Abstract
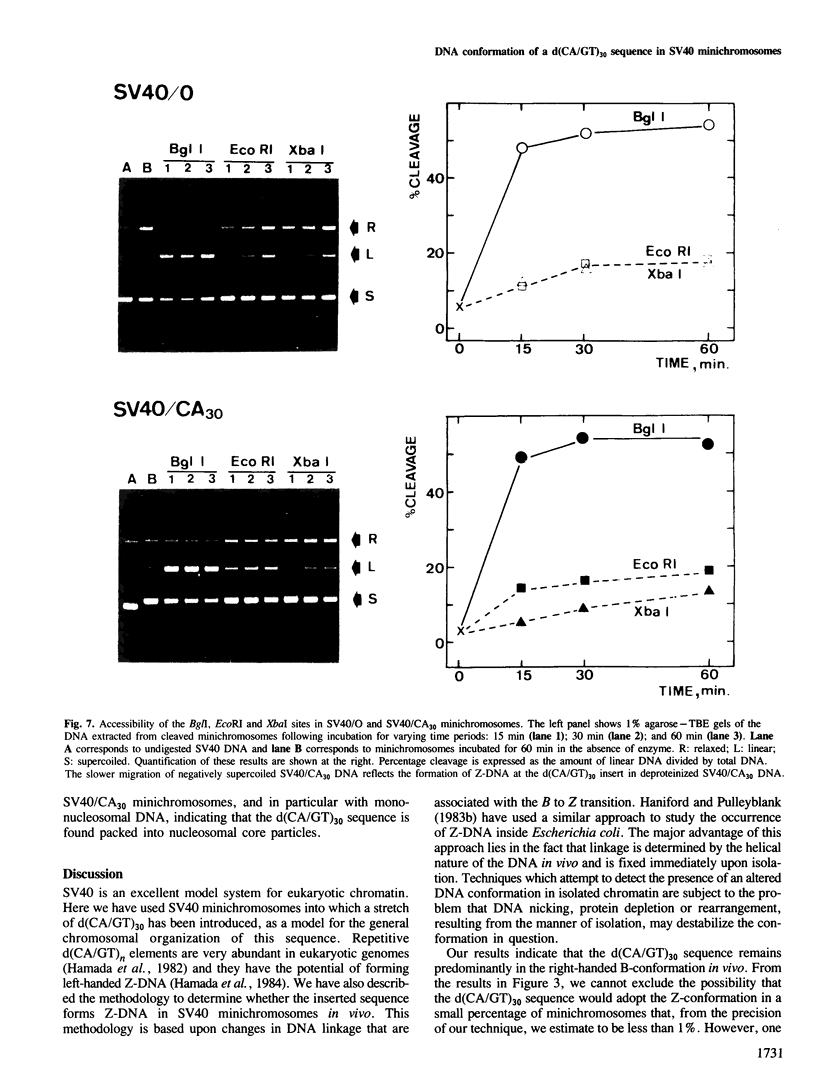
The alternating DNA sequence d(CA/GT)n is known to adopt the left-handed Z-form in negatively supercoiled DNA in vitro. This element represents a significant fraction of the highly repeated DNA sequences found in eukaryotic genomes. We have cloned an alternating d(CA/GT)30 sequence into SV40 minichromosomes at the unique HpaII restriction site which occurs in the transcriptional leader region of the viral late genes. By comparing the linkage differences of topoisomers obtained from viral DNA with or without the d(CA/GT)30 insert, at various stages of the lytic cycle, we conclude that this sequence does not predominate in the Z-form in vivo. Furthermore, we find that the d(CA/GT)30 sequence is packaged into nucleosomal core particles and the region of the minichromosomes which contain the d(CA/GT)30 sequence is organized as nucleosomes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azorin F., Nordheim A., Rich A. Formation of Z-DNA in negatively supercoiled plasmids is sensitive to small changes in salt concentration within the physiological range. EMBO J. 1983;2(5):649–655. doi: 10.1002/j.1460-2075.1983.tb01479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azorin F., Rich A. Isolation of Z-DNA binding proteins from SV40 minichromosomes: evidence for binding to the viral control region. Cell. 1985 Jun;41(2):365–374. doi: 10.1016/s0092-8674(85)80009-x. [DOI] [PubMed] [Google Scholar]
- Barsoum J., Berg P. Simian virus 40 minichromosomes contain torsionally strained DNA molecules. Mol Cell Biol. 1985 Nov;5(11):3048–3057. doi: 10.1128/mcb.5.11.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. S., Hsu M. T. Evidence for variation of supercoil densities among simian virus 40 nucleoprotein complexes and for higher supercoil density in replicating complexes. J Virol. 1984 Jul;51(1):14–19. doi: 10.1128/jvi.51.1.14-19.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Capua E., Stasiak A., Koller T., Brahms S., Thomae R., Pohl F. M. Torsional stress induces left-handed helical stretches in DNA of natural base sequence: circular dichroism and antibody binding. EMBO J. 1983;2(9):1531–1535. doi: 10.1002/j.1460-2075.1983.tb01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Huang S. Y., Garrard W. T. Chromatin structure of the potential Z-forming sequence (dT-dG)n X (dC-dA)n. Evidence for an "alternating-B" conformation. J Mol Biol. 1985 May 25;183(2):251–265. doi: 10.1016/0022-2836(85)90218-9. [DOI] [PubMed] [Google Scholar]
- Hamada H., Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982 Jul 22;298(5872):396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T., Seidman M., Stollar B. D. Characterization of genomic poly(dT-dG).poly(dC-dA) sequences: structure, organization, and conformation. Mol Cell Biol. 1984 Dec;4(12):2610–2621. doi: 10.1128/mcb.4.12.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. The in-vivo occurrence of Z DNA. J Biomol Struct Dyn. 1983 Dec;1(3):593–609. doi: 10.1080/07391102.1983.10507467. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. A nucleosome-free region in SV40 minichromosomes. Nature. 1980 May 22;285(5762):263–265. doi: 10.1038/285263a0. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Angelides K. J., Holloman W. K. Left-handed DNA and the synaptic pairing reaction promoted by Ustilago rec1 protein. Cell. 1985 Jan;40(1):139–145. doi: 10.1016/0092-8674(85)90317-4. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Synapsis promoted by Ustilago rec1 protein. Cell. 1984 Mar;36(3):593–598. doi: 10.1016/0092-8674(84)90338-6. [DOI] [PubMed] [Google Scholar]
- Kmiec E., Holloman W. K. Homologous pairing of DNA molecules promoted by a protein from Ustilago. Cell. 1982 Jun;29(2):367–374. doi: 10.1016/0092-8674(82)90153-2. [DOI] [PubMed] [Google Scholar]
- Kłysik J., Stirdivant S. M., Larson J. E., Hart P. A., Wells R. D. Left-handed DNA in restriction fragments and a recombinant plasmid. Nature. 1981 Apr 23;290(5808):672–677. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- Lafer E. M., Sousa R., Rich A. Anti-Z-DNA antibody binding can stabilize Z-DNA in relaxed and linear plasmids under physiological conditions. EMBO J. 1985 Dec 30;4(13B):3655–3660. doi: 10.1002/j.1460-2075.1985.tb04131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer E. M., Sousa R., Rosen B., Hsu A., Rich A. Isolation and characterization of Z-DNA binding proteins from wheat germ. Biochemistry. 1985 Sep 10;24(19):5070–5076. doi: 10.1021/bi00340a017. [DOI] [PubMed] [Google Scholar]
- Lai C. J. Mapping simian virus 40 mutants by marker rescue. Methods Enzymol. 1980;65(1):811–816. doi: 10.1016/s0076-6879(80)65075-7. [DOI] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller F. D., Dixon G. H., Rattner J. B., van de Sande J. H. Assembly and characterization of nucleosomal cores on B- vs. Z-form DNA. Biochemistry. 1985 Jan 1;24(1):102–109. doi: 10.1021/bi00322a015. [DOI] [PubMed] [Google Scholar]
- Nickol J., Behe M., Felsenfeld G. Effect of the B--Z transition in poly(dG-m5dC) . poly(dG-m5dC) on nucleosome formation. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1771–1775. doi: 10.1073/pnas.79.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Lafer E. M., Peck L. J., Wang J. C., Stollar B. D., Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982 Dec;31(2 Pt 1):309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Tesser P., Azorin F., Kwon Y. H., Möller A., Rich A. Isolation of Drosophila proteins that bind selectively to left-handed Z-DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7729–7733. doi: 10.1073/pnas.79.24.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revet B., Zarling D. A., Jovin T. M., Delain E. Different Z DNA forming sequences are revealed in phi X174 RFI by high resolution darkfield immuno-electron microscopy. EMBO J. 1984 Dec 20;3(13):3353–3358. doi: 10.1002/j.1460-2075.1984.tb02303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Stockton J. F., Miller F. D., Jorgenson K. F., Zarling D. A., Morgan A. R., Rattner J. B., van de Sande J. H. Left-handed Z-DNA regions are present in negatively supercoiled bacteriophage PM2 DNA. EMBO J. 1983;2(12):2123–2128. doi: 10.1002/j.1460-2075.1983.tb01712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R. T., DePamphilis M. L. Simian virus 40 DNA replication in isolated replicating viral chromosomes. J Virol. 1978 Oct;28(1):53–65. doi: 10.1128/jvi.28.1.53-65.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O. H., Bohn M. J. SV40 viral minichromosome: preferential exposure of the origin of replication as probed by restriction endonucleases. Nucleic Acids Res. 1978 Oct;5(10):3469–3477. doi: 10.1093/nar/5.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Weiss E., Ghose D., Schultz P., Oudet P. T-antigen is the only detectable protein on the nucleosome-free origin region of isolated simian virus 40 minichromosomes. Chromosoma. 1985;92(5):391–400. doi: 10.1007/BF00327472. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B. The three-dimensional structure of DNA. Annu Rev Biochem. 1982;51:395–427. doi: 10.1146/annurev.bi.51.070182.002143. [DOI] [PubMed] [Google Scholar]