<0.001.
RESULTS
Overview of sequencing data
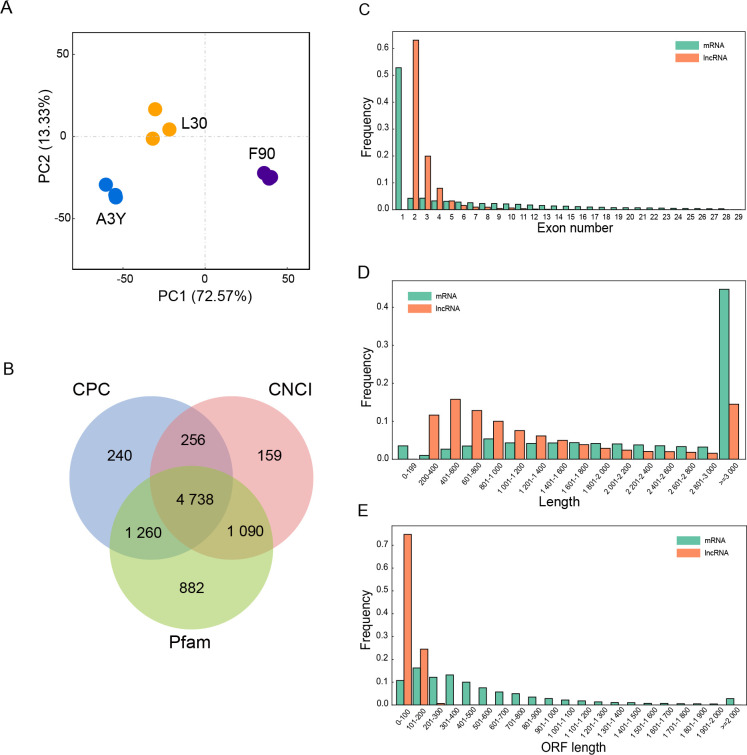
To systematically characterize lncRNA expression and identify potentially functional lncRNAs involved in ovine longissimus dorsi muscle development, the expression profiles of lncRNAs were generated from skeletal muscle samples of 90-day-old embryos (F90), 1-month-old lambs (L30), and 3-year-old adult sheep (A3Y) using ribosome-depleted, strand-specific transcriptome sequencing. Each sample yielded more than 10 Gb of raw paired-end reads. The average GC content of clean reads was 49.08%, and Q30 percentages exceeded 87% for all samples (Supplementary Table S2). Over 83% of the clean reads from each sample were aligned to the Ovis aries reference genome (Oar v.4.0). Principal component analysis (PCA) of the lncRNA expression profiles ( Figure 1A) revealed clear separation of the nine samples into three distinct groups. The qPCR results corroborated the transcriptome sequencing data (Supplementary Figure S1), indicating high reliability of the dataset for subsequent analysis.
Figure 1.
Characterization of lncRNAs during muscle development
A: Principal component analysis (PCA) of lncRNAs in nine samples. B: Venn diagram of lncRNA transcripts identified by Pfam, CPC, and CNCI. C: Distribution of mRNA and lncRNA exon numbers. D: Distribution of mRNA and lncRNA lengths. E: Distribution of mRNA and lncRNA ORF lengths.
Characterization of mRNAs and lncRNAs
In total, 4 738 lncRNAs were identified across the three developmental stages ( Figure 1B). Analysis of exon characteristics indicated that the vast majority of lncRNAs contained two exons, while most mRNAs contained one exon ( Figure 1C), consistent with previous reports on exon characteristics ( Wei et al., 2023). Length distribution analysis indicated that most lncRNAs ranged from 200 bp to 1 600 bp or exceeded 3 000 bp, while most mRNAs were longer than 3 000 bp ( Figure 1D). The open reading frames (ORFs) of lncRNAs were shorter than those of mRNAs, ranging from 0 bp to 200 bp for lncRNAs compared to 0 bp to 1 000 bp for mRNAs ( Figure 1E).
DE lncRNAs during ovine muscle development
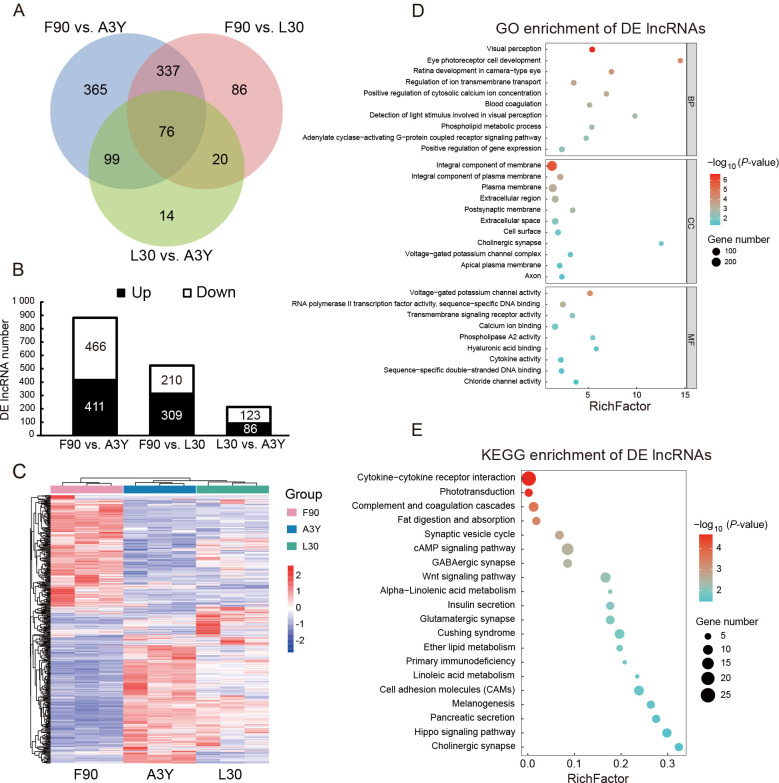
A total of 997 DE lncRNAs were identified across the three developmental stages, with 365, 86, and 14 stage-specific lncRNAs found in the F90 vs. A3Y, F90 vs. L30, and L30 vs. A3Y comparisons, respectively ( Figure 2A). The numbers of up-regulated and down-regulated lncRNAs in the three groups are shown in Figure 2B. Hierarchical clustering revealed that the expression of lncRNAs displayed strong temporal specificity across developmental stages ( Figure 2C).
Figure 2.
Differentially expressed (DE) lncRNAs during muscle development
A: Venn diagram of number of DE lncRNAs. B: Stacked histogram of number of DE lncRNAs across three comparison groups. C: Heatmap of 76 overlapping DE lncRNAs across three groups. D: GO enrichment analysis of genes potentially regulated by 76 overlapping DE lncRNAs. E: KEGG enrichment analysis of genes potentially regulated by 76 overlapping DE lncRNAs.
To investigate the biological functions of the genes potentially regulated by the overlapping DE lncRNAs across the three groups, GO and KEGG analyses were performed for their cis- and trans-targeted genes ( Kopp & Mendell, 2018). The top 20 significantly enriched GO terms and KEGG pathways were identified. GO terms were classified into biological process (BP), cellular component (CC), and molecular function (MF) categories. As illustrated in Figure 2D, genes were enriched in BP terms associated with adenylate cyclase-activating G-protein-coupled receptor signaling pathway, phospholipid metabolic process, and positive regulation of cytosolic calcium ion concentration. In the CC category, genes were enriched in terms related to integral component of membrane, integral component of plasma membrane, plasma membrane, extracellular region, and cell surface. In the MF category, genes were enriched in terms associated with transcriptional activator activity, calcium ion binding, and RNA polymerase II transcription factor activity. These findings suggest that these genes may be involved in cell fate determination and cellular signal transduction. KEGG analysis further revealed gene enrichment in the cAMP signaling pathway, Wnt signaling pathway, fat digestion and absorption, and insulin secretion ( Figure 2E).
STEM analysis
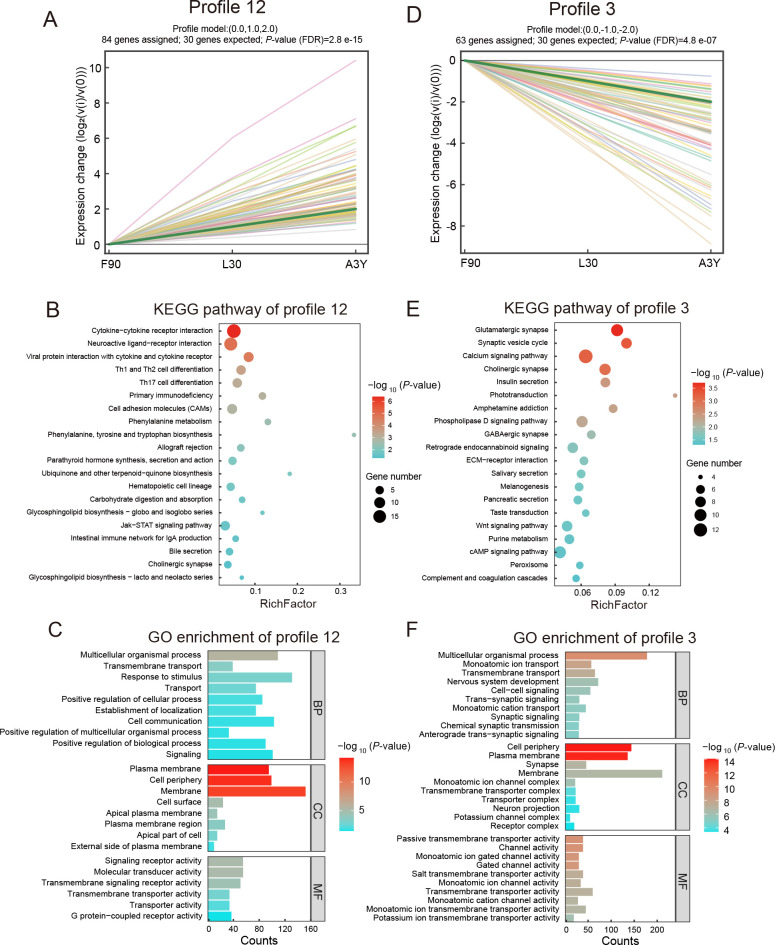
As the transcriptomic data were obtained from ovine skeletal muscle at three developmental stages, the expression profiles of DE lncRNAs were determined by STEM analysis. To explore the persistent expression changes of lncRNAs, 16 clustering profiles were generated, eight of which were significant (Supplementary Figure S2). Among these, we focused on profile 12, which displayed continuous up-regulation, and profile 3, which exhibited continuous down-regulation without inflection points. In profile 12, 84 DE lncRNAs were significantly up-regulated ( Figure 3A; Supplementary Table S3). KEGG enrichment analysis showed that the predicted target genes of these DE lncRNAs were associated with carbohydrate digestion, absorption signaling pathways, and disease-related pathways ( Figure 3B). GO analysis indicated that the predicted target genes were associated with transmembrane transport, cell periphery, membrane, and plasma membrane ( Figure 3C). The top five DE lncRNAs from profile 12 (TCONS_00 330 276, TCONS_01 105 309, TCONS_00 783 895, TCONS_00 389 861, and TCONS_00 838 251) and their predicted target genes, including TNNC2, MyoM1, ACTN4, and PPP1R3A ( Table 1), are reportedly related to muscle development. In profile 3, 63 DE lncRNAs were significantly down-regulated ( Figure 3D; Supplementary Table S4). KEGG results showed that the predicted target genes of these DE lncRNAs were enriched in the calcium, Wnt, and cAMP signaling pathways ( Figure 3E). GO analysis indicated that the predicted target genes were associated with nervous system development, cell periphery, synaptic signaling, and passive transmembrane transporter activity ( Figure 3F). The top five DE lncRNAs and corresponding potentially regulated genes in profile 3 are shown in Table 2. Notably, TCONS_00 790 974, TCONS_00 774 517, TCONS_00 685 284, TCONS_00 164 122, and TCONS_01 405 904 showed abundant expression, with their predicted target genes, such as FOXM1, implicated in myogenesis.
Figure 3.
Short time-series expression miner (STEM) clustering analysis of differentially expressed (DE) lncRNAs
A: Continuously up-regulated profile 12 ( P<0.05), where green line represents overall trend, and each colored line represents individual DE lncRNAs. B: KEGG analysis of genes potentially regulated by DE lncRNAs in profile 12. C: GO analysis of genes potentially regulated by DE lncRNAs in profile 12. D: Continuously down-regulated profile 3 ( P<0.05), where green line represents overall trend, and each colored line represents individual DE lncRNAs. E: KEGG analysis of genes potentially regulated by DE lncRNAs in profile 3. F: GO analysis of genes potentially regulated by DE lncRNAs in profile 3. Enriched terms and pathways are shown according to P-values.
Table 1. Top five DE lncRNAs and their potentially regulated genes in continuously up-regulated profile 12 based on short time-series expression miner (STEM) analysis.
| LncRNA | F90 (FPKM) | L30 (FPKM) | A3Y (FPKM) | Target gene of lncRNA |
| TCONS_00 330 276 | 746.26 | 3 311.85 | 26 624.87 | TNNC2, SPINT4, ZNF335, PCIF1, WFDC3 |
| TCONS_00 783 895 | 482.22 | 816.04 | 2 801.51 | FMO1, SALL4, NOD2, IL7R, TARSL2 |
| TCONS_00 838 251 | 105.18 | 254.69 | 965.04 | LPIN2, MYOM1 |
| TCONS_00 389 861 | 95.90 | 340.59 | 2 750.72 | ACTN4, MAP4K1, EIF3K, RYR1, RASGRP4 |
| TCONS_01 105 309 | 42.83 | 169.98 | 1 076.83 | PPP1R3A |
Table 2. Top five DE lncRNAs and their potentially regulated genes in continuously down-regulated profile 3 based on short time-series expression miner (STEM) analysis.
| LncRNA | F90 (FPKM) | L30 (FPKM) | A3Y (FPKM) | Target gene of lncRNA |
| TCONS_00 790 974 | 2 017.67 | 391.16 | 101.99 | MAGEL2, SNAP25, NGFR, PDYN, KCNV1 |
| TCONS_00 774 517 | 85.13 | 8.21 | 1.19 | FOXM1, CIITA, TMEM121, EXO1 |
| TCONS_00 685 284 | 74.02 | 34.02 | 17.46 | SCNN1B, SPATS2L, KCTD18, FOXM1 |
| TCONS_00 164 122 | 34.68 | 17.84 | 8.01 | TGDS, GPR180, SOX21 |
| TCONS_01 405 904 | 29.06 | 16.96 | 8.24 | A2ML1, STK32C, ADAMTS13, ASIC3, CCNA1 |
LncRNA-mRNA network analysis
A network of 2 526 lncRNA-mRNA interactions was identified between the 76 overlapping DE lncRNAs and 995 potentially regulated genes. To further explore the roles and regulatory mechanisms of these lncRNAs, a correlation analysis between mRNAs and lncRNAs was conducted using PCCs, with |PCC|≥0.99 as the threshold for constructing the interactive network. As shown in Figure 4 (Supplementary Figure S3), the network revealed 25 lncRNAs and 77 mRNAs. Notably, TCONS_00 544 451 was found to regulate a significant number of genes enriched in pathways related to muscle development, such as the cAMP, Wnt, and PI3K-Akt signaling pathways. In the STEM analysis, TCONS_00 544 451 was significantly enriched in expression profile 0, showing a marked decrease after birth (Supplementary Figure S2). Therefore, TCONS_00 544 451 was selected for further functional verification.
Figure 4.
Co-expression network of LncRNAs and their potentially regulated genes
Red nodes represent lncRNAs, blue nodes represent their potentially regulated genes.
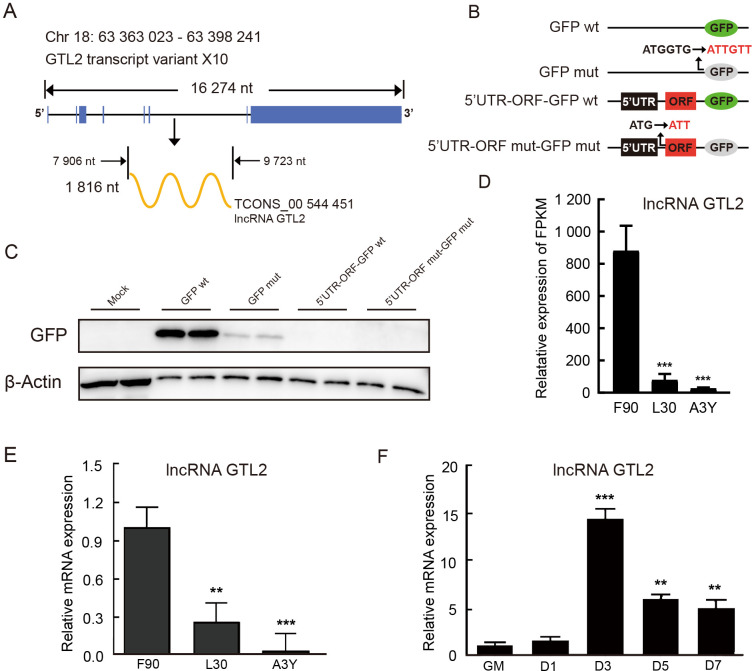
Identification of lncRNA GTL2 as a ncRNA
TCONS_00 544 451 was located on ovine chromosome 18 and overlapped with GTL2. We identified transcript variant X10 of GTL2 ( Figure 5A; Supplementary Table S5) and named it lncRNA GTL2 in subsequent research. To confirm whether the start codon of lncRNA GTL2 is active, a series of plasmid expression vectors were constructed, including GFPwt, GFPmut (with the start codon ATGGTG mutated to ATTGTT), 5’UTR-ORFmut-GFPwt (with the start codon ATG mutated to ATT), and 5’UTR-ORFmut-GFPmut (expressing a GFPmut ORF fusion at the C-terminus of 5’UTR-ORFmut) ( Figure 5B). Western blot analysis clearly demonstrated a lack of 5’UTR-ORFmut-GFPwt and 5’UTR-ORFmut-GFPmut fusion protein expression ( Figure 5C). RNA-seq showed that lncRNA GTL2 was highly expressed in the longissimus dorsi muscle of F90, with its expression levels gradually decreasing postnatally ( Figure 5D). This trend was validated by qPCR, which confirmed significantly higher expression of lncRNA GTL2 in F90 compared to L30 and A3Y ( Figure 5E). Furthermore, the expression of lncRNA GTL2 increased and then decreased during the differentiation of ovine SCs ( Figure 5F). Taken together, these results suggest that lncRNA GTL2 is a ncRNA, with high expression in prenatal skeletal muscle and during the early stages of SC differentiation.
Figure 5.
Identification of lncRNA GTL2 as a non-coding RNA
A: Chromosomal location and length of lncRNA GTL2. Blue represents exon of GTL2, and yellow represents lncRNA GTL2. nt: nucleotide. B: GFP fusion plasmid construction for transfection, showing wild-type GFP gene (GFPwt) with the start codon ATGGTG and mutant GFP gene (GFPmut) with the start codon ATTGTT; lncRNA GTL2 ORF start codon ATG is mutated to ATT. C: Western blot analysis of GFP protein expression levels in 293T cells. D: RNA-seq analysis of lncRNA GTL2 expression in longissimus dorsi muscle across F90, L30, and A3Y stages. E: LncRNA GTL2 expression in longissimus dorsi muscle across three development stages (F90, L30, and A3Y). F: qPCR detection of lncRNA GTL2 in SCs during proliferation (GM) and differentiation (D1, D3, D5, and D7). Results are presented as mean±SEM, **: P<0.01; ***: P<0.001.
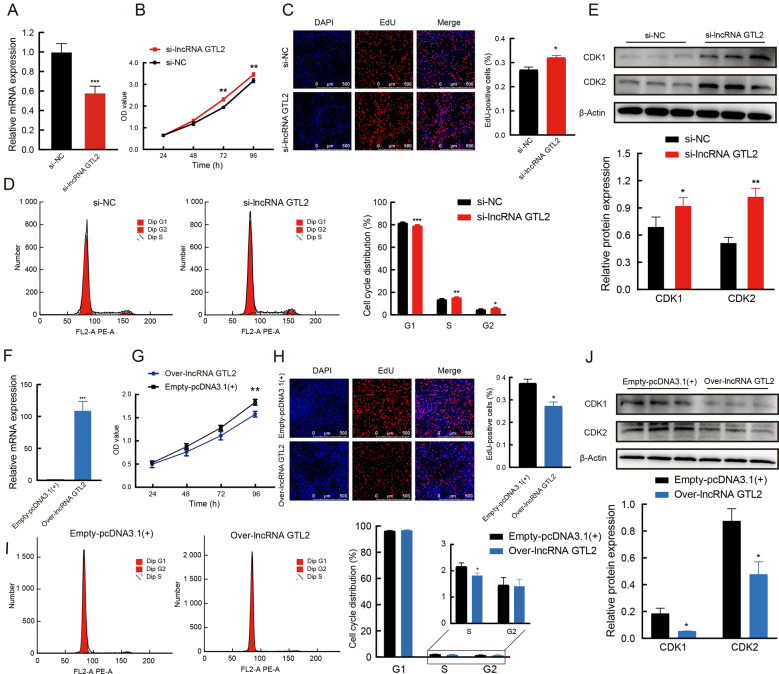
LncRNA GTL2 inhibits proliferation of ovine skeletal muscle SCs
To better understand the function of lncRNA GTL2, its expression levels were modulated in ovine skeletal muscle SCs through RNA interference (RNAi) strategies and overexpression vector constructs for lncRNA GTL2 ( Figure 6A, F). The CCK-8 assay demonstrated that lncRNA GTL2 knockdown significantly increased cell activity compared to the negative control ( Figure 6B). Furthermore, the EdU assay showed that depletion of lncRNA GTL2 increased the number of EdU-positive cells ( Figure 6C). Flow cytometry revealed that lncRNA GTL2 knockdown stimulated the transition from the G1 to S phase of the cell cycle ( Figure 6D). Western blot analysis further confirmed that lncRNA GTL2 knockdown significantly increased the protein levels of cyclin-dependent kinase 1 (CDK1) and cyclin-dependent kinase 2 (CDK2), known markers of cell proliferation ( Figure 6E). Conversely, overexpression of lncRNA GTL2 suppressed ovine SC proliferation compared to the empty-pcDNA3.1(+) group ( Figure 6C–J).
Figure 6.
LncRNA GTL2 inhibits proliferation of ovine skeletal muscle satellite cells (SCs)
A: Knockdown of lncRNA GTL2 utilizing RNA interference (RNAi). B: CCK-8 assay showing cell vitality of SCs transfected with negative control or si-lncRNA GTL2. C: EdU assay detecting proliferation of SCs after knockdown of lncRNA GTL2. D: Numbers of cells in Gap 1 phase (G1), synthesis phase (S), and Gap 2 phase (G2) were calculated by flow cytometry of si-lncRNA GTL2. E: Western blot assay of CDK1 and CDK2 in SCs transfected with si-lncRNA GTL2. F: Cell transfection efficiency of over-lncRNA GTL2. G: CCK-8 assay showing cell vitality of SCs transfected with empty-pcDNA3.1(+) or over-lncRNA GTL2. H: EdU assay detecting proliferation of SCs after overexpression of lncRNA GTL2. I: Numbers of cells in Gap 1 phase (G1), synthesis phase (S), and Gap 2 phase (G2) were calculated by flow cytometry of over-lncRNA GTL2. J: Western blot assay of CDK1 and CDK2 in SCs transfected with over-lncRNA GTL2. Results are presented as mean±SEM, *: P<0.05; **: P<0.01; ***: P<0.001.
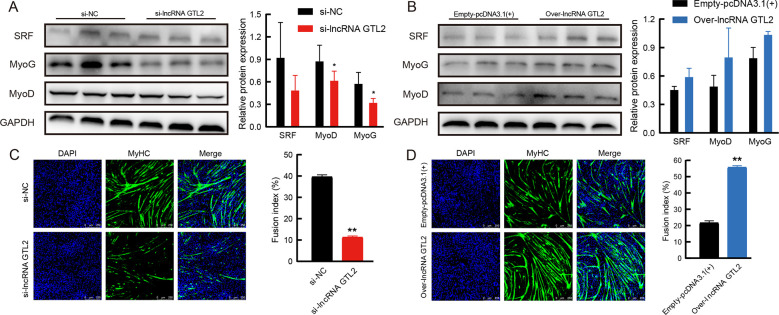
LncRNA GTL2 promotes differentiation of ovine skeletal muscle SCs
Next, the role of lncRNA GTL2 in SC myogenic differentiation was examined. Knockdown of lncRNA GTL2 down-regulated the protein expression of myogenin (MyoG), myogenic differentiation (MyoD), and serum response factor (SRF) ( Figure 7A). Conversely, western blot analysis showed that overexpression of lncRNA GTL2 resulted in an increase in MyoG, MyoD, and SRF protein expression ( Figure 7B). Furthermore, lncRNA GTL2 depletion markedly suppressed myotube formation during differentiation ( Figure 7C), while overexpression of lncRNA GTL2 promoted myotube formation ( Figure 7D). Together, these findings suggest that lncRNA GTL2 promotes the differentiation of ovine muscle SC in vitro.
Figure 7.
LncRNA GTL2 promotes differentiation of ovine skeletal muscle satellite cells (SCs)
A: Western blot assay of MyoG, MyoD, and SRF in SCs transfected with negative control or lncRNA GTL2 siRNA. B: Western blot assay of MyoG, MyoD, and SRF in SCs transfected with empty-pcDNA3.1(+) or over-lncRNA GTL2. C: Representative myotube staining of differentiated SCs transfected with si-lncRNA GTL2. Cells were stained with DAPI (blue) and MyHC (green) to visualize nuclei and myotubes, respectively. D: Immunofluorescence after transfection with over-lncRNA GTL2. SCs were differentiated for 3 days in differentiation medium. Results are presented as mean±SEM, *: P<0.05; **: P<0.01.
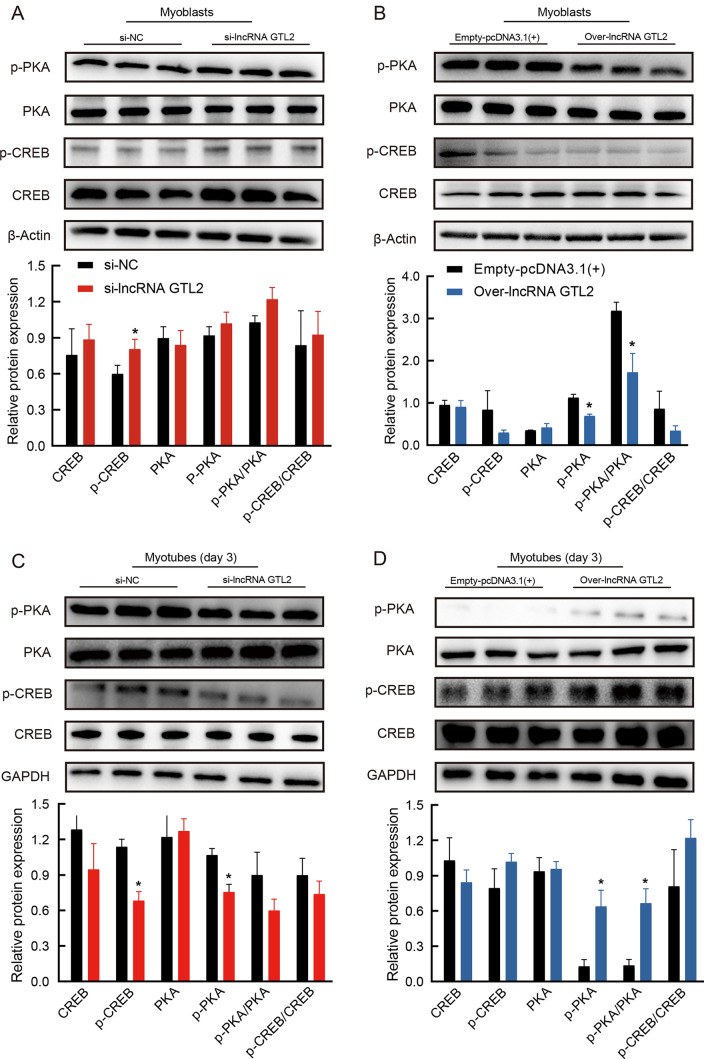
LncRNA GTL2 regulates proliferation and differentiation of ovine SCs via the PKA-CREB signaling pathway
To explore the regulatory mechanism of lncRNA GTL2 in myogenesis, we focused on the cAMP signaling pathway, as indicated by KEGG enrichment analysis of genes potentially regulated by lncRNA GTL2. Previous studies have shown that cAMP regulates various cellular processes primarily through protein kinase A (PKA) and its downstream effectors, such as the transcription factor cAMP responsive element binding protein (CREB) ( Zhang et al., 2020a). Furthermore, activation of the cAMP signaling pathway has been shown to promote myogenic differentiation of C2C12 cells ( Marco-Bonilla et al., 2023). Thus, we hypothesized that lncRNA GTL2 may regulate ovine myoblast differentiation through the PKA-CREB signaling pathway.
To test this, we explored whether lncRNA GTL2 modulates SC proliferation and differentiation via the PKA-CREB signaling pathway. Results showed that overexpression of lncRNA GTL2 significantly reduced the phosphorylation level of PKA, while knockdown of lncRNA GTL2 did not significantly affect SC proliferation ( Figure 8A, B). Compared to the control group, both p-CREB and p-PKA expression levels were decreased in the lncRNA GTL2 knockdown group ( Figure 8C). Conversely, overexpression of lncRNA GTL2 increased the protein expression levels of p-CREB and p-PKA ( Figure 8D). Collectively, these findings demonstrate that lncRNA GTL2 regulates SC proliferation and differentiation through the PKA-CREB signaling pathway.
Figure 8.
LncRNA GTL2 regulates proliferation and differentiation of ovine SCs via the PKA-CREB signaling pathway
A: Western blot assay of PKA, p-PKA, CREB, and p-CREB in proliferating SCs transfected with negative control or lncRNA GTL2 siRNA. B: Western blot assay of PKA, p-PKA, CREB, and p-CREB in proliferating SCs transfected with empty-pcDNA3.1(+) or over-lncRNA GTL2. C: Western blot assay of PKA, p-PKA, CREB, and p-CREB in differentiating SCs transfected with negative control or lncRNA GTL2 siRNA. D: Western blot assay of PKA, p-PKA, CREB, and p-CREB in differentiating SCs transfected with empty-pcDNA3.1(+) or over-lncRNA GTL2. SCs were differentiated for 3 days in differentiation medium. Results are presented as mean±SEM, *: P<0.05.
DISCUSSION
Increasing evidence highlights the pivotal role of lncRNAs in regulating muscle growth and development ( Sui et al., 2019; Wang et al., 2019b; Yu et al., 2021). Although several studies have explored lncRNA expression profiles in ovine skeletal muscle, the precise mechanisms by which lncRNAs regulate skeletal muscle development and growth in sheep remain unclear. In this study, 4 738 lncRNAs were identified in ovine skeletal muscle tissue across three key developmental stages using RNA-seq. Similarly, previous studies on goat skeletal muscle identified 19 880 mRNAs and 5 966 lncRNAs at two different developmental stages ( Huang et al., 2023). Our results demonstrated that the predicted targeted genes of overlapping DE lncRNAs were primarily involved in the cAMP and Wnt signaling pathways, both of which are well-established regulators of muscle development and myogenesis ( Chung et al., 2022; Da Silva et al., 2023; Klemm et al., 2001; Russell et al., 2023), with the Wnt signaling pathway also shown to influence the expression of myogenic regulatory factors (MRFs) ( Tajbakhsh et al., 1998; Takata et al., 2007; Von Maltzahn et al., 2012). Additionally, pathways involved in fat digestion and absorption, insulin secretion, and ether lipid metabolism were also highly enriched, further suggesting that these lncRNAs play important roles in a wide range of biological processes.
STEM analysis is widely used to study dynamic biological processes ( Ernst & Bar-Joseph, 2006). In this study, we applied STEM analysis to 997 DE lncRNAs involved in sheep skeletal muscle development at three developmental stages (F90, L30, and A3Y). Analysis revealed significant gene expression changes across eight profiles (15, 12, 11, 13, 0, 3, 4, and 2) ( P<0.05). We focused on two significant profiles (12 and 3), which exhibited contrasting expression patterns. In profile 3, forkhead box M1 ( FoxM1), a transcriptional factor critical for regulating muscle cell proliferation ( Chen et al., 2018; Wang et al., 2022a), was identified as a potentially regulated gene of TCONS_00 685 284 and TCONS_00 774 517. CIITA, a major histocompatibility complex (MHC) class II transactivator that inhibits myogenesis by repressing MyoG in differentiating myoblasts and reducing myogenin activity in myotubes ( Adhikari et al., 2020; Londhe & Davie, 2011), was identified as being regulated by TCONS_00 774 517. In profile 12, bioinformatic predictions highlighted a large number of lncRNAs potentially involved in myogenesis. For example, myomesin-1 ( MYOM1) was identified as a target gene of TCONS_00 838 251, with its knockout in human cardiomyocytes reported to result in myocardial atrophy ( Hang et al., 2021). PPP1R3A, the target gene of TCONS_01 105 309, has been linked to reduced muscle glycogen content in humans and mice ( Savage et al., 2008). α-Actinin-4 ( ACTN4), identified as a target gene of TCONS_00 389 861, increases the expression of muscle-specific proteins via interactions with MEF2 ( An et al., 2014; Chakraborty et al., 2006). These findings suggest that the identified lncRNAs contribute to skeletal muscle development in sheep.
In this study, genes potentially regulated by ovine lncRNAs were predicted using the LncTar tool. Based on lncRNA-mRNA interaction network analysis, we identified 76 overlapping DE lncRNAs with potentially regulated genes involved in muscle development, suggesting that these lncRNAs play vital roles in ovine muscle growth. For instance, NEK5, a gene potentially regulated by TCONS_01 396 701, promotes myogenic differentiation through up-regulation of caspase activity ( Shimizu & Sawasaki, 2013). Similarly, P2RX3, potentially regulated by TCONS_00 790 974, is implicated in neuromuscular junction development in mice ( Carré et al., 2022; Hui et al., 2021). Among the down-regulated lncRNAs, GTL2 was identified as a key regulator, interacting with 17 potentially regulated genes in the network. LncRNA GTL2 was highly expressed in embryonic stages, with peak expression observed on the third day of SC differentiation. Notably, KCNH7 (also known as EGR3), a gene potentially regulated by lncRNA GTL2, has been reported to promote the differentiation of normal intrafusal muscle fibers ( Belengeanu et al., 2014; Fernandes & Tourtellotte, 2015). KEGG pathway analysis further revealed that all genes potentially regulated by lncRNA GTL2 were enriched in the Wnt and cAMP signaling pathways, both of which participate in myogenesis and skeletal muscle development. These findings enhance our understanding of lncRNA GTL2 functions and suggest that it may regulate muscle development through multiple signaling pathways.
Fan et al. ( 2022) reported that the expression of lncRNA GTL2 is regulated by DNA methylation in muscle tissue, suggesting potential involvement in ovine skeletal muscle development and growth. LncRNA GTL2 has also been reported to regulate muscle development through the ceRNA mechanism, where lncRNAs compete with miRNAs to promote the expression of target genes ( Mi et al., 2023; Wang et al., 2019a; Yao et al., 2023). Our results showed that lncRNA GTL2 decreased the proliferation of ovine SCs while enhancing their differentiation. This is consistent with previous studies reporting that lncRNA GTL2 promotes the differentiation of skeletal muscle SCs in pigs ( Cheng et al., 2020; Liu et al., 2023) and cattle ( Liu et al., 2019). Furthermore, our results indicated that lncRNA GTL2 participated in myogenesis via the PKA-CREB signaling pathway. Specifically, lncRNA GTL2 inhibited the proliferation of SCs and promoted their differentiation by affecting the protein expression levels of phospho-PKA and phospho-CREB. Previous studies have shown that activation of PKA expression can reverse the inhibitory effects of isoprenaline on C2C12 cell differentiation and myoblast fusion ( Chen et al., 2019). Additionally, CREB depletion has been reported to accelerate the proliferation of smooth muscle cells ( Klemm et al., 2001). Inhibition of adenosine triphosphate (ATP) release leads to reduced myotube fusion and decreased expression of p-CREB during C2C12 differentiation ( Marco-Bonilla et al., 2023). In addition, p-CREB is highly expressed in Pax7-expressing SCs and nascent myofibers during muscle regeneration ( Stewart et al., 2011). In conclusion, these findings suggest that lncRNA GTL2 may have a profound effect on myogenesis by regulating the PKA-CREB signaling pathway.
In conclusion, we systematically characterized the lncRNA expression profiles of sheep skeletal muscle across different developmental stages and constructed a comprehensive lncRNA-mRNA network for the identified DE lncRNAs. Several lncRNAs with potential roles in muscle development were discovered. Functional analyses revealed that key signaling pathways, including the cAMP and Wnt signaling pathways, were involved in sheep muscle development. Mechanistically, lncRNA GTL2 was identified as a critical regulator of ovine skeletal muscle development, acting through the modulation of the PKA-CREB signaling pathway. This study not only provides a valuable resource for understanding lncRNAs in sheep muscle but also offers new insights into the molecular mechanisms driving muscle development in sheep, laying the groundwork for future studies in muscle biology and livestock production.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Q.J.Z. conceived and designed the experiments. Q.J.Z. and Y.H.M. revised the manuscript. Q.C. and J.J.B. performed the experiments. Q.C. wrote the manuscript. J.J.B., C.H., and Q.Z. analyzed the data. A.H.M.I. and T.H. revised the manuscript. H.C.Z., Y.B.P., X.H.H., and L.J. contributed to sample collection. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Key R&D Program of China (2 021YFD1 300 901), Natural Science Foundation of China (32 172 701), and Modern Wool Sheep Industry System (CARS-39-01)
DATA AVAILABILITY
The sequencing data obtained in this study have been uploaded to the Science Data Bank database (SDB, Data DOI: 10.57760/sciencedb.j00139.00092), Genome Sequence Archive database (GSA, accession number PRJCA030094), and National Center for Biotechnology Information database (NCBI, BioProjectID PRJNA883616).
References
- Adhikari A, Cobb B, Eddington S, et al IFN-γ and CIITA modulate IL-6 expression in skeletal muscle. Cytokine: X. 2020;2(2):100023. doi: 10.1016/j.cytox.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An HT, Kim J, Yoo S, et al Small leucine zipper protein (sLZIP) negatively regulates skeletal muscle differentiation via interaction with α-actinin-4. Journal of Biological Chemistry. 2014;289(8):4969–4979. doi: 10.1074/jbc.M113.515395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczak W, Carr SM, Liu G, et al Long non-coding RNA-derived peptides are immunogenic and drive a potent anti-tumour response. Nature Communications. 2023;14(1):1078. doi: 10.1038/s41467-023-36826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belengeanu V, Gamage TH, Farcas S, et al A de novo 2.3 Mb deletion in 2q24.2q24.3 in a 20-month-old developmentally delayed girl . Gene. 2014;539(1):168–172. doi: 10.1016/j.gene.2014.01.060. [DOI] [PubMed] [Google Scholar]
- Brown J, Pirrung M, Mccue LA FQC dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 2017;33(19):3137–3139. doi: 10.1093/bioinformatics/btx373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun CE, Sincennes MC, Lin AYT, et al GLI3 regulates muscle stem cell entry into G Alert and self-renewal . Nature Communications. 2022;13(1):3961. doi: 10.1038/s41467-022-31695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubner B, Baldwin IT Use of real-time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Reports. 2004;23(5):263–271. doi: 10.1007/s00299-004-0859-y. [DOI] [PubMed] [Google Scholar]
- Butchart LC, Fox A, Shavlakadze T, et al The long and short of non-coding RNAs during post-natal growth and differentiation of skeletal muscles: focus on lncRNA and miRNAs. Differentiation. 2016;92(5):237–248. doi: 10.1016/j.diff.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Cao YT, Ai Y, Zhang XS, et al Genome-wide epigenetic dynamics during postnatal skeletal muscle growth in Hu sheep. Communications Biology. 2023;6(1):1077. doi: 10.1038/s42003-023-05439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré D, Martin V, Kouidri Y, et al The distribution of neuromuscular junctions depends on muscle pennation, when botulinum neurotoxin receptors and SNAREs expression are uniform in the rat. Toxicon. 2022;212:34–41. doi: 10.1016/j.toxicon.2022.04.003. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Reineke EL, Lam M, et al α-Actinin 4 potentiates myocyte enhancer factor-2 transcription activity by antagonizing histone deacetylase 7. Journal of Biological Chemistry. 2006;281(46):35070–35080. doi: 10.1074/jbc.M602474200. [DOI] [PubMed] [Google Scholar]
- Charlier C, Segers K, Wagenaar D, et al. 2001. Human–ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge ( clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Research, 11(5): 850–862.
- Chen BJ, Cai HF, Niu YF, et al. 2024. Whole transcriptome profiling reveals a lncMDP1 that regulates myogenesis by adsorbing miR-301a-5p targeting CHAC1. Communications Biology, 7(1): 518.
- Chen MM, Zhang LL, Guo YW, et al A novel lncRNA promotes myogenesis of bovine skeletal muscle satellite cells via PFN1-RhoA/Rac1. Journal of Cellular and Molecular Medicine. 2021;25(13):5988–6005. doi: 10.1111/jcmm.16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SJ, Yue J, Zhang JX, et al Continuous exposure of isoprenaline inhibits myoblast differentiation and fusion through PKA/ERK1/2-FOXO1 signaling pathway. Stem Cell Research & Therapy. 2019;10(1):70. doi: 10.1186/s13287-019-1160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XN, Xue G, Zhao JY, et al Lockd promotes myoblast proliferation and muscle regeneration via binding with DHX36 to facilitate 5′ UTR rG4 unwinding and Anp32e translation. Cell Reports. 2022;39(10):110927. doi: 10.1016/j.celrep.2022.110927. [DOI] [PubMed] [Google Scholar]
- Chen Z, Bu NP, Qiao XH, et al Forkhead box M1 transcriptionally regulates the expression of long noncoding RNAs Snhg8 and Gm26917 to promote proliferation and survival of muscle satellite cells. Stem Cells. 2018;36(7):1097–1108. doi: 10.1002/stem.2824. [DOI] [PubMed] [Google Scholar]
- Cheng XF, Li L, Shi GL, et al MEG3 promotes differentiation of porcine satellite cells by sponging miR-423-5p to relieve inhibiting effect on SRF. Cells. 2020;9(2):449. doi: 10.3390/cells9020449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Choi HJ, Kang YJ, et al MHY4571, a novel diarylcyclohexanone derivative, exerts anti-cancer activity by regulating the PKA-cAMP-response element-binding protein pathway in squamous cell lung cancer. Experimental Hematology & Oncology. 2022;11(1):68. doi: 10.1186/s40164-022-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TC, Gionbelli MP, Duarte MDS Fetal programming in ruminant animals: understanding the skeletal muscle development to improve meat quality. Animal Frontiers. 2021;11(6):66–73. doi: 10.1093/af/vfab061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva MT, Joshi AS, Castillo MB, et al Fn14 promotes myoblast fusion during regenerative myogenesis. Life Science Alliance. 2023;6(12):e202302312. doi: 10.26508/lsa.202302312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill TL, Carroll A, Pinheiro A, et al The long noncoding RNA Meg3 regulates myoblast plasticity and muscle regeneration through epithelial-mesenchymal transition . Development. 2021;148(2):dev194027. doi: 10.1242/dev.194027. [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Nau GJ, Bar-Joseph Z Clustering short time series gene expression data. Bioinformatics. 2005;21(suppl_1):i159–i168. doi: 10.1093/bioinformatics/bti1022. [DOI] [PubMed] [Google Scholar]
- Fan YX, Ren CF, Deng KP, et al The regulation of LncRNA GTL2 expression by DNA methylation during sheep skeletal muscle development. Genomics. 2022;114(5):110453. doi: 10.1016/j.ygeno.2022.110453. [DOI] [PubMed] [Google Scholar]
- Fernandes MO, Tourtellotte WG Egr3-dependent muscle spindle stretch receptor intrafusal muscle fiber differentiation and fusimotor innervation homeostasis. Journal of Neuroscience. 2015;35(14):5566–5578. doi: 10.1523/JNEUROSCI.0241-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer J, Dimitrova N Transcription regulation by long non-coding RNAs: mechanisms and disease relevance. Nature Reviews Molecular Cell Biology. 2024;25(5):396–415. doi: 10.1038/s41580-023-00694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, et al Pfam: the protein families database. Nucleic Acids Research. 2014;42(D1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming-Waddell JN, Olbricht GR, Taxis TM, et al Effect of DLK1 and RTL1 but not MEG3 or MEG8 on muscle gene expression in Callipyge lambs . PLoS One. 2009;4(10):e7399. doi: 10.1371/journal.pone.0007399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang CW, Song YX, Li YN, et al Knockout of MYOM1 in human cardiomyocytes leads to myocardial atrophy via impairing calcium homeostasis. Journal of Cellular and Molecular Medicine. 2021;25(3):1661–1676. doi: 10.1111/jcmm.16268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CN, Liu CL, Zeng SQ, et al Identification of differentially expressed long non-coding RNAs and messenger RNAs involved with muscle development in Dazu black goats through RNA sequencing. Animal Biotechnology. 2023;34(4):1305–1313. doi: 10.1080/10495398.2021.2020804. [DOI] [PubMed] [Google Scholar]
- Huang XW, Fu CH, Liu WH, et al Chemerin-induced angiogenesis and adipogenesis in 3 T3-L1 preadipocytes is mediated by lncRNA Meg3 through regulating Dickkopf-3 by sponging miR-217. Toxicology and Applied Pharmacology. 2019;385:114815. doi: 10.1016/j.taap.2019.114815. [DOI] [PubMed] [Google Scholar]
- Hui TK, Jing HY, Lai XS Neuromuscular junction-specific genes screening by deep RNA-seq analysis. Cell & Bioscience. 2021;11(1):81. doi: 10.1186/s13578-021-00590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm DJ, Watson PA, Frid MG, et al cAMP response element-binding protein content is a molecular determinant of smooth muscle cell proliferation and migration. Journal of Biological Chemistry. 2001;276(49):46132–46141. doi: 10.1074/jbc.M104769200. [DOI] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, et al CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research. 2007;35(suppl_2):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Mendell JT Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JW, Ma W, Zeng P, et al LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Briefings in Bioinformatics. 2015;16(5):806–812. doi: 10.1093/bib/bbu048. [DOI] [PubMed] [Google Scholar]
- Li RY, Li BJ, Shen M, et al LncRNA 2310043L19Rik inhibits differentiation and promotes proliferation of myoblast by sponging miR-125a-5p. Aging. 2020;12(7):5625–5639. doi: 10.18632/aging.102905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Liu CN, Yuan XY, et al Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic Acids Research. 2011;39(9):3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Xiao MH, Chen HX, et al A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death & Disease. 2019;10(7):528. doi: 10.1038/s41419-019-1767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Li B, Peng WW, et al LncRNA-MEG3 promotes bovine myoblast differentiation by sponging miR-135. Journal of Cellular Physiology. 2019;234(10):18361–18370. doi: 10.1002/jcp.28469. [DOI] [PubMed] [Google Scholar]
- Liu Q, Li MX, Xie S, et al MYOD induced lnc-MEG3 promotes porcine satellite cell differentiation via interacting with DLST. Epigenetics. 2023;18(1):2237789. doi: 10.1080/15592294.2023.2237789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londhe P, Davie JK Gamma interferon modulates myogenesis through the major histocompatibility complex class II transactivator, CIITA. Molecular and Cellular Biology. 2011;31(14):2854–2866. doi: 10.1128/MCB.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XH, Yang XR, Zhang DQ, et al RNA-seq analysis reveals the critical role of the novel lncRNA BIANCR in intramuscular adipogenesis through the ERK1/2 signaling pathway . Journal of Animal Science and Biotechnology. 2023;14(1):21. doi: 10.1186/s40104-022-00820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco-Bonilla M, Herencia R, Fresnadillo M, et al Dipyridamole activates adenosine A2B receptor and AMPK/cAMP signaling and promotes myogenic differentiation of myoblastic C2C12 cells. Frontiers in Pharmacology. 2023;14:1247664. doi: 10.3389/fphar.2023.1247664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Pasut A, Matsumoto M, et al mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541(7636):228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- Mi S, Huang F, Jiao ML, et al Inhibition of MEG3 ameliorates cardiomyocyte apoptosis and autophagy by regulating the expression of miRNA-129-5p in a mouse model of heart failure. Redox Report. 2023;28(1):2224607. doi: 10.1080/13510002.2023.2224607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Gries KJ, Marcotte GR, et al Human myofiber-enriched aging-induced lncRNA FRAIL1 promotes loss of skeletal muscle function . Aging Cell. 2024;23(4):e14097. doi: 10.1111/acel.14097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning XH, Han B, Peng Y, et al. 2024. LncRNA pol-lnc78 as a ceRNA regulates antibacterial responses via suppression of pol-miR-n199-3p-mediated SARM down-regulation in Paralichthys olivaceus. Zoological Research, 45(1): 25–35.
- Papaioannou D, Petri A, Dovey OM, et al The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia . Nature Communications. 2019;10(1):5351. doi: 10.1038/s41467-019-13259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelló-Amorós M, Otero-Tarrazón A, Jorge-Pedraza V, et al Myomaker and myomixer characterization in gilthead sea bream under different myogenesis conditions. International Journal of Molecular Sciences. 2022;23(23):14639. doi: 10.3390/ijms232314639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, et al StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza SHA, Khan R, Cheng G, et al RNA-Seq reveals the potential molecular mechanisms of bovine KLF6 gene in the regulation of adipogenesis . International Journal of Biological Macromolecules. 2022;195:198–206. doi: 10.1016/j.ijbiomac.2021.11.202. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Mccarthy DJ, Smyth GK edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell NX, Burra K, Shah RM, et al Wnt signaling regulates ion channel expression to promote smooth muscle and cartilage formation in developing mouse trachea. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2023;325(6):L788–L802. doi: 10.1152/ajplung.00024.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DB, Zhai LM, Ravikumar B, et al A prevalent variant in PPP1R3A impairs glycogen synthesis and reduces muscle glycogen content in humans and mice . PLoS Medicine. 2008;5(1):e27. doi: 10.1371/journal.pmed.0050027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Sawasaki T Nek5, a novel substrate for caspase-3, promotes skeletal muscle differentiation by up-regulating caspase activity. FEBS Letters. 2013;587(14):2219–2225. doi: 10.1016/j.febslet.2013.05.049. [DOI] [PubMed] [Google Scholar]
- Simionescu-Bankston A, Kumar A Noncoding RNAs in the regulation of skeletal muscle biology in health and disease. Journal of Molecular Medicine. 2016;94(8):853–866. doi: 10.1007/s00109-016-1443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CC, Yang ZX, Jiang R, et al LncRNA IGF2 AS regulates bovine myogenesis through different pathways . Molecular Therapy - Nucleic Acids. 2020;21:874–884. doi: 10.1016/j.omtn.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R, Flechner L, Montminy M, et al CREB is activated by muscle injury and promotes muscle regeneration. PLoS One. 2011;6(9):e24714. doi: 10.1371/journal.pone.0024714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui YT, Han Y, Zhao XY, et al Long non-coding RNA Irm enhances myogenic differentiation by interacting with MEF2D . Cell Death & Disease. 2019;10(3):181. doi: 10.1038/s41419-019-1399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Zhang F, Luo X, et al Long noncoding RNA AC092155 facilitates osteogenic differentiation of adipose-derived stem cells through the miR-143-3p/STMN1 axis. The Journal of Gene Medicine. 2021;23(8):e3363. doi: 10.1002/jgm.3363. [DOI] [PubMed] [Google Scholar]
- Sun L, Luo HT, Bu DC, et al Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research. 2013;41(17):e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Borello U, Vivarelli E, et al. 1998. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development, 125(21): 4155–4162.
- Takata H, Terada K, Oka H, et al Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Developmental Dynamics. 2007;236(10):2800–2807. doi: 10.1002/dvdy.21327. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Maltzahn J, Chang NC, Bentzinger CF, et al Wnt signaling in myogenesis. Trends in Cell Biology. 2012;22(11):602–609. doi: 10.1016/j.tcb.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu MR, Ding ZX, et al Phosphoglycerate dehydrogenase positively regulates the proliferation of chicken muscle cells. Poultry Science. 2022a;101(5):101805. doi: 10.1016/j.psj.2022.101805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen MY, Chen JF, et al LncRNA IMFlnc1 promotes porcine intramuscular adipocyte adipogenesis by sponging miR-199a-5p to up-regulate CAV-1. BMC Molecular and Cell Biology. 2020a;21(1):77. doi: 10.1186/s12860-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LT, Fan J, Han LL, et al The micropeptide LEMP plays an evolutionarily conserved role in myogenesis. Cell Death & Disease. 2020b;11(5):357. doi: 10.1038/s41419-020-2570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MH, Li C, Zhang Y, et al LncRNA MEG3-derived miR-361-5p regulate vascular smooth muscle cells proliferation and apoptosis by targeting ABCA1. American Journal of Translational Research. 2019a;11(6):3600–3609. [PMC free article] [PubMed] [Google Scholar]
- Wang RT, Chen FL, Chen Q, et al MyoD is a 3D genome structure organizer for muscle cell identity. Nature Communications. 2022b;13(1):205. doi: 10.1038/s41467-021-27865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Tan BH, Xiao LY, et al Long non-coding RNA Gm10561 promotes myogenesis by sponging miR-432 . Epigenetics. 2022c;17(13):2039–2055. doi: 10.1080/15592294.2022.2105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Zuo H, Jin JJ, et al Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2 . Cell Death & Disease. 2019b;10(7):505. doi: 10.1038/s41419-019-1742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Gu MF, Wang ZF, et al Let-7a-5p regulated by lncRNA-MEG3 promotes functional differentiation to Schwann cells from adipose derived stem cells via directly inhibiting RBPJ-mediating notch pathway. Apoptosis. 2021;26(9-10):548–560. doi: 10.1007/s10495-021-01685-x. [DOI] [PubMed] [Google Scholar]
- Wei CH, Li L, Su HW, et al Identification of the crucial molecular events during the large-scale myoblast fusion in sheep. Physiological Genomics. 2014;46(12):429–440. doi: 10.1152/physiolgenomics.00184.2013. [DOI] [PubMed] [Google Scholar]
- Wei CH, Wu MM, Wang CD, et al Long noncoding RNA Lnc-SEMT modulates IGF2 expression by sponging miR-125b to promote sheep muscle development and growth. Cellular Physiology and Biochemistry. 2018;49(2):447–462. doi: 10.1159/000492979. [DOI] [PubMed] [Google Scholar]
- Wei Y, Guo DS, Bai YB, et al Transcriptome analysis of mRNA and lncRNA related to muscle growth and development in Gannan yak and Jeryak. International Journal of Molecular Sciences. 2023;24(23):16991. doi: 10.3390/ijms242316991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HQ, Ren Y, Li S, et al In vitro culture and induced differentiation of sheep skeletal muscle satellite cells. Cell Biology International. 2012;36(6):579–587. doi: 10.1042/CBI20110487. [DOI] [PubMed] [Google Scholar]
- Wu TY, Wang SH, Wang LH, et al Long noncoding RNA (lncRNA) CTTN-IT1 elevates skeletal muscle satellite cell proliferation and differentiation by acting as ceRNA for YAP1 through absorbing miR-29a in Hu sheep . Frontiers in Genetics. 2020a;11:843. doi: 10.3389/fgene.2020.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YX, Lian KQ, Sun C LncRNA LEF1-AS1 promotes osteogenic differentiation of dental pulp stem cells via sponging miR-24-3p. Molecular and Cellular Biochemistry. 2020b;475(1-2):161–169. doi: 10.1007/s11010-020-03868-7. [DOI] [PubMed] [Google Scholar]
- Xu XC, Ji SY, Li WL, et al LncRNA H19 promotes the differentiation of bovine skeletal muscle satellite cells by suppressing Sirt1/FoxO1. Cellular & Molecular Biology Letters. 2017;22:10. doi: 10.1186/s11658-017-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YL, Wang ZS, Chen Y, et al Single-cell analysis reveals the lncRNA-MEG3/miRNA-133a-3p/PRRT2 axis regulates skeletal muscle regeneration and myogenesis. Genes & Diseases. 2023;10(2):359–362. doi: 10.1016/j.gendis.2022.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JA, Wang ZJ, Yang X, et al LncRNA-FKBP1C regulates muscle fiber type switching by affecting the stability of MYH1B. Cell Death Discovery. 2021;7(1):73. doi: 10.1038/s41420-021-00463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Zheng YJ, et al LncRNA TUG1 promoted osteogenic differentiation through promoting bFGF ubiquitination. In Vitro Cellular & Developmental Biology - Animal. 2020;56(1):42–48. doi: 10.1007/s11626-019-00410-y. [DOI] [PubMed] [Google Scholar]
- Yue YQ, Yue YR, Fan ZY, et al The long noncoding RNA lnc-H19 is important for endurance exercise by maintaining slow muscle fiber types. Journal of Biological Chemistry. 2023;299(11):105281. doi: 10.1016/j.jbc.2023.105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HJ, Yu YH, Jiake C Expression signatures of lncRNAs in skeletal muscles at the early flow phase revealed by microarray in burned rats. Turkish Journal of Trauma and Emergency Surgery. 2016;22(3):224–232. doi: 10.5505/tjtes.2015.04831. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Kong QB, Wang J, et al Complex roles of cAMP–PKA–CREB signaling in cancer. Experimental Hematology & Oncology. 2020a;9(1):32. doi: 10.1186/s40164-020-00191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Chen MM, Liu XF, et al A novel lncRNA, lnc403, involved in bovine skeletal muscle myogenesis by mediating KRAS/Myf6. Gene. 2020b;751:144706. doi: 10.1016/j.gene.2020.144706. [DOI] [PubMed] [Google Scholar]
- Zhang XR, Zhang YN, Geng GF, et al lncRNA NEAT1 is required for splenic erythroid differentiation . Journal of Genetics and Genomics. 2023;50(6):454–457. doi: 10.1016/j.jgg.2023.01.012. [DOI] [PubMed] [Google Scholar]
- Zhao W, Chen L, Zhong T, et al The differential proliferation and differentiation ability of skeletal muscle satellite cell in Boer and Nanjiang brown goats. Small Ruminant Research. 2018;169:99–107. doi: 10.1016/j.smallrumres.2018.07.006. [DOI] [Google Scholar]
- Zhou CN, Li MT, Sun YY, et al Systematic identification of long noncoding RNAs during three key organogenesis stages in zebrafish. International Journal of Molecular Sciences. 2024;25(6):3440. doi: 10.3390/ijms25063440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Zhang SY, Wang HT, et al LncFunNet: an integrated computational framework for identification of functional long noncoding RNAs in mouse skeletal muscle cells. Nucleic Acids Research. 2017;45(12):e108. doi: 10.1093/nar/gkx232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The sequencing data obtained in this study have been uploaded to the Science Data Bank database (SDB, Data DOI: 10.57760/sciencedb.j00139.00092), Genome Sequence Archive database (GSA, accession number PRJCA030094), and National Center for Biotechnology Information database (NCBI, BioProjectID PRJNA883616).