Abstract
Background
Periodontitis is among the most prevalent inflammatory conditions and greatly impacts oral health. This study aimed to elucidate the role of basement membrane-related genes in the pathogenesis and diagnosis of periodontitis.
Methods
GSE10334 was used for identification of hub genes via the differential analysis, protein-protein interaction network, MCC and DMNC algorithms, and evaluation via LASSO regression and support vector machine analysis to identify basement membrane-related markers in patients with periodontitis. Findings were validated by analysis of the GSE16134 dataset and quantitative reverse transcription PCR. The regulatory interplay among lncRNAs, miRNAs, and mRNAs was investigated through multiple databases. Immune infiltration analysis was performed to assess the immune landscape in periodontitis.
Results
ITGA7 was identified as a key gene for periodontitis, as supported by machine learning analysis, validation of expression, and receiver operating characteristic analysis from external datasets. Immune infiltration analysis revealed significant associations between ITGA7 expression and the infiltration of numerous immune cells implicated in periodontitis. Additionally, our findings suggest that the expression of the lncRNA LINC-PINT is significantly increased in periodontitis, and that it can modulate ITGA7 expression through hsa-miR-1293.
Conclusion
ITGA7 is a potential diagnostic and therapeutic target for periodontitis. The LINC-PINT/hsa-miR-1293/ITGA7 axis and the relationship between ITGA7 and immune infiltration provide new insights into the molecular mechanisms underlying periodontitis and highlight potential avenues for clinical intervention.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12903-024-05201-w.
Keywords: Periodontitis, Basement membranes, Immune, Machine learning, Biological marker
Introduction
Periodontitis, the leading cause of tooth loss in adults, is a chronic immunoinflammatory disease affecting both soft and hard oral tissues. It arises from the host’s interaction with bacterial biofilms around the teeth and is influenced by genetic, behavioural, social, and environmental factors [1–4]. The development of periodontitis is closely related to the infection of periodontal tissue with specific bacteria, which can cause inflammation, weaken the patient’s immune system and dysregulate the oral microbiota. Periodontitis destroys the structures supporting the teeth and progressively destroys the alveolar bone and periodontal ligaments, leading to periodontal pocket formation, bone resorption, and progressive attachment loss. Ultimately, oral health is adversely affected, leading to tooth loss [5–9]. Currently, some clinical parameters, such as the probing pocket depth, clinical attachment loss, and the presence and extent of angular bone defects, are the primary factors used to evaluate periodontal disease, determine subsequent treatment protocols and predict prognosis [10]. However, the role of these parameters in early diagnosis and treatment is limited. At the same time, the current treatment of periodontitis mainly focuses on oral hygiene guidance, plaque control and scaling [11]. However, there are also disadvantages, such as frequent administration, poor patient compliance, and systemic adverse reactions, which increase the risk of bacterial resistance and other problems [12].Therefore, the identification of more biomarkers for the diagnosis of periodontitis is helpful for its early diagnosis, and these biomarkers could also be used as therapeutic targets, which is helpful for early intervention and treatment with periodontitis and the prevention of some serious sequelae.
The basement membrane (BM), the earliest extracellular matrix identified in animals, was first observed in skeletal muscle in 1840. The BM is a sheet-like extracellular matrix and is implicated in various diseases, including cancer, congenital muscular dystrophies, and Pierson syndrome [13]. Recent studies highlight the critical role of the BM in the development, organization, and preservation of oral tissues and its function as a barrier against infections [14]. For example, Groeger SE demonstrated the protective role of the BM in oral health [15], and James P Simmer emphasized its importance in dental enamel formation [16]. Additionally, Nurcan Gurses reported a wider distribution of BM-associated proteins in the connective tissue of periodontitis-affected tissues [17]. Recently, advancements in high-throughput genetic microarray technologies have enabled genetic-level analysis of disease onset [18, 19]. For example, Ryan T Demmer identified 61 differentially expressed genes (DEGs) in periodontitis tissues via high-throughput sequencing [20], whereas Hansong Lee discovered an immunological link between periodontitis and type 2 diabetes through single-cell sequencing [21]. Furthermore, Qi Xie used bioinformatics to identify dysregulated BM-related genes, such as MMP2, in periodontitis [22]. Machine learning, as a crucial algorithmic approach, has also been extensively applied in periodontitis research. Protein interaction networks rely heavily on interactomic hub genes, which can be predicted using machine learning. By utilizing network analysis, feature selection, and predictive modeling, machine learning algorithms are able to identify and rank hub genes according to their possible significance in biological pathways. Pradeep Kumar Yadalam utilized machine learning to identify key genes involved in periodontitis, offering new insights for further investigation [23]. Recently, Haoran Yang employed machine learning to identify nine key genes associated with mitochondrial extracellular vesicles with high diagnostic value, demonstrating their strong correlation with immune cell infiltration [24]. Despite these advancements, the potential contributions of numerous genes to the diagnosis and management of periodontitis remain unclear.
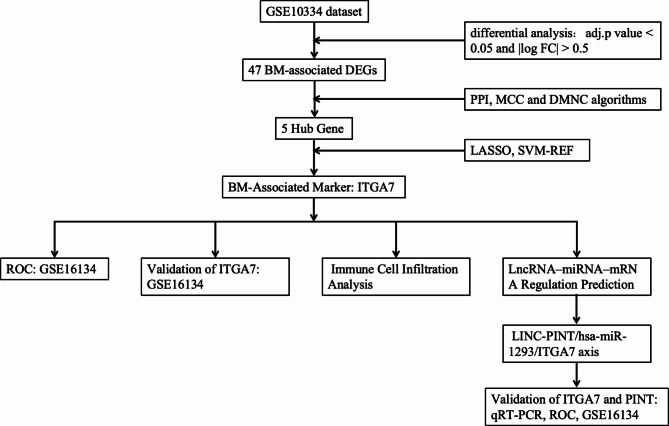
To enhance our understanding and improve the diagnostic and therapeutic approaches for periodontitis, this study aimed to explore the relationships between BM genes and periodontitis. The dataset GSE10334 was used for analysis in this study. The expression and diagnostic utility of these genes were validated by analysis of the validation dataset GSE16134 and by quantitative reverse transcription PCR (qRT‒PCR) (Fig. 1).
Fig. 1.
The flowchart of this study
Materials and methods
Identification of BM-associated DEGs in periodontitis
The GSE10334 dataset (platform: GPL570, Affymetrix Human Genome U133 Plus 2.0 Array) was obtained from the Gene Expression Omnibus (GEO) database and contains 247 gingival tissue samples (183 diseased sites and 64 healthy sites) from 90 patients (63 patients with chronic periodontitis and 27 patients with aggressive periodontitis). The 2017 classification of periodontal and peri-implant diseases and conditions introduced substantial changes, categorizing periodontitis into three main types: necrotizing periodontitis, periodontitis as a manifestation of systemic disease, and periodontitis. The forms of the disease previously recognized as ‘chronic’ or ‘aggressive’ are now grouped under a single category, ‘periodontitis,’ according to the new classification technique [4, 25]. Therefore, this study analyzed the two types of patients in this dataset together.
The datasets were retrieved via the GEOquery package (version 2.64.2) [26] and statistical analysis was performed with R (version 4.2.1). Probes corresponding to multiple molecules were eliminated, retaining only the probe with the highest signal value. The data were normalized using the normalizeBetweenArrays function of the limma package (version 3.52.2) [27]. Differential analysis between the two groups was conducted using the limma package, and the results were visualized with volcano plots and heatmaps.
The significance threshold was set at an adjusted p value < 0.05 and |log FC| > 0.5 for generating the volcano plot, which was performed via the ggplot2 package (version 2.3.6). Genes associated with the BM were identified from prior research [28], and BM-associated DEGs were identified through Venn diagrams. The BM-associated DEGs were visualized in heatmaps generated with the ComplexHeatmap package (version 2.13.1) [29].
Hub gene determination
The DEGs related to the BM were used to construct a protein‒protein interaction (PPI) network via the STRING online analysis tool (https://string-db.org/) with a minimum required interaction score of 0.400 [30]. Hub genes were identified using the MCC and DMNC algorithms in the cytoHubba plugin of Cytoscape (version 3.9.1), and the top 10 hub genes were determined.
Identification of BM-associated markers
Two types of machine learning were used to analyze the BM-Associated DEGs obtained previously, and the prediction results were intersected with the hub genes to determine the final BM-Associated Markers. Using the R glmnet package, the power of genes to distinguish periodontitis samples from healthy samples was assessed using the LASSO regression approach with the optimal lambda identified. Tenfold cross-validation validated the parameter selection to ensure the partial likelihood deviation met the minimum criteria. Support vector machine (SVM) analysis, combined with recursive feature elimination (RFE), was used to filter optimal genes. Significant potential BM-associated markers were further evaluated using the GSE16134 dataset (Appendix 1).
Receiver operating characteristic (ROC) analysis
Following the acquisition of the GSE16134 dataset from the GEO database, a logistic model was constructed using the R glm function. Analysis was performed with the pROC package, and results were visualized using the ggplot2 package.
Immune cell infiltration analysis
Based on the ssGSEA algorithm in the R package -GSVA [31], the markers for 24 discrete types of immune cells reported in an article published in Immunity were used to calculate immune infiltration scores [32]. Then, the Wilcoxon rank–sum test was used for statistical analysis of immune cell infiltration in the disease group and the healthy group, and the data were visualized with the ggplot2 package.
LncRNA‒miRNA‒mRNA regulation prediction
Five databases (TargetMiner, miRDB, RNAInter, miRWalk, and RNA22) were used to predict miRNAs that were related to the candidate diagnostic markers. The final miRNAs were identified by comparing those that interacted with each marker. From the HGNC database, we obtained 9090 lncRNAs (retrieved on October 5, 2023), and 19 differentially expressed lncRNAs were identified in the GSE10334 dataset (|logFC| > 0.5, adj. P < 0.05). The DIANA database (https://diana.e-ce.uth.gr/lncbasev3) was used to predict the relationship between differentially expressed lncRNAs and miRNAs [33]. The data were visualized using Cytoscape (version 3.9.1).
In vitro cell experiment
Human gingival fibroblasts (HGF-1, ATCC, USA, #CRL-2014) were cultured in DMEM supplemented with 10% foetal bovine serum (Gibco, USA). HGF-1 cells were seeded in plastic culture dishes and incubated at 37 °C with 5% CO2. At 80% confluence, cells were subcultured and divided into control and experimental group. The cells in the experimental group were processed with Porphyromonas gingivalis LPS (InvivoGen, USA) at a final concentration of 10 µg/mL for 12 h, while the cells in the control group were treated with an equal volume of PBS [34].
qRT‒PCR
Total RNA was extracted using TRIzol. cDNA was synthesized by reverse transcription at 95 °C for 5 min followed by incubation at 37 °C for 15 min. qRT‒PCR was performed for 5 min at 95 °C, followed by an annealing stage of 40 cycles for 10 s at 95 °C and 30 s at 60 °C. Relative gene expression was normalized to that of ACTB (anti-actin) and calculated via the 2−△△CT method. The primer sequences used are listed in Table 1.
Table 1.
Specific sequences of the primers used in the polymerase chain reaction (PCR) assay
| Gene | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| ACTB | GTTGTCGACGACGAGCG | GCACAGAGCCTCGCCTT |
| ITGA7 | TTGAGCTGCCACTGTCCATTGC | CCGTGACCTCATACTTGACCTTGC |
| PINT | TGCCATCTGGATTTCTCTGCC | TGTTCAGTGGTTTATTCTGCTTCA |
Statistical analysis
Statistical comparisons between periodontitis and control specimens were performed using the student’s t test in R. Differences were considered statistically significant at ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, or ∗p < 0.05.
Results
Identification of DEGs
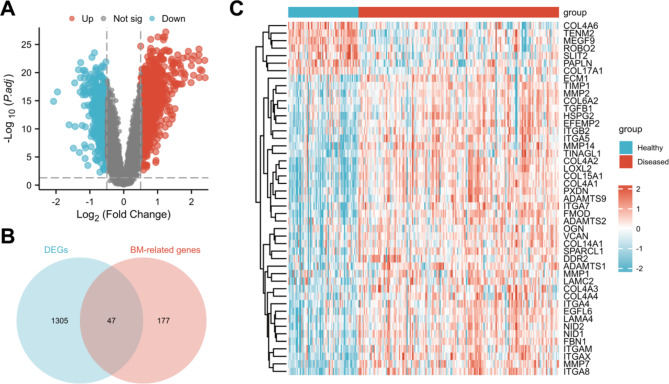
BM-associated DEGs of periodontitis patients and healthy controls were identified using the GSE10334 dataset. A total of 813 genes were significantly upregulated (log FC > 0.5), and 540 genes were significantly downregulated (log FC < -0.5) in patients with periodontitis (Fig. 2A). A Venn diagram was generated to determine the intersection between the BM-related genes and the DEGs, and 47 BM-associated DEGs were identified (Fig. 2B). Furthermore, a heatmap of these genes was generated. The results showed that 6 genes were downregulated and 41 genes were upregulated in the disease group (Fig. 2C).
Fig. 2.
A: A total of 813 genes were significantly upregulated (log FC > 0.5, red) and 540 genes were significantly downregulated (log FC < -0.5, blue) in patients with periodontitis. B: A total of 47 BM-associated DEGs were identified by taking the intersection of the BM-related genes and the DEGs. C: Heatmap of the 47 BM-associated DEGs
Hub gene determination
A PPI network for the 47 BM-associated DEGs was constructed using the STRING database (Fig. 3A). The top 10 hub genes were identified using the MCC and DMNC algorithms (Fig. 3B-C). Finally, 5 hub genes, COL4A6, ITGA7, COL4A4, LAMA4, and ITGA8, were identified by taking the intersection of the genes identified by the two algorithms (Fig. 3D).
Fig. 3.
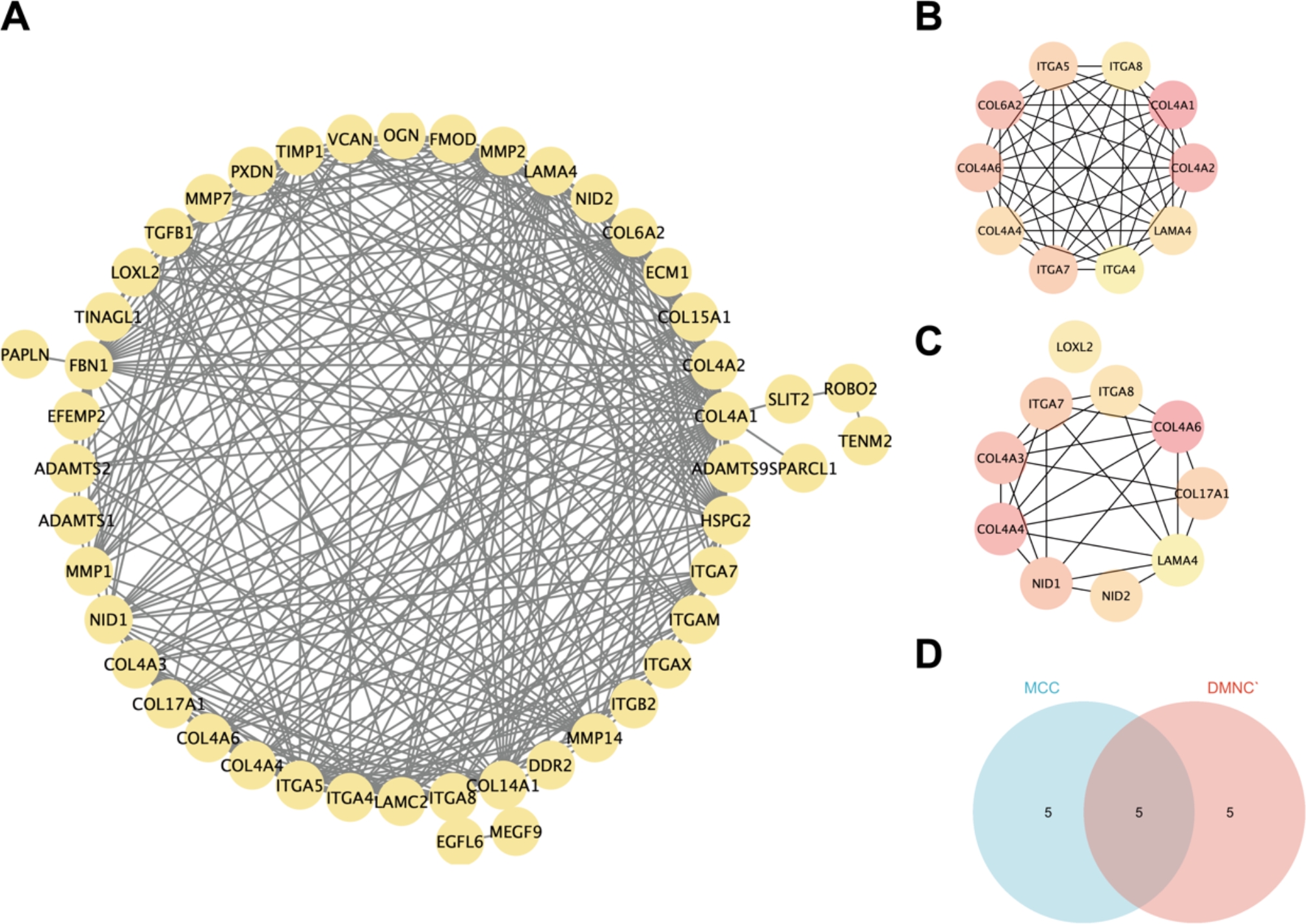
A: PPI network relationships of the 47 BM-associated DEGs. B: The top 10 BM-associated DEGs identified with the MCC algorithm. C: The top 10 BM-associated DEGs identified by the DMNC algorithm. D: The intersection of the results of the MMC and DMNC algorithms was determined to identify five hub genes
Identification of a BM-associated marker for periodontitis
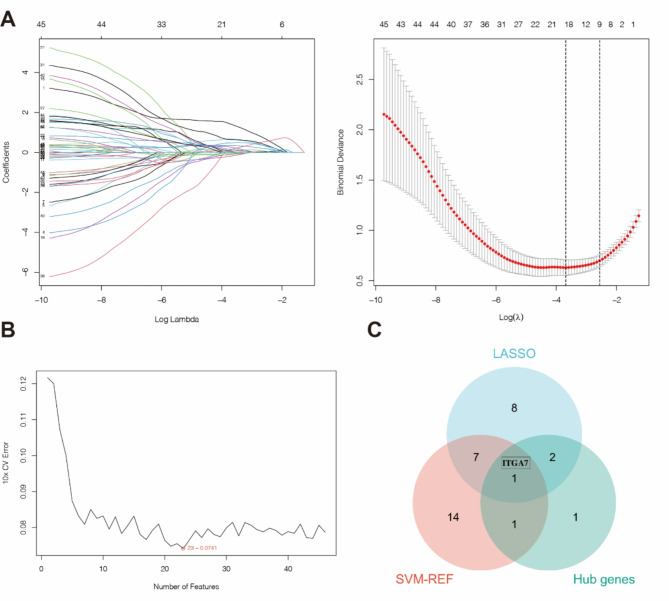
Based on the 47 BM-associated DEGs, important BM-associated genes were identified using LASSO regression analysis and the SVM-RFE method. A total of 17 important BM-associated genes were identified by LASSO regression analysis (Fig. 4A), and 23 important BM-associated genes were identified with the SVM-RFE algorithm (Fig. 4B). The final BM-associated marker, ITGA7, was identified by comparing these genes with the 5 hub genes (Fig. 4C).
Fig. 4.
A: LASSO regression analysis was used to identify 18 important BM-associated genes with the minimum error (left line in the right panel of Fig. 3A). (The numbers on the top horizontal axis in both graphs in Fig. 3A are the numbers of nonzero coefficients corresponding to this λ in the model.) B: The SVM–RFE algorithm was used to identify 23 important BM-associated genes. C: The genes identified by LASSO regression analysis and SVM-REF analysis were intersected with the hub genes to identify the BM-associated marker, ITGA7
Validation of the BM-associated marker
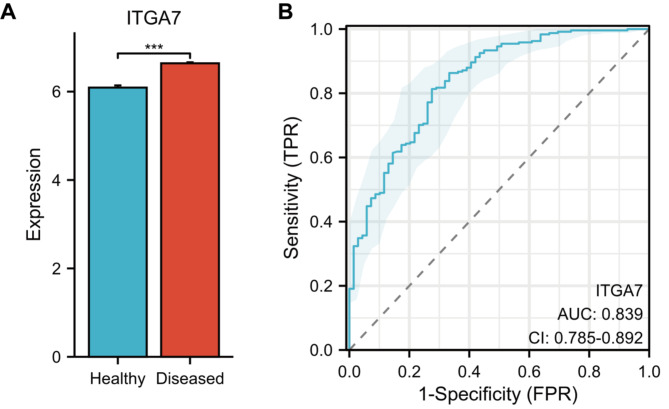
The expression of ITGA7 was validated using the GSE16134 dataset, and the results showed that ITGA7 expression was significantly higher in samples from periodontitis patients than healthy individuals (Fig. 5A). ROC analysis also demonstrated its high diagnostic value (Fig. 5B).
Fig. 5.
A: Validated the expression of ITGA7 in samples from the GSE16134 dataset. B: The ROC curve showed that ITGA7 had a high diagnostic value
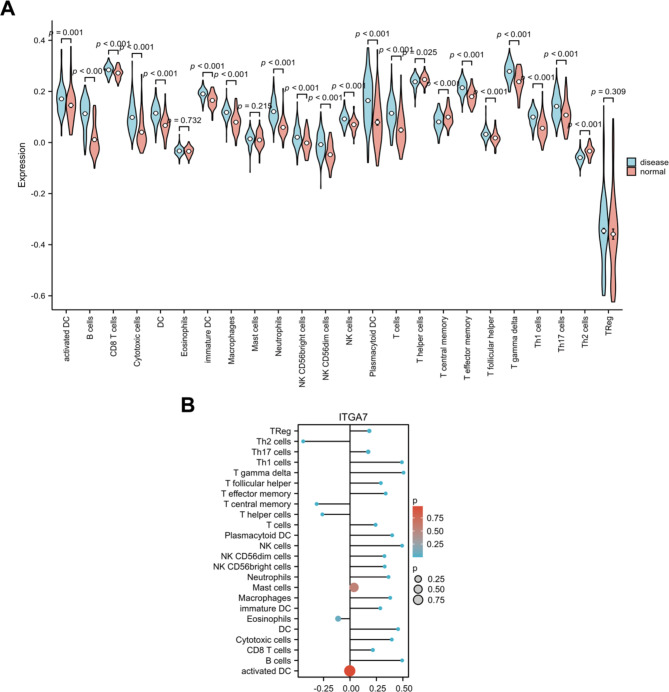
Immune infiltration analysis
The research also investigated the role of immune infiltration in periodontitis using GSEA. The analysis revealed significant differences in the abundances of various types of immune cells. Among them, activated DCs, B cells, CD8 + T cells, cytotoxic cells, DCs, immature DCs, macrophages, neutrophils, CD56bright NK cells, CD56dim NK cells, NK cells, plasmacytoid DCs, T cells, T effector memory cells, T follicular helper cells, T gamma delta cells, Th1 cells and Th17 cells were more abundant in the disease group, while T helper cells, T central memory and Th2 cells were less abundant in the disease group (Fig. 6A). Moreover, our results showed strong associations between ITGA7 expression and immune cell infiltration. Specifically, the infiltration of B cells, CD8 + T cells, cytotoxic cells, DCs, immature DCs, macrophages, neutrophils, CD56bright NK cells, CD56dim NK cells, NK cells, plasmacytoid DCs, T cells, T effector memory cells, T follicular helper cells, T gamma delta cells, Th1 cells, Th17 cells and Tregs was significantly positively correlated with ITGA7 expression, while the infiltration of T helper cells, T central memory cells and Th2 cells was significantly negatively correlated with ITGA7 expression. However, there was no statistically significant correlation between the infiltration of activated DCs, eosinophils, or mast cells and the expression of ITGA7 (Fig. 6B). The results showed a surprising association among ITGA7, immune cells and disease. When ITGA7 was upregulated, the abundances of positively related immune cells were increased in the disease group, and the abundances of negatively related immune cells were decreased in the disease group. Therefore, ITGA7 may be involved in the occurrence and development of periodontitis by altering immune cell infiltration.
Fig. 6.
A: Differences in immune infiltration between the disease and control groups. B: Correlations between ITGA7 expression and immune cell infiltration
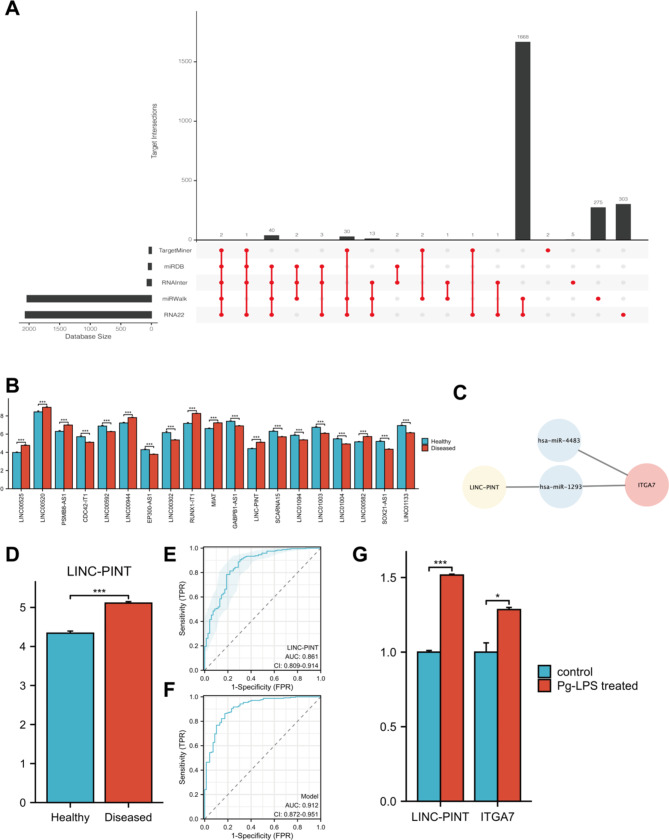
LncRNA‒miRNA‒mRNA regulation
ITGA7-related miRNAs were predicted using five databases, leasing to the identification of two miRNAs (Fig. 7A). From the GSE10334 dataset, 19 differentially expressed lncRNAs were identified between the diseased and healthy groups (Fig. 7B). The regulatory relationship between LINC-PINT, hsa-miR-1293 and ITGA7 was established using the DIANA database (Fig. 7C). Analysis of the GSE16134 dataset showed elevated expression of LINC-PINT in periodontitis samples (Fig. 7D). ROC analysis indicated that LINC-PINT has high diagnostic value (Fig. 7E). Using both ITGA7 and LINC-PINT enhanced diagnostic accuracy compared to that of ITGA7 or LINC-PINT alone (Fig. 7F). Cell experiments further confirmed the upregulation of LINC-PINT and ITGA7 in the experimental group, consistent with our data analysis (Fig. 7G).
Fig. 7.
A: 2 ITGA7-relevated miRNAs were predicted using five databases. B: The expression of 19 differentially expressed lncRNAs in GSE10334. C: The LncRNA‒miRNA‒mRNA network predicted that LINC-PINT/hsa-miR-1293/ITGA7 had a regulatory relationship. D: LINC-PINT expression in GSE16134. E: ROC of LINC-PINT. F: ROC of ITGA7 and LINC-PINT as a shared indication. G: The expression of LINC-PINT and ITGA7 in the experimental group
Discussion
The BM-associated marker, ITGA7, was identified by differential expression analysis, PPI network analysis, the identification of hub genes via the MCC and DMNC algorithms, and evaluation by LASSO regression and support vector machine analysis. In addition, an unexpected association among disease, immune cell infiltration, and ITGA7 expression was identified. When ITGA7 expression was upregulated, the abundances of positively related immune cells were increased and the abundances of negatively related immune cells were decreased in the disease group. This pattern suggests that ITGA7 may affect the occurrence and development of diseases by regulating the infiltration of immune cells. Then, using the lncRNA‒miRNA‒mRNA network, we found that LINC-PINT was highly likely to upregulate ITGA7 expression through hsa-miR-1293, thereby affecting the development and progression of periodontitis. Finally, the diagnostic value of LINC-PINT and ITGA7 was verified by ROC curve analysis, and their expression levels were validated in an in vitro cell experiment. The findings suggested that the BM-associated marker ITGA7 plays an important role in periodontitis and is a potential diagnostic and therapeutic target.
ITGA7 encodes integrin alpha 7, a protein that mediates cell-matrix and cell‒cell interactions, influencing cell morphology, migration, differentiation, and invasion. This protein acts as a receptor for the BM protein laminin 1 and is mainly expressed in cardiac and skeletal muscles, playing a role in myogenesis [35–38]. ITGA7 has been associated with congenital muscular dystrophy and congenital myopathy [39, 40]. It has been shown that myogenic differentiation is regulated by integrin α7β1 signaling [41]. We found that the expression of ITGA7 was significantly upregulated in periodontitis, consistent with the findings of Ulvi K. Gürsoy [42]. Additionally, immunosuppressive drugs, linked to gingival overgrowth, have been shown to upregulate ITGA7 expression in fibroblasts [43]. Lauritano demonstrated that amlodipine exacerbates fibrotic responses and gingival overgrowth via ITGA7 [44]. Li found that p75NTR + cells with enhanced osteogenic capacity involve ITGA7 as a receptor for laminin [45]. Meanwhile, LINC-PINT is associated with various diseases, including chronic obstructive pulmonary disease and colorectal cancer [46, 47]. Although its role in periodontitis is not well understood, our study showed that LINC-PINT may upregulates ITGA7 through hsa-miR-1293, impacting periodontitis progression via the BM. In general, the components of the LINC-PINT/hsa-miR-1293/ITGA7 axis, especially ITGA7, are potentially important diagnostic and therapeutic targets for periodontitis. Further research is needed to elucidate their specific regulatory mechanisms in periodontitis.
Periodontitis is closely linked to immune responses [8, 48, 49]. Immune infiltration analysis revealed that the infiltration of B cells, CD8 + T cells, cytotoxic cells, DCs, immature DCs, macrophages, neutrophils, CD56bright NK cells, CD56dim NK cells, NK cells, plasmacytoid DCs, T cells, T effector memory cells, T follicular helper cells, T gamma delta cells, Th1 cells, Th17 cells and Tregs all was significantly positively correlated with the expression of ITGA7, while the infiltration of T helper cells, T central memory cells and Th2 cells was significantly negatively correlated with ITGA7 expression. Accumulating studies have shown that excessive activation of the immune response can activate osteoclasts and lead to alveolar bone loss. The inflammatory factors produced by Th17 cells play an important role in the occurrence and development of periodontitis [50–53]. However, whether Th17 cells and Th17 cell-secreted IL-17 play a promoting or inhibitory role in the development of periodontitis remains controversial. IL-17 has anti-infective properties and can function as an immunological surveillance factor. However, because IL-17 can play a direct role in Rank/Rankl pathway-mediated periodontal bone loss, Th17 cells can contribute to bone degradation in the context of periodontitis in addition to their function in mucosal immunity [54]. Our study showed that ITGA7 expression is significantly positively correlated with Th17 cell infiltration. Therefore, the upregulation of ITGA7 may promote the infiltration of Th17 cells and aggravate periodontitis. This hypothesis is consistent with our observation that Th17 cells were significantly more abundant in the periodontitis group than in the control group. Moreover, some studies have shown that B cells play a dual role in the development and progression of periodontitis [55–57]. B cells can play a protective role by promoting the clearance of bacteria and preventing the further development of periodontitis [56]. B cells can also mediate the destruction of alveolar bone by supporting the differentiation of osteoclasts, further exacerbating periodontitis [55]. However, the role of B cells in periodontal progression remains incompletely understood. Our study showed that the expression of the BM-associated marker ITGA7 was significantly associated with B cell infiltration, suggesting its potential role in regulating the function of B cells in periodontitis. However, the detailed mechanism by which ITGA7 regulates the progression of periodontitis by regulating the function of B cells requires further investigation. Moreover, our findings revealed an unexpected correlation between disease, immune cells, and ITGA7. When ITGA7 was upregulated, the abundances of positively related immune cells were increased and the abundances of negatively related immune cells were decreased in the disease group. Overall, our findings indicated that ITGA7 expression was positively correlated with the infiltration of the majority of immune cells, suggesting that ITGA7 may play an important role in the immune processes associated with periodontitis.
The most important finding of this study is the identification of an important BM-associated marker, ITGA7, and its related regulatory axis (LINC-PINT/hsa-miR-1293/ITGA7) and relationship with immune cells. ITGA7 is a potential diagnostic and therapeutic target for periodontitis. Moreover, our findings provide new insights for studying the mechanisms underlying the occurrence and development of periodontitis.
However, this study has limitations. First, the analysis used data from online databases that lacked clinical information necessary for analyses such as site severity. Second, this retrospective study requires prospective studies to confirm our findings. Third, further verification of ITGA7, LINC-PINT, and hsa-miR-1293 expression and regulatory relationships in human tissues is needed. For example, the gingival tissues of periodontitis patients and healthy individuals could be obtained and the expression of related genes evaluated by qRT-PCR. In addition, the expression of hsa-miR-1293 and ITGA7 and the severity of periodontitis could be evaluated by silencing or knocking down LINC-PINT in mouse and cell models of periodontitis and comparing the control group with the model group. Finally, the mechanism by which ITGA7 influences local immune cell infiltration and periodontitis progression, including the role of LINC-PINT via hsa-miR-1293, requires further study.
Conclusion
Here, we identified ITGA7, a BM-associated marker, as a potential diagnostic and therapeutic target for periodontitis. We also showed that ITGA7 expression a is closely related to immune cell infiltration. Moreover, we further discovered the regulatory role of the LINC-PINT/hsa-miR-1293/ITGA7 axis. These insights offer a new perspective on the diagnosis and treatment of periodontitis, emphasizing the importance of BM-related genes and their regulatory networks in the disease’s pathogenesis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- BM
Basement membrane
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- ROC
Receiver operating characteristic
- DEGs
Differentially expressed genes
- PPI
Protein-protein interaction network
- GEO
Gene Expression Omnibus
- BP
Biological process
- CC
Cellular component
- MF
Molecular function
- MCC
Maximal clique centrality
- DMNC
Density of Maximum Neighbourhood Component
- SVM
Support vector machine
- LASSO
Least-Absolute Shrinkage and Selection Operator
- qRT-PCR
Quantitative reverse transcription PCR
Author contributions
YH and ZZ conducted the study design. YH and GX performed the data analysis. YH, GX, MY and HS performed chart production. YH and ZZ wrote the main manuscript text. All authors reviewed the manuscript.
Funding
None.
Data availability
The datasets generated and analyzed during the current study are available in the GEO repository(https://www.ncbi.nlm.nih.gov/geo/).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Montenegro SCL, Retamal-Valdes B, Bueno-Silva B, Duarte PM, Faveri M, Figueiredo LC, et al. Do patients with aggressive and chronic periodontitis exhibit specific differences in the subgingival microbial composition? A systematic review. J Periodontol. 2020;91(11):1503–20. [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee S, Chopra A, Karmakar S, Bhat SG. Periodontitis increases the risk of gastrointestinal dysfunction: an update on the plausible pathogenic molecuar mechanisms. Crit Rev Microbiol. 2024;1–31. https://www.tandfonline.com/doi/full/10.1080/1040841X.2024.2339260. [DOI] [PubMed]
- 3.Augimeri G, Caparello G, Caputo I, Reda R, Testarelli L, Bonofiglio D. Mediterranean diet: a potential player in the link between oral microbiome and oral diseases. J Oral Microbiol. 2024;16(1):2329474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papapanou PN, Sanz M, Buduneli N, Dietrich T, Feres M, Fine DH, et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World workshop on the classification of Periodontal and Peri-implant diseases and conditions. J Periodontol. 2018;89(Suppl 1):S173–82. [DOI] [PubMed] [Google Scholar]
- 5.Pardo A, Fiorini V, Zangani A, Faccioni P, Signoriello A, Albanese M et al. Topical agents in Biofilm disaggregation: a systematic review and Meta-analysis. J Clin Med. 2024;13(8). [DOI] [PMC free article] [PubMed]
- 6.Ray RR. Periodontitis: an oral disease with severe consequences. Appl Biochem Biotechnol. 2023;195(1):17–32. [DOI] [PubMed] [Google Scholar]
- 7.Gopinath D, Kunnath Menon R, George Botelho SKV, Johnson M. NW. Periodontal diseases as putative risk factors for Head and Neck Cancer: systematic review and Meta-analysis. Cancers (Basel). 2020;12(7). [DOI] [PMC free article] [PubMed]
- 8.Pan S, Li Y, He H, Cheng S, Li J, Pathak JL. Identification of ferroptosis, necroptosis, and pyroptosis-associated genes in periodontitis-affected human periodontal tissue using integrated bioinformatic analysis. Front Pharmacol. 2022;13:1098851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arigbede AO, Babatope BO, Bamidele MK. Periodontitis and systemic diseases: a literature review. J Indian Soc Periodontol. 2012;16(4):487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosin-Villanueva M, Alminana-Pastor PJ, Garcia-Gimenez JL, Lopez-Roldan A. Study of microRNAs in Gingival Crevicular Fluid as Periodontal diseases biomarkers: systematic review. Int J Mol Sci. 2024;25(15). [DOI] [PMC free article] [PubMed]
- 11.Kucukturkmen B, Oz UC, Toptas M, Devrim B, Saka OM, Bilgili H, et al. Development of Zoledronic Acid containing biomaterials for enhanced guided bone regeneration. J Pharm Sci. 2021;110(9):3200–7. [DOI] [PubMed] [Google Scholar]
- 12.Zeng J, Mamitimin M, Song Y, Sun WB, Wu ZH, Qi XL. Chairside administrated antibacterial hydrogels containing berberine as dental temporary stopping for alveolar ridge preservation. Eur Polym J. 2021;160.
- 13.Pozzi A, Yurchenco PD, Iozzo RV. The nature and biology of basement membranes. Matrix Biol. 2017;57–58:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marre ATO, Domingues R, Lobo LA. Adhesion of anaerobic periodontal pathogens to extracellular matrix proteins. Braz J Microbiol. 2020;51(4):1483–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groeger SE, Meyle J. Epithelial barrier and oral bacterial infection. Periodontol 2000. 2015;69(1):46–67. [DOI] [PubMed] [Google Scholar]
- 16.Simmer JP, Hu JC, Hu Y, Zhang S, Liang T, Wang SK, et al. A genetic model for the secretory stage of dental enamel formation. J Struct Biol. 2021;213(4):107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurses N, Thorup AK, Reibel J, Carter WG, Holmstrup P. Expression of VLA-integrins and their related basement membrane ligands in gingiva from patients of various periodontitis categories. J Clin Periodontol. 1999;26(4):217–24. [DOI] [PubMed] [Google Scholar]
- 18.Parola C, Neumeier D, Reddy ST. Integrating high-throughput screening and sequencing for monoclonal antibody discovery and engineering. Immunology. 2018;153(1):31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14(6):671–83. [DOI] [PubMed] [Google Scholar]
- 20.Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79(11):2112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Joo JY, Song JM, Kim HJ, Kim YH, Park HR. Immunological link between periodontitis and type 2 diabetes deciphered by single-cell RNA analysis. Clin Transl Med. 2023;13(12):e1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Q, Lv H, Wang T, Sun J, Li Y, Niu Y, et al. Identifying common genes and pathways Associated with Periodontitis and Aging by Bioinformatics Analysis. Dis Markers. 2022;2022:4199440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadalam PK, Arumuganainar D, Ronsivalle V, Di Blasio M, Badnjevic A, Marrapodi MM, et al. Prediction of interactomic hub genes in PBMC cells in type 2 diabetes mellitus, dyslipidemia, and periodontitis. BMC Oral Health. 2024;24(1):385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, Zhao A, Chen Y, Cheng T, Zhou J, Li Z. Exploring the potential link between MitoEVs and the immune microenvironment of periodontitis based on machine learning and bioinformatics methods. BMC Oral Health. 2024;24(1):169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caton JG, Armitage G, Berglundh T, Chapple ILC, Jepsen S, Kornman KS, et al. A new classification scheme for periodontal and peri-implant diseases and conditions - introduction and key changes from the 1999 classification. J Periodontol. 2018;89(Suppl 1):S1–8. [DOI] [PubMed] [Google Scholar]
- 26.Davis S, Meltzer PS. GEOquery: a bridge between the Gene expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–7. [DOI] [PubMed] [Google Scholar]
- 27.Dicken SJ, Batterham RL. The role of Diet Quality in Mediating the Association between Ultra-processed Food Intake, obesity and health-related outcomes: a review of prospective cohort studies. Nutrients. 2021;14(1). [DOI] [PMC free article] [PubMed]
- 28.Jayadev R, Morais M, Ellingford JM, Srinivasan S, Naylor RW, Lawless C, et al. A basement membrane discovery pipeline uncovers network complexity, regulators, and human disease associations. Sci Adv. 2022;8(20):eabn2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. [DOI] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–95. [DOI] [PubMed] [Google Scholar]
- 33.Karagkouni D, Paraskevopoulou MD, Tastsoglou S, Skoufos G, Karavangeli A, Pierros V, et al. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020;48(D1):D101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiroch J, Dunkel A, Sterneder S, Zehentner S, Behrens M, Di Pizio A, et al. Human gingival fibroblasts as a Novel Cell Model describing the association between bitter taste thresholds and Interleukin-6 release. J Agric Food Chem. 2023;71(13):5314–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flintoff-Dye NL, Welser J, Rooney J, Scowen P, Tamowski S, Hatton W, et al. Role for the alpha7beta1 integrin in vascular development and integrity. Dev Dyn. 2005;234(1):11–21. [DOI] [PubMed] [Google Scholar]
- 36.Wu Z, Kong X, Wang Z. Integrin alpha7 knockdown suppresses cell proliferation, migration, invasion and EMT in hepatocellular carcinoma. Exp Ther Med. 2021;21(4):309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10(1):9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heller KN, Montgomery CL, Shontz KM, Clark KR, Mendell JR, Rodino-Klapac LR. Human alpha7 integrin gene (ITGA7) delivered by Adeno-Associated Virus extends survival of severely affected Dystrophin/Utrophin-Deficient mice. Hum Gene Ther. 2015;26(10):647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42(10):737–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esposito T, Sampaolo S, Limongelli G, Varone A, Formicola D, Diodato D, et al. Digenic mutational inheritance of the integrin alpha 7 and the myosin heavy chain 7B genes causes congenital myopathy with left ventricular non-compact cardiomyopathy. Orphanet J Rare Dis. 2013;8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClure MJ, Clark NM, Hyzy SL, Chalfant CE, Olivares-Navarrete R, Boyan BD, et al. Role of integrin alpha7beta1 signaling in myoblast differentiation on aligned polydioxanone scaffolds. Acta Biomater. 2016;39:44–54. [DOI] [PubMed] [Google Scholar]
- 42.Gursoy UK, Zeidan-Chulia F, Yilmaz D, Ozdemir V, Maki-Petays J, Neves de Oliveira BH, et al. Analyses of Gingival Adhesion Molecules in Periodontitis: theoretical in Silico, comparative in vivo, and explanatory in Vitro models. J Periodontol. 2016;87(2):193–202. [DOI] [PubMed] [Google Scholar]
- 43.Lauritano D, Moreo G, Limongelli L, Palmieri A, Carinci F. Drug-Induced Gingival Overgrowth: the Effect of Cyclosporin A and Mycophenolate Mophetil on Human Gingival fibroblasts. Biomedicines. 2020;8(7). [DOI] [PMC free article] [PubMed]
- 44.Lauritano D, Lucchese A, Di Stasio D, Della Vella F, Cura F, Palmieri A et al. Molecular aspects of Drug-Induced Gingival Overgrowth: an in Vitro Study on Amlodipine and Gingival fibroblasts. Int J Mol Sci. 2019;20(8). [DOI] [PMC free article] [PubMed]
- 45.Li J, Zhao M, Wang Y, Shen M, Wang S, Tang M, et al. p75NTR optimizes the osteogenic potential of human periodontal ligament stem cells by up-regulating alpha1 integrin expression. J Cell Mol Med. 2020;24(13):7563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the polycomb repressive complex 2. Genome Biol. 2013;14(9):R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang W, Shen Z, Guo J, Sun S. Screening of long non-coding RNA and TUG1 inhibits proliferation with TGF-beta induction in patients with COPD. Int J Chron Obstruct Pulmon Dis. 2016;11:2951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bunte K, Beikler T. Th17 cells and the IL-23/IL-17 Axis in the Pathogenesis of Periodontitis and Immune-mediated inflammatory diseases. Int J Mol Sci. 2019;20(14). [DOI] [PMC free article] [PubMed]
- 49.Liu J, Dan R, Zhou X, Xiang J, Wang J, Liu J. Immune senescence and periodontitis: from mechanism to therapy. J Leukoc Biol. 2022;112(5):1025–40. [DOI] [PubMed] [Google Scholar]
- 50.Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG 3rd, Hayashi A, Imai J, et al. The intermucosal connection between the Mouth and Gut in Commensal Pathobiont-Driven colitis. Cell. 2020;182(2):447–62. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leng S, Xu W, Wu L, Liu L, Du J, Yang F, et al. NLRP3 disturbs Treg/Th17 Cell Balance to aggravate apical periodontitis. J Dent Res. 2023;102(6):656–66. [DOI] [PubMed] [Google Scholar]
- 52.Pan W, Wang Q, Chen Q. The cytokine network involved in the host immune response to periodontitis. Int J Oral Sci. 2019;11(3):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.An Y, Zhao R, Liu W, Wei C, Jin L, Zhang M, et al. Quercetin through miR-147-5p/Clip3 axis reducing Th17 cell differentiation to alleviate periodontitis. Regen Ther. 2024;27:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Xue N, Wang Z, Zeng X, Ji N, Chen Q. Targeting Th17 cells: a promising strategy to treat oral mucosal inflammatory diseases. Front Immunol. 2023;14:1236856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hetta HF, Mwafey IM, Batiha GE, Alomar SY, Mohamed NA, Ibrahim MA et al. CD19(+) CD24(hi) CD38(hi) Regulatory B cells and memory B cells in Periodontitis: Association with pro-inflammatory and anti-inflammatory cytokines. Vaccines (Basel). 2020;8(2). [DOI] [PMC free article] [PubMed]
- 56.Zeng W, Liu G, Luan Q, Yang C, Li S, Yu X, et al. B-Cell Deficiency exacerbates inflammation and bone loss in Ligature-Induced Experimental Periodontitis in mice. J Inflamm Res. 2021;14:5367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun X, Wu T, Yang Z, Chen S, Zhao Z, Hu C, et al. Regulatory role of PDK1 via integrated gene analysis of mitochondria-immune response in periodontitis. Gene. 2024;918:148476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the GEO repository(https://www.ncbi.nlm.nih.gov/geo/).