Abstract
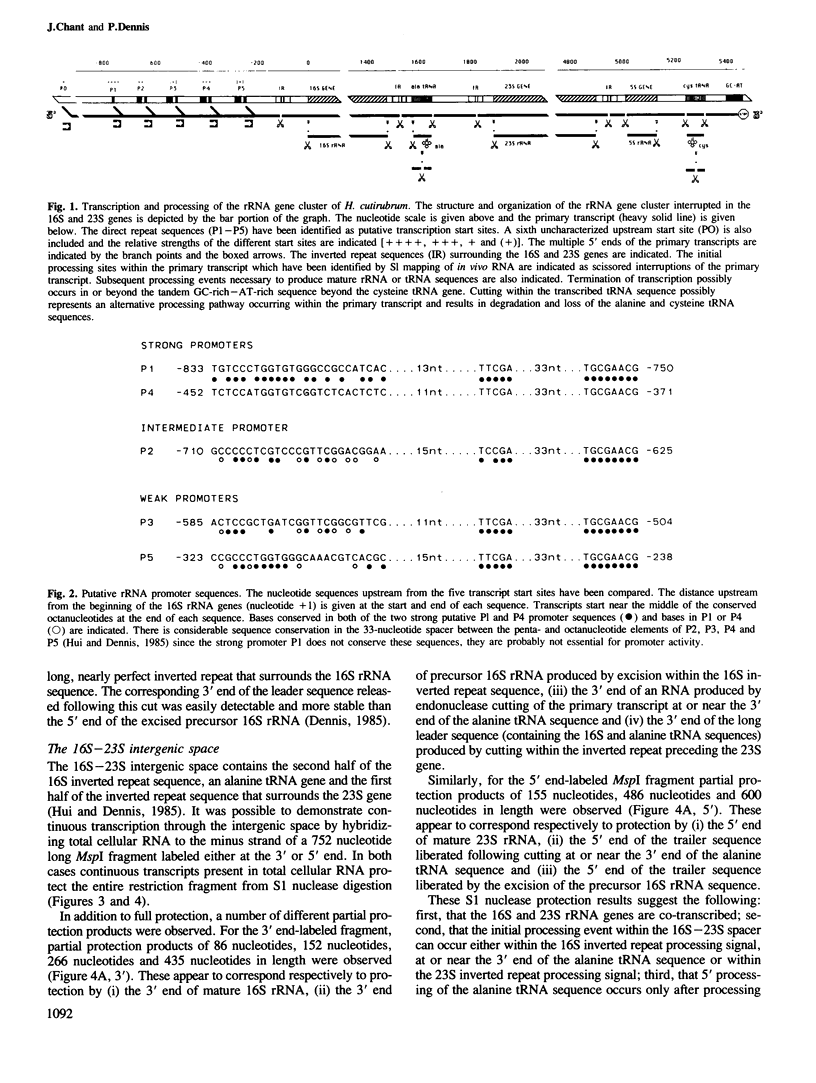
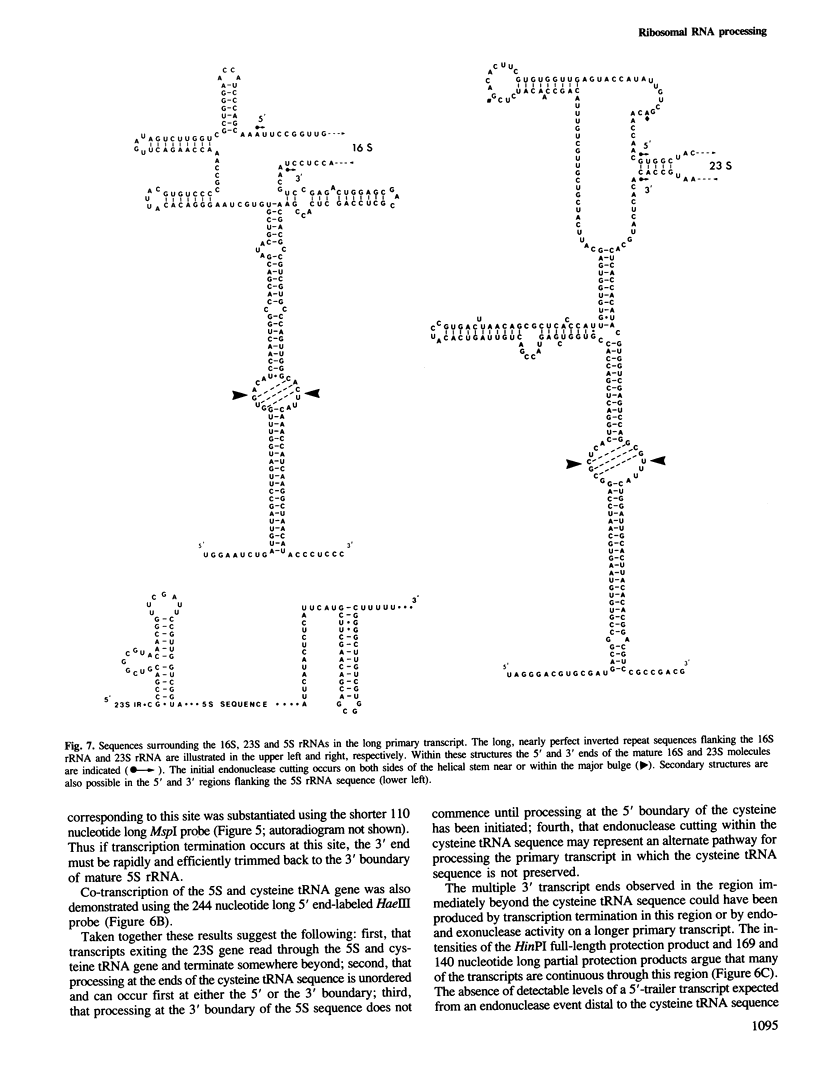
The chromosome of Halobacterium cutirubum contains a single ribosomal RNA gene cluster. The 5' to 3' organization of genes within this 6-kbp region is: 16S, alanine tRNA, 23S, 5S, cysteine tRNA. The entire gene cluster is transcribed as a single long primary transcript; processing of mature RNA sequences from the 5' region of the transcript begins prior to the completion of synthesis at the 3' end. There are five conserved octanucleotide direct repeats (TGCGAACG) in the 900-bp 5'-flanking sequence in front of the 16S gene. The positions of these repeat sequences correspond to the different 5' ends of the primary transcript and probably represent the RNA polymerase start sites. The 16S and 23S rRNA genes are surrounded by long nearly perfect inverted repeat sequences. These sequences probably form duplex structures in the primary transcript and are recognized by an RNaseIII-like endonuclease activity that carries out the initial excision of the precursor 16S and 23S rRNA sequences. These precursors are rapidly trimmed to the mature 16S and 23S molecules and assembled into ribosomal particles. The processing sites for 5S rRNA appear to be at or very near to the mature ends of the 5S molecule. The tRNA sequences are processed with reduced efficiency from the primary transcript. Nuclease cuts have been detected at the ends as well as in the middle of the cysteine tRNA sequence suggesting that there may be alternative processing pathways, one resulting in proper excision of the mature tRNA sequence and the other resulting in improper excision and degradation of the tRNA sequence. The transcription termination sequence is believed to be at or beyond an AT-rich sequence preceded by a GC-rich sequence located distal to the cysteine tRNA gene.
Keywords: archaebacteria, Halobacterium, rRNA processing
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Daniels C. J., Gupta R., Doolittle W. F. Transcription and excision of a large intron in the tRNATrp gene of an archaebacterium, Halobacterium volcanii. J Biol Chem. 1985 Mar 10;260(5):3132–3134. [PubMed] [Google Scholar]
- Daniels C. J., Hofman J. D., MacWilliam J. G., Doolittle W. F., Woese C. R., Luehrsen K. R., Fox G. E. Sequence of 5S ribosomal RNA gene regions and their products in the archaebacterium Halobacterium volcanii. Mol Gen Genet. 1985;198(2):270–274. doi: 10.1007/BF00383005. [DOI] [PubMed] [Google Scholar]
- Dennis P. P. Multiple promoters for the transcription of the ribosomal RNA gene cluster in Halobacterium cutirubrum. J Mol Biol. 1985 Nov 20;186(2):457–461. doi: 10.1016/0022-2836(85)90117-2. [DOI] [PubMed] [Google Scholar]
- Dennis P. P. Transcription patterns of adjacent segments on the chromosome of Escherichia coli containing genes coding for four 50S ribosomal proteins and the beta and beta' subunits of RNA polymerase. J Mol Biol. 1977 Oct 5;115(4):603–625. doi: 10.1016/0022-2836(77)90105-x. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of procaryotic ribonucleic acid. Microbiol Rev. 1981 Dec;45(4):502–541. doi: 10.1128/mr.45.4.502-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman J. D., Lau R. H., Doolittle W. F. The number, physical organization and transcription of ribosomal RNA cistrons in an archaebacterium: Halobacterium halobium. Nucleic Acids Res. 1979 Nov 10;7(5):1321–1333. doi: 10.1093/nar/7.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Schnabel R., Sentenac A., Zillig W. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J. 1983;2(8):1291–1294. doi: 10.1002/j.1460-2075.1983.tb01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui I., Dennis P. P. Characterization of the ribosomal RNA gene clusters in Halobacterium cutirubrum. J Biol Chem. 1985 Jan 25;260(2):899–906. [PubMed] [Google Scholar]
- Kaine B. P., Gupta R., Woese C. R. Putative introns in tRNA genes of prokaryotes. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3309–3312. doi: 10.1073/pnas.80.11.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin A. S., Teterina N. L., Rubtsov P. M., Baratova L. A., Kagramanova V. K. Putative promoter region of rRNA operon from archaebacterium Halobacterium halobium. Nucleic Acids Res. 1984 Aug 24;12(16):6537–6546. doi: 10.1093/nar/12.16.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]