Abstract
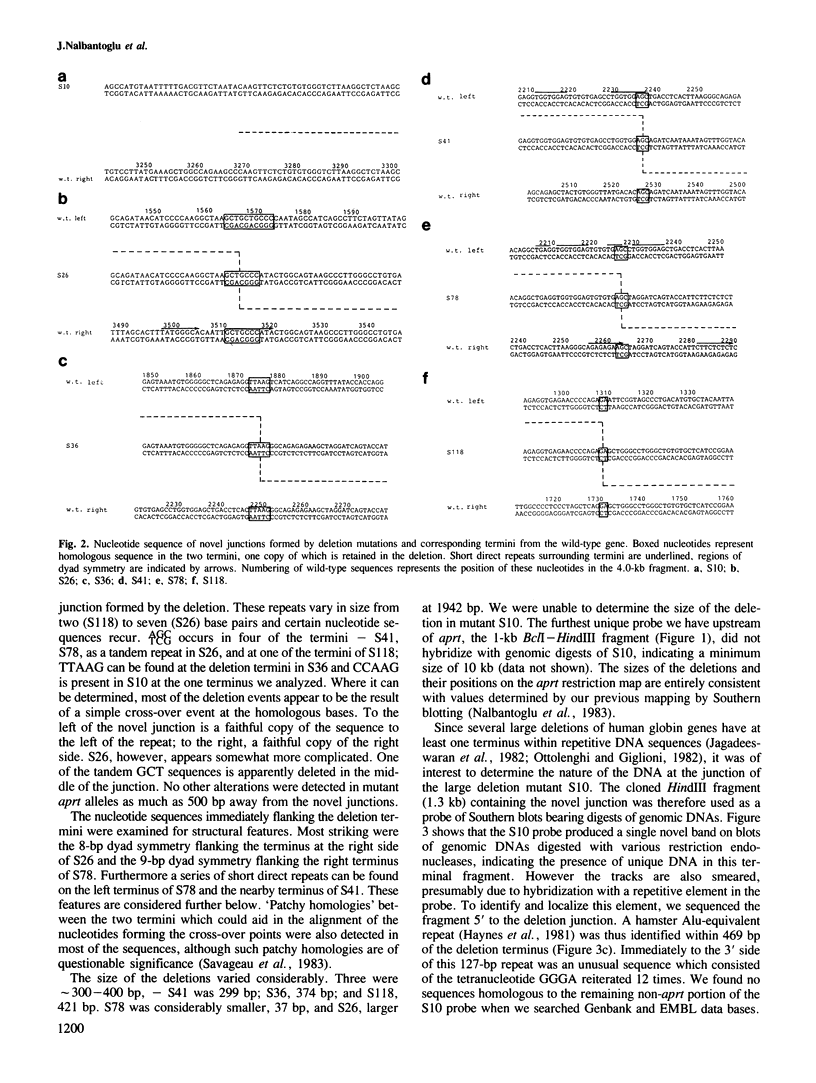
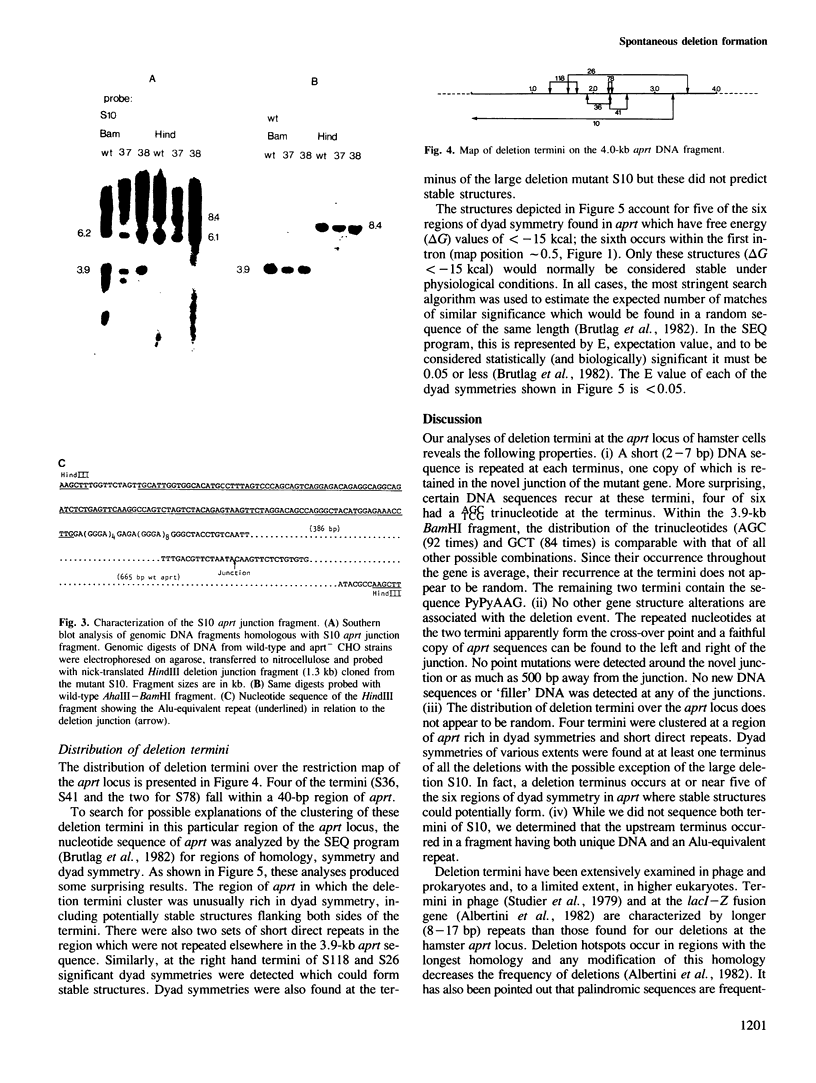
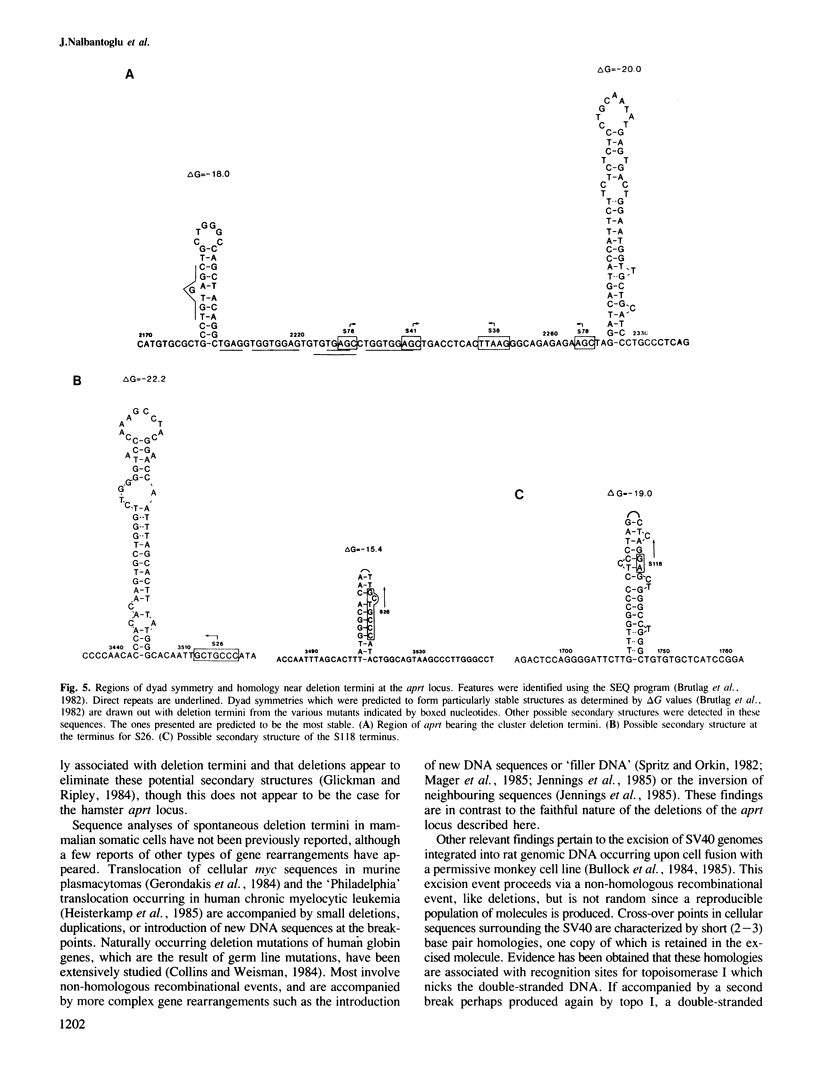
To examine the factors governing the generation of DNA sequence rearrangements in mammalian somatic cells, we have cloned and sequenced novel junctions produced by six spontaneous deletion mutations at the aprt locus of Chinese hamster ovary cells. Our analyses indicate that these rearrangements were produced by non-homologous recombinational events occurring between short (2-7 bp) sequence repeats at the two termini of the deletion which leave one copy of the repeat in the mutant gene. Certain tri- and tetranucleotides recur at the deletion termini, suggesting that these may possibly be a recognition sequence for an enzyme involved in the event. No other gene structural alterations were found at the novel junctions or in neighbouring sequences. The deletions are not randomly distributed over the aprt gene; four termini clustered in a 40-bp sequence. This region of aprt is unusual as it contains both significant stretches of dyad symmetry which could potentially form stable DNA secondary structures and short direct repeats. Regions of dyad symmetry were also found at at least one terminus of all the deletions. In view of the similar properties of this set of deletions, possible mechanisms for the formation of this type of gene rearrangement are considered.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley W. E., Letovanec D. High-frequency nonrandom mutational event at the adenine phosphoribosyltransferase (aprt) locus of sib-selected CHO variants heterozygous for aprt. Somatic Cell Genet. 1982 Jan;8(1):51–66. doi: 10.1007/BF01538650. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Lebkowski J. S., Botchan M. R. High mutation frequency in DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3015–3019. doi: 10.1073/pnas.80.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. S., Weissman S. M. The molecular genetics of human hemoglobin. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. doi: 10.1016/s0079-6603(08)60382-7. [DOI] [PubMed] [Google Scholar]
- Dush M. K., Sikela J. M., Khan S. A., Tischfield J. A., Stambrook P. J. Nucleotide sequence and organization of the mouse adenine phosphoribosyltransferase gene: presence of a coding region common to animal and bacterial phosphoribosyltransferases that has a variable intron/exon arrangement. Proc Natl Acad Sci U S A. 1985 May;82(9):2731–2735. doi: 10.1073/pnas.82.9.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Gerondakis S., Cory S., Adams J. M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell. 1984 Apr;36(4):973–982. doi: 10.1016/0092-8674(84)90047-3. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Toomey T. P., Leinwand L., Jelinek W. R. The Chinese hamster Alu-equivalent sequence: a conserved highly repetitious, interspersed deoxyribonucleic acid sequence in mammals has a structure suggestive of a transposable element. Mol Cell Biol. 1981 Jul;1(7):573–583. doi: 10.1128/mcb.1.7.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp N., Stam K., Groffen J., de Klein A., Grosveld G. Structural organization of the bcr gene and its role in the Ph' translocation. 1985 Jun 27-Jul 3Nature. 315(6022):758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran P., Tuan D., Forget B. G., Weissman S. M. A gene deletion ending at the midpoint of a repetitive DNA sequence in one form of hereditary persistence of fetal haemoglobin. Nature. 1982 Apr 1;296(5856):469–470. doi: 10.1038/296469a0. [DOI] [PubMed] [Google Scholar]
- Jennings M. W., Jones R. W., Wood W. G., Weatherall D. J. Analysis of an inversion within the human beta globin gene cluster. Nucleic Acids Res. 1985 Apr 25;13(8):2897–2906. doi: 10.1093/nar/13.8.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. E., Sargent P. A. Mutants of cultured chinese hamster cells deficient in adenine phosphoribosyl transferase. Cell. 1974 May;2(1):43–54. doi: 10.1016/0092-8674(74)90007-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lowy I., Pellicer A., Jackson J. F., Sim G. K., Silverstein S., Axel R. Isolation of transforming DNA: cloning the hamster aprt gene. Cell. 1980 Dec;22(3):817–823. doi: 10.1016/0092-8674(80)90558-9. [DOI] [PubMed] [Google Scholar]
- Mager D. L., Henthorn P. S., Smithies O. A Chinese G gamma + (A gamma delta beta)zero thalassemia deletion: comparison to other deletions in the human beta-globin gene cluster and sequence analysis of the breakpoints. Nucleic Acids Res. 1985 Sep 25;13(18):6559–6575. doi: 10.1093/nar/13.18.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Goncalves O., Meuth M. Structure of mutant alleles at the aprt locus of Chinese hamster ovary cells. J Mol Biol. 1983 Jul 5;167(3):575–594. doi: 10.1016/s0022-2836(83)80099-0. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Phear G. A., Meuth M. Nucleotide sequence of hamster adenine phosphoribosyl transferase (aprt) gene. Nucleic Acids Res. 1986 Feb 25;14(4):1914–1914. doi: 10.1093/nar/14.4.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ottolenghi S., Giglioni B. The deletion in a type of delta 0-beta 0-thalassaemia begins in an inverted AluI repeat. Nature. 1982 Dec 23;300(5894):770–771. doi: 10.1038/300770a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savageau M. A., Metter R., Brockman W. W. Statistical significance of partial base-pairing potential: implications for recombination of SV40 DNA in eukaryotic cells. Nucleic Acids Res. 1983 Sep 24;11(18):6559–6570. doi: 10.1093/nar/11.18.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalenghe F., Buscaglia M., Steinheil C., Crippa M. Large scale isolation of nuclei and nucleoli from vitellogenic oocytes of Xenopus laevis. Chromosoma. 1978 May 16;66(4):299–308. doi: 10.1007/BF00328531. [DOI] [PubMed] [Google Scholar]
- Spritz R. A., Orkin S. H. Duplication followed by deletion accounts for the structure of an Indian deletion beta (0)-thalassemia gene. Nucleic Acids Res. 1982 Dec 20;10(24):8025–8029. doi: 10.1093/nar/10.24.8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Simon M. N., Dunn J. J. Genetic and physical mapping in the early region of bacteriophage T7 DNA. J Mol Biol. 1979 Dec 25;135(4):917–937. doi: 10.1016/0022-2836(79)90520-5. [DOI] [PubMed] [Google Scholar]
- Tindall K. R., Stankowski L. F., Jr, Machanoff R., Hsie A. W. Detection of deletion mutations in pSV2gpt-transformed cells. Mol Cell Biol. 1984 Jul;4(7):1411–1415. doi: 10.1128/mcb.4.7.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin E. F., Henthorn P. S., Kioussis D., Grosveld F., Smithies O. Unexpected relationships between four large deletions in the human beta-globin gene cluster. Cell. 1983 Dec;35(3 Pt 2):701–709. doi: 10.1016/0092-8674(83)90103-4. [DOI] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]



