Abstract
Background
Wastewater-based epidemiology (WBE) is already being adopted for the surveillance of health conditions of communities and shows great potential for the monitoring of infectious pathogens of public health importance. There is however paucity of robust data to support extensive WBE in Nigeria. This study evaluated the prevalence of clinically relevant infectious pathogens and provided antimicrobial resistance profiles of bacteria pathogens in wastewater canals in Lagos State at a single point in time.
Methods
This is a cross-sectional survey of wastewater canals in 20 Local Government Areas (LGAs) in Lagos State for detection of bacteria pathogens of public health importance including non-tuberculous mycobacteria and SARS-Cov-2 virus using cultural analysis and conventional Polymerase Chain Reaction (PCR) techniques. Descriptive epidemiological survey of communities around the canals was done using questionnaires to assess exposure pathways. Statistical analysis was done using SPSS version 27 while P value of < 0.05 was considered as significant.
Results
Three thousand and fifty-four (3054) questionnaires were administered to 1215 (39.8%) females and 1658 (54.3%) males in communities situated around 40 canals in 20 LGAs. Although majority (81.8%) reported using water closet toilet system and pit latrine (12.5%), a few of them admitted to open defaecation [101 (3.3%)] while 299 (9.8%) engaged in open field waste disposal. SARS-CoV-2 was not detected from wastewater in this study. Two mycobacterial species that included Mycobacterium fortitium group (13, 32.5%) and Mycobacterium kansasii (11, 27.5%) were identified in 15 out of 20 LGAs sampled. A total of 123 bacteria pathogens were isolated across the 40 canals. Prominent enteropathogens isolated included Escheriachia coli (28.5%), Salmonella spp (16.3%), Vibro cholerae (10.6%) and Shigella spp (5.7%). Extended spectrum beta-lactamase genes were prominent (87.5%) in the wastewater samples with almost a half (42.5%) of the canals containing both SHV and CTX-M.
Conclusion
This study highlights the presence of pathogens with potential to cause epidemic in wastewater canals in Lagos State and provides evidence to inform policy and strategies for wastewater monitoring and treatment. Further studies involving longitudinal monitoring of time-based variations is needed to identify trends in pathogen loads and AMR patterns over time.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12889-024-21157-6.
Keywords: Wastewater-based epidemiology, Escherichia coli, Vibrio cholera, Mycobacterium kansasii, Mycobacterium fortitium, Antimicrobial resistance, Antimicrobial resistance genes, Public health surveillance, Pathogen detection, Lagos
Introduction
Experience has shown that monitoring sewage for traces of a pathogen enables effective surveillance of entire communities, providing a sensitive signal of whether the pathogen is present in the population [1]. Wastewaters generated by domestic users, have been largely investigated in the last years [1–4] with several studies providing strong evidence for wastewater monitoring as a powerful tool in tracking the spread of infectious diseases including antimicrobial resistant bacteria (ARB) [5–9]. All these suggests wastewater-based epidemiology (WBE) for the early detection of presence of pathogen and for the surveillance and monitoring purposes.
Over the years, the overuse and abuse of antibiotics and other antimicrobial agents has led to antimicrobial-resistant bacteria and their antimicrobial resistance genes (ARGs) being widespread in the environment, including in wastewater [10–12]. They can persist for long periods of time in the wastewater posing great risk to public health. Furthermore, widespread use of broad-spectrum antibiotics and increased community consumption of antibiotics reported following the advent of Corona virus disease 2019 [13, 14] has heightened concerns on antimicrobial resistance (AMR), although carefully collected data is still limited. Consequently, there is possibility of further emergence and selection of new AMR traits in bacteria, including pathogenic bacteria of human health concern [15, 16], with loss of antimicrobial treatment options for humans. High morbidity and mortality due to the high prevalence of AMR in the environment is the biggest threat to public health [17].
Tuberculosis (TB) disease caused by non-tuberculous mycobacteria (NTM) are increasing globally in the context of immunosuppression [18, 19] and are highly resistant to common antibiotics regimens, thus, retrogressing achieving TB elimination by year 2030. Available information on the role of environment especially the wastewater as reservoir for mycobacteria and possible transmission to human in Nigeria remains limited. It is clinically important to identify NTM at the species level to distinguish pathogenic and non-pathogenic species with implication for public health and patient management [20].
Antimicrobial resistance and COVID-19 are two high profile public health challenges threatening the attainment of sustainable development goals of good health for all. Wastewater surveillance for pathogens of public health importance could have many benefits. It is a cost-effective way to survey transmission dynamics of entire communities. It avoids the biases of other epidemiological indicators [21] and collects data from people who lack access to healthcare. Also, it has the potential of revealing infection dynamics earlier than diagnostic testing thereby providing public-health officials with near-real-time information on disease prevalence. Canals in Lagos State serve as a confluent for domestic wastewater and for sewage disposal while people have been known to directly defecate into the canal.
In Nigeria, little is known about the role of NTM disease and there is no diagnostic algorithms or treatment guidelines established for NTM. In addition, species identification of NTM is largely unavailable in TB laboratories, hence NTM is frequently misdiagnosed as TB. More so, the recent covid-19 pandemic was believed to have led to a surge in the use of azithromycin, a key drug in the treatment of NTM [14]. Inappropriate use of azithromycin and other macrolides may lead to acquired resistance thus impacting treatment. The high population density in Lagos and poor urban communities increase the risk of infections through exposure to contaminated water, soil, and medical equipment [22]. Similarly, Lagos has a history of cholera outbreaks, driven by poor sanitation, inadequate clean water supply, and climate variability affecting water systems [23, 24]. Additionally, Lagos, as a global trade hub, faces the risk of accelerating AMR due to unregulated antibiotic use and high human mobility [25]. Investigating pathogens of public health importance including Vibrio cholerae in Lagos wastewater is crucial to safeguarding public health. It will provide opportunity for early identification of immediate risks of infections with the ultimate goal of mitigating future outbreak of water and food borne diseases and contributing to the long-term goal of achieving sustainable health and environmental safety in the region. Unfortunately, wastewater-based epidemiological monitoring has not been fully explored for infectious disease surveillance in Lagos State. This study therefore collected data and evaluated the prevalence of pathogens of public health importance in the wastewater canals in Lagos State for epidemiological purposes.
Methodology
Study site/design
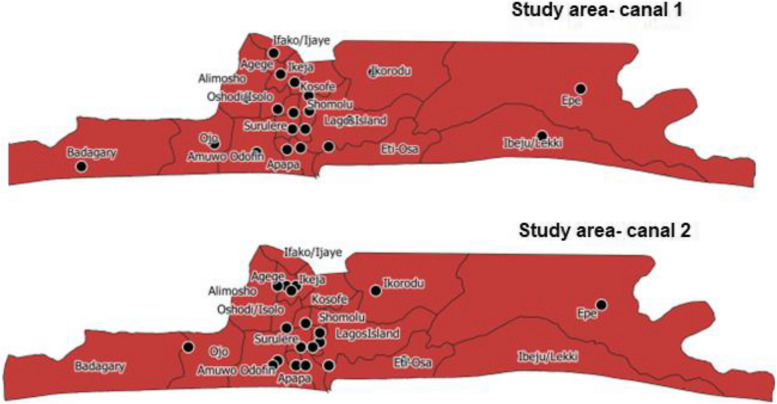
This study was a cross-sectional survey of wastewater canals in Lagos State for infectious pathogens including SARS-CoV-2, Vibrio cholerae, Escherichia coli, Salmonella spp., Shigella spp., Mycobacterium tuberculosis complex and non-tuberculous mycobacteria (NTM). The study was conducted in Lagos state, Nigeria. Lagos is the commercial nerve centre and former administrative capital of Nigeria. It is a populous melting pot of culture and diversity, with over 20 million people from all the major tribes in the country largely represented. It has 20 Local Government Areas (LGAs) and 37 Local Council Development Areas (LCDAs) [22]. We mapped Lagos State according to the 20 LGAs and two major canals were identified in each LGA and included in the study (Fig. 1, Suppl-2 Figure S1A&B). The selection criteria considered canals closer to the communities with relatively heavy human activity. The study was carried out over a period of 4-months from April to July 2022.
Fig. 1.
Map of Lagos State showing the location of the canals that were sampled
Epidemiological survey
A descriptive epidemiology study via questionnaire survey was done to assess exposure pathway using communities around the selected canals. The sample size was calculated based on the number of residents across the Local Government Areas (LGAs) in Lagos State [22]. The objective was to ensure a representative sample for investigating exposure pathways. We used the sample size estimation formula from Sharma et al. [26] to calculate the minimum required sample size, n, based on the prevalence of the outcome and desired precision. To account for potential non-response, we increased the sample size by 10%. Additionally, given that the study involved complex sampling across multiple LGAs, a design effect of 4.0 was applied to ensure statistical reliability across the different regions. The final adjusted sample size was 2881. The final sample size was allocated proportionally across the LGAs based on their population sizes, as shown in Table 2. This ensured that each LGA is represented according to its contribution to the overall population of Lagos State. We slightly exceeded the calculated sample size, resulting in a total of 3054 questionnaires distributed to increase precision and account for any discrepancies in response rates.
Table 2.
Distribution of questionnaires across the different LGAs
| SN | LGAs | Sample sizes (Questionnaire) | No of Bacteria Isolated |
|---|---|---|---|
| 1 | LAGOS ISLAND (LI) | 82 | 8 |
| 2 | LAGOS MAINLAND (LM) | 110 | 6 |
| 3 | SURULERE (SU) | 163 | 9 |
| 4 | APAPA (AP) | 80 | 5 |
| 5 | ETI-OSA (EO) | 89 | 6 |
| 6 | SHOMOLU (SH) | 142 | 10 |
| 7 | KOSOFE (KO) | 220 | 3 |
| 8 | EPE (EP) | 70 | 6 |
| 9 | IBEJU-LEKKI (IL) | 50 | 3 |
| 10 | IKORODU (IK) | 180 | 5 |
| 11 | AGEGE (AG) | 160 | 6 |
| 12 | IFAKO-IJAYE (II) | 150 | 7 |
| 13 | ALIMOSHO (AL) | 410 | 7 |
| 14 | BADAGRY (BA) | 90 | 4 |
| 15 | OJO (OJ) | 201 | 5 |
| 16 | AJEROMI/IFELODUN (AI) | 210 | 9 |
| 17 | AMUWO-ODOFIN (AO) | 111 | 3 |
| 18 | OSHODI/ISOLO (OI) | 210 | 4 |
| 19 | IKEJA (IKJ) | 111 | 6 |
| 20 | MUSHIN (MU) | 214 | 11 |
| Total | 3054 | 123 |
The questionnaire was designed through consultation with relevant previous literature [13, 25, 27] and extensive discussions among the research team on what questions would be relevant given the diversity of the investigation. It consisted of 39 questions divided into four sections to capture information on demographics (8 questions), households-environment-water and sanitation (14 questions), COVID-19 exposure (6 questions) and Antibiotic use behaviour (11 questions). The questionnaire was initially validated among the research team to ensure that the questions are reliable and effectively capture the intent of the investigation. Then it was pretested among the non-research community in the NIMR staff quarters (35 people) and analysed. The outcome of analysis of the pilot data and all feedback were incorporated into the final questionnaire. The validated questionnaires (Suppl. 1) were interviewer-administered to residents of communities close to the selected canals in the 20 LGAs to assess risk factors responsible for observed pathogens.
Sampling methodology
Two major wastewater canal were identified per LGA and included in the study (Fig. 1). Wastewater samples were collected from each of the 40 selected canals from 20 LGAs. Wastewater samples were collected using wide-mouthed sterile bottles (200 ml). Five samples, 200 ml each were collected at different points per canal and pooled together for a total volume of 1 L (using a 1 L bottle). Pooled samples were later aliquoted into four places, one placed in Viral transport media (VTM) to be used for SARS CoV-2 detection, another to be used for bacteria culture, while the remaining two were reserved for molecular NTM analysis and ARG detection respectively. The VTM used in this study was an in-house preparation containing 10 g veal infusion broth and 2 g bovine albumin fraction V in 400 ml sterile distilled water. Then 0.8 ml gentamicin sulfate solution (50 mg/ml) and 3.2 ml amphotericin B (250 µg/ml) were added, and the solution was sterilized by filtration and stored at 2–8 °C for use. All four sample bottles were placed in an ice box (4°-8 °C) and transported to the Microbiology Laboratory of Nigerian Institute of Medical Research for storage and analyses.
Detection of SARS-CoV-2
Wastewater samples from the canals were concentrated by an adsorption-elution method using an electronegative filter as described by Katayama et al. [28]. Viral nucleic acids were extracted from 200 μL of the final elute obtained from the concentration step using a NIMR BIOTECH RNA Extraction Kit according to the manufacturer’s protocol. The extracted nucleic acid was kept at − 80 °C until additional downstream analysis was carried out.
The presence of SARS-CoV-2 RNA in wastewater canal was detected using SARS-CoV-2 Detection Assay (SCODA) [targets ORF1ab and N-gene] on Quantstudio5 Real Time PCR machine (Thermo Fisher Scientific). For quality control, the assay was run with a positive control, negative control and a housekeeping internal control according to the manufacturer’s instructions. Negative control was nuclease free water while positive control was synthetic gene from the target region. Samples with a cycle threshold of ≤ 38 was considered positive.
Molecular detection of MTB and NTM
Multiplex PCR and hybridization were performed using GenoType CMdirect kits to detect NTM and Mycobacterium tuberculosis complex (MTBC) respectively. The GenoType CMdirect Ver 1.0 is a qualitative in-vitro test based on the DNA strip technology for identification of the MTBC as well as species of nontuberculous mycobacteria from clinical specimens. It permits the rapid and reliable differentiation of relevant mycobacteria. The whole procedure is divided into three steps: DNA extraction from decontaminated sample; multiplex amplification with biotinylated primers; and a reverse hybridization. Line probe assay technology (GenoType CMdirect ver 1.0) was used in identification of mycobacteria from wastewater. Briefly, 15 ml of wastewater was centrifuged at 3000 X G for 15 min and the pellets were re-suspended in 5 ml distilled water. The re-suspended pellets were decontaminated using 4% NaOH [29] and DNA was extracted using the Genolyse kit according to manufacturer’s protocol (Hain Lifescience, Nehren, Germany). Then, 5 µl of the extracted DNA was added to the PCR reaction mix and amplified using multiplex PCR with biotinylated primers. The amplification was performed as follows: enzyme activation for 15 min at 950C, 10 cycles of 30 s at and 2 min at 580C; and 30 cycles of 25 s at 950C, and 40 s at 530C 40 s at 700C. The final cycle consisted of 8 min at 700C. A 20 µl aliquot of amplified PCR product was hybridized to DNA probe-labeled strips provided in the GenoType CMdirect assay kit for detection of mycobacteria. All hybridization and detection steps were performed as per the manufacturer’s instructions. The American Type Culture Collection (ATCC) reference strains, Mycobacterium tuberculosis 27294 H37Rv and Mycobacterium terrae 15755 were used as quality control (QC) for the assay.
Detection of Enteropathogens
Isolation and identification of bacterial isolates from wastewater samples was carried out.
aseptically using standard microbiological techniques as described by Sarkar et al. [30].Waste-water samples were passed through membrane filters (0.45 µm) and the filters were incubated in liquid Cary Blair medium (20 ml) prepared in MacCartney bottles and incubated overnight (18–24 h) at 37 °C to stabilize the entero-pathogens in the medium. Thereafter, enrichment, enriched and selective media were inoculated for targeted pathogen Isolation. MacConkey agar was inoculated for isolation of potential coliforms including Escherichia coli, klebsiella spp., Citrobacter spp. and Enterobacter spp. Alkaline peptone water (enrichment) was inoculated (200 µl), incubated overnight at 37 °C and subsequently plated on Thiosulphate citrate bile salt sucrose agar (TCBS) for selective isolation of Vibrio cholerae and related species. Similarly, an aliquot (200 µl) was enriched in Selenite F broth and subsequently plated on Salmonella Shigella agar (SSA) for selective isolation of Salmonella and Shigella species. Blood agar was also inoculated as an enriched media to facilitate the growth of other nutritionally fastidious pathogens and incubated aerobically at 37 °C for 24—48 h. Other selective media including mannitol salt agar and cetrimide agar were also inoculated to target Staphylococcus aureus and Pseudomonas aeruginosa respectively. All media used in this study were sourced from OXOID ThermoFisher Scientific. The above bacteria pathogens were targeted due to their relevance either as diarrhoea pathogens (Enterobacteriaceae) or their role as leading pathogens implicated in deaths associated with antimicrobial resistance (Staphylococcus aureus and Pseudomonas aeruginosa) [31]. Pure colonies were identified using colonial morphology, Gram reaction, biochemical tests and further confirmed using BD BioMic V3 biochemical rating Identification system while pure isolates were stored at -80 °C for further analysis. Identified Vibrio cholerae were further serotyped using Vibrio cholerae polyvalent and monovalent antisera to determine the serotypes. All bacteriological and molecular analyses were done at the Microbiology Department of the Nigerian Institute of Medical Research with ISO 15189 accredited laboratories (Facility Accreditation Number: M0588).
Antibiotic susceptibility testing
In vitro susceptibility testing of the isolated bacteria strains was performed using the Kirby-Bauer disk diffusion method with 12 antimicrobial agents (OXOID): ampicillin (AM, 10 μg), amoxicillin/clavulanic acid (AMC-CLA, AMC 20 μg / CLA 10 μg), cefotaxime (CTX, 30 μg), ceftazidime (CAZ, 30 μg), imipenem (IPM, 10 μg), tetracycline (TC, 30 μg), ciprofloxacin (CIP, 5 μg), trimethoprim/sulfamethoxazole (SMZ-TMP, SMZ 1.25 μg / TMP 23.75 μg), Amikacin (AK, 30 μg), Levofloxacin (LEV, 5 μg), Erythromycin (E, 15 μg), Ceftriaxone (CRO, 30 μg). These antibiotics were selected to represent eight major classes (Penicillin, Carbapenems, Fluoroquinolones, Macrolides, Aminoglycosides, Cephalosporins, Tetracyclines and Sulphonamides) of antibiotics commonly used for treatment of bacterial infections and have been shown to be prone to abuse. The AST was carried out in accordance with the recommendation of the Clinical and Laboratory Standards Institute (CLSI), 2023 [32]. The reference strain Escherichia coli ATCC®25,922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms.
Detection of Antibiotic-resistant genes (ARG)
Metagenomic DNA was extracted using ZymoBIOMICS™ DNA Miniprep Kit (ZYMO RESEARCH) for microbiome/ metagenomic analysis and the extracted DNA was used for resistant gene detection. Presence of extended spectrum beta-lactamase (ESBL), methicillin and macrolide resistance genes were assessed by PCR using the sets of primers listed in Table 1. Thermal cycling was conducted in a PTC 200 gradient thermal Cycler for an initial denaturation of 95 °C for 5 min followed by 30 amplification cycles of 30 s at 95 °C; 30 s at (refer to annealing temperature for respective target gene in Table 1) and for the respective extension time for each gene (Table 1) at 72 °C. This was followed by a final extension step of 10 min at 72 °C. The resulting amplicons were separated on a 1.5% agarose gel and electrophoresis was carried out at 80 V for 1 h 30 min. After electrophoresis, DNA bands were visualized by ethidium bromide staining and a UV- Trans-illuminator. A 100 bp DNA ladder was used as DNA molecular weight standard.
Table 1.
List of Primers used for Antimicrobial resistant genes detection PCR
| Primers | Forward | Annealing temp. (oC) | Extension time | Band size (bp) | References |
|---|---|---|---|---|---|
| ermF |
F- CGA CAC AGC TTT GGT TGA AC R- QQA CCT ACC TCA TAG ACA AG |
60 | 30 s | 309 | [33] |
| ermX |
F- GAG ATC GGR CCA GGA AGC R- GTG TGC ACC ATC GCC TGA |
60 | 30 s | 488 | [33] |
| ereA |
F- GCC GGT GCT CAT CAT GAA CTT GAG R- CGA CTC TAT TCG ATC AGA GGC |
60 | 30 s | 420 | [34] |
| MsrA |
F- GCA CTT ATT GGG GGT AAT GG R- GTC TAT AAG TGC TCT ATC GTG |
58 | 30 s | 384 | [34] |
| CTX-M |
F- CGC TGT TGT TAG GAA GTG TG R- GGC TGG GTG AAG TAA GTG AC |
56 | 1 min | 754 | [35] |
| SHV |
F- CGC CTG TGT ATT ATC TCC CT R- CGA GTA CTC CAC GAG ATC CT |
56 | 30 s | 293 | [35] |
| TEM |
F- TTT CGT GTC GCC CTT ATT CC R- ATC GTT GTC AGA AGT AAG TTG |
56 | 30 s | 403 | [35] |
| VEB |
F- GAT GGT GTT TGG TCG CAT ATC GCA AC R- CAT CGC TGT TGG GGT TGC CCA ATT TT |
58 | 1 min | 391 | [36] |
| MecA |
F- AAA ATC GAT GGT AAA GGT TGG R- AGT TCT GCA GTA CCG GAT TTG |
58 | 45 s | 532 | [37] |
| IMP |
F- TTG ACA CTC CAT TTA CDG R- GAT YGA GAA TTA AGC CAC YCT |
54 | 30 s | 183 | [38] |
| KPC |
F- CAT TCA AGG TTC TTG CTG C R- ACG ACG GCA TAG TCA TTT GC |
56 | 30 s | 340 | [38] |
Statistical analysis
The statistical package for social sciences (SPSS) version 27 was used for analysis, figure preparation and report generation. Quantitative data were summarized using descriptive statistics. Pearson’s correlation was used to examine association between outcome variables. A P value of < 0.05 was considered as significant.
Results
Epidemiological survey
Three thousand and fifty-four (3054) questionnaires were administered to people in the communities around the canals (Table 2). Respondents’ demographics can be seen on Table 3. Although majority (81.8%) reported using water closet toilet system and pit latrine (12.5), a few of them admitted to open defecation [101 (3.3%)] (Suppl-2 Figure S2). Twenty-nine-point nine percent (29.9%) of the respondents had been screened previously for SARS-CoV-2 prior to this study of which 2.5% tested positive for the virus. About 29.2% were vaccinated for COVID-19 with 21.2% completing double dose while 9.7% reported taking a booster dose.
Table 3.
Socio-demographics of respondents
| Variable | Number (%) |
|---|---|
| Gender | |
| Male | 1658 (54.3) |
| Female | 1215 (39.8) |
| Not provided | 181 (5.9) |
| Total | 3054 (100) |
| Mean age 35.4 ± 12.9 (18- 94) | |
| Marital Status | |
| Single | 1223 (40) |
| Married | 1620 (53.0) |
| Widowed | 51 (1.7) |
| Divorced | 17 (0.6) |
| Education | |
| No education | 83 (2.7) |
| Informal education | 28 (0.9) |
| Primary education | 250 (8.2) |
| Secondary education | 1504 (49.2) |
| Tertiary education | 842 (27.6) |
| Family type | |
| Polygamous | 440 (14.4) |
| Monogamous | 2036 (66.7) |
| Not provided | 578 (18,9) |
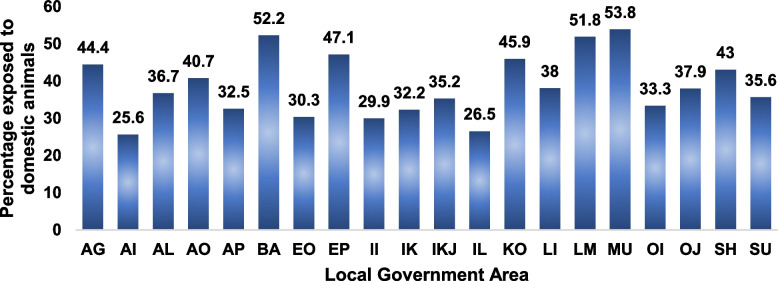
Out of all the respondents, 38.7% were exposed to domestic animals in their residents while 299 (9.8%) engaged in open field waste disposal. Self-reported exposure to domestic animals varied across the LGAs (Fig. 2). The domestic animals mentioned included dog (23.9%), cat (10.5%), poultry (10.5%), goat (10.3%), cow (2.7%), pig (1.5%), rat (0.4%) and others (1.0%).
Fig. 2.
Percentage of respondents exposed to domestic animals disaggregated across the LGAs
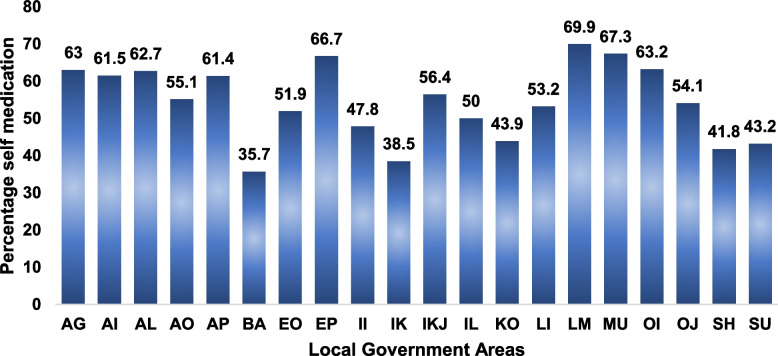
There was a substantial knowledge of antibiotics (70.1%) among the respondents, however, 54.7% of the respondent’s reported self-medication in the last 6 months and 31.6% would stop their antibiotic medication when they feel better. Figure 3 shows the distribution of reported self-medication across the LGAs.
Fig. 3.
Percentage distribution of self-reported antibiotic use (without prescription) among respondents from the different LGAs
SARS-CoV-2 detection
Analysis of the 40 wastewater samples from the 20 LGAs did not detect any SARS-CoV-2 RNA positive result. Study was carried out at a non-peak period.
NTB detection
Two mycobacterial species that included Mycobacterium fortitium group (13, 54.2%) and Mycobacterium kansasii (11; 45.8%) were identified in 15 out of 20 LGAs sampled, Table 4. Mycobacterium fortitium group was solely identified in 7 LGAs (LI, KO, AI, OJ, AP, IKJ, and SH) while Mycobacterium kansasii was identified solely in IK, AL, SU, AO and BA respectively. Furthermore, Mycobacterium fortitium group and Mycobacterium kansasii were equally distributed in 3 LGAs (LM, EP and II). There was no detection of any member of the Mycobacterium tuberculosis complex.
Table 4.
Distribution of Mycobacterial species obtained with GenoType Cmdirect
| LGA | Mycobacterium fortitium group | Mycobacterium kansasii | Total |
|---|---|---|---|
| EO | 0 | 0 | 0 |
| LI | 1 | 0 | 1 |
| KO | 2 | 0 | 2 |
| AG | 0 | 0 | 0 |
| IK | 0 | 2 | 2 |
| LM | 1 | 1 | 2 |
| AI | 2 | 0 | 2 |
| EP | 1 | 1 | 2 |
| OI | 0 | 0 | 0 |
| OJ | 1 | 0 | 1 |
| IC | 0 | 0 | 0 |
| AL | 0 | 2 | 2 |
| II | 1 | 1 | 2 |
| SU | 0 | 1 | 1 |
| AP | 2 | 0 | 2 |
| IKJ | 1 | 0 | 1 |
| AO | 0 | 2 | 2 |
| MU | 0 | 0 | 0 |
| BA | 0 | 1 | 1 |
| SH | 1 | 0 | 1 |
| Total | 13 | 11 | 24 |
Bacteria pathogen detection
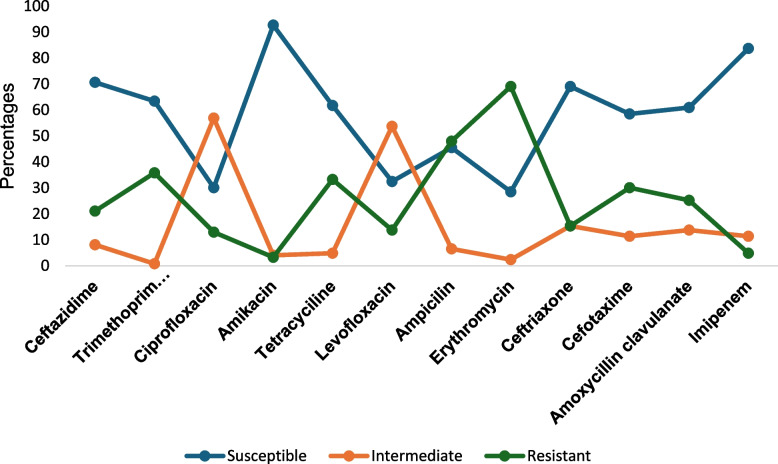
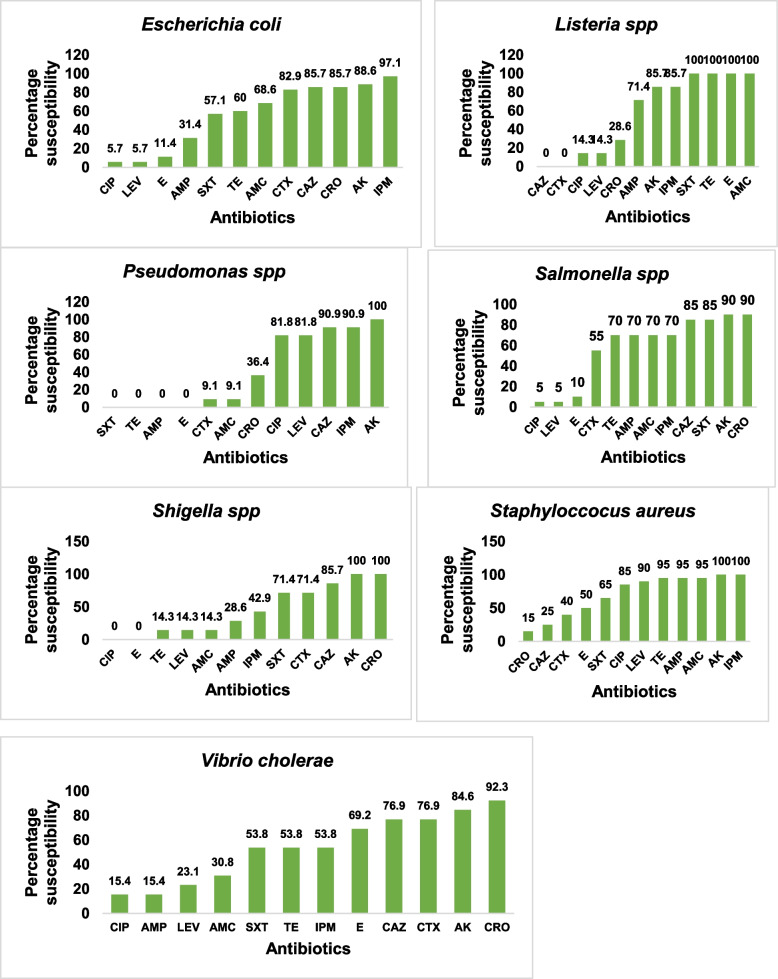
A total of 123 bacteria pathogens were isolated from 40 canals in 20 LGAs. Prominent bacteria pathogens isolated included Escherichia coli (28.5%), Staphylococcus aureus (16,3%), Salmonella spp. (16.3%), Vibro cholerae (10.6%) and Shigella spp. (5.7%) see Table 5. Of major concern is the detection of V. cholerae O1 in 13 canals located in nine (45%) LGAs in Lagos. The LGAs implicated included Ajeromi Ifelodun, Ikorodu, Mushin, Oshodi/Isolo, Lagos Island, Surulere, Shomolu, Agege and Epe LGAs. (See Suppl-2 Table S1). In general, the isolates were susceptible to Amikacin (92.7%), Imipenem (83.7%) and Ceftazidime (70.7%) and least susceptible to Erythromycin (28.5%), Ciprofloxacin (30.1%), Levofloxacin (32.5%) and Ampicillin (45.5%). See Fig. 4
Table 5.
List of Bacteria species isolated
| Bacteria taxa | Number of isolates (%) |
|---|---|
| Escherichia coli | 35 (28.5) |
| Staphylococcus aureus | 20 (16.3) |
| Salmonella spp | 20 (16.3) |
| Psuedomonas spp | 11 (8.9) |
| Vibro cholerae | 13 (10.6) |
| Shigella spp | 7 (5.7) |
| Listeria spp | 7 (5.7) |
| Aeromonas hydrophila | 4 (3.3) |
| Enterobacter cloacae | 2 (1.6) |
| Citrobacter freundi | 1 (0.8) |
| Coagulase negative Staph | 1 (0.8) |
| Proteus vulgaris | 1 (0.8) |
| Pantoea agglomerans | 1 (0.8) |
| Total | 123 (100) |
Fig. 4.
Overall Antibiotic susceptibility profile of bacteria isolates from wastewater canals in Lagos State
Figure 5 shows the susceptibility profile of key priority bacteria pathogens isolated. There was overall low susceptibility to Erythromycin and Ciprofloxacin across all isolates. The Pseudomonas spp. isolated were resistant to Tetracycline, Ampicillin, Erythromycin and trimethoprim/sulfamethoxazole (Fig. 5). The V. cholerae species were susceptible to Ceftriaxone (92.3%), Amikacin (84.6%) and Ceftazidime (76.9%) but resistant to Ampicillin (69.2%), Cotrimoxazole (46.2%) and Tetracycline (38.5%). Carbapenem resistance was recorded in Shigella spp. (42.9%) and Vibrio cholerae (15.4%).
Fig. 5.
Suceptibility profile of key bacteria pathogens isolated from wastewater canals in Lagos State, to the Antibiotics tested. Key: AMP-ampicillin, AMC- amoxicillin/clavulanic acid, CTX-cefotaxime, CAZ-ceftazidime, IPM- imipenem, TE-tetracycline, CIP- ciprofloxacin, SXT-trimethoprim/sulfamethoxazole, AK- amikacin, LEV-Levofloxacin, E- erythromycin, CRO- Ceftriaxone
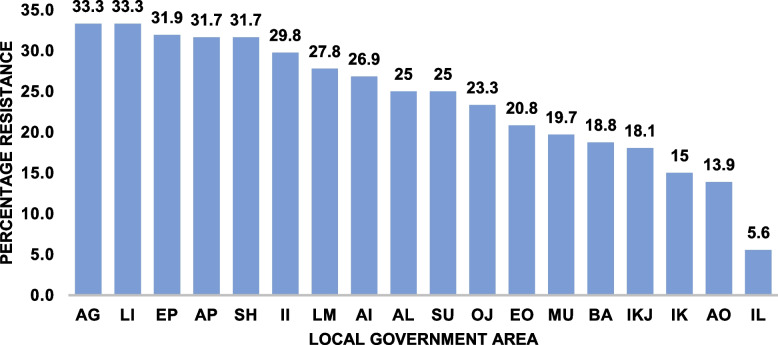
The prevalence of antibiotic resistance varied across the LGAs with highest level observed in Agege (33.3%) and lowest seen in Ibeju-lekki (5.6%) Fig. 6. The results showed that there was a positive correlation between community antibiotic resistance level and self-reported use of antibiotics without prescription r [18] = 0.24, p = 0.31 and community antibiotic resistance level and exposure to domestic animals r [18] = 0.18, p = 0.44, although the correlation was weak.
Fig. 6.
Overall antibiotic resistance profile across the 20 LGAs studied. Calculated as the percentage total resistance to all antibiotic tested weighted against the total number of isolates. KEY: AP = Apapa, EO = Eti-osa (EO), SH = Shomolu, KO = Kosofe, EP = Epe, IL = Ibeju-Lekki, IK = Ikorodu, AG = Agege, II = Ifako-Ijaye, AL = Alimosho, BA = Badagry, OJ = Ojo, AI = Ajeromi/Ifelodun, AO = Amuwo-Odofin, OI = Oshodi/Isolo, IKJ = Ikeja, MU = Mushin
Antibiotic resistant genes (ARG) detection
Extended spectrum betalactamse (ESBL) genes were the most abundant with a majority (87.5%) of the samples harboring the SHV genes (Suppl-2 Figure S3A&B) while 42.5% harbored the CTX-M gene (Table 6; Suppl-2 Figure S4 A&B). Only one sample (2.5%) was positive for macrolide efflux pump gene MsrA (Suppl-2 Figure S5) and none of the sampels habored macrolide resistant genes (ermF, ermX, ereA), methicillin resistant gene (MecA) and ESBL resistant genes (IMP, KPC, TEM and VEB).
Table 6.
Distribution of the Antimicrobial resistance genes detected across the different canals in Lagos State
| S/N | Canal ID | ARGs |
|---|---|---|
| 1 | AG C1 | SHV, CTX-M |
| 2 | AG C2 | SHV, CTX-M |
| 3 | AI C1 | SHV |
| 4 | AI C2 | SHV |
| 5 | AL C1 | SHV, MsrA |
| 6 | AL C2 | SHV |
| 7 | AO C1 | SHV, CTX-M |
| 8 | AO C2 | SHV, CTX-M |
| 9 | AP C1 | SHV, CTX-M |
| 10 | AP C2 | SHV, CTX-M |
| 11 | BA C1 | SHV |
| 12 | BA C2 | - |
| 13 | EO C1 | - |
| 14 | EO C2 | SHV |
| 15 | EP C1 | SHV, CTX-M |
| 16 | EP C2 | - |
| 17 | II C1 | SHV, CTX-M |
| 18 | II C2 | SHV |
| 19 | IK C1 | SHV, CTX-M |
| 20 | IK C2 | SHV, CTX-M |
| 21 | IKJ C1 | SHV |
| 22 | IKJ C2 | SHV, CTX-M |
| 23 | IL C1 | - |
| 24 | IL C2 | SHV |
| 25 | KO C1 | SHV, CTX-M |
| 26 | KO C2 | SHV |
| 27 | LI C1 | SHV, CTX-M |
| 28 | LI C2 | SHV |
| 29 | LM C1 | SHV |
| 30 | LM C2 | SHV |
| 31 | MU C1 | SHV |
| 32 | MU C2 | SHV, CTX-M |
| 33 | OI C1 | SHV, CTX-M |
| 34 | OI C2 | SHV |
| 35 | OJ C1 | SHV |
| 36 | OJ C2 | SHV |
| 37 | SH C1 | - |
| 38 | SH C2 | SHV, CTX-M |
| 39 | SU C1 | SHV |
| 40 | SU C2 | SHV, CTX-M |
Key: C1- Canal 1, C2- Canal 2
Discussions
Wastewater epidemiology is the future of disease surveillance for protecting and safeguarding public health [39]. Worldwide, before its use in tracking COVID-19, WBE has been used to track the spread of Vibro cholerae, Escherichia coli, Staphylococcus aureus, Psuedomonas species, Salmonella species and many other micro-organisms [7, 8, 40, 41]. SARS-CoV-2 RNA is able to persist in faeces due to the ability of the virus to infect Angiotensin-converting enzyme 2 (ACE2) -expressing cells in the small intestine [42] and faecal shedding of the virus by both symptomatic and asymptomatic COVID-19 patients has been extensively reported [39, 43, 44]. The quantities of the virus found in wastewater have been shown to directly correlate with the number of persons infected in the corresponding area [2, 39, 45]. Pecca and team measured SARS-CoV-2 RNA concentrations in primary sewage sludge and demonstrated the continued detection during COVID-19 outbreak in 2020 [5]. The authors further noted that when adjusted for time lags, SARS-CoV-2 RNA detection in wastewater can be used to track the rise and fall of cases seen in clinical test results and local COVID-19 hospital admissions [5]. In our study, SARS-CoV-2 RNA was not detected in the untreated wastewater canals of Lagos, which was consistent with the national report of few active cases at the time of sample collection [46]. Nonetheless, we recognise that effective wastewater-based disease monitoring must be carried out systematically, constantly and longitudinally to be effective in tracking and monitoring disease spread.
Epidemiological survey
Poor antibiotic stewardship culture including overuse, underuse and misuse have inevitably increased the environmental burden of antibiotic resistance among bacteria [11, 47]. Although a substantial number of the respondents in this study understood what an antibiotic was, 54.7% of the respondents admitted to using antibiotics without prescription over the last six months and 31.6% would stop their antibiotic medication when they feel better. These results show a significant increase when compared with our previous report of 31.3% self-medication rate among the general Nigerian public [25]. Unfortunately, these risky behaviours constitute major drivers of AMR in the communities and have continued to fester, re-enforcing the need for evidence-based behavioural change intervention. This was weakly supported by a positive correlation recorded between self-reported use of antibiotics without prescription and community level of antibiotic resistance. The low correlation coefficient recorded in this study, however, limited our ability to make equivocal statements on the relationship between exposure to animals and community AMR rate.
Detection of Enteropathogens in wastewater
The Lagos State government has estimated that the state generates around 2.2 billion cubic meters of wastewater every day. Unfortunately, only a small fraction of the wastewater is being treated and majority of the treatment plants are non-functional [48]. The traversing wastewater canals are prominent landmarks in Lagos State, and have become like huge funnels dispensing, in most cases, untreated effluents, from varied sources, into the lagoon [49]. These canals, which may be structured or unstructured, tend to overflow during heavy rainfalls due to blockages posing a public health threat to millions of residents [49, 50]. This overflowing of wastewater canal has been fingered to be responsible for the seasonal outbreak of infectious diarrhoeas diseases in Lagos and environs notably Cholera. Diarrhoea prevalence rate in Nigeria is currently one of the worst in Sub Saharan Africa (18.8%) and is above the 16% global average, accounting for over 16% of child deaths mainly amongst children under five years [51, 52] and responsible for 9% of childhood deaths in Lagos State [53].
Prominent bacterial enteropathogens isolated from the canals included Escheriachia coli, Salmonella spp, Vibrio cholerae and Shigella spp. A similar study by Dungeni et al. [41] reported Vibrio cholerae among other related pathogenic bacteria such as Escherichia coli and Salmonella typhimuriam on four wastewater treatment plants located in Gauteng Province, South Africa. The abundance of faecal indicator bacteria (coliforms) in the wastewater canals in Lagos State is a clear indication of faecal contamination and correlates with our descriptive survey findings with some respondents admitting to open defaecation and open-field waste disposal. Most of these wastes are washed directly into the canals during the heavy rainfalls and find their way into surrounding households contaminating drinking water and food. Other bacteria pathogens isolated in this study included staphylococcus aureus and Pseudomonas aeruginosa detected in approximately 50% and 25% of the canals respectively. These bacteria species exhibited high level of resistance (over 50%) to cephalosporins tested (Fig. 5) and has been listed by WHO as part of bacterial priority pathogens for AMR surveillance [54].
Cholerae is caused by the release of enterotoxins by the aetiological agent, Vibrio cholerae, resulting in an efflux of important cell nutrients including sodium and water, leading to diarrhoea and dehydration [55]. The current detection of Vibrio cholera in canals in Lagos State is a major concern and serves as a sensitive signal of a potential Cholera outbreak. Vibrio cholerae 01 has been implicated in Cholera out breaks in Nigeria especially in the North-eastern part with drinking of unsafe water, a major risk factor [56]. Several studies have reiterated the need for bacteria organisms causing illnesses of outbreak potential, such as Vibrio cholerae, with their characteristic environmental resilience, to be monitored in wastewater [8, 40]. Our findings align with the body of evidence for wastewater-based epidemiology. A subsequent cholera outbreak in Nigeria reported about 1,141 suspected cholera cases and over 30 deaths as of 11th June 2024 [24]. Lagos State declared an outbreak on the 15 June 2024 with 436 suspected cases across the 20 LGAs and a 5.4% case fatality rate [23]. This further gives credence to our findings and amplifies our concern on the role of these canals as reservoirs for infectious pathogens and the need for constant wastewater monitoring and treatment. As of the 21st of July 2024, total suspected cases rose to 4,809 (CFR 3.2%) from 35 states in Nigeria with Lagos State accounting for 65% (3,126 cases) of all suspected cases in the country and Lagos Island LGA (496 cases) in Lagos State accounting for 10% of all suspected cases reported in the country [57]. The World Health Organisation is currently lending its support through provision of vaccines and community outreach support aimed at halting community transmission with reasonable progress [23]. It is apparent that curbing future cholera outbreaks will require multifaceted approach ranging from vaccination to water, sanitation and hygiene measures, adequate and proper drainage system, and wastewater treatment. Nonetheless, wastewater surveillance can provide warning signals to warrant proactive measures to mitigate future outbreaks.
Detection of ARBs and ARGs
The reported prevalence of ARBs and ARGs in wastewater canals in Lagos State has several critical implications for both human health and environmental safety. The high levels of macrolide resistance observed in Listeria spp., Vibrio cholerae, and Staphylococcus aureus align with patterns in clinical isolates [58], signalling a concerning overlap between environmental and clinical resistance profiles. This indicates that wastewater may serve as a reservoir for resistant pathogens capable of reinfecting humans. The presence of MsrA in only one canal sample suggests that phenotypic resistance to macrolides could be mediated by alternative resistance mechanisms, which may complicate treatment strategies in clinical settings. Similar resistance pattern has also been reported in wastewater bacteria pathogens [7]. Infectious diseases caused by antibiotic resistance bacteria are major public health threat for humans and animals. Surveillance of wastewater for ARBs and ARGs has been advocated for as a means of mitigating the emergence and spread of AMR [7]. There is a global rise of bacteria with the potential to produce ESBL with the most common classes including blaTEM, blaSHV and blaCTX-M [59]. The widespread presence of ESBL genes (blaSHV and blaCTX-M) in 87.5% of wastewater samples underscores the role of wastewater as a reservoir for ARGs (antibiotic resistance genes). [59–61]. ESBL-producing bacteria are associated with limited treatment options and increased healthcare costs, especially in nosocomial infections [62]. Wastewater harbouring ARGs and ARBs (antibiotic-resistant bacteria) may facilitate the evolution and dissemination of multi-drug-resistant pathogens, posing a direct threat to public health. Although resistance to erythromycin was generally high across the isolates, the macrolide genes tested in this study were negative except for one sample harbouring MsrA implying that the observed phenotypic resistance to erythromycin maybe via other mechanism of resistance. These findings support assertions that wastewater is a major repository for ARGs and ARBs, creating opportunities for horizontal gene transfer between bacteria, potentially amplifying resistance in the broader environment. Furthermore, resistant bacteria from wastewater can contaminate agricultural irrigation systems and aquaculture, leading to the transfer of ARGs into the food chain.
Detection of non-tuberculous mycobacteria
The prevalence of non-tuberculous mycobacteria and associated disease is increasing across the globe. The source of non-tuberculous mycobacteria is wide and varied and they have been detected in soil, water, vegetation, and community environment [19]. In this study, 13 canals were positive for Mycobacterium kansasii while 11 were positive for Mycobacterium fortitium. This suggests that the risk of water contamination with these organisms is high. There are reports on the clinical significance of Mycobacterium kansasii and Mycobacterium fortitium as the most common rapidly growing mycobacteria and water as their main source of infection [19, 63]. Immune-compromised individuals are at high risk of being infected with Mycobacterium kansasii and Mycobacterium fortitium from water source [9]. Emerging infections with these organisms include skin and soft tissue infection, pulmonary infections and disseminated infection [9, 64]. Non-tuberculous mycobacteria are involved in causing different infections and some of them cause diseases similar to TB. Treatment of individuals infected with NTM is different from those infected with TB. Therefore, correct and rapid identification of mycobacteria at species level is important for effective treatment. Diseases caused by these bacteria are often indistinguishable from that caused by MTBC, thus, making accurate diagnosis challenging. Infections with MTBC is known to occur mainly through air-borne transmission, however, the role of wastewater in feaco-oral transmission of tuberculosis is poorly understood. There are evidence supporting the detection of MTBC in both treated and untreated wastewater [65, 66]. Although members of the MTBC were not detected in this study, there is rising concerns that wastewater containing MTBC could potentially contribute to indirect transmission of pulmonary or extra-pulmonary infections in humans and animals [67]. The current survey of wastewater canals provided sensitive signal of pathogens present in the population and outlined the different NTM present in the wastewater samples. This data can be used as a complement to epidemiological data on infectious diseases of public health importance.
Implication for health policy and interventions
This study provided useful information that can be harnessed to inform local public health policies and to Strengthen existing policies especially for antimicrobial stewardship and restricting the indiscriminate use of antibiotics. We, therefore, recommend regular and sustained monitoring of wastewater for ARGs and ARBs to track emerging resistance trends and inform public health strategies. Advanced wastewater treatment methods capable of reducing ARGs and ARBs, such as membrane filtration or advanced oxidation processes can be implemented [68]. Additionally, community education on the risk posed by improper waste disposal and wastewater management through awareness campaigns advocating for the responsible use of antibiotics will go a long way to mitigate the spread of antibiotic resistance.
Limitations
The study had several limitations. One major one being that the study was a cross-sectional survey intended to provide a one-time point indication of the prevalent pathogens in the wastewater canals. This limited the ability to track temporal trends in pathogen prevalence and resistance patterns. Future studies would involve longitudinal monitoring of seasonal or time-based variations with goal of identifying trends in pathogen loads and resistance patterns over time. Effective surveillance will entail periodic monitoring of wastewater canals for pathogens of public health importance which will avert huge humanitarian disaster. We present here, the findings of our pilot survey of wastewater canals in Lagos State, Nigeria. The study did not detect resistant genes from individual bacteria isolates but attempted to demonstrate the role of wastewater canals as a reservoir of ARGs that may be of significant health risk to humans if exposed. The authors are also mindful that relating wastewater ARBs and ARGs with clinical infection is problematic as the source of ARB/ ARG in wastewater cannot be only from symptomatic human individuals but can also be from asymptomatic carriers as well as from animal sources. The use of antibiotics without prescription (in the last six months) reported in this study were purely self-reported by the respondents as a YES or No question which was categorically analysed. The ARGs reported in this study were not comprehensive but targeted macrolides due to the perceived prevalent Azithromycin use for COVID-19 related symptoms and clinical significance of ESBL producing bacteria as a public health threat. The descriptive epidemiological survey was not a focus of this study and was minimally reported. However, it will be more comprehensively addressed as part of a subsequent community based-epidemiological report.
Conclusions
This study highlights importance of wastewater-based epidemiology (WBE) as an early warning system for infectious disease surveillance and exposes the presence of antibiotic resistant enteropathogens in canals in Lagos State with potential to cause diarrhoea epidemic. The detection of Vibrio cholerae O1 in 9 out of the 20 LGAs in wastewater canals in Lagos and subsequent outbreak of Cholera further reiterates the utility of wastewater monitoring as a sensitive, warning signal of impending outbreak. Wastewater monitoring can be used for effective surveillance of communities with high risk of disease outbreak as well as transmission. Although our study was limited by cross-sectional nature of the design, we provide evidence to inform proactive measures to mitigate future outbreaks of cholera and other diarrheal diseases in the state. Future longitudinal monitoring will enable a more comprehensive understanding of seasonal or time-based trends in infectious pathogens prevalence and spread of AMR genes, facilitating targeted interventions to combat future outbreaks. Additionally, diagnostic algorithm and treatment guidelines for NTM infections should be developed by the National TB Control programme in addition to awareness creation in the communities on the presence of mycobacteria species in environmental samples such as canals. The study therefore provides evidence-based recommendations for more stringent measures to be taken by health authorities to disinfect the wastewater at community level. Targeted behavioural change interventions are crucial to curb community risk behaviours like open defaecation, open field waste disposal and self-medication which drives emergence and spread of antimicrobial resistance. Wastewater monitoring has emerged as a valuable public health tool with potential for providing robust data for disease prevention, outbreak management, and environmental protection. Its standardization and implementation should be prioritized to build resilient, data-driven public health systems capable of addressing current and emerging challenges. Rehabilitation of wastewater treatment plants in Lagos State with effective monitoring of outbreak-prone pathogenic organisms is highly recommended.
Supplementary Information
Acknowledgements
The authors wish to acknowledge the management of the Nigerian Institute of Medical Research for providing the funding and enabling environment for this project. We are also grateful to the Industrial training (IT) students at the Centre for Infectious Diseases Research for their secretarial and technical roles. We want to specifically acknowledge Ms Zainab Abdulganniyu, Esther Momoh and Ahmed Tajudeen for technical support. We also express our gratitude to the Lagos State Ministry of Health that gave approval to conduct this study at the communities where samples were collected. Sincere thanks to the study participants at the community level, Environmental Health Officers and Local Government Area management team.
Authors’ contributions
All listed authors collaborated to carry out this work. Author EEC Conceptualised and designed the study and wrote the protocol. Authors EEC, AO, CK, NO, JS, MA and RAA revised the protocol, designed the data collection tool. Authors WO, DA, EA, OA and KO were involved in the sample collection. EEC, OA, KO, TI, JS and NO performed the laboratory analysis, entered and helped in interpreting data. EEC wrote the draft of the manuscript. Authors KO and EEC performed data analysis and visualization. Authors AM and RAA supervised the work, reviewed the drafts and provided suggestions. All authors contributed to the literature searches and approved the final manuscript. All authors read and approved the final manuscript.
Funding
This study received funding support from the Microbiology Department of the Nigerian Institute of Medical Research but did not receive any Grant from local or international body.
Data availability
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Declarations
Ethics approval and consent to participate
Ethical clearance was obtained from Nigerian Institute of Medical Research Institutional Review Board (IRB/21/068) and the Lagos Ministry of Health (LSMH/9117/1/98). Also, participants in the descriptive epidemiological survey were required to sign informed consent form and questionnaires were used to obtain demographic information and data on the predictors and risk behaviours. All procedures were performed in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ Sci Technol Lett. 2020;7(7):511–6. [DOI] [PubMed] [Google Scholar]
- 2.Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;15(181):115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Núñez-Delgado A. What do we know about the SARS-CoV-2 coronavirus in the environment? Sci Total Environ. 2020;20(727):138647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;1(728):138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol. 2020;38(10):1164–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas K, Hillary LS, Malham SK, McDonald JE, Jones DL. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr Opin Environ Sci Health. 2020;17:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiwari A, Kurittu P, Al-Mustapha AI, Heljanko V, Johansson V, Thakali O, et al. Wastewater surveillance of antibiotic-resistant bacterial pathogens: A systematic review. Front Microbiol. 2022;13. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2022.977106. Cited 2023 Jul 16. [DOI] [PMC free article] [PubMed]
- 8.Okeyo AN, Nontongana N, Fadare TO, Okoh AI. Vibrio Species in Wastewater Final Effluents and Receiving Watershed in South Africa: Implications for Public Health. Int J Environ Res Public Health. 2018;15(6):1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falsafi S, Bostanabad SZ, Feizabadi MM, Ali R, Shahraki A, Sheikhi N, et al. Isolation and Molecular Identification of Mycobacterium fortuitum isolates from Environmental water and clinical samples at different regions of Iran. Bull Env Pharmacol LifeSci. 2015;4(10):63–8.
- 10.Global action plan on antimicrobial resistance. Available from: https://www.who.int/publications-detail-redirect/9789241509763. Cited 2023 Aug 21.
- 11.Mao D, Yu S, Rysz M, Luo Y, Yang F, Li F, et al. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015;85:458–66. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen AQ, Vu HP, Nguyen LN, Wang Q, Djordjevic SP, Donner E, et al. Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci Total Environ. 2021;783:146964. [DOI] [PubMed] [Google Scholar]
- 13.Chukwu EE, Musa AZ, Enwuru C, Ohihion A, Bamidele T, Olukosi A, et al. Determinants of Antimicrobial Use for Covid-19 Related Symptoms among Nigerians. West Afr J Med. 2021;38(3):213–21. [PubMed] [Google Scholar]
- 14.Abdelmalek SM, Mousa A. Azithromycin Misuse During the COVID-19 Pandemic: A Cross-Sectional Study from Jordan. Infect Drug Resist. 2022;15:747–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut Barking Essex 1987. 2009;157(11):2893–902. [DOI] [PubMed]
- 16.Wright GD. Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol. 2010;13(5):589–94. [DOI] [PubMed] [Google Scholar]
- 17.Matthiessen L, Bergström R, Dustdar S, Meulien P, Draghia-Akli R. Increased momentum in antimicrobial resistance research. The Lancet. 2016;388(10047):865. [DOI] [PubMed] [Google Scholar]
- 18.Cook JL. Nontuberculous mycobacteria: opportunistic environmental pathogens for predisposed hosts. Br Med Bull. 2010;96:45–59. [DOI] [PubMed] [Google Scholar]
- 19.Holton J. Non-Tuberculous Mycobacteria: An Emerging Clinical Problem. SPG BioMed. 2019;1(2).
- 20.Cook VJ, Turenne CY, Wolfe J, Pauls R, Kabani A. Conventional Methods versus 16S Ribosomal DNA Sequencing for Identification of Nontuberculous Mycobacteria: Cost Analysis. J Clin Microbiol. 2003;41(3):1010–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe AK, Kachur SP, Yoon SS, Lynch M, Slutsker L, Steketee RW. Caution is required when using health facility-based data to evaluate the health impact of malaria control efforts in Africa. Malar J. 2009;8(1):209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagos State Government_LGA Statistics. 2020. LAGOS BUREAU OF STATISTICS: Local Government Statistics. Available from: https://mepb.lagosstate.gov.ng/wp-content/uploads/sites/29/2018/06/Abstract-of-LG-Statistics-2017editted.pdf
- 23.WHO | Regional Office for Africa. 2024. Rallying action to curb cholera in Nigeria’s Lagos state. Available from: https://www.afro.who.int/countries/nigeria/news/rallying-action-curb-cholera-nigerias-lagos-state
- 24.Reporters O. Cholera outbreak: Nigeria runs out of vaccine as death toll hits 40. Punch Newspapers. 2024. Available from: https://punchng.com/cholera-outbreak-nigeria-runs-out-of-vaccine-as-death-toll-hits-40/. Cited 2024 Aug 2.
- 25.Chukwu EE, Oladele DA, Awoderu OB, Afocha EE, Lawal RG, Abdus-salam I, et al. A national survey of public awareness of antimicrobial resistance in Nigeria. Antimicrob Resist Infect Control. 2020;9(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma SK, Mudgal SK, Thakur K, Gaur R. How to calculate sample size for observational and experimental nursing research studies?. Nat J Physiol, Pharm and Pharmacol. 2020;10(01).
- 27.National Population Commission - NPC, ICF. Nigeria Demographic and Health Survey 2018 - Final Report. Abuja, Nigeria: NPC and ICF; 2019. Available from: http://dhsprogram.com/pubs/pdf/FR359/FR359.pdf
- 28.Katayama H, Shimasaki A, Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Appl Environ Microbiol. 2002;68(3):1033–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi K, Tripathi PC, Nema S, Shrivastava AK, Dwiwedi K, Dhanvijay AK. Modified Petroff’s Method: an excellent simplified decontamination technique in comparison with Petroff’s Method. IJRTSAT. 2014;10:461–4. [Google Scholar]
- 30.Sarkar M, Chakraborty S, Kundu D, Ghosh S, Khan A, Karmakar D, et al. Isolation and Characterization of Bacteria from Sewage and Pond Water, Malda. India Acta Sci Microbiol. 2019;2(9):28–34. [Google Scholar]
- 31.Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet. 2022;399(10325):629–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical & Laboratory Standards Institute. M100Ed33 | Performance Standards for Antimicrobial Susceptibility Testing, 33rd Edition. Available from: https://clsi.org/standards/products/microbiology/documents/m100/. Cited 2023 Jul 16.
- 33.Chen J, Michel FC, Sreevatsan S, Morrison M, Yu Z. Occurrence and Persistence of Erythromycin Resistance Genes (erm) and Tetracycline Resistance Genes (tet) in Waste Treatment Systems on Swine Farms. Microb Ecol. 2010;60(3):479–86. [DOI] [PubMed] [Google Scholar]
- 34.De R, Mukhopadhyay AK, Dutta S. Molecular Analysis of Selected Resistance Determinants in Diarrheal Fecal Samples Collected From Kolkata, India Reveals an Abundance of Resistance Genes and the Potential Role of the Microbiota in Its Dissemination. Front Public Health. 2020;8. Available from: https://www.frontiersin.org/journals/public-health/articles/10.3389/fpubh.2020.00061/full. Cited 2024 Nov 16. [DOI] [PMC free article] [PubMed]
- 35.Worku S, Abebe T, Seyoum B, Alemu B, Denkayehu G, Seyoum T, et al. Molecular characterization of carbapenemase and extended spectrum beta-lactamase producing Acinetobacter baumannii isolates causing surgical site infections in Ethiopia. BMC Infect Dis. 2024;24(1):459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trung NT, Hien TTT, Huyen TTT, Quyen DT, Binh MT, Hoan PQ, et al. Simple multiplex PCR assays to detect common pathogens and associated genes encoding for acquired extended spectrum betalactamases (ESBL) or carbapenemases from surgical site specimens in Vietnam. Ann Clin Microbiol Antimicrob. 2015;14(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-Specific and Ubiquitous-DNA-Based Assays for Rapid Identification of Staphylococcus aureus. J Clin Microbiol. 1998;36(3):618–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Characterization of carbapenemases. ESBLs, and plasmid-mediated quinolone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J Infect Public Health. 2018;11(1):64–8. [DOI] [PubMed] [Google Scholar]
- 39.Ladyzhets B. What toilets can reveal about COVID, cancer and other health threats. Nature. 2024;628(8008):492–4. [DOI] [PubMed] [Google Scholar]
- 40.Zohra T, Ikram A, Salman M, Amir A, Saeed A, Ashraf Z, et al. Wastewater based environmental surveillance of toxigenic Vibrio cholerae in Pakistan. PLoS ONE. 2021;16(9):e0257414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dungeni M, Merwe RR van der, Momba M. Abundance of pathogenic bacteria and viral indicators in chlorinated effluents produced by four wastewater treatment plants in the Gauteng Province, South Africa. Water SA. 2010;36(5). Available from: https://www.ajol.info/index.php/wsa/article/view/61994. Cited 2023 Jul 18.
- 42.Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5(47):eabc3582. [DOI] [PMC free article] [PubMed]
- 43.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–5. [DOI] [PubMed] [Google Scholar]
- 44.Wang XW, Li JS, Guo TK, Zhen B, Kong QX, Yi B, et al. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J Gastroenterol WJG. 2005;11(28):4390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graham KE, Loeb SK, Wolfe MK, Catoe D, Sinnott-Armstrong N, Kim S, et al. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ Sci Technol. 2021;55(1):488–98. [DOI] [PubMed] [Google Scholar]
- 46.Nigeria: WHO Coronavirus Disease (COVID-19) Dashboard With Vaccination Data. Available from: https://covid19.who.int. Cited 2023 Sep 21.
- 47.Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sànchez-Melsió A, et al. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015;69:234–42. [DOI] [PubMed] [Google Scholar]
- 48.Asemota L, Alkhaddar R, Sertyesilisik B, Tunstall A. Wastewater management in Lagos State: Moving toward a more sustainable approach. Environ Qual Manag. 2011;20(4):63–72. [Google Scholar]
- 49.Nigeria G. The Guardian Nigeria News - Nigeria and World News. 2016. Lagos Canals: Polluted city’s waterways. Available from: https://guardian.ng/sunday-magazine/lagos-canals-polluted-citys-waterways/. Cited 2024 Nov 12.
- 50.Pulitzer Center. Public Health Impact of the Drainage System in Lagos: A Photo Essay. Available from: https://pulitzercenter.org/stories/public-health-impact-drainage-system-lagos-photo-essay. Cited 2024 Nov 12.
- 51.Okafor IP, Akinyemi OT, Wika-Kobani BN, Olubodun T, Eze UT. Childhood diarrhoea: a cross-sectional survey on maternal knowledge, hygienic practices and use of oral zinc for home management in a Nigerian community. Pan Afr Med J. 2022;42:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The United Nations Children’s Fund (UNICEF)/World Health Organization (WHO), 2009. Diarrhoea : Why children are still dying and what can be done. Available from: https://iris.who.int/bitstream/handle/10665/44174/9789241598415_eng.pdf?sequence=1
- 53.Akinyemi YC. Spatial pattern and determinants of diarrhoea morbidity among under-five-aged children in Lagos State. Nigeria Cities Health. 2022;6(1):180–91. [Google Scholar]
- 54.WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Available from: https://www.who.int/publications-detail-redirect/9789240093461. Cited 2024 May 30.
- 55.Chatterjee SN, Maiti M. Vibriophages And Vibriocins: Physical, Chemical, And Biological Properties. In: Lauffer MA, Maramorosch K, editors. Advances in Virus Research. Academic Press; 1984. p. 263–312. Available from: https://www.sciencedirect.com/science/article/pii/S006535270860411X. Cited 2023 Jul 18. [DOI] [PubMed]
- 56.Fagbamila IO, Abdulkarim MA, Aworh MK, Uba B, Balogun MS, Nguku P, et al. Cholera outbreak in some communities in North-East Nigeria, 2019: an unmatched case–control study. BMC Public Health. 2023;23(1):446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nigeria Centre for Disease Control and Prevention. Available from: https://ncdc.gov.ng/diseases/sitreps/?cat=7&name=An%20update%20of%20Cholera%20outbreak%20in%20Nigeria. Cited 2024 Aug 14.
- 58.Chukwu EE, Awoderu OB, Enwuru CA, Afocha EE, Lawal RG, Ahmed RA, et al. High prevalence of resistance to third-generation cephalosporins detected among clinical isolates from sentinel healthcare facilities in Lagos, Nigeria. Antimicrob Resist Infect Control. 2022;11(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nzima B, Adegoke AA, Ofon UA, Al-Dahmoshi HOM, Saki M, Ndubuisi-Nnaji UU, et al. Resistotyping and extended-spectrum beta-lactamase genes among Escherichia coli from wastewater treatment plants and recipient surface water for reuse in South Africa. New Microbes New Infect. 2020;31(38):100803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oliveira M, Truchado P, Cordero-García R, Gil MI, Soler MA, Rancaño A, et al. Surveillance on ESBL-Escherichia coli and Indicator ARG in Wastewater and Reclaimed Water of Four Regions of Spain: Impact of Different Disinfection Treatments. Antibiotics. 2023;12(2):400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fadare FT, Okoh AI. The Abundance of Genes Encoding ESBL, pAmpC and Non-β-Lactam Resistance in Multidrug-Resistant Enterobacteriaceae Recovered From Wastewater Effluents. Front Environ Sci. 2021;9. Available from: https://www.frontiersin.org/journals/environmental-science/articles/10.3389/fenvs.2021.711950/full.Cited 2024 Aug 16.
- 62.Bouassida K, Jaidane M, Bouallegue O, Tlili G, Naija H, Mosbah AT. Nosocomial urinary tract infections caused by extended-spectrum beta-lactamase uropathogens: Prevalence, pathogens, risk factors, and strategies for infection control. Can Urol Assoc J. 2016;10(3–4):E87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steadham JE. High-catalase strains of Mycobacterium kansasii isolated from water in Texas. J Clin Microbiol. 1980;11(5):496–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown-Elliott BA, Wallace RJ. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev. 2002;15(4):716–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Exploring the role of wastewater-based epidemiology in understanding tuberculosis burdens in Africa. Environ Res. 2023;231:115911. [DOI] [PMC free article] [PubMed]
- 66.Mtetwa HN, Amoah ID, Kumari S, Bux F, Reddy P. Molecular surveillance of tuberculosis-causing mycobacteria in wastewater. Heliyon. 2022;8(2):e08910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mtetwa HN, Amoah ID, Kumari S, Bux F, Reddy P. The source and fate of Mycobacterium tuberculosis complex in wastewater and possible routes of transmission. BMC Public Health. 2022;22:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oladele O. Advanced Wastewater Treatment Systems: Emerging Technologies and Innovations. 2024. Available from https://www.researchgate.net/publication/385214693.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].