Abstract
Dynamic 3D covalent organic frameworks (COFs) have shown a concerted structural transformation upon adaptive guest inclusion. However, the origin of the conformational mobility and the host–guest adaptivity remain conjecture of the pedal motions of revolving imine linkages, often without considering the steric hindrance from the interwoven frameworks. Here, we present atomic-level observation of the rotational and translational dynamics in single-crystal COF-300 upon adaptive guest inclusion of various organic molecules, featuring multiple rotamers of covalent linkages and switchable interframework noncovalent interactions. Specifically, we developed a diffusion gradient transimination protocol to facilitate the growth of COF single crystals, enabling a high-resolution X-ray diffraction structural analysis. We uncovered metastable and low-symmetry intermediate phases from contracted to expanded phases during structural evolution. We identified torsion angles in the terephthalaldehyde diimine motifs that switch from anti-periplanar to syn-periplanar/anticlinal conformations. Moreover, the rotational dynamics of the imine linkage were concurrent with the translational dynamics of tetraphenylmethane units, which tend to form the translational quadruple phenyl embrace. Such conformational mobility allows the frameworks to adapt to various guest molecules, such as alcohols, esters, phenols, and diols, featuring double linear, herringbone, zigzag chains, triple helix, and tubular alignments. Quantitative energy analyses revealed that such dynamic structure transformations are not arbitrary but follow specific pathways that resemble protein folding. The work is paving the way to developing robust, dynamic, and crystalline molecular sponges for studying the condensed structure of liquids without the need for further crystallization.
Introduction
Covalent organic frameworks (COFs) are robust porous crystals constructed by stitching organic building units into extended networks through strong covalent bonds.1,2 Featuring various conformations without breaking and remaking bonds upon adaptive guest inclusion, the dynamics responses of COFs have attracted great attention for their high functions in gas storage, adsorptive separation, heterogeneous catalysis, and molecular sensing.3−8 Thus, understanding their dynamic mechanisms at the atomic-level is crucial for correlating their conformational geometry variations with their dynamic responses coupled with their physicochemical properties. In this work, we unraveled the rotational and translational dynamics for adaptive guest alignments tracking by single-crystal X-ray crystallography with atomic resolution in COF-300,9 enabling computational analyses on the energy pathway of structural transformation.
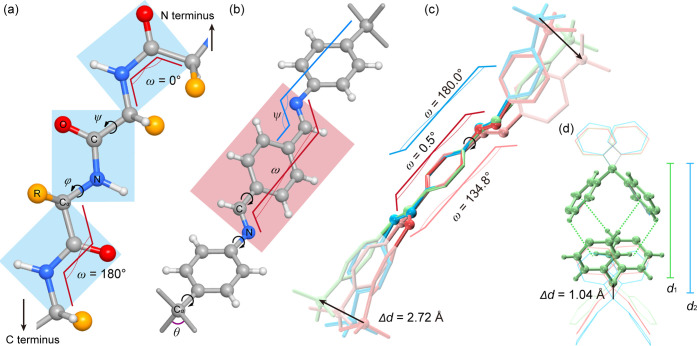
Inspired by the protein folding, conformations of the polypeptide backbones are determined by the degrees of freedom at each peptide linkage (Figure 1a; ω, ψ, and φ): With the partial conjugation of the amide bond, the Cα–CO–N–Cα motifs are generally coplanar, leading to ω torsion angles of either 180 or 0°, which dictate the formation of α-helices/β-strands or big turns in their folded structures to dictate their biological functions.10a Levinthal’s paradox highlights the contradiction between the rapid folding of proteins, often within seconds or less, and the astronomical time theoretically required to randomly explore all possible conformations (i.e., 3100 = 5 × 1043 for a protein containing 100 peptide linkages).10b This suggests that protein folding is not random but instead follows specific pathways, as described by the “folding energy funnel model.″ In contrast to the 1D backbones in protein folding, the structural transformation of dynamic 3D COFs (Figure 1b) remains elusive due to the steric hindrance imposed by their interwoven networks.3d Therefore, atomic understanding of the conformation diversity and quantitative analyses of their host–host, host–guest, and guest–guest interactions are essential to portray their energy pathway of structural transformation. For describing the rotational dynamics of COF-300, we identified the critical conformational geometry variation as the torsion angles of its terephthalaldehyde diimine (TPDI) motifs (ω in Figure 1b) adopting anti-periplanar, syn-periplanar, and anticlinal conformations, leading to crystallography symmetry-breaking. Another key conformational geometry variation is the torsion angle of the diphenylimine (ψ in Figure 1b), which determines the contraction and expansion of frameworks without changing the ω angle and crystallography symmetry. Overlaying these conformations (Figure 1c) reveals the rotational freedom of the imine linkage within the COF’s closely interwoven structures. Moreover, the steric hindrance imposed by the interwoven frameworks is mitigated by the translational dynamics between tetraphenylmethane (TPM) motifs, whose interframework noncovalent interactions switched from π–π interaction between TPM and TPDI to the formation of translational quadruple phenylene embrace (TQPE)11 between TPMs (Figure 1d).
Figure 1.
Conformational mobility of dynamic 3D COFs akin to protein folding. (a) Degrees of freedom in the polypeptide backbone, defined by the coplanar torsion angle Cα–N–C–Cα (ω = 0 or 180°) and the rotational torsion angles N–Cα–C–N (ψ) and C–N–Cα–C (φ) for each peptide linkage. (b) Key parameters for the conformation of dynamic 3D COFs, highlighting the torsion angles of diimine (ω = 0, 134, or 180°) and biphenyl imine (ψ = 160, −163, 176.88, or −179°). (c) Superimposed view of covalent linkages in anti-periplanar, syn-periplanar, and anticlinal conformations for COF-300-cp, -ip, and -op, demonstrating the pedal motion by revolving imine bonds and shift of the tetrahedral nodes. (d) Shuttle movement of tetraphenylene methane leads to the formation of the translation quadruple phenylene embrace.
Such extraordinary conformational mobility, coupled with structural regularity, enables adaptive guest alignments within the dynamic confinement space with atomic precision. Adaptive guest alignments of organic molecules such as alcohols, esters, and diols, with varying sizes, functionality, and complexity, have shown rich guest–guest interactions to perform double linear, herringbone, zigzag chains, triple helix, and tubular alignments, manifesting their exceptional mutual host–guest adaptivity, precise molecular recognition, and multimodule self-organization. This work is paving the way to developing robust, dynamic, and crystalline molecular sponges for studying the condensed structure of liquids without the need for further crystallization.
Experimental Section
Rapid Growth of COF Single Crystals for High-Resolution X-ray Structural Analysis
Single crystals of COF-300, suitable for high-resolution X-ray diffraction analysis, were synthesized using a diffusion gradient transimination protocol (Section S1 in the Supporting Information).12 Terephthalaldehyde (TPA, 12.0 mg, 0.089 mmol) is dissolved in 1,4-dioxane (0.5 mL, ultradry), added with aniline (0.12 mL, 15 equiv.) and aqueous acetic acid (AcOH; 6 mol L–1, 0.2 mL), resulting in white flack crystal precipitated out at the bottom of the glass diffusion tube (inner diameter ∼4.24 mm) standing vertically. Then, a solution of tetra-(p-aminophenyl)-methane (TAM, 20.0 mg, 0.052 mmol) dissolved in 1,4-dioxane (0.5 mL, ultradry) was then added drop by drop onto the first layer to form stratification. The diffusion gradient transimination protocol controls the nucleation and crystal growth, yielding prismatic crystals of COF-300 with a size of ∼50 μm within 6 days (denoted as COF-300-DC). For comparison of crystallinity, imine-exchange and solvent-ventilation controlled crystal growth are also implemented to yield single crystals of COF-300 with sizes of ∼40 μm within 30 or 14 days (denoted as COF-300-MC and -VC), respectively.
Size and Morphology Control of Single-Crystal COF Bulk Sample for In Situ Powder X-ray Diffraction (PXRD) Analysis
To improve the adsorption kinetics and achieve coherent transformation,3f we have developed the ventilation-vial synthetic protocol to obtain bulk samples with uniform size and morphology (Section S1 in the Supporting Information).3b TPA (60.0 mg, 0.445 mmol) was dissolved in 1,4-dioxane (1 mL, ultradry) in a 20 mL ventilation glass vial, which was added with aniline (0.20 mL) and AcOH (6 mol L–1, 1 mL). A solution of TAM (100.0 mg, 0.26 mmol) dissolved in 1,4-dioxane (5 mL, ultradry) was then added to the vial, which was heated at 65 °C for 5 days. Single crystals of COF-300 with a size of 10 μm (donated as COF-300–10 μm) were then isolated and washed with fresh 1,4-dioxane and fully exchanged with tetrahydrofuran (THF). The phase purity and crystallinity of samples were confirmed by PXRD on a Bruker D8 advance diffractometer with Cu Kα radiation. The crystal size and morphology were examined through scanning electron microscopy (SEM) on a JEOL JSM 7800F Prime microscope.
Single-Crystal Structural Analysis
The single crystal X-ray diffraction (SXRD) data were collected at the Shanghai Synchrotron Light Source Line BL17B1/BL10U2 station. The structure was solved by direct methods using the SHELXTL software package and further developed with different Fourier syntheses. All non-hydrogen atoms on the framework were refined anisotropically; all hydrogen atoms were generated geometrically and refined in the riding mode. All of the phenyl rings are treated with rigid constraints.
Gas/Vapor Adsorption Isotherms
The organic vapor adsorption isotherms of the COF-300-DC samples were collected using a BELSORP-MAX and MicrotracBELSopr-Aqua3 adsorption apparatus with a water circulator bath. Anhydrous solvents were used for vapor adsorption, which degassed at least five times before the isotherm collection.
Solid-State Nuclear Magnetic Resonance Spectroscopy
Solid-state nuclear magnetic resonance (ssNMR) spectroscopy was performed with cross-polarization with magic angle spinning (CP/MAS) techniques on a Bruker AVANCE III HD 400 MHz wide-bore solid-state NMR spectrometer at a magnetic field of 9.4 T equipped with a standard Bruker MAS probe with a 3.2 mm (o.d.) zirconia rotor. 13C CP/MAS NMR spectra were acquired at a Larmor frequency of 100.6 MHz. 13C chemical shifts were referenced to tetramethyl silane (TMS) at 0 ppm and were further calibrated using the carboxylic carbon of the glycine assigned to 176.2 ppm as a secondary reference. All the 13C CP/MAS experiments were carried out on a standard 3.2 mm double-resonance probe with a sample spinning rate of 12 kHz, and 13C experiments were carried out with a 1H π/2 pulse length of 3.5 μs, a contact time of 2 ms, a pulse delay of 5 s, and a SPINAL-64 at a decoupling frequency of 81 kHz.
Dynamic Vapor Sorption In Situ PXRD Measurement
The organic vapor concentration in the working gas was regulated by blending vapor-saturated nitrogen (N2) from an inlet connected to a solvent reservoir with dry N2, using two 100-sccm mass flow controllers (MFCs).8c This working gas was directed into the Anton Paar in situ PXRD chamber on a Bruker D8 diffractometer. PXRD patterns were collected sequentially under a stable vapor concentration until the reflection positions and intensities reached equilibrium, after which the conditions were adjusted to the next set points.
Results and Discussion
Optimizing Crystallinity and Efficiency in COF Single Crystal Growth
The crystallization problem of COFs has long posed a significant challenge, particularly in growing sufficiently large single crystals (∼50 μm) suitable for X-ray diffraction analysis. Several synthetic strategies, such as introducing amine modulators,10 amine transimination,13 crystal growth in ionic liquids,14 and amphiphilic micelle reaction vessels,15 have yielded larger single crystals. However, large-sized single crystals do not guarantee high-resolution X-ray diffraction for structure analysis of COFs.16 While the aniline-modulated strategies have led to decent crystallinity and X-ray diffraction resolution up to 0.83 Å, the process has been time-consuming (30–80 days).
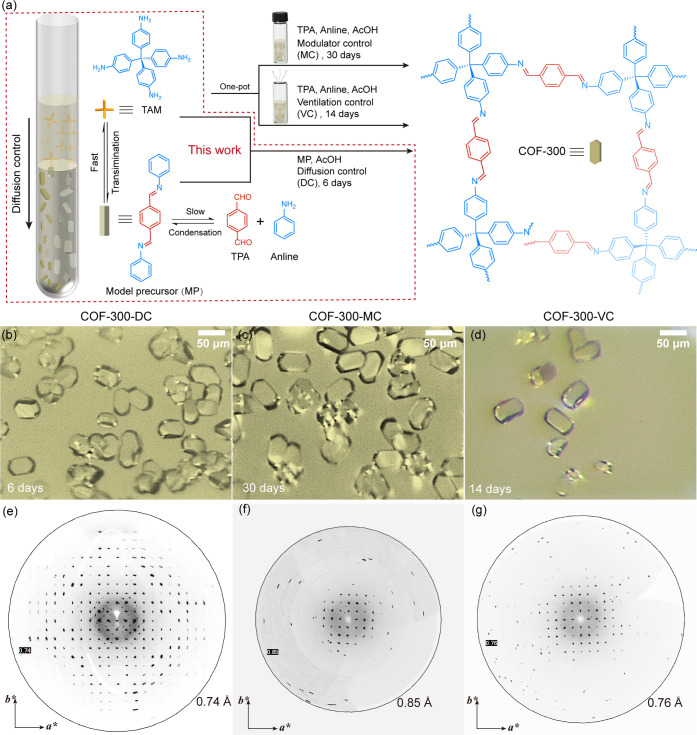
To enable precise host–guest structure analysis, we developed a diffusion gradient transimination protocol that facilitates the growth of COF single crystals (∼50 μm) within just 6 d or less, achieving X-ray diffraction with resolutions of up to 0.68 Å (Figure S24 in the Supporting Information). This process involves the imine condensation to form model precursor (MP, Figure S22 and Table S1 in the Supporting Information) immediately precipitated out at the bottom of the diffusion tube, alongside a transimination with TAM diffused from its top layer, resulting in nucleation control and a reactant diffusion ingredient conducive to the single crystal growth. In general, the reaction kinetics of imine exchange is faster than that of imine condensation, such sequential transimination bypassing the slower imine condensation competition between TAM and aniline in a one-pot reaction, accelerating the crystal growth. Indeed, we observe the full conversion of the white crystal of the model precursor into the yellow crystal of COF-300-DC (Figure 2b), resulting in an X-ray diffraction resolution of 0.74 Å (Figures 2e, S23 and Table S2 in the Supporting Information).
Figure 2.
Rapid growth of COF single crystals for high-resolution X-ray diffraction. (a) Gradient diffusion protocol showcasing sequential COF construction through imine condensation of TPA and aniline to form a modulated molecular precursor, followed by imine exchange with TAM to form COF-300 with diffusion-controlled kinetics (COF-300-DC), compared to one-pot reactions with kinetics controlled by modulator-controlled in sealed vials (-MC) and solvent ventilation in vented vials (-VC). (b–g) Optical images of single crystals grown via -DC (6 days), -MC (30 days), and -VC (14 days) approaches, respectively. (e–g) Reconstructed single-crystal X-ray diffraction projected along the reciprocal c* direction, with resolutions of 0.74, 0.85, and 0.76 Å for COF-300-DC, -MC, and -VC, respectively.
Fourier-transform infrared spectroscopy (FT-IR, Figure S1 in the Supporting Information) confirmed the formation of imine bonds (−C=N–stretching at 1626 cm–1), and thermal gravimetric analysis (TGA, Figure S2 in the Supporting Information) indicated thermal stability up to 520 °C. Moreover, PXRD patterns (Figure S3 in the Supporting Information) displayed sharp reflection peaks with a full width at half-maximum (fwhm) of 0.123° (200), suggesting high crystallinity. The 13C ssNMR spectra revealed a homogeneous local chemical environment of COF-300-DC with significantly improved resolution (fwhm ∼0.52 ppm, Figure S4 in the Supporting Information). The CO2 (195 K), N2 (77 K), CH4 (112 K) and n-Butane (298 K) adsorption isotherms have shown their dynamic responses upon gas inclusion, consistent with the crystal-size effect observed in dynamic 3D COFs3f (Figures S7 and S10 in the Supporting Information).
In comparison, single crystal growth through the traditional amine modulated protocols controlled by imine exchange (COF-300-MC) and solvent ventilation controlled (COF-300-VC) requires 30 and 14 days, respectively, resulting in X-ray diffraction resolutions of 0.86 and 0.76 Å (Figure 2d–g, Table S3 in the Supporting Information).
Suitable Probe for Ex Situ Single-Crystal Structural Evolution of COF-300
To identify a suitable probe molecule for monitoring the dynamic structural transformations of COF-300, we collected adsorption isotherms for various organic vapors, including alcohols, esters, ethers, and alkanes (Figures S12 and S20 in the Supporting Information). These experiments allowed us to screen for anomalous adsorption behavior accompanied by dynamic structural transformation. Among the tested vapors, n-butanol (n-BuOH) emerged as the most effective probe, owing to its broad pressure response range (P/P0: 0–0.50) and high heat of adsorption, ranging from 65 to 81 kJ/mol (Figure S21 in the Supporting Information). These characteristics suggest strong host–guest and guest–guest interactions, making n-BuOH particularly well suited for capturing subtle phase transitions.
Rotational Dynamics of Covalent Linkages Visualized by the COF Rotamers
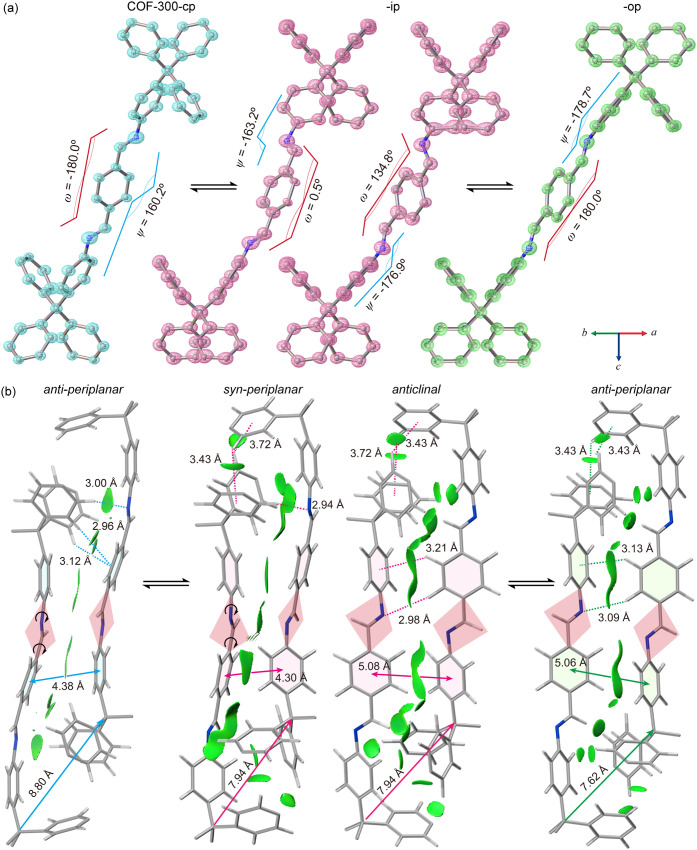
High-resolution synchrotron SXRD analysis (Figures 3; S25–S29 and Tables S4–S6 in the Supporting Information) provided atomic-resolution structures of COF-300 across different guest stoichiometries, including the contracted phase (-cp, tetragonal I41/a, a = b = 20.49 Å, c = 8.82 Å, V = 3703.0 Å3), the intermediated phase (-ip, orthorhombic I212121, a = 24.30 Å, b = 20.76 Å, c = 8.17 Å, V = 4123.0 Å3), and the open phase (-op, tetragonal I41/a, a = b = 25.41 Å, c = 7.78 Å, V = 5021.6 Å3). The structural transformation was highlighted by the conformational changes in the covalent linkages, as observed from the electron density maps calculated from the SXRD data (Figure 3a). Both COF-300-cp and -op shared the global symmetry of space group I41/a (No. 88), with the diimine structure units exhibiting an anti-periplanar conformation (ω = 180°). Notably, COF-300-ip crystallized in lower symmetry, I212121(No. 24), and displayed a doubled asymmetric unit compared to that of COF-300-cp and -op, resulting in a syn-periplanar conformation (ω = 0.5°) and an anticlinal conformation (ω = 134.5°). This geometric variation of covalent linkages between anti-periplanar (180°), syn-periplanar (0.5°)/anticlinal (134.5°), and anti-periplanar (180°) conformations of ω is direct evidence of the rotational dynamics of the COF. Despite both COF-300-cp and -op adopting the same anti-periplanar conformations, the structural differences are characterized by the torsion angles Cα-C=N–C (denoted as ψ, Figure 3a), which shift from 160.2 to −176.9°. By overlaying the phenyl rings of the TPDI for comparison (Figure S59 in the Supporting Information), it becomes clear that the ψ angle significantly impacts the distance between the tetrahedral nodes. This conformational mobility is further enabled by a concerted “hinge” movement of adjacent phenyl groups in TPM, where the angle θ varies between 101 and 113° (Figure S58 and Table S21 in the Supporting Information).
Figure 3.
X-ray diffraction single-crystal structural evolution during guest removal illustrated the COF-300-cp (left column), -ip (middle column), and -op (right column) phases observed through ex-situ desorption of n-BuOH guest molecules. (a) Atomic-level conformational variations highlighting the pedal motion of covalent linkages, characterized by changes in torsion angles (ω and ψ). (b) Interframework displacement is driven by shifts in π–π interactions and rotation of the imine linkage, transitioning from the node (TPM)–to-edge (TPDI) interactions to node–to-node and edge–to-node interactions, mitigating the steric hindrance in the closely interwoven frameworks. (Color codes: C, gray; N, blue; and H, white.).
The free energy landscape17 of conformational search of TPDI shows dependences of both ω and ψ (Figure S64 in the Supporting Information). Energy minima are located at ψ = 168° and ω = 0 or 180°. The observed conformers of COF-300-cp (ψ = 160° and ω = −180°), the syn-periplanar of -ip (ψ = −163.2° and ω = 0.5°;) and -op (ψ = −178.7° and ω = 180°) approximate the ideal conformational geometry, while the anticlinal of -ip (ψ = −176.9° and ω = 134.8°) is of energy penalty.
Translational Dynamics Enabled by Switching Interframework π–π Interactions
The conformational mobility of dynamic 3D COFs is further facilitated by the displacement of interwoven dia networks. Comparing their simplified networks, the angle of the tetrahedral nodes varied from 67° to 83.4 and 87.4°, with the displacement between subnets decreasing from 8.80 to 7.94 and 7.62 Å (Figure S58 in the Supporting Information). As shown in Figure 3b, the dominant interaction in the COF-300-cp involves the phenyl edge-to-face π–π interaction, featuring the distance between TPM and TPDI (2.96 and 3.12 Å). In the COF-300-ip and -op conformations, the TPM phenyls form the 4-fold edge-to-face interactions known as phenyl embrace (Figure 1d). The distances between TPM phenyl are 3.43 and 3.72 Å in COF-300-ip and only 3.43 Å in COF-300-op. In addition to the phenyl embrace, edge-to-face π–π interaction is observed between TPDI and TPM in both the COF-300-ip phase anticlinal and COF-300-op anti-periplanar conformation, with distances of 3.21 and 3.13 Å, respectively. Throughout the conformational transition, the dominant interaction within the framework changes from node (TPM)–edge (TPDI) to both node–node and edge–node interactions, demonstrating the flexibility of the moieties within the dynamic framework. These moieties within this dynamic framework can rotate, bend, and deform freely in a concerted manner through interwoven displacement.
Further analysis of framework interaction was conducted using an independent gradient model Hirshfeld partition of molecular density (IGMH)18 via Multiwfn software19 (Section S6 in the Supporting Information). Focusing on the δginter (isosurfaces of 0.0045 a.u.) intermolecular interaction region (Figure 3b), the results revealed that the van der Waals interactions between the frameworks are consistent with the single-crystal structure analysis. Moreover, the dominant π–π interaction evolves from TPM-TPDI to TPM-TPM and TPDI–TPDI upon guest inclusion. Benefiting from precise SXRD analysis and computational calculations, we have elucidated the dynamic mechanism governing the COF-300 response upon guest inclusion and removal.
Coherent Structural Transformation Upon Adaptive Guest Inclusion within Dynamic Confinement Space in COF-300
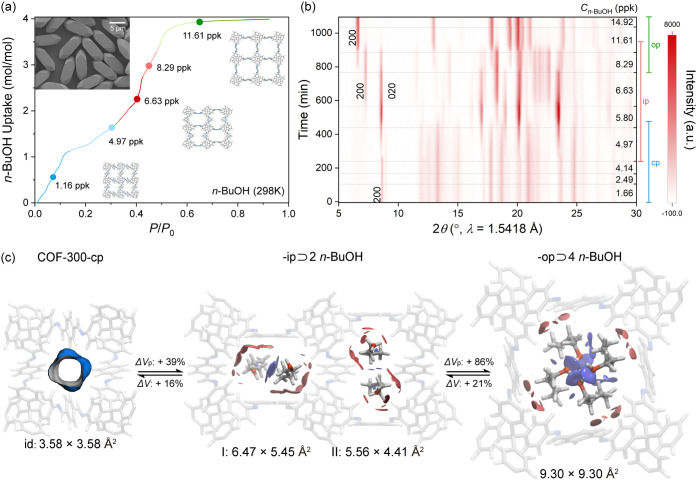
The coherent structural transformation of COF-300 was investigated using COF-300-10-μm crystals with size and morphology control (inset in Figure 4a, Section S1 and Figures S1–S6, S19), whose transition kinetics for a bulk sample has been optimized with the consideration of crystal size effect.3f By combining the organic vapor adsorption isotherm with in situ PXRD under various concentrations of organic vapor (e.g., n-BuOH carried by N2 gas), the host-guest adaptivity can be, therefore, correlated with their anomalous adsorption behavior (Section S5; Figures S60-S63 in the Supporting Information). The n-BuOH (298 K) vapor adsorption isotherm of COF-300 (Figure 4a) shows an initial plateau upon uptake over 1 mol mol-1, followed by approaching 4 mol mol–1. Partial vapor pressures with anomalous adsorption points were therefore selected as representative vapor concentrations (e.g., 1.66, 2.49, 4.14, 4.97, 5.80, 6.63, 8.29, 11.61 and 14.92 ppk) for in situ PXRD patterns reveals multistep coherent structural transformation upon n-BuOH inclusion: (i) at low vapor pressure (–4.14 ‰ n-BuOH), the lattice constants remained unchanged with a peak at 8.73°, attributed to 200 reflection of COF-300-cp (Figure S62 in the Supporting Information); (ii) a detectable shift for a peak at 8.67° attributed to the 020 reflection of COF-300-ip, along with the emergence of new peaks, indicates the onset of structural transformation at 4.97‰ n-BuOH; (iii) a new significant peak at 7.32°, corresponding to the 200 reflection of COF-300-op, marks the transition from COF-300-cp to COF-300-ip at 6.63‰ n-BuOH (upon 2 mol mol–1 uptake); and (iv) the complete transformation is observed at 14.92‰ n-BuOH, with a peak at 6.66° corresponding to the 200 reflection of COF-300-op. Remarkably, the structural transformation is fully reversible upon heating, with the desorption of n-BuOH restoring the pristine COF-300-cp (Figure S63 in the Supporting Information).
Figure 4.
Concerted structural transformation of COF-300 upon adaptive guest inclusion with controlled single crystal size and morphology. (a) Anomalous adsorption behavior of COF-300, correlated with guest-induced structure transformations at representative n-BuOH vapor uptakes at 298 K. Inset, SEM image of the bulk sample of COF-300 single crystals with controlled size and morphology. (b) Contour plot of in situ PXRD patterns of COF-300, tracing its intermediate phase at varying concentrations of n-BuOH vapor. (c) Adaptive guest alignments of n-BuOH within the dynamic confinement space in COF-300 from -cp to -ip showing pore enlargement and geometry diversification for quantitative host–guest (red isosurface) and guest–guest (blue isosurface) interactions (sign(λ2)ρ colored isosurfaces of δginter = 0.005 a.u.). Color codes: C, gray; N, blue; and H, white.
Notably, the d200 or d020 value progressively changes from 10.13 to 12.24 Å and finally to 13.32 Å during the inclusion of n-BuOH, suggesting a stepwise increase in pore volume—39% for COF-300-ip and 86% for -op (Figure 4c), as well as the dynamic confinement space within COF-300 as determined from SXRD analysis (Figures S26–S29; Tables S5–S6 in the Supporting Information). The enlargement of pore diameters from COF-300-cp (3.58 × 3.58 Å2) and COF-300-op (9.30 × 9.30 Å2) is therefore evident with square pore geometry.
Notably, COF-300-ip exhibits symmetry-breaking dynamics, featuring pore geometry deformation into rectangular shapes and diversification of pore environments in the channels I (6.47 × 5.45 Å2) and II (5.56 × 4.41 Å2). IGMH analysis (Figure 4c) indicated that host–guest and guest–guest interaction regions are more prevalent in channel I than in channel II. Upon the adsorption of n-BuOH molecules, the guest alignment undergoes reorganization, and within the COF-300-op, the guest–guest interactions become stronger than the host–guest interactions.
Adaptive Molecular Alignments within the Dynamic Confinement Space of COF-300
Such a dynamic confinement space within COF-300 provides exceptional host–guest adaptivity tolerance for various organic molecules. Thus, we further extended our investigation for other guest molecules of varying sizes, complexities, and functionalities, such as alcohols, phenols, esters, and diols (Figure 5a), leading to diverse adaptive guest alignments within the dynamic confinement spaces of COF-300-ip and -op determined through high-resolution X-ray crystallography (Section S4 in the Supporting Information).
Figure 5.
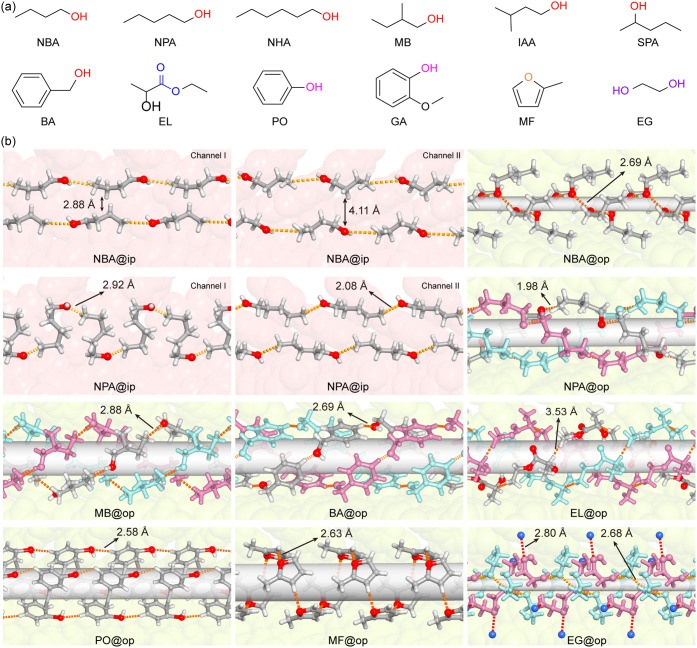
Diverse spatial arrangements of organic molecules within the dynamic confinement space of COF-300. (a) Chemical formulas and abbreviations of representative organic guest molecules (n-Butanol, NBA; n-Pentanol, NPA; n-Hexanol, NHA; 2-Methyl-n-butanol, MB; Isoamyl alcohol, IAA; 2-Pentanol, SPA; Benzyl alcohol, BA; Ethyl lactate, EL; Phenol, PO; Guaiacol, GA; 2-Methylfuran, MF; Ethylene glycol, EG) with functionalities highlighted with colors: red, alcoholic hydroxyl; violet, phenolic hydroxyl; blue, ester group; purple, hydrate glycol. (b) Diverse arrangements of organic guest molecules—such as double linear, herringbone, zigzag chain, triple helix, and tubular structures—within the adaptive channels of COF-300. The channel walls of COF-300-ip and COF-300-op are depicted with red and green space-filling models to represent the distinct confined environments in these phases.
Remarkably, the formation of distinct and intricate alignments of organic guest molecules mainly relies on the partial pressure of organic vapor and their guest–guest and host–guest, noncovalent interactions. Upon 2 mol mol–1 uptake of n-butanol (NBA, Figures S26–S29, and Tables S5–S6 in the Supporting Information), the NBA molecules align head-to-tail to form double linear chains within two types of channels of the COF-300-ip, featuring distinct chain-to-chain distances (2.88 and 4.11 Å within channels I and II, respectively, Figure 5b). Driving by the intermolecular hydrogen bonding of NBA guest molecules, NBA guest molecules rearranged, upon 4 mol mol–1 uptake, to form head-to-head herringbone alignment, having their alkyl chains face toward the pore wall. At the same time, the hydroxy groups aggregated inward within the expanded channels. With larger molecular size, the n-pentanol (NPA) molecules adopt head-to-tail zigzag chain alignment within the contracted channel I of COF-300-ip and further align head-to-tail to form a triple helix fashion within the expanded channel of COF-300-op upon 3 mol mol–1 uptake (Figures S30–S33, S65a,b and Tables S7–S8 in the Supporting Information). Such triple helix alignments can also be observed for n-hexanol (NHA), 2-methyl-n-butanol (MB), isoamyl alcohol (IAA), and 2-phenol (SPA), as well as benzyl alcohol (BA) and ethyl lactate (EL), showing the excellent host–guest adaptivity of varying molecular sizes and varieties (Figures 5b, S34–S47, S65c–h and Tables S9–S15 in the Supporting Information).
Different from aliphatic molecules, adaptive guest alignments of aromatic guest molecules, such as phenol (PO, Figure 5b, S48–S49, S65i–g, and Table S16 in the Supporting Information), are driven by two types of noncovalent interactions—interphenylene π–π interactions and interhydroxyl hydrogen bonding —resulting in a pinwheel tubular arrangement within the expanded channel of COF-300-op. With an additional methoxy group, Guaiacol (GA) forms helix alignment through intermolecular hydrogen bonding by inhibiting interphenylene π–π interactions (Figures S50–S51, and Table S17 in the Supporting Information). Single helix arrangements of 2-methylfuran (MF, Figures 5b, S52–S53, S65k-i, and Table S18 in the Supporting Information) within the expanded channels of COF-300-op were observed, driven by guest–guest hydrogen bonding.
For an aliphatic organic molecule with bifunctionality, intricate alignment of ethylene glycol (EG) was observed (Figure 5b, Figures S54–S56, S65m, and Table S19 in the Supporting Information), driven by guest–guest and host–guest interactions. One type of EG guest molecule (EG-I, colored with cyan, with a torsion angle of OCCO at 31.9°) aligned head-to-tail to form a helix arrangement within the expanded channel of COF-300-op. Host–guest interactions were observed in another type of guest molecule (EG-II, colored purple, with a torsion angle of OCCO at 10.4°) docking on the pore wall through O–H···N hydrogen bonding (O···N, distance: 2.80 Å), with the other, ends attaching to the helix mentioned above with O···O distances of 2.83 and 2.88 Å (Figure S55 in the Supporting Information), respectively. The rotational and translational dynamics in COF-300 enabled such adaptive guest alignments, which is evident by the atomic-level conformational changes of TPM and TPDI, as detailed in Tables S20–S21 in Supporting Information and Figure S57 in Supporting Information. The quantitative energy analyses of host–guest and guest–guest interactions for these adaptive guest alignments are provided in Figure S65 in the Supporting Information.
Dynamic 3D COFs represent a new form of soft porous crystals (SPCs)20 with covalent linkage for a higher level of robustness than crystalline sponges,21 and the adaptive guest alignment exempts the requirement of strong coordination ability compared to the coordinative alignment (CAL) method22 using robust metal–organic frameworks (MOFs). Furthermore, the adaptive guest alignment can work for gas-phase molecules without needing further recrystallization compared with crystal mates through a cocrystal approach.23 By leveraging the guest–guest interactions within the dynamic confinement space, the adaptive guest alignment approach has the potential to unveil the condensed structures of organic liquids, which are usually dynamic and intricate.24
Conclusions
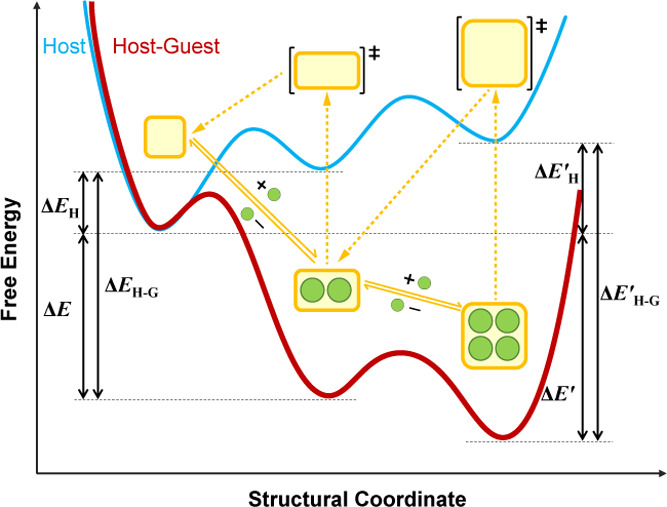
In conclusion, we have uncovered the rotational and translational dynamics of single-crystal dynamic 3D COFs through high-resolution X-ray crystallography, featuring conformation diversity of anti-periplanar, syn-periplanar, and anticlinal forms and switchable interframework π–π interactions. Such exceptional conformational mobility coupled with precise host–guest adaptability allows for adaptive guest alignments within the dynamic confinement space for various guest molecules varying in size, functionality, and complexity. With these precise adaptive guest alignment structures in hand, we are able to perform quantitative analyses of their host–host, host–guest, and guest–guest interactions to portray their energy pathway of structural transformation (Figure S66 and Table S22 in the Supporting Information). Inspired by the observed diverse and intricate alignments, this work is paving the way to robust yet dynamic porous crystals for studying the condensed-state chemistry of liquids at atomic-level precision.24
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 22271189, 92356301, and 21522105), the Science and Technology Commission of Shanghai Municipality (Nos. 21XD1402300, 22QC1401500, 21JC1401700, and 21DZ2260400), and the Double First-Class Initiative Fund of ShanghaiTech University (SYLDX0052022). The authors thank the staffs at the BL17B1 and BL10U2 beamlines from the National Facility for Protein Science in Shanghai and Shanghai Synchrotron Radiation Facility of Shanghai Advanced Research Institute, Chinese Academy of Sciences, respectively, for providing beam time and technical support during X-ray diffraction data collection. The authors thank Dr. Q. Zhang for the support of SEM facilities at CℏEM (#EM02161943); Dr. M. Peng, Dr. N. Yu, and Ms. L. Long at the Analytical Instrumentation Center of the School of Physical Science and Technology (#SPST-AIC10112914) for technical support for XRD, solid-state NMR spectroscopy, and gas adsorption measurements; and the HPC Platform of ShanghaiTech University for theoretical calculation. This work is dedicated to the 100th anniversary of Sun Yat-Sen University (SYSU) and the centennial celebration of its chemistry discipline. We thank Prof. J.-P. Zhang at SYSU for the beneficial discussion on flexibility.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.4c13377.
Synthetic details, single-crystal structural analysis, static gas/vapor adsorption isotherms, theoretical calculation, ssNMR spectroscopy, FT-IR, PXRD, TGA, in situ, and time-dependent PXRD patterns (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- a Yaghi O. M.; Kalmutzki M. J.; Diercks C. S.. Introduction to Reticular Chemistry: Metal–Organic Frameworks and Covalent Organic Frameworks; Wiley-VCH: Weinheim, 2019. [Google Scholar]; b Diercks C. S.; Yaghi O. M. The Atom, the Molecule, and the Covalent Organic Framework. Science 2017, 355 (6328), eaal1585 10.1126/science.aal1585. [DOI] [PubMed] [Google Scholar]
- a Kandambeth S.; Dey K.; Banerjee R. Covalent Organic Frameworks: Chemistry beyond the Structure. J. Am. Chem. Soc. 2019, 141, 1807–1822. 10.1021/jacs.8b10334. [DOI] [PubMed] [Google Scholar]; b Haase F.; Lotsch B. V. Solving the COF Trilemma: Towards Crystalline, Stable and Functional Covalent Organic Frameworks. Chem. Soc. Rev. 2020, 49, 8469–8500. 10.1039/D0CS01027H. [DOI] [PubMed] [Google Scholar]; c Huang N.; Wang P.; Jiang D. Covalent Organic Frameworks: A Materials Platform for Structural and Functional Designs. Nat. Rev. Mater. 2016, 1 (10), 16068. 10.1038/natrevmats.2016.68. [DOI] [Google Scholar]; d Guan X.; Chen F.; Fang Q.; Qiu S. Design and Applications of Three Dimensional Covalent Organic Frameworks. Chem. Soc. Rev. 2020, 49, 1357–1384. 10.1039/C9CS00911F. [DOI] [PubMed] [Google Scholar]; e Gui B.; Lin G.; Ding H.; Gao C.; Mal A.; Wang C. Three-Dimensional Covalent Organic Frameworks: From Topology Design to Applications. Acc. Chem. Res. 2020, 53, 2225–2234. 10.1021/acs.accounts.0c00357. [DOI] [PubMed] [Google Scholar]
- a Uribe-Romo F. J.; Hunt J. R.; Furukawa H.; Klock C.; O’Keeffe M.; Yaghi O. M. A Crystalline Imine-Linked 3-D Porous Covalent Organic Framework. J. Am. Chem. Soc. 2009, 131, 4570–4571. 10.1021/ja8096256. [DOI] [PubMed] [Google Scholar]; b Chen Y.; Shi Z.-L.; Wei L.; Zhou B.; Tan J.; Zhou H.-L.; Zhang Y.-B. Guest-Dependent Dynamics in a 3D Covalent Organic Framework. J. Am. Chem. Soc. 2019, 141, 3298–3303. 10.1021/jacs.8b13691. [DOI] [PubMed] [Google Scholar]; c Sun T.; Wei L.; Chen Y.; Ma Y.; Zhang Y.-B. Atomic-Level Characterization of Dynamics of a 3D Covalent Organic Framework by Cryo-Electron Diffraction Tomography. J. Am. Chem. Soc. 2019, 141, 10962–10966. 10.1021/jacs.9b04895. [DOI] [PubMed] [Google Scholar]; d Chi H.; Liu Y.; Li Z.; Chen W.; He Y. Direct Visual Observation of Pedal Motion-Dependent Flexibility of Single Covalent Organic Frameworks. Nat. Commun. 2023, 14, 5061. 10.1038/s41467-023-40831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Wang M.; Zeng T.; Yu Y.; Wang X.; Zhao Y.; Xi H.; Zhang Y.-B. Flexibility On-Demand: Multivariate 3D Covalent Organic Frameworks. J. Am. Chem. Soc. 2024, 146, 1035–1041. 10.1021/jacs.3c11944. [DOI] [PubMed] [Google Scholar]; f Ma T.; Wei L.; Liang L.; Yin S.; Xu L.; Niu J.; Xue H.; Wang X.; Sun J.; Zhang Y.-B.; Wang W. Diverse crystal size effects in covalent organic frameworks. Nat. Commun. 2020, 11, 6128. 10.1038/s41467-020-19858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zhang Y.-B.; Su J.; Furukawa H.; Yun Y.; Felipe G.; Doung A.; Zou X.; Yaghi O. M. Single-Crystal Structure of a Covalent Organic Framework. J. Am. Chem. Soc. 2013, 135, 16336–16339. 10.1021/ja409033p. [DOI] [PubMed] [Google Scholar]; b Ma Y.-X.; Li Z.-J.; Wei L.; Ding S.-Y.; Zhang Y.-B.; Wang W. A Dynamic Three-Dimensional Covalent Organic Framework. J. Am. Chem. Soc. 2017, 139, 4995–4998. 10.1021/jacs.7b01097. [DOI] [PubMed] [Google Scholar]; c Kang C.; Zhang Z.; Kusaka S.; Negita K.; Usadi A. K.; Calabro D. C.; Baugh L. S.; Wang Y.; Zou X.; Huang Z.; Matsuda R.; Zhao D. Covalent Organic Framework Atropisomers with Multiple Gas-Triggered Structural Flexibilities. Nat. Mater. 2023, 22, 636–643. 10.1038/s41563-023-01523-2. [DOI] [PubMed] [Google Scholar]; d Zeng T.; Ling Y.; Jiang W.; Yao X.; Tao Y.; Liu S.; Liu H.; Yang T.; Wen W.; Jiang S.; Zhao Y.; Ma Y.; Zhang Y.-B. Atomic Observation and Structural Evolution of Covalent Organic Framework Rotamers. Proc. Natl. Acad. Sci. U. S. A. 2024, 121, e2320237121 10.1073/pnas.2320237121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Liu X.; Li J.; Gui B.; Lin G.; Fu Q.; Yin S.; Liu X.; Sun J.; Wang C. A Crystalline Three-Dimensional Covalent Organic Framework with Flexible Building Blocks. J. Am. Chem. Soc. 2021, 143, 2123–2129. 10.1021/jacs.0c12505. [DOI] [PubMed] [Google Scholar]; b Liu X.; Wang Z.; Zhang Y.; Gui B.; Sun J.; Wang C. Gas-Triggered Gate-Opening in a Flexible Three-Dimensional Covalent Organic Framework. J. Am. Chem. Soc. 2024, 146, 11411–11417. 10.1021/jacs.4c01331. [DOI] [PubMed] [Google Scholar]
- a Zhu Q.; Wang X.; Clowes R.; Cui P.; Chen L.; Little M. A.; Cooper A. I. 3D Cage COFs: A Dynamic Three-Dimensional Covalent Organic Framework with High-Connectivity Organic Cage Nodes. J. Am. Chem. Soc. 2020, 142, 16842–16848. 10.1021/jacs.0c07732. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ma J.-X.; Li J.; Chen Y.-F.; Ning R.; Ao Y.-F.; Liu J.-M.; Sun J.; Wang D.-X.; Wang Q.-Q. Cage Based Crystalline Covalent Organic Frameworks. J. Am. Chem. Soc. 2019, 141, 3843–3848. 10.1021/jacs.9b00665. [DOI] [PubMed] [Google Scholar]; c Ji C.; Su K.; Wang W.; Chang J.; El-Sayed M.; Zhang L.; Yuan D. Tunable Cage-Based Three-Dimensional Covalent Organic Frameworks. CCS. Chem. 2022, 4, 3095–3105. 10.31635/ccschem.021.202101453. [DOI] [Google Scholar]
- a Li Y.; Sui J.; Cui L.-S.; Jiang H.-L. Hydrogen Bonding Regulated Flexibility and Disorder in Hydrazone-Linked Covalent Organic Frameworks. J. Am. Chem. Soc. 2023, 145, 1359–1366. 10.1021/jacs.2c11926. [DOI] [PubMed] [Google Scholar]; b Auras F.; Ascherl L.; Bon V.; Vornholt S. M.; Krause S.; Döblinger M.; Bessinger D.; Reuter S.; Chapman K. W.; Kaskel S.; Friend R. H.; Bein T. Dynamic Two-Dimensional Covalent Organic Frameworks. Nat. Chem. 2024, 16, 1373–1380. 10.1038/s41557-024-01527-8. [DOI] [PubMed] [Google Scholar]; c Yi L.; Gao Y.; Luo S.; Wang T.; Deng H. Structure Evolution of 2D Covalent Organic Frameworks Unveiled by Single-Crystal X-ray Diffraction. J. Am. Chem. Soc. 2024, 146, 19643–19648. 10.1021/jacs.4c05705. [DOI] [PubMed] [Google Scholar]
- a Hu Y.; Sengupta B.; Long H.; Wayment L. J.; Ciora R.; Jin Y.; Wu J.; Lei Z.; Friedman K.; Chen H.; Yu M.; Zhang W. Molecular Recognition with Resolution Below 0.2 Angstroms through Thermoregulatory Oscillations in Covalent Organic Frameworks. Science 2024, 384, 1441–1447. 10.1126/science.adj8791. [DOI] [PubMed] [Google Scholar]; b Yin Y.; Zhang Y.; Zhou X.; Gui B.; Wang W.; Jiang W.; Zhang Y.-B.; Sun J.; Wang C. Ultrahigh-Surface Area Covalent Organic Frameworks for Methane Adsorption. Science 2024, 386, 693–696. 10.1126/science.adr0936. [DOI] [PubMed] [Google Scholar]; c Wei L.; Sun T.; Shi Z.; Xu Z.; Wen W.; Jiang S.; Zhao Y.; Ma Y.; Zhang Y.-B. Guest-Adaptive Molecular Sensing in a Dynamic 3D Covalent Organic Framework. Nat. Commun. 2022, 13, 7936. 10.1038/s41467-022-35674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Xu Y.; Sun T.; Zeng T.; Zhang X.; Yao X.; Liu S.; Shi Z.; Wen W.; Zhao Y.; Jiang S.; Ma Y.; Zhang Y.-B. Symmetry-Breaking Dynamics in a Tautomeric 3D Covalent Organic Framework. Nat. Commun. 2023, 14, 4215. 10.1038/s41467-023-39998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Gao C.; Li J.; Yin S.; Sun J.; Wang C. Redox-Triggered Switching in Three-Dimensional Covalent Organic Frameworks. Nat. Commun. 2020, 11, 4919. 10.1038/s41467-020-18588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Fang J.; Fu Z.; Chen X.; Liu Y.; Chen F.; Wang Y.; Li H.; Yusran Y.; Wang K.; Valtchev V.; Qiu S.; Zou B.; Fang Q. Piezochromism in Dynamic Three-Dimensional Covalent Organic Frameworks. Angew. Chem., Int. Ed. 2023, 62, e202304234 10.1002/anie.202304234. [DOI] [PubMed] [Google Scholar]
- a Ma T.; Kapustin E. A.; Yin S. X.; Liang L.; Zhou Z.; Niu J.; Li L.-H.; Wang Y.; Su J.; Li J.; Wang X.; Wang W. D.; Wang W.; Sun J.; Yaghi O. M. Single-Crystal X-Ray Diffraction Structures of Covalent Organic Frameworks. Science 2018, 361, 48–52. 10.1126/science.aat7679. [DOI] [PubMed] [Google Scholar]; b Han J.; Feng J.; Kang J.; Chen J.-M.; Du X.-Y.; Ding S.-Y.; Liang L.; Wang W. Fast Growth of Single-Crystal Covalent Organic Frameworks for Laboratory X-Ray Diffraction. Science 2024, 383 (6686), 1014–1019. 10.1126/science.adk8680. [DOI] [PubMed] [Google Scholar]
- a Kuriyan J.; Konforti B.; Wemmer D.. The Molecules of Life: Physical and Chemical Properties; Garland Science: New York, 2012. [Google Scholar]; b Zwanzig R.; Szabo A.; Bagchi B. Levinthal’s Paradox. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 20–22. 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Dance I.; Scudder M. Supramolecular Motifs: Concerted Multiple Phenyl Embraces Between Ph4P+ Cations Are Attractive and Ubiquitous. Chem.—Eur. J. 1996, 2, 481–486. 10.1002/chem.19960020505. [DOI] [PubMed] [Google Scholar]; b Dance I.; Scudder M. Concerted Supramolecular Motifs: Linear Columns and Zigzag Chains Of Multiple Phenyl Embraces Involving Ph4P+ Cations in Crystals. J. Chem. Soc., Dalton Trans. 1996, 3755–3769. 10.1039/dt9960003755. [DOI] [Google Scholar]
- Heinz K. H.Crystals in Gels and Liesegang Ring; Cambridge University Press: Cambridge, 1988, 1–197. [Google Scholar]
- a Vitaku E.; Dichtel W. R. Synthesis of 2D Imine-Linked Covalent Organic Frameworks through Formal Transimination Reactions. J. Am. Chem. Soc. 2017, 139, 12911–12914. 10.1021/jacs.7b06913. [DOI] [PubMed] [Google Scholar]; b Corbett P. T.; Leclaire J.; Vial L.; West K. R.; Wietor J.-L.; Sanders J. K. M.; Otto S. Dynamic Combinatorial Chemistry. Chem. Rev. 2006, 106, 3652–3711. 10.1021/cr020452p. [DOI] [PubMed] [Google Scholar]; c Wang L.-M.; Yue J.-Y.; Cao X.; Wang D. Insight into the Transimination Process in the Fabrication of Surface Schiff-Based Covalent Organic Frameworks. Langmuir 2019, 35, 6333–6339. 10.1021/acs.langmuir.9b00565. [DOI] [PubMed] [Google Scholar]; d Han X.; Zhang J.; Huang J.; Wu X.; Yuan D.; Liu Y.; Cui Y. Chiral induction in covalent organic frameworks. Nat. Commun. 2018, 9, 1294. 10.1038/s41467-018-03689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Vitaku E.; Gannett C. N.; Carpenter K. L.; Shen L.; Abruña H. D.; Dichtel W. R. Phenazine-Based Covalent Organic Framework Cathode Materials with High Energy and Power Densities. J. Am. Chem. Soc. 2020, 142, 16–20. 10.1021/jacs.9b08147. [DOI] [PubMed] [Google Scholar]; f Zhong H.; Wang M.; Ghorbani-Asl M.; Zhang J.; Ly K. H.; Liao Z.; Chen G.; Wei Y.; Biswal B. P.; Zschech E.; Weidinger I. M.; Krasheninnikov A. V.; Dong R.; Feng X. Boosting the Electrocatalytic Conversion of Nitrogen to Ammonia on Metal-Phthalocyanine-Based Two-Dimensional Conjugated Covalent Organic Frameworks. J. Am. Chem. Soc. 2021, 143, 19992–20000. 10.1021/jacs.1c11158. [DOI] [PubMed] [Google Scholar]; g Zhang Q.; Zhang F.; Dong J.; Shao M.; Zhu M.; Wang D.; Guo Y.; Zhang J.; Liu Y. Controlling the Nucleation Process to Prepare a Family of Crystalline Tribenzimidazole-Based Covalent Organic Frameworks. Chem. Mater. 2022, 34, 6977–6984. 10.1021/acs.chemmater.2c01401. [DOI] [Google Scholar]; h Sprachmann J.; Wachsmuth T.; Bhosale M.; Burmeister D.; Smales G. J.; Schmidt M.; Kochovski Z.; Grabicki N.; Wessling R.; List-Kratochvil E. J. W.; Esser B.; Dumele O. Antiaromatic Covalent Organic Frameworks Based on Dibenzopentalenes. J. Am. Chem. Soc. 2023, 145, 2840–2851. 10.1021/jacs.2c10501. [DOI] [PubMed] [Google Scholar]
- Wang X.; Enomoto R.; Murakami Y. Ionic Additive Strategy to Control Nucleation and Generate Larger Single Crystals of 3D Covalent Organic Frameworks. Chem. Commun. 2021, 57, 6656–6659. 10.1039/D1CC01857D. [DOI] [PubMed] [Google Scholar]
- a Zhou Z.; Zhang L.; Yang Y.; Vitorica-Yrezabal I. J.; Wang H.; Tan F.; Gong L.; Li Y.; Chen P.; Dong X.; Liang Z.; Yang J.; Wang C.; Hong Y.; Qiu Y.; Gölzhäuser A.; Chen X.; Qi H.; Yang S.; Liu W.; Sun J.; Zheng Z. Growth of Single-Crystal Imine-Linked Covalent Organic Frameworks Using Amphiphilic Amino-Acid Derivatives in Water. Nat. Chem. 2023, 15, 841–847. 10.1038/s41557-023-01181-6. [DOI] [PubMed] [Google Scholar]; b Hong Y.; Zhang M.; Zhou Z.; Zhu Z.; Lu J.; Sun J.; Zheng Z. Synthesis and Structure of the Inclusion Compound COF-300-ALA. Chin. J. Chem. 2023, 41, 2957–2962. 10.1002/cjoc.202300393. [DOI] [Google Scholar]; c Zhou Z.; Xiong X.-H.; Zhang L.; Li Y.; Yang Y.; Ding X.; Luo D.; Wei Z.; Liu W.; Su C.-Y.; Sun J.; Zheng Z. Linker-Guided Growth of Single-Crystal Covalent Organic Frameworks. J. Am. Chem. Soc. 2024, 146, 3449–3457. 10.1021/jacs.3c13069. [DOI] [PubMed] [Google Scholar]
- a Yu B.; Lin R.-B.; Xu G.; Fu Z.-H.; Wu H.; Zhou W.; Lu S.; Li Q.-W.; Jin Y.; Li J.-H.; Zhang Z.; Wang H.; Yan Z.; Liu X.; Wang K.; Chen B.; Jiang J. Linkage Conversions in Single-Crystalline Covalent Organic Frameworks. Nat. Chem. 2024, 16, 114–121. 10.1038/s41557-023-01334-7. [DOI] [PubMed] [Google Scholar]; b Yu B.; Li W.; Wang X.; Li J.-H.; Lin R.-B.; Wang H.; Ding X.; Jin Y.; Yang X.; Wu H.; Zhou W.; Zhang J.; Jiang J. Observation of Interpenetrated Topology Isomerism for Covalent Organic Frameworks with Atom-Resolution Single Crystal Structures. J. Am. Chem. Soc. 2023, 145, 25332–25340. 10.1021/jacs.3c09001. [DOI] [PubMed] [Google Scholar]; c Yu B.; Tao Y.; Yao X.; Jin Y.; Liu S.; Xu T.; Wang H.; Wu H.; Zhou W.; Zhou X.; Ding X.; Wang X.; Xiao X.; Zhang Y.-B.; Jiang J. Single-Crystalline 3D Covalent Organic Frameworks with Exceptionally High Specific Surface Areas and Gas Storage Capacities. J. Am. Chem. Soc. 2024, 146, 28932–28940. 10.1021/jacs.4c09680. [DOI] [PubMed] [Google Scholar]
- Lu T.Molclus Program, Version 1.12, http://www.keinsci.com/research/molclus.html (accessed Nov 6, 2024).
- a Lu T.; Chen Q. Independent Gradient Model Based on Hirshfeld Partition: A New Method for Visual Study of Interactions in Chemical Systems. J. Comput. Chem. 2022, 43, 539–555. 10.1002/jcc.26812. [DOI] [PubMed] [Google Scholar]; b Lefebvre C.; Rubez G.; Khartabil H.; Boisson J.-C.; Contreras-García J.; Hénon E. Accurately Extracting the Signature of Intermolecular Interactions Present in The NCI Plot of The Reduced Density Gradient Versus Electron Density. Phys. Chem. Chem. Phys. 2017, 19, 17928–17936. 10.1039/C7CP02110K. [DOI] [PubMed] [Google Scholar]
- Lu T.; Chen F. Multiwfn: A Multifunctional Wavefunction Analyzer. J. Comput. Chem. 2012, 33, 580–592. 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- a Horike S.; Shimomura S.; Kitagawa S. Soft Porous Crystals. Nat. Chem. 2009, 1, 695–704. 10.1038/nchem.444. [DOI] [PubMed] [Google Scholar]; b Schneemann A.; Bon V.; Schwedler I.; Senkovska I.; Kaskel S.; Fischer R. A. Flexible Metal-Organic Frameworks. Chem. Soc. Rev. 2014, 43, 6062–6096. 10.1039/C4CS00101J. [DOI] [PubMed] [Google Scholar]; c Zhang J. P.; Liao P. Q.; Zhou H. L.; Lin R. B.; Chen X. M. Single-Crystal X-Ray Diffraction Studies on Structural Transformations of Porous Coordination Polymers. Chem. Soc. Rev. 2014, 43, 5789–5814. 10.1039/C4CS00129J. [DOI] [PubMed] [Google Scholar]; d Katsoulidis A. P.; Antypov D.; Whitehead G. F. S.; Carrington E. J.; Adams D. J.; Berry N. G.; Darling G. R.; Dyer M. S.; Rosseinsky M. J. Chemical Control of Structure and Guest Uptake by a Conformationally Mobile Porous Material. Nature 2019, 565, 213–217. 10.1038/s41586-018-0820-9. [DOI] [PubMed] [Google Scholar]; e Krause S.; Bon V.; Senkovska I.; Stoeck U.; Wallacher D.; Tobbens D. M.; Zander S.; Pillai R. S.; Maurin G.; Coudert F. X.; Kaskel S. A Pressure-Amplifying Framework Material with Negative Gas Adsorption Transitions. Nature 2016, 532, 348–352. 10.1038/nature17430. [DOI] [PubMed] [Google Scholar]; f Zhou D.-D.; Zhang J.-P. On the Role of Flexibility for Adsorptive Separation. Acc. Chem. Res. 2022, 55, 2966–2977. 10.1021/acs.accounts.2c00418. [DOI] [PubMed] [Google Scholar]
- a Inokuma Y.; Yoshioka S.; Ariyoshi J.; Arai T.; Hitora Y.; Takada K.; Matsunaga S.; Rissanen K.; Fujita M. X-Ray Analysis on The Nanogram to Microgram Scale Using Porous Complexes. Nature 2013, 495, 461–466. 10.1038/nature11990. [DOI] [PubMed] [Google Scholar]; b Yan K.; Dubey R.; Arai T.; Inokuma Y.; Fujita M. Chiral Crystalline Sponges for the Absolute Structure Determination of Chiral Guests. J. Am. Chem. Soc. 2017, 139, 11341–11344. 10.1021/jacs.7b06607. [DOI] [PubMed] [Google Scholar]
- a Lee S.; Kapustin E.; Yaghi O. M. Coordinative Alignment of Molecules in Chiral Metal-Organic Frameworks. Science 2016, 353, 808–811. 10.1126/science.aaf9135. [DOI] [PubMed] [Google Scholar]; b Pei X.; Bürgi H.-B.; Kapustin E. A.; Liu Y.; Yaghi O. M. Coordinative Alignment in the Pores of MOFs for the Structural Determination of N-, S-, and P-Containing Organic Compounds Including Complex Chiral Molecules. J. Am. Chem. Soc. 2019, 141, 18862–18869. 10.1021/jacs.9b10501. [DOI] [PubMed] [Google Scholar]
- Song J.-G.; Zheng J.; Wei R.-J.; Huang Y.-L.; Jiang J.; Ning G.-H.; Wang Y.; Lu W.; Ye W.-C.; Li D. Crystalline Mate for Structure Elucidation of Organic Molecules. Chem. 2024, 10, 924–937. 10.1016/j.chempr.2023.12.004. [DOI] [Google Scholar]
- a Xu R. Towards a New Discipline of Condensed Matter Chemistry. Natl. Sci. Rev. 2018, 5, 1. 10.1093/nsr/nwx155. [DOI] [Google Scholar]; b Xu R.; Wang K.; Chen G.; Yan W. Condensed-Matter Chemistry: From Materials to Living Organisms. Natl. Sci. Rev. 2019, 6, 191–194. 10.1093/nsr/nwy128. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Xu R.; Yu J.; Yan W.. Introduction to Condensed Matter Chemistry; Elsevier, 2024, 1–446. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.