Abstract
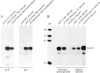
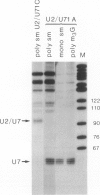
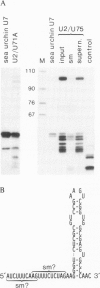
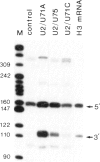
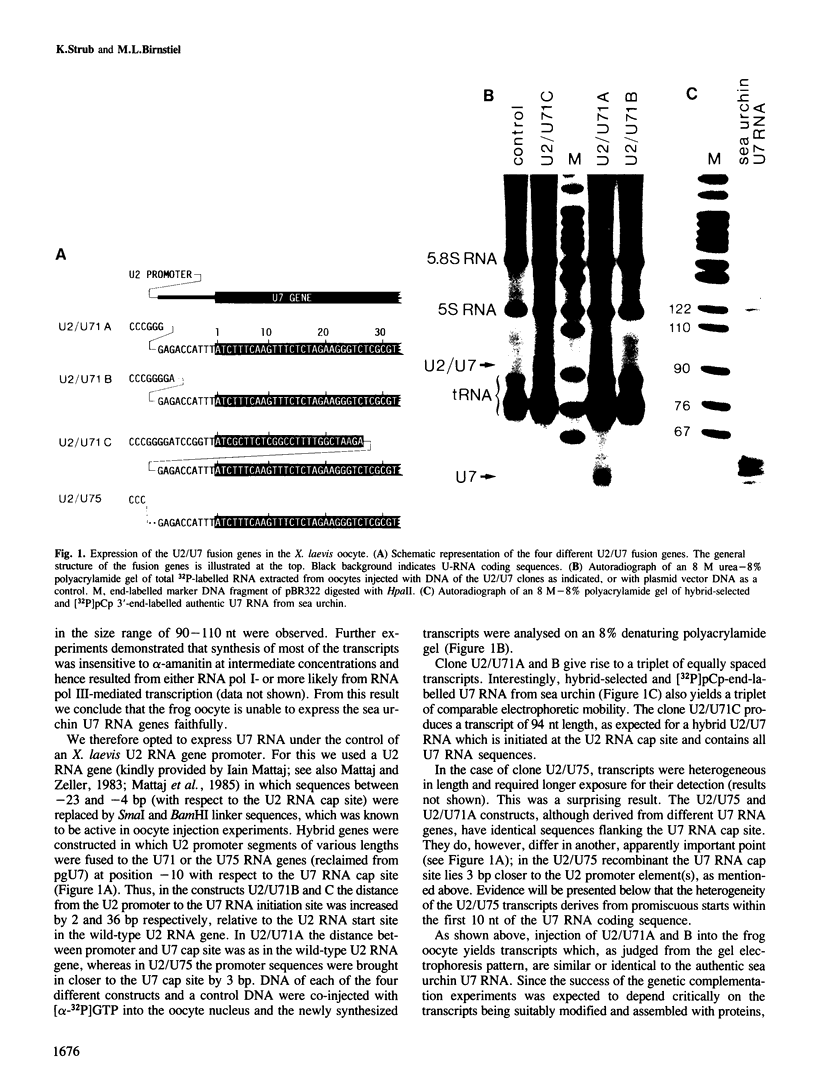
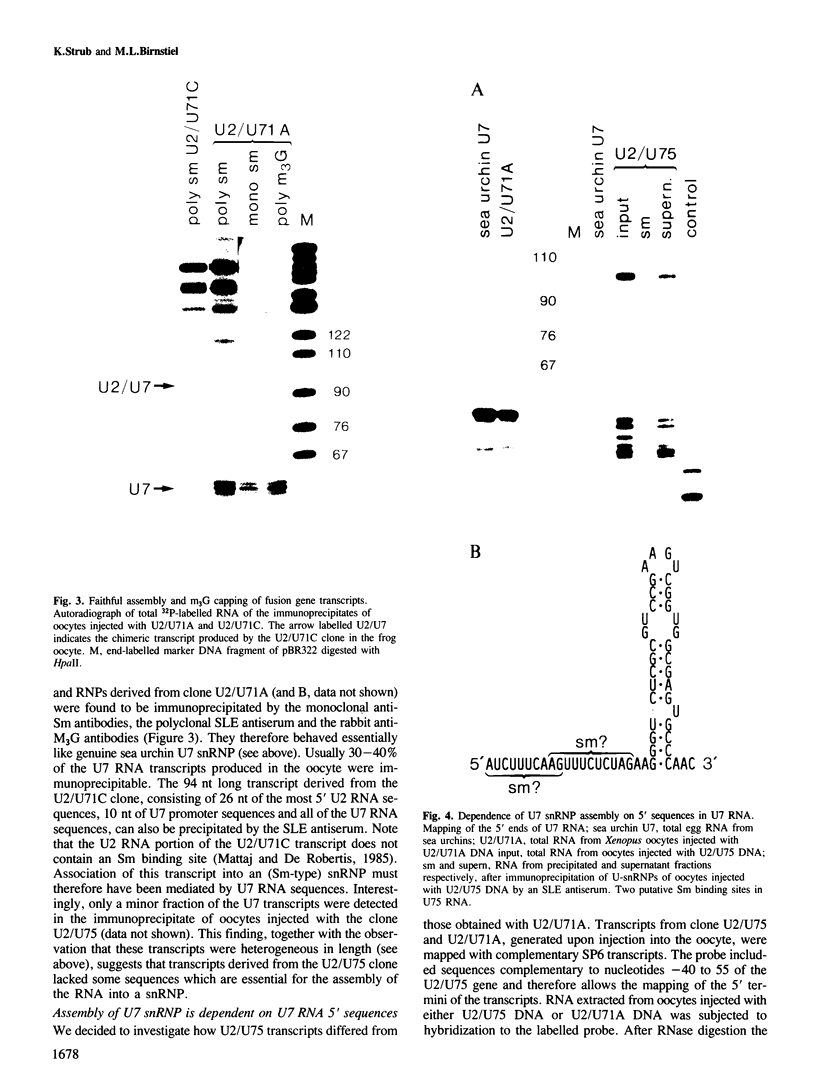
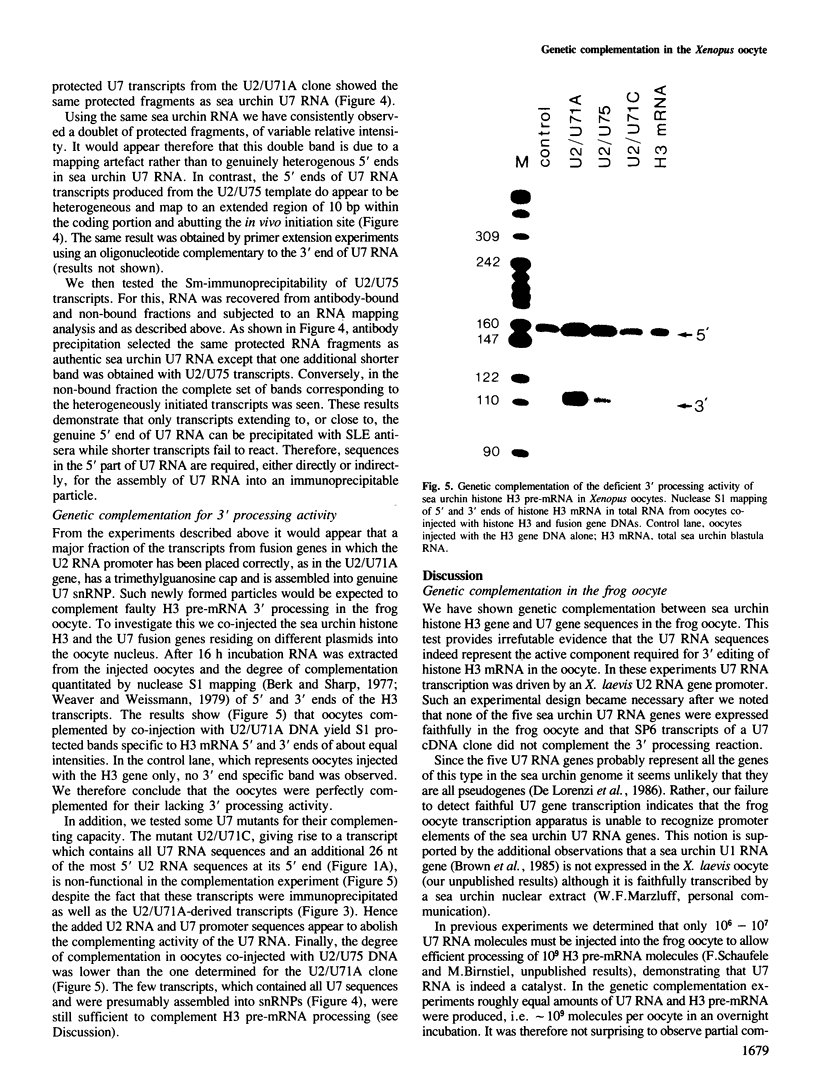
Efficient 3' processing of sea urchin histone H3 pre-mRNA in the Xenopus oocyte is dependent on co-injection of sea urchin U7 RNA or U7 RNP. We show that U7 RNP is a genuine member of the (Sm-type) U-snRNP family as judged by immunoprecipitation of the U7 RNA and U7 RNPs with polyclonal anti-trimethylguanosine and monoclonal anti-Sm antibodies. Sea urchin U7 RNA can be produced in the frog oocyte from RNA genes driven by a Xenopus U2 promoter. The transcripts thus synthesized have all the known characteristics of authentic U7 RNA. Our experiments further suggest that sequences near the 5' end of the U7 RNA mediate Sm-immunoprecipitability. Co-expression of sea urchin U7 RNA gene and H3 histone gene sequences in the frog oocyte complements faulty 3' maturation of histone pre-mRNA. This provides irrefutable evidence that the sea urchin U7 RNA identified earlier is indeed an essential component of the 3' processing of histone mRNAs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. Aspects of biochemical catalysis. Cell. 1984 Feb;36(2):237–239. doi: 10.1016/0092-8674(84)90216-2. [DOI] [PubMed] [Google Scholar]
- Ares M., Jr, Mangin M., Weiner A. M. Orientation-dependent transcriptional activator upstream of a human U2 snRNA gene. Mol Cell Biol. 1985 Jul;5(7):1560–1570. doi: 10.1128/mcb.5.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Brown D. T., Morris G. F., Chodchoy N., Sprecher C., Marzluff W. F. Structure of the sea urchin U1 RNA repeat. Nucleic Acids Res. 1985 Jan 25;13(2):537–556. doi: 10.1093/nar/13.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Chabot B., Black D. L., LeMaster D. M., Steitz J. A. The 3' splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science. 1985 Dec 20;230(4732):1344–1349. doi: 10.1126/science.2933810. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzi M., Rohrer U., Birnstiel M. L. Analysis of a sea urchin gene cluster coding for the small nuclear U7 RNA, a rare RNA species implicated in the 3' editing of histone precursor mRNAs. Proc Natl Acad Sci U S A. 1986 May;83(10):3243–3247. doi: 10.1073/pnas.83.10.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis E. M., Lienhard S., Parisot R. F. Intracellular transport of microinjected 5S and small nuclear RNAs. Nature. 1982 Feb 18;295(5850):572–577. doi: 10.1038/295572a0. [DOI] [PubMed] [Google Scholar]
- Frederiksen S., Hellung-Larsen P., Gram Jensen E. The differential inhibitory effect of alpha-amanitin on the synthesis of low molecular weight RNA components in BHK cells. FEBS Lett. 1978 Mar 15;87(2):227–231. doi: 10.1016/0014-5793(78)80338-x. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Stunnenberg H. G., Birnstiel M. L. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3' histone mRNA termini. Cell. 1983 Oct;34(3):823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- Georgiev O., Birnstiel M. L. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3' termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 1985 Feb;4(2):481–489. doi: 10.1002/j.1460-2075.1985.tb03654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., Shewmaker C. K., Jat P., Flavell R. A. Localization of DNA sequences necessary for transcription of the rabbit beta-globin gene in vitro. Cell. 1981 Jul;25(1):215–226. doi: 10.1016/0092-8674(81)90246-4. [DOI] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985 Oct;42(3):725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- Kunkel G. R., Pederson T. Transcription boundaries of U1 small nuclear RNA. Mol Cell Biol. 1985 Sep;5(9):2332–2340. doi: 10.1128/mcb.5.9.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E. A., Lerner M. R., Janeway C. A., Jr, Steitz J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981 May;78(5):2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., De Robertis E. M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985 Jan;40(1):111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Lienhard S., Jiricny J., De Robertis E. M. An enhancer-like sequence within the Xenopus U2 gene promoter facilitates the formation of stable transcription complexes. Nature. 1985 Jul 11;316(6024):163–167. doi: 10.1038/316163a0. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Zeller R. Xenopus laevis U2 snRNA genes: tandemly repeated transcription units sharing 5' and 3' flanking homology with other RNA polymerase II transcribed genes. EMBO J. 1983;2(11):1883–1891. doi: 10.1002/j.1460-2075.1983.tb01675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Busch H. Small nuclear RNAs and RNA processing. Prog Nucleic Acid Res Mol Biol. 1983;30:127–162. doi: 10.1016/s0079-6603(08)60685-6. [DOI] [PubMed] [Google Scholar]
- Reddy R., Henning D., Busch H. Primary and secondary structure of U8 small nuclear RNA. J Biol Chem. 1985 Sep 15;260(20):10930–10935. [PubMed] [Google Scholar]
- Reddy R., Henning D., Busch H. Substitutions, insertions, and deletions in two highly conserved U3 RNA species. J Biol Chem. 1980 Jul 25;255(14):7029–7033. [PubMed] [Google Scholar]
- Rogers J., Wall R. A mechanism for RNA splicing. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1877–1879. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop D. R., Kristo P., Stumph W. E., Tsai M. J., O'Malley B. W. Structure and expression of a chicken gene coding for U1 RNA. Cell. 1981 Mar;23(3):671–680. doi: 10.1016/0092-8674(81)90430-x. [DOI] [PubMed] [Google Scholar]
- Skuzeski J. M., Lund E., Murphy J. T., Steinberg T. H., Burgess R. R., Dahlberg J. E. Synthesis of human U1 RNA. II. Identification of two regions of the promoter essential for transcription initiation at position +1. J Biol Chem. 1984 Jul 10;259(13):8345–8352. [PubMed] [Google Scholar]
- Strub K., Galli G., Busslinger M., Birnstiel M. L. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984 Dec 1;3(12):2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G., Lund E., Murphy J. T., Pettersson U., Dahlberg J. E. Human U2 and U1 RNA genes use similar transcription signals. EMBO J. 1984 Dec 20;3(13):3295–3301. doi: 10.1002/j.1460-2075.1984.tb02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo C. Y., Ares M., Jr, Weiner A. M. Sequences required for 3' end formation of human U2 small nuclear RNA. Cell. 1985 Aug;42(1):193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]