Highlights
-
•
Edwardsiella tarda induced a RIPK3-dependent necroptosis in RAW264.7 cells.
-
•
E. tarda infection activated the RIG-I/MDA5 pathway in RAW264.7 cells.
-
•
RIG-I and MDA5 were required for E. tarda-induced necroptosis in RAW264.7 cells.
-
•
Our results unveiled a RIG-I/MDA-mediated necroptosis upon bacterial infection.
Keywords: RIG-I, MDA5, Necroptosis, Edwardsiella tarda, Infection
Abstract
RIG-I and MDA5 are members of RIG-I-like receptors (RLRs) that detect viral RNA within the cytoplasm and subsequently initiate antiviral immune responses. Necroptosis is a form of programmed cell death (PCD) executed by mixed lineage kinase domain-like (MLKL), which, upon phosphorylation by receptor-interacting protein kinase 3 (RIPK3), causes necrotic cell death. To date, no link between RLRs and necroptosis has been observed during bacterial infection. Edwardsiella tarda is a zoonotic bacterial pathogen that can thrive in host macrophages. In a previous study, we identified RIG-I and MDA5 as two hub factors of RAW264.7 cells responsive to E. tarda infection. The present study aimed to determine the specific form of cell death triggered by E. tarda and explore the association between RIG-I/MDA5 and PCD in the context of bacterial infection. Our results showed that E. tarda infection induced RIPK3-MLKL-mediated necroptosis, rather than pyroptosis or apoptosis, in RAW264.7 cells. Meanwhile, E. tarda promoted RIG-I/MDA5 production and activated the RIG-I/MDA5 pathways that led to IRF3 phosphorylation, IFN-β secretion, and interferon-stimulated gene (ISG) and cytokine expression. Both RIG-I and MDA5 were essential for E. tarda-triggered necroptosis and required for effective inhibition of intracellular bacterial replication. Furthermore, the regulatory effect of RIG-I/MDA5 on necroptosis was not affected by type I IFN or TNF-α signaling blockage. Together these results revealed that necroptosis could be triggered by intracellular bacterial infection through the RIG-I/MDA5 pathways, and that there existed intricate interplays between PCD and RLRs induced by bacterial pathogen.
Graphical abstract
1. Introduction
Retinoic acid-inducible gene I (RIG-I) like receptors (RLRs) are a family of DExD/H box RNA helicases, including RIG-I, melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (Loo and Gale, 2011; Yoneyama and Fujita, 2009). During viral infection, RIG-I and MDA5 can sense viral double-stranded RNA (dsRNA) and interact with mitochondrial antiviral signaling protein (MAVS) via the caspase activation and recruitment domain (CARD), thereby eliciting the production of type I interferons (IFN1) through pathways mediated by interferon regulatory factor 3 (IRF3) and IRF7 (Schweibenz et al., 2022; Seth et al., 2005; Thoresen et al., 2021). IFN1 binds to its receptor (IFNAR) and induces downstream signaling, resulting in the expression of a variety of interferon-stimulated genes (ISGs) to facilitate pathogen clearance (Brisse and Ly, 2019). While viruses are well known to be inducers of varied RLR cascades, only three intracellular bacteria, i.e., Listeria monocytogenes, Mycobacterium tuberculosis, and Salmonella enterica serovar Typhimurium, have been reported to activate the RLR signaling (Abdullah et al., 2012; Bullen et al., 2023; Hagmann et al., 2013; Schmolke et al., 2014). Furthermore, the immunological consequences and signaling processes of RLR activation during bacterial infection are largely unclear.
Necroptosis is an inflammatory form of programmed cell death (PCD) executed by the pore-forming protein mixed lineage kinase domain-like (MLKL). MLKL exists as an inactive protein that can be activated by receptor-interacting protein kinase (RIPK) 3, which phosphorylates MLKL at the C-terminal region, the hallmark of necroptosis (He et al., 2016; Linkermann and Green, 2014). The phosphorylated MLKL forms oligomers, which translocate to the plasma membrane and form channels in the membrane, leading to membrane rupture and the release of cytoplasmic contents, including damage-associated molecular patterns (DAMPs) (Humphries et al., 2015; Seo et al., 2021; Sethi et al., 2022). Necroptosis is known to be triggered by bacterial pathogens, but no reports on bacteria-induced connection between necroptosis and the RLR pathways have been documented.
Edwardsiella tarda is a Gram-negative, facultative intracellular bacterial pathogen with a wide range of hosts, including fish and humans (Leung et al., 2012; Li et al., 2019; Sun et al., 2022). E. tarda is equipped with an array of virulence factors that enable bacterial survival and proliferation within host cells, including macrophages (Chen et al., 2017; Li et al., 2021; Merrifield et al., 2012; Sun et al., 2012; Wei et al., 2019; Zhou et al., 2015). In a recent study, we found that E. tarda infection promoted the expression of MLKL and RIPK3 in the teleost fish Paralichthys olivaceus (Hao et al., 2024), but the mechanism is unclear. No other association of E. tarda and necroptosis has been reported. In this study, we discovered that E. tarda induced necroptosis of murine macrophages in a manner that depended on activation of the RIG-I and MDA5 pathways. Our results added new insights into the roles of programmed cell death and RLRs in the mechanism of host-bacterial pathogen interaction.
2. Materials and methods
2.1. Cell lines and bacteria strains
HEK293T cells (American Type Culture Collection) and RAW264.7 cells (American Type Culture Collection) were cultured in DMEM medium (Corning, NY, USA) supplemented with 10 % fetal bovine serum at 37 °C with 5 % CO2. RAW264.7 derived cells of RAW-Lucia™ ISG (wild type), RAW-Lucia™ ISG-KO-RIG-I (RIG-I-/-), and RAW-Lucia™ ISG-KO-MDA5 (MDA5-/-) were purchased from InvivoGen (San Diego, CA, USA) and cultured in DMEM with 200 µg/mL Zeocin (InvivoGen, San Diego, CA, USA). To determine the luciferase activity of these cells, the cell culture supernatants were mixed with QUANTI-Luc™−4 Reagent (InvivoGen, San Diego, CA, USA), and the luciferase activity was measured using a microplate reader (Biotek, Winooski, USA). The Edwardsiella tarda strain TX1 and the GFP-expressing TX1 (TX1 G) were grown as reported previously (Sui et al., 2017; Zhang et al., 2008).
2.2. Cellular infection
RAW264.7 cells were infected with E. tarda as reported previously (Li et al., 2019). In brief, the cells were infected with E. tarda at a multiplicity of infection (MOI) of 3:1 at 30 °C for 1 h The extracellular bacteria were killed by gentamicin (300 µg/mL). The cells were washed with PBS and incubated for various hours in Opti-MEM containing 30 µg/mL gentamicin for subsequent assays. The control group was treated in the same manner with PBS.
2.3. Cell death observation
RAW264.7 cells were plated into 24-well plate or 35 mm glass-bottom culture dish (Nest Biotechnology, Wuxi, China) at about 60 % confluency and subjected to the indicated treatment. PI (Invitrogen, Carlsbad, CA, USA) was added to the culture medium to monitor cell death. To visualize the cell death process, the cells were infected with GFP-expressing E. tarda in the presence of Annexin V-Alexa Fluor 647 (Yeasen, Shanghai, China), and the process of cell death were recorded using confocal microscope (Carl Zeiss, Jena, Germany) as described previously (Jiang et al., 2023; Xu et al., 2022a). Lactate dehydrogenase (LDH) release was perfromed as reported previously (Xu et al., 2022a).
2.4. Immunoblotting and ELISA
RAW 264.7 and derived cells were infected with E. tarda as above. The preparation of cell lysates and culture supernatants, and subsequent immunoblotting were conducted as reported previously (Xu et al., 2022a). In brief, the lysates and supernatants were collected and subjected to 12 % SDS-PAGE (GenScript, Piscataway, NJ). The proteins were transferred to nitrocellulose blotting membranes (Millipore, MA, USA), and the membranes were blocked with 5 % BSA or 5 % skimmed milk for 1 h and treated with appropriate primary antibodies. The membranes were then incubated with HRP-conjugated secondary antibody for 1 h, and the target proteins were visualized using an ECL kit (Sparkjade Biotechnology Co. Ltd, Shandong, China). Anti-phospho-RIPK3 antibody (ab222320), anti-phospho-MLKL antibody (ab196436), anti-GSDMD (ab219800) antibody, anti-caspase-3 (ab219800) antibody, and anti-MAVS antibody (ab31334) were purchased from Abcam (Cambridge, UK). Anti-phosphp-IRF3 antibody (D601 M), anti-MDA5 antibody (D74E4), and anti-RIG-I antibody (D14G6) were purchased from Cell Signaling Technology (Beverly, MA, USA). Anti-IRF3 antibody (bs-2993R) was purchased from Bioss (Beijing, China). Anti-RIPK3 antibody (A5431), anti-MLKL antibody (A17312), anti-IFN-β antibody, (A16223), anti-GAPDH antibody (AC001), and anti-β-actin antibody (AC026) were purchased from ABclonal (Wuhan, China). Anti-ISG54 antibody (12604-1-AP) was purchased from Proteintech (Wuhan, China).
The secretion of IFN-β was quantified with a Mouse IFN-β ELISA kit (Finetest, Wuhan, China) following the manufacturer's protocol.
2.5. Co-immunoprecipitation (Co-IP)
Co-IP was performed as reported previously (Wu et al., 2024). Briefly, HEK293T cells were transfected with plasmids expressing the indicated Myc- or Flag-tagged proteins for 24 h and lysed with lysis buffer. The cells lysates were loaded onto anti-Myc affinity gels (Beyotime, Shanghai, China), and the gels were incubated at room temperature for 4 h, followed by washing with PBS for five times. The samples were analyzed by immunoblotting with anti-Myc/Flag antibodies as described above.
2.6. Nitric oxide (NO) measurement
RAW264.7 cells were incubated with 2′,7′-dichlorofluorescein diacetate (DCFH-DA) (Beyotime, Shanghai, China) at 37 °C for 2 h and then washed with PBS. The cells were infected with E. tarda as above for different hours. NO production was measured at 495 nm excitation and 515 nm emission using a fluorescence spectrophotometer (Infinite M1000, Tecan, Switzerland).
2.7. Antibody neutralization
Antibody blocking of IFNAR1 was performed using InVivoMab anti-mouse IFNAR1 (BioXCell, New Hampshire, USA) and InVivoMab anti-mouse IgG1 isotype control (BioXCell, New Hampshire, USA). Antibodies were added to RAW264.7 cells at a final concentration of 100 ng/ml, followed by incubation for 2 h. For antibody blocking of TNF-α, different doses of InVivoMAb anti-mouse TNF-α (BioXCell, New Hampshire, USA) or isotype control were added to E. tarda-infected RAW264.7 cell.
2.8. Quantitative real-time PCR (qRT–PCR)
RAW264.7 cells were infected with E. tarda as above. Total RNA was extracted from the cells with a FastPure Cell/Tissue Total RNA Isolation Kit V2 (Vazyme Biotech Co. Ltd., Nanjing, China). The total RNA was used for cDNA synthesis with a First Strand cDNA Synthesis Kit (TOYOBO, Osaka, Japan). qRT-PCR was conducted in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) as reported previously (Xu et al., 2022a). The PCR primers used are listed in Table S1.
2.9. RNA interference (RNAi) and RNA transfection
RNAi was performed as described previously (Xu et al., 2022b). Briefly, RAW264.7 cells were transfected with or without (control) small interfering RNA (siRNA) targeting RIG-1 (siRIG-I), MDA5 (siMDA5), MAVS (siMAVS), or the negative control siRNA (siNC) by Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. After transfection for 24 h, the efficiency of gene knockdown was verified by qRT-PCR and immunoblotting. The siRNAs used were synthesized by GenePharma (Shanghai, China) and listed in Table S2. For RNA transfection, E. tarda RNA was extracted as described above and transfected into RAW264.7 cells with Lipofectamine RNAiMAX.
2.10. Statistical analysis
Data were analyzed with Student's t-test (for Fig. 1C and Fig. 1, Fig. 2), one-way analysis of variance (ANOVA) (for Fig. 3), or two-way ANOVA (for all others). Statistical analysis was performed with GraphPad Prism 7 (www.graphpad.com/) software. Statistical significance was defined as p < 0.05.
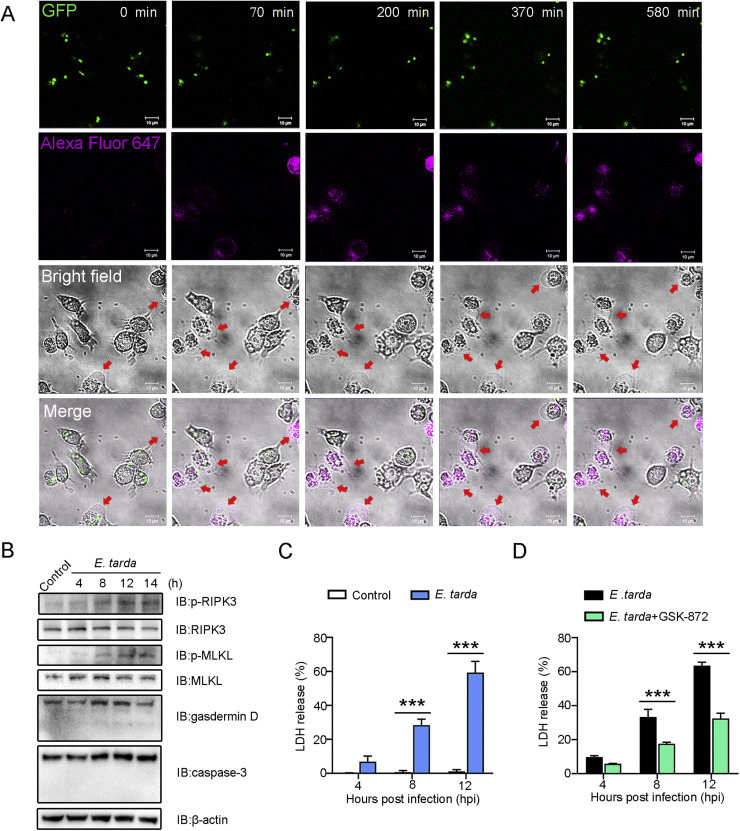
Fig. 1.
Edwardsiella tarda-induced death of RAW264.7 cells. (A) Representative time-lapse microscopic images showing Annexin V-Alexa Fluor 647 stained RAW264.7 cells infected with GFP-tagged E. tarda. Scale bar, 10 μm. The red arrows indicate dying cells. (B) RAW264.7 cells were infected with E. tarda for 4, 8, 12, or 14 h, and the cell lysate was immunoblotted with antibody against the indicated proteins. (C) RAW264.7 cells were infected with E. tarda for 4, 8, or 12 h, and LDH release was measured. (D) RAW264.7 cells were infected with E. tarda as above in the absence or presence of the RIPK3 inhibitor, GSK-872. LDH release was then determined. For (C) and (D), values are the means of three experimental replicates and shown as means ± SD. ***p < 0.001; **p < 0.01; *p < 0.05.
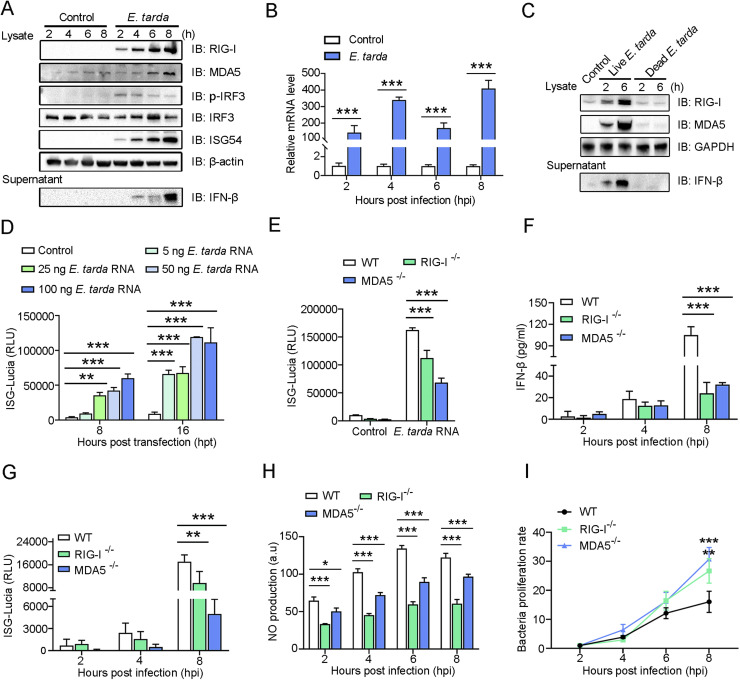
Fig. 2.
Effect of Edwardsiella tarda on RIG-I/MDA5 pathway activation. (A) RAW264.7 cells were infected with or without (control) E. tarda for indicated hours. The cell lysates and supernatants were immunoblotted (IB) with antibodies against the indicated proteins. (B) RAW264.7 cells were treated as above, and the IFN-β mRNA level was determined by qRT-PCR. (C) RAW264.7 cells were treated with live or dead E. tarda for different hours. RIG-I/MDA5 production and FN-β secretion were determined as above. (D) RAW264.7 cells were transfected with different doses of E. tarda RNA for different hours. ISG-Lucia activity was then assessed. (E) RIG-I-/-, MDA5-/-, or wild type (WT) RAW264.7 cells were treated with 50 ng E. tarda RNA for 16 h, and ISG-Lucia activity was then determined. (F-G) IFN-β secretion (F) and ISG-Lucia activity (G) in RIG-I-/-, MDA5-/-, or wild type (WT) RAW264.7 cells following E. tarda infection were determined. (H) RIG-I-/-, MDA5-/-, or WT RAW264.7 cells were incubated with a NO probe for 1 h and then infected with E. tarda. The production of NO was measured at different hours. (I) RIG-I-/-, MDA5-/-, or WT RAW264.7 cells were infected with E. tarda for indicated hours, and the intracellular bacterial proliferation rate was determined. For (B, and d-I), values are the means of three experimental replicates and shown as means ± SD. ***p < 0.001; **p < 0.01.
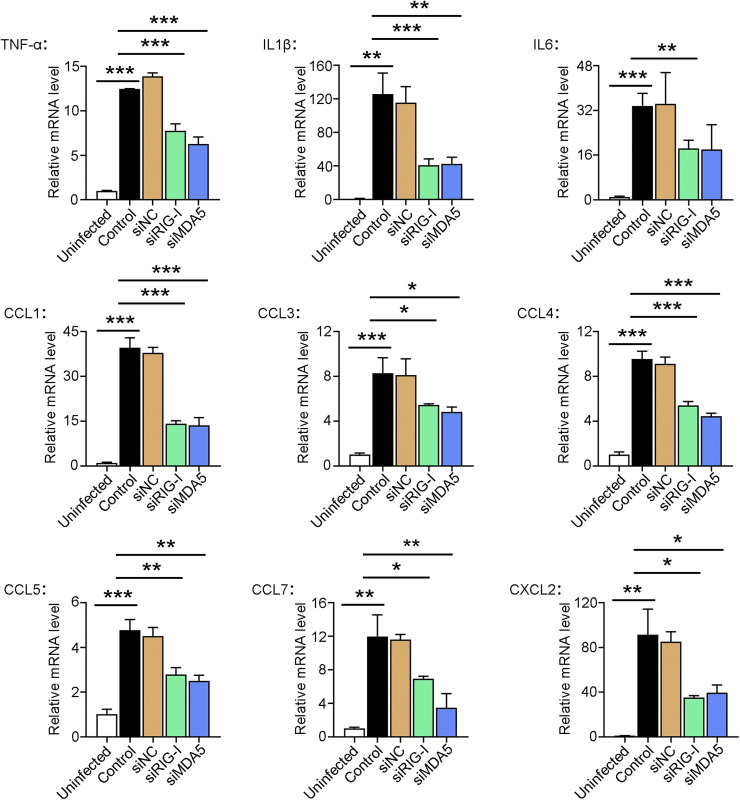
Fig. 3.
The effect of RIG-I/MDA5 knockdown on cytokine expression in Edwardsiella tarda-infected RAW264.7 cells. RAW264.7 cells were treated with or without (control) siRIG-I, siMDA5, or siNC. The cells were incubated with E. tarda or PBS (“Uninfected” group) for 4 h The mRNA levels of the inflammatory cytokines and chemokines in the cells were determined by qRT-PCR. Values are the means of three experimental replicates and shown as means ± SD. ***p < 0.001; **p < 0.01; *p < 0.05.
3. Results
3.1. E. tarda induces RIPK3-MLKL-mediated necroptosis
Time-lapse imaging showed that RAW264.7 cells infected with GFP-expressing E. tarda exhibited morphological changes over time, including cell membrane swelling and phosphatidylserine exposure (Annexin V-Alexa Fluor 647-positive), which ultimately led to membrane rupture (Fig. 1A, Video S1). These observations indicated a necrotic form of cell death induced by E. tarda. To further examine the molecular characteristics of the cell death, the activations of MLKL and gasdermin D (GSDMD), which are indicators of necroptosis and pyroptosis, respectively, were determined. The result showed that E. tarda induced a time-dependent increase in the phosphorylation of MLKL, along with the phosphorylation of RIPK3, the activating kinase of MLKL (Fig. 1B), suggesting the occurrence of necroptosis. In contrast, E. tarda infection had no apparent effect on the cleavage and activation of either GSDMD or caspase-3 (the effector of apoptosis) (Fig. 1B), suggesting the lack of pyroptosis and apoptosis. Furthermore, in support of the occurrence of necroptosis, the release of LDH was significantly elevated in E. tarda-infected cells. Both LDH release and MLKL phosphorylation were significantly attenuated in E. tarda-infected cells treated with GSK-872, a specific inhibitor of RIPK3 (Fig. 1C,D and Figure S1). The specific inhibitors of RIPK1 and MLKL, i.e., Nec-1 s and NSA, respectively, also significantly inhibited E. tarda-triggered cell death (Figure S2).
3.2. E. tarda activates the RIG-I/MDA5 pathways that inhibit intracellular bacterial replication
In a previous transcriptome analysis, we identified the genes encoding RIG-I (DDX58) and MDA5 (IFIH1) as the hub genes in the immune network of RAW264.7 cells in response to E. tarda infection (Li et al., 2019). In the present work, we found that E. tarda infection induced time-dependent productions of RIG-I and MDA5, which were accompanied by IRF3 phosphorylation and enhanced ISG54 production, IFN-β expression, and IFN-β secretion (Fig. 2A,B). In contrast, inactivated (dead) E. tarda had no apparent effect on RIG-I or MDA5 production (Fig. 2C). RAW264.7 cells transfected with E. tarda RNA exhibited a significant increase in ISG production in a manner that depended on the dose of the RNA (Fig. 2D). However, the ability of E. tarda RNA to induce ISG production was significantly reduced in RIG-I-/- and MDA5-/- RAW264.7 cells (Fig. 2E). Upon E. tarda infection, RIG-I-/- and MDA5-/- RAW264.7 cells exhibited significantly decreased IFN-β secretion, ISG production, and NO synthesis, than the wild type cells. (Fig. 2F-H). As a result, the bacterial proliferation rates in RIG-I-/- and MDA5-/- cells were significantly higher than that in the wild type cells (Fig. 2I). To further examine the importance of the RLR pathways in E. tarda infection, RIG-I and MDA5 expressions in RAW264.7 cells were knocked down by RNAi (Figure S3). Comparing with normal cells, in which E. tarda stimulated the expression of inflammatory cytokines (IL1β, IL6, and TNF-α) and a wide range of chemokines (CCL1/3/4/5/7 and CXCL2), cells with RIG-I and MDA5 knockdown exhibited significantly decreased expressions of these cytokines during E. tarda infection (Fig. 3).
3.3. RIG-I and MDA5 are essential for E. tarda-induced necroptosis
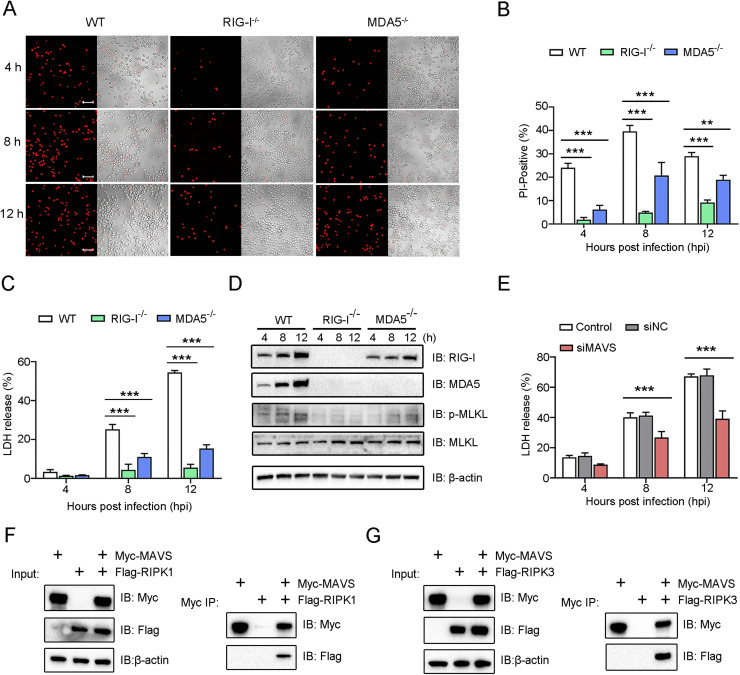
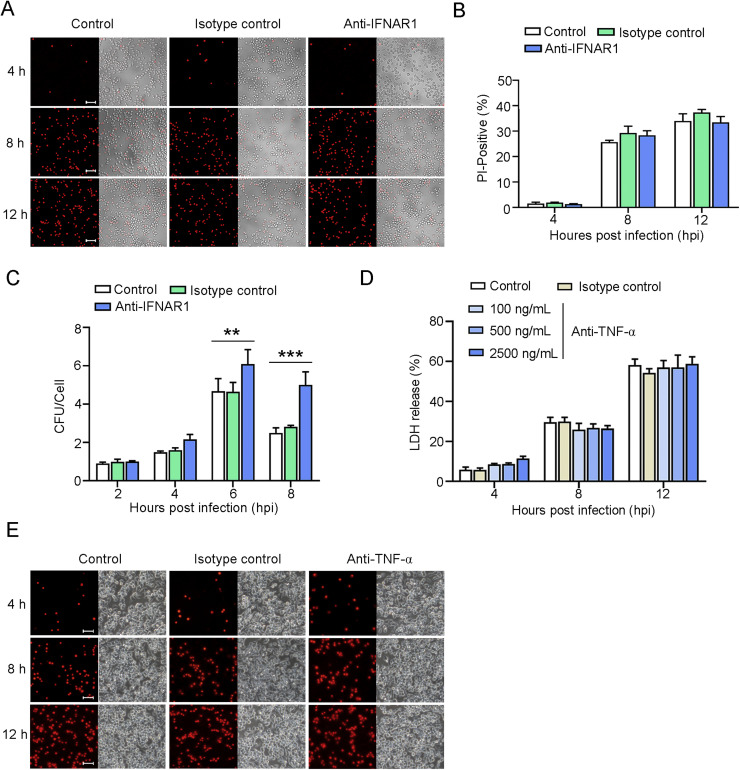
Since E. tarda infection, on the one hand, induced necroptosis and, on the other hand, activated RIG-I/MDA5, we investigated the potential link between these two cellular processes. The results showed that, following E. tarda infection, the death of RIG-I-/- and MDA5-/- RAW264.7 cells was much less than that of the wild type cells, as evidenced by the significantly lower amounts of PI-positive cells and LDH release exhibited by the RIG-I/MDA5 knockout cells (Fig. 4A-C). Similarly, in RIG-I knockdown and MDA5 knockdown cells, LDH release during E. tarda infection significantly decreased (Figure S4). RIG-I and MDA5 were also required for E. tarda-induced MLKL phosphorylation, which was markedly diminished in RIG-I-/- and MDA5-/- cells (Fig. 4D). In line with these observations, when MAVS, a key adaptor in the RIG-I/MDA5 pathways, was knocked down by siRNA, the release of LDH from E. tarda-infected cells significantly decreased (Figure S5 and Fig. 4E). Furthermore, Co-IP analysis detected MAVS interaction with RIPK1 and RIPK3 (Fig. 4F,G). To examine whether RIG-I/MDA5 functioned via IFN1, E. tarda infection was performed in the presence of anti-IFNAR1 antibody. The result showed that antibody blocking of IFNAR1 had no apparent effect on the cell death of RAW264.7 cells caused by E. tarda (Fig. 5A,B), suggesting that the RIG/MDA5-mediated necroptosis was independent of IFN1 signaling. However, IFNAR1 blockage significantly enhanced intracellular bacterial proliferation (Fig. 5C). Antibody blocking of TNF-α had no apparent effect on the death of RAW264.7 cells infected with E. tarda (Fig. 5, Fig. 5).
Fig. 4.
RIG-I and MDA5 are required for Edwardsiella tarda-induced necroptosis. (A-D) RIG-I-/-, MDA5-/-, or wild type (WT) RAW264.7 cells were infected with E. tarda for 1 h After killing the extracellular bacteria, the cells were cultured for 4, 8, or 12 h and then treated as follows. (A) The cells were stained with PI and then observed using a confocal microscope at different time points. Scale bar, 50 μm. (B) The percentages of PI-positive cells were calculated. (C) LDH release from the cells was measured. (D) The cells were lysed and then subjected to immunoblotting using the indicated antibodies. (E) RAW264.7 cells were transfected with or without (control) siMAVS or siNC for 24 h The cells were infected with E. tarda for various hours and then determined for LDH release. (F, G) HEK293T cells were transfected with plasmids expressing Myc-tagged MAVS and/or Flag-tagged RIPK1 (F) or Myc-tagged MAVS and/or Flag-tagged RIPK3 (G). The cell lysates were immunoprecipitated (IP), and the input and IP samples were immunoblotted (IB) with anti-Myc/Flag antibodies. For panels B, C and E, values are the means of three experimental replicates and shown as means ± SD. ***p < 0.001; **p < 0.01.
Fig. 5.
The effect of IFNAR1 and TNF-α neutralization on Edwardsiella tarda-induced cell death. (A-C) RAW264.7 cells were treated with or without (control) anti-IFNAR1 antibody or isotype control for 4 h, and then infected with E. tarda for indicated hours. The cells were then treated as follows. (A) The cells were stained with PI and observed with a confocal microscope at different time points. Scale bar, 50 μm. (B) The percentages of PI-positive cells were calculated. (C) The intracellular bacterial number (shown as Colony Forming Unit, CFU) was determined by plate count. Values are the means of triplicates and shown as means ± SD. ***p < 0.001; **p < 0.01. (D) RAW264.7 cells were incubated with or without (control) different doses of anti-TNF-α antibody or isotype control, and then infected with E. tarda for various hours. LDH release from the cells was determined. (E) RAW264.7 were treated with anti-TNF-α antibody and infected with E. tarda as above. The cells were stained with PI and observed with a microscope at different time points.
4. Discussion
Intracellular pathogens are able to thrive inside host cells. As a countermeasure, the hosts have evolved a variety of molecular machines that carry out PCD, such as apoptosis, pyroptosis, and necroptosis, to destroy the replication niches of the pathogens (Bertheloot et al., 2021; Humphries et al., 2015; Yuan and Ofengeim, 2023). On the other hand, pathogens may utilize PCD to eliminate the host immune cells or to break the physical barriers for bacterial dissemination (Bleriot and Lecuit, 2016; Zhang et al., 2022). E. tarda infection has been reported to be associated with pyroptosis and apoptosis (Chen et al., 2017; He et al., 2020; Niu et al., 2023; Wang et al., 2021; Zhou and Sun, 2016), but its association with necroptosis remains to be explored. In the present work, we found that E. tarda induced RAW264.7 cells to exhibit necrotic cell death with enhanced phosphorylation of RIPK3 and MLKL, suggesting the occurrence of RIPK3-MLKL-mediated necroptosis. This conclusion was further supported by the observation that the cell death was significantly blocked by RIPK1, RIPK3 or MLKL inhibition. E. tarda has been shown to prevent apoptosis in various hosts and cells, including murine macrophages, zebrafish cells and larvae, and blue gourami fish (He et al., 2020; Zhou and Sun, 2016). In line with these reports, we found that E. tarda-infected cells exhibited no apparent apoptosis morphology. This observation, together with the lack of caspase-3 activation, indicated the absence of apoptosis. Similarly, gasdermin D, which executes pyroptosis, was not activated by E. tarda. It is possible that the silence of the pyroptosis and apoptosis pathways may be the result of some immune evasion mechanisms of E. tarda.
RLRs are best known as the key sensors of viral infection that act by direct interaction with viral RNAs, whereby promoting the production of type I IFN and other antiviral molecules to establish antiviral immunity (Chow et al., 2018). In this study, we found that E. tarda infection significantly augmented RIG-I/MDA5 production, IRF3 phosphorylation, IFN-β expression/secretion, and ISG production, which together indicated activation of the RIG-I and MDA5 pathways. Consistently, E. tarda-induced IFN-β expression and secretion in RIG-I/MDA5 knockdown cells and RIG-I/MDA5 knockout cells significantly decreased, implying that the observed IFN-β production in the wild type cells was indeed due to RIG-I/MDA5 signaling. In line with the fact that RIG-I and MDA5 are RNA sensors, we found that E. tarda RNA stimulated ISG production in a RIG-I/MDA5-dependent manner, suggesting that during intracellular infection, E. tarda RNA may serve as a ligand for RIG-I and MDA5. Previous studies showed that RIG-I and MDA5 played important roles in eliminating cytoplasmic viruses (Chow et al., 2018). Here, we found that in RIG-I-/- and MDA5-/- cells, E. tarda proliferation significantly increased compared to that in the wild type cells, suggesting that the RIG-I and MDA5 pathways were required for optimal inhibition of the intracellular infection of E. tarda. This result is similar to that of a previous report, which showed that RIG-I and MDA5 were essential for intracellular bacterial removal during septicemia (Asadpour-Behzadi and Kariminik, 2018). It is known that RIG-I and MDA5 signaling can also lead to NF-κB activation. Likewise, in our study, we found that in addition to type I IFN, the expressions of inflammatory cytokines and chemokines were also enhanced in a RIG-I/MDA5-dependent manner, suggesting activation of the NF-κB pathway. It is likely that the enhanced expression of these cytokines, as well as NO production, accounted for the effect of RIG-I/MDA5 against E. tarda replication.
Pathogen-induced RLR-mediated necroptosis has been observed only with Sendai virus, which induced necroptosis in the fibrosarcoma L929 cell line by activating the RIG-I pathway (Schock et al., 2017). Necroptosis mediated by RLRs has not been reported in the context of bacterial infection. In this study, we found that E. tarda-mediated necroptosis, including RIPK3/MLKL activation and cell death, was markedly weakened in RIG-I-/- and MDA5-/- cells, indicating a vital contribution of RIG-I/MDA5 to necroptosis. Since the effects of RIG-I/MDA5 were not significantly affected by IFNAR1 neutralization, the functions of RIG-I/MDA5 in necroptosis were probably independent of type I IFN. TNF-α has been reported to be involved in the onset of necroptosis (Ting and Bertrand, 2016). In this work, we found that E. tarda-triggered RIG-I/MDA5 signaling stimulated the expression of TNF-α. However, antibody blocking of TNF-α had no apparent effect on the death of E. tarda-infected cells, implying that E. tarda-induced necroptosis was likely not a downstream event of TNF-α signaling. In contrast, MAVS knockdown significantly inhibited E. tarda-induced cell death, indicating an important role of MAVS in the signaling cascade of E. tarda-induced necroptosis. MAVS has been reported to interact with RIPK1/3 to regulate innate anti-viral immunity against Influenza A virus (Downey et al., 2017). In accordance with this report, in this study we found interactions between MAVS and RIPK1/3. It is possible that these interactions may contribute to the activation of necroptosis by the RIG-I/MDA5 pathway during E. tarda infection of RAW264.7 cells.
5. Conclusions
In this study, we discovered that the intracellular bacterial pathogen E. tarda specifically activated RIPK3-MLKL-mediated necroptosis and the RIG-I/MDA5 pathways. Furthermore, both RIG-I and MDA5 were essential for necroptosis induction, likely in a type I IFN-independent manner, and effective inhibition of intracellular infection. These findings indicated the existence of complex interactions between RLRs and necroptosis, and provided new insights into the host immune response against intracellular bacterial pathogen.
Declaration of competing interest
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the grants of the National Natural Science Foundation of China (31330081, 41576150, and 31730100).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2024.100318.
Contributor Information
Boguang Sun, Email: sunboguang@qdio.ac.cn.
Li Sun, Email: lsun@qdio.ac.cn.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Abdullah Z., Schlee M., Roth S., Abu Mraheil M., Barchet W., Boettcher J., Hain T., Geiger S., Hayakawa Y., Fritz J.H., Civril F., Hopfner K.-P., Kurts C., Ruland J., Hartmann G., Chakraborty T., Knolle P.A. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. Embo J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadpour-Behzadi A., Kariminik A. RIG-1 and MDA5 are the important intracellular sensors against bacteria in septicemia suffering patients. J. Appl. Biomed. 2018;16:358–361. [Google Scholar]
- Bertheloot D., Latz E., Franklin B.S. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell Mol. Immunol. 2021;18:1106–1121. doi: 10.1038/s41423-020-00630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleriot C., Lecuit M. The interplay between regulated necrosis and bacterial infection. Cell Mol. Life Sci. 2016;73:2369–2378. doi: 10.1007/s00018-016-2206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse M., Ly H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen C.K., Singh A.K., Krug S., Lun S., Thakur P., Srikrishna G., Bishai W.R. MDA5 RNA-sensing pathway activation by Mycobacterium tuberculosis promotes innate immune subversion and pathogen survival. JCI. Insight. 2023;8 doi: 10.1172/jci.insight.166242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Yang D., Han F., Tan J., Zhang L., Xiao J., Zhang Y., Liu Q. The Bacterial T6SS Effector EvpP Prevents NLRP3 Inflammasome Activation by Inhibiting the Ca2+-Dependent MAPK-Jnk Pathway. Cell Host. Microbe. 2017;21:47–58. doi: 10.1016/j.chom.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Chow K.T., Gale M., Jr., Loo Y.M. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu. Rev. Immunol. 2018;36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- Downey J., Pernet E., Coulombe F., Allard B., Meunier I., Jaworska J., Qureshi S., Vinh D.C., Martin J.G., Joubert P., Divangahi M. RIPK3 interacts with MAVS to regulate type I IFN-mediated immunity to Influenza A virus infection. PLoS. Pathog. 2017;13 doi: 10.1371/journal.ppat.1006326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann C.A., Herzner A.M., Abdullah Z., Zillinger T., Jakobs C., Schuberth C., Coch C., Higgins P.G., Wisplinghoff H., Barchet W., Hornung V., Hartmann G., Schlee M. RIG-I Detects Triphosphorylated RNA of Listeria monocytogenes during Infection in Non-Immune Cells. PLoS. One. 2013;8 doi: 10.1371/journal.pone.0062872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K., Xu H., Jiang S., Sun L. Paralichthys olivaceus MLKL-mediated necroptosis is activated by RIPK1/3 and involved in anti-microbial immunity. Front. Immunol. 2024;15 doi: 10.3389/fimmu.2024.1348866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Huang S., Shen Z. Biomarkers for the detection of necroptosis. Cell Mol. Life Sci. 2016;73:2177–2181. doi: 10.1007/s00018-016-2192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.T., Zhou Y., Liu Y.L., Li D.Y., Nie P., Li A.H., Xie H.X. Edwardsiella piscicida type III protein EseJ suppresses apoptosis through down regulating type 1 fimbriae, which stimulate the cleavage of caspase-8. Cell Microbiol. 2020;22:e13193. doi: 10.1111/cmi.13193. [DOI] [PubMed] [Google Scholar]
- Humphries F., Yang S., Wang B., Moynagh P.N. RIP kinases: key decision makers in cell death and innate immunity. Cell Death. Differ. 2015;22:225–236. doi: 10.1038/cdd.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Qin K., Sun L. Time-lapse live-cell imaging of pyroptosis by confocal microscopy. STAR. Protoc. 2023;4 doi: 10.1016/j.xpro.2023.102708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K.Y., Siame B.A., Tenkink B.J., Noort R.J., Mok Y.K. Edwardsiella tarda - virulence mechanisms of an emerging gastroenteritis pathogen. Microbes. Infect. 2012;14:26–34. doi: 10.1016/j.micinf.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Li D.Y., Liu Y.L., Liao X.J., He T.T., Sun S.S., Nie P., Xie H.X. Identification and Characterization of EvpQ, a Novel T6SS Effector Encoded on a Mobile Genetic Element in Edwardsiella piscicida. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.643498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Sun B., Ning X., Jiang S., Sun L. A Comparative Analysis of Edwardsiella tarda-Induced Transcriptome Profiles in RAW264.7 Cells Reveals New Insights into the Strategy of Bacterial Immune Evasion. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20225724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkermann A., Green D.R. Necroptosis. N. Engl. J. Med. 2014;370:455–465. doi: 10.1056/NEJMra1310050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield D., He Y., Xu T., Fossheim L.E., Zhang X.-H. FliC, a Flagellin Protein, Is Essential for the Growth and Virulence of Fish Pathogen Edwardsiella tarda. PLoS. One. 2012;7 doi: 10.1371/journal.pone.0045070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M., Sui Z., Jiang G., Wang L., Yao X., Hu Y. The Mutation of the DNA-Binding Domain of Fur Protein Enhances the Pathogenicity of Edwardsiella piscicida via Inducing Overpowering Pyroptosis. Microorganisms. 2023;12 doi: 10.3390/microorganisms12010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmolke M., Patel J.R., de Castro E., Sanchez-Aparicio M.T., Uccellini M.B., Miller J.C., Manicassamy B., Satoh T., Kawai T., Akira S., Merad M., Garcia-Sastre A. RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection. mBio. 2014;5:e01006–e01014. doi: 10.1128/mBio.01006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schock S.N., Chandra N.V., Sun Y., Irie T., Kitagawa Y., Gotoh B., Coscoy L., Winoto A. Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death. Differ. 2017;24:615–625. doi: 10.1038/cdd.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweibenz B.D., Devarkar S.C., Solotchi M., Craig C., Zheng J., Pascal B.D., Gokhale S., Xie P., Griffin P.R., Patel S.S. The intrinsically disordered CARDs-Helicase linker in RIG-I is a molecular gate for RNA proofreading. EMBO J. 2022;41 doi: 10.15252/embj.2021109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J., Nam Y.W., Kim S., Oh D.B., Song J. Necroptosis molecular mechanisms: recent findings regarding novel necroptosis regulators. Exp. Mol. Med. 2021;53:1007–1017. doi: 10.1038/s12276-021-00634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sethi A., Horne C.R., Fitzgibbon C., Wilde K., Davies K.A., Garnish S.E., Jacobsen A.V., Samson A.L., Hildebrand J.M., Wardak A., Czabotar P.E., Petrie E.J., Gooley P.R., Murphy J.M. Membrane permeabilization is mediated by distinct epitopes in mouse and human orthologs of the necroptosis effector. MLKL. Cell Death Differ. 2022;29:1804–1815. doi: 10.1038/s41418-022-00965-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Z.H., Xu H., Wang H., Jiang S., Chi H., Sun L. Intracellular Trafficking Pathways of Edwardsiella tarda: from Clathrin- and Caveolin-Mediated Endocytosis to Endosome and Lysosome. Front. Cell Infect. Microbiol. 2017;7:400. doi: 10.3389/fcimb.2017.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Sun B., Zhang B., Sun L. Temperature induces metabolic reprogramming in fish during bacterial infection. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1010948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zheng W.J., Hu Y.H., Sun B.G., Sun L. Edwardsiella tarda Eta1, an in vivo-induced antigen that is involved in host infection. Infect. Immun. 2012;80:2948–2955. doi: 10.1128/IAI.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen D., Wang W., Galls D., Guo R., Xu L., Pyle A.M. The molecular mechanism of RIG-I activation and signaling. Immunol. Rev. 2021;304:154–168. doi: 10.1111/imr.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting A.T., Bertrand M.J.M. More to Life than NF-kappaB in TNFR1 Signaling. Trends. Immunol. 2016;37:535–545. doi: 10.1016/j.it.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kong X., Liu X., Wang X., Wang Z., Liu J., Zhang Q., Yu H. Edwardsiella tarda triggers the pyroptosis of the macrophage of Japanese flounder (Paralichthys olivaceus) Aquaculture. 2021;533 [Google Scholar]
- Wei L., Qiao H., Sit B., Yin K., Yang G., Ma R., Ma J., Yang C., Yao J., Ma Y., Xiao J., Liu X., Zhang Y., Waldor M.K., Wang Q. A Bacterial Pathogen Senses Host Mannose to Coordinate Virulence. iScience. 2019;20:310–323. doi: 10.1016/j.isci.2019.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Chen Y., Yuan Z., Xu H., Sun L. CRADD and cIAP1 antagonistically regulate caspase-9-mediated apoptosis in teleost. Int. J. Biol. Macromol. 2024;279 doi: 10.1016/j.ijbiomac.2024.135265. [DOI] [PubMed] [Google Scholar]
- Xu H., Jiang S., Yu C., Yuan Z., Sun L. GSDMEa-mediated pyroptosis is bi-directionally regulated by caspase and required for effective bacterial clearance in teleost. Cell Death. Dis. 2022;13:491. doi: 10.1038/s41419-022-04896-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yuan Z., Sun L. A Non-Canonical Teleost NK-Lysin: antimicrobial Activity via Multiple Mechanisms. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232112722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Yuan J., Ofengeim D. A guide to cell death pathways. Nat. Rev. Mol. Cell Biol. 2023 doi: 10.1038/s41580-023-00689-6. [DOI] [PubMed] [Google Scholar]
- Zhang G., Wang J., Zhao Z., Xin T., Fan X., Shen Q., Raheem A., Lee C.R., Jiang H., Ding J. Regulated necrosis, a proinflammatory cell death, potentially counteracts pathogenic infections. Cell Death. Dis. 2022;13:637. doi: 10.1038/s41419-022-05066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Sun K., Sun L. Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain. Microbiology (Reading) 2008;154:2060–2069. doi: 10.1099/mic.0.2008/017343-0. [DOI] [PubMed] [Google Scholar]
- Zhou Z.-j., Sun B.-g., Sun L. Edwardsiella tarda Sip1: a serum-induced zinc metalloprotease that is essential to serum resistance and host infection. Vet. Microbiol. 2015;177:332–340. doi: 10.1016/j.vetmic.2015.03.030. [DOI] [PubMed] [Google Scholar]
- Zhou Z.J., Sun L. Edwardsiella tarda-Induced Inhibition of Apoptosis: a Strategy for Intracellular Survival. Front. Cell Infect. Microbiol. 2016;6:76. doi: 10.3389/fcimb.2016.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.