Abstract
Background
Silicon has an important role in regulating water management in plants. It is deposited in cell walls and creates a mechanical barrier against external factors. The aim of this study was to determine the role of silicon supplementation in the synthesis and distribution of callose in oilseed rape roots and to characterize the modifications of cell wall structure of these organs after exposure to drought stress. Histological and ultrastructural analyses were performed to determine the changes in the distribution of arabinogalactan proteins, pectins, and extensin in roots of Brassica napus growing under drought and supplemented with silicon. Callose deposition and the accumulation of callose synthase protein were assessed, followed by transcriptional analysis of callose synthase genes.
Results
The results showed that silicon supplementation under drought conditions alter the direction of cortex cell differentiation, promoting fiber formation and proliferation of callose-depositing cells in the roots of the tested plants. This was reflected in an increase in the level of callose synthase and a decrease in the transcriptional activity of the gene encoding this enzyme, indicating regulation based on negative feedback under drought stress. The changes in abundance and distribution of investigated arabinogalactan proteins, pectins and extensin in roots of Si supplemented plants growing under drought stress were observed, indicating cell walls remodeling.
Conclusion
Silicon supplementation in oilseed rape roots induced significant changes in cell wall composition, including increased callose deposition and altered pectins and arabinogalactan proteins distribution. These modifications, along with the formation of fibres in the root cortex, likely contribute to enhanced cell wall strength providing a physical barrier against water loss and mechanical stress, as a probable defence mechanism induced during drought stress.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05967-9.
Keywords: Brassica napus var napus L., Silicon, Drought stress, Callose synthase accumulation, Roots structure, Cells walls, Roots tissues
Introduction
Silicon (Si) is an element occurring naturally in significant amounts in the Earth’s crust but, due to the high instability of silicic acid and its easy conversion into forms unavailable to plants [1], it was not recognized for a long time as a microelement that may have a significant role in the growth and development of plants. However, over the years, it has been shown that some plants can absorb Si from the environment and accumulate it in significant amounts—high accumulators, and others in small amounts—low accumulators [2]. Moreover, it has been observed that it plays an important role in plant defense reactions against the negative effects of various stress factors, including drought [3]. Our research to date has proven that Si supplementation in Brassica napus var napus brings benefits in the regulation of physiological processes [4–6]; however, further studies are required to explain the mechanism of its action. Si accumulation in oilseed rape proved to be significantly higher in the above-ground part of the plant compared to the roots [4, 6] but despite that Si stimulates the root development. It has been shown to promote the growth of the root system, thereby increasing its absorbent surface, improving the water balance of plants by influencing water uptake by the roots, and increasing water retention in cells, which is strictly regulated by tonoplast aquaporins [6]. Moreover, Si stimulates catalase activity in the roots of plants grown under both well-watered and drought conditions, and this effect is especially pronounced under stress conditions, which emphasizes the role of Si in mitigating the effects of oxidative stress by activating the antioxidant system [6]. Si is deposited in plants, creating a mechanical barrier [7], mainly in the cell walls of the covering, conducting, or strengthening tissue [8], where it combines with its polysaccharide components and lignin or forms silica [9]. Even very small amounts of this element can change the structure of the cell wall as it was shown in the case of rice, but also in plants that poorly accumulate Si, such as oilseed rape [10]. This process occurs by the formation of covalent bonds with plant cell wall components such as hemicelluloses, pectin, and lignin, which may be crucial in plant cell wall structure and remodeling [8, 11].
Callose mechanically strengthens the cell wall, and the timing and location of its accumulation in the cell wall may be crucial for the proper progression of processes such as megasporogenesis, microsporogenesis, embryogenesis [12], cytokinesis [13], transport of assimilates, intercellular communication [14] and plant responses to environmental stresses [15]. The levels of callose in plants are regulated by the coordinated activities of callose synthase (CalS, also referred to as GSL for glucan-synthase-like) and β-1,3-glucanase (BG). Callose synthase facilitates the biosynthesis of callose, whereas β-1,3-glucanase is responsible for its breakdown. The finding of the CalS gene family in many plant species revealed its significance in plant development and responses to environmental stress. So far, 15 CalS genes have been identified in Brassica rapa, 7 in Hordeum vulgare, 12 in Zea mays, 8 in Vitis vinifera, 32 in Brassica napus, 12 in Citrus sinensis, and 12 in Arabidopsis thaliana [16]. In Arabidopsis thaliana, the callose synthases are grouped into two functional categories. The first group, which includes CalS1, CalS2, CalS6, CalS8 and CalS10, is mainly involved in callose synthesis during pollen development and cell division. The second group, consisting of CalS3, CalS4, CalS5, CalS7 and CalS12, is primarily responsible for producing callose in the cell wall as part of the plant’s defense mechanism against phytopathogen infections. The roles of other members, such as CalS9 and CalS11 are still not well understood [17]. Studies on callose synthesis induced by plant silicon supplementation have primarily focused on conditions of biotic stress. For example, increased callose accumulation triggered by a compound mimicking fungal infection has been shown to coincide significantly with elevated silica deposition in leaf trichomes and mesophyll tissues [18]. In oilseed rape (Brassica napus), research on callose accumulation and the callose synthase gene family responsible for its synthesis has so far been limited to responses to infection by the pathogen Leptosphaeria maculans [11]. However, the effects of Si supplementation and drought stress on callose synthesis and its related mechanisms have not been yet explored. Therefore, this study aimed to investigate the role of Si supplementation in the synthesis and distribution of callose in the roots of oilseed rape plants. Additionally, it sought to characterize modifications to the cell wall structure of these organs, focusing on the localization of arabinogalactan proteins, pectins and extensin.
Materials and methods
Plant material, experimental design and sampling
The experiment was planned according to Saja-Garbarz et al. (2022) [5]. In the experiment plants of Brassica napus var napus cv. Markus (from the Institute of Plant Protection – National Research Institute, Poznań, Poland) were used. The design of the experiment and information about conditions of growth are presented in Fig. 1.
Fig. 1.
Experimental design. In the experiment seeds of oilseed rape which were not subjected to sterilization and stratification were used. Samples were taken from the middle part of the root (root hair zone and lateral root zone). The roots were washed several times with distilled water and excess soil was removed with a paintbrush
Preparation of roots for histological and immunohistological analysis
Samples from roots were fixed in a mixture of 4% paraformaldehyde (PFA), 1% glutaraldehyde (GA) in phosphate-buffered saline (PBS, pH = 7.2; overnight at 4 ◦C), dehydrated in increasing ethanol concentrations and gradually embedded in Steedman’s wax as described previously by Sala et al., (2019) [19]. Cross Sect. (6-µm thick) were cut using a HYRAX M40 rotary microtome (Zeiss, Germany) and collected on microscopic slides covered with Mayer’s albumin.
Callose detection
Callose (Cal) was detected by staining with 0.1% (w/v) aniline blue (AppliChem) in phosphate buffer, pH 7.2 for 30 min, rinsed several times in PBS and mounted in a Fluoromount anti-fading medium [20]. Aniline blue was observed using epifluorescence microscopy (Nikon Eclipse Ni) in UV light. The average number of cells with callose was determined based on the number of cells with callose per 10 cells in radial rows selected randomly around the entire root circumference.
Ultrastructural analysis of roots
For transmission electron microscopy (TEM), the material was prepared according to the procedure described by Brzezicka and Kozieradzka-Kiszkurno (2021) [21]. The samples were fixed in 2.5% (w/v) formaldehyde and 2.5% (v/v) glutaraldehyde in a 0.05 M cacodylate buffer (pH 7.0) for 4 h at room temperature. Next, were rinsed in the same buffer and post-fixed in 1% (w/v) osmium tetroxide in a cacodylate buffer at 4 °C overnight, dehydrated in a series of graded acetone (25–100%) and embedded in Spurr’s low-viscosity resin (Polysciences, Germany). Ultrathin Sects. (50–90 nm) were cut using a diamond knife on a Leica EM UC7 ultramicrotome (Leica, Germany). The sections were stained with Uranyless (Delta Microscopies) and Reynold’s lead citrate (Delta Microscopies), and examined on a Tecnai G2 Spirit BioTWIN (FEI, United States) transmission electron microscope at 120 kV. TEM analyses were done at the Bioimaging Laboratory, Faculty of Biology, University of Gdańsk, Poland.
Protein concentration in crude root extract
Protein extracts were prepared from roots following Saja-Garbarz et al. (2024) [6]. Protein concentration was determined spectrometrically using the Bio-Rad Protein Assay, based on the Bradford method [22]. Absorbance changes were measured with Synergy 2 BioTek. Three independent biological replicates were used, each consisting of root fragments from five plants, with two technical replicates per sample.
Accumulation of CalS12 protein in roots
The identification of the callose synthase protein (CalS12) was performed using immunodetection on nitrocellulose membranes. Protein fractions (5.5 µg) were diluted in loading buffer, denatured at 70 °C for 10 min, and separated on 10% polyacrylamide gels according to Laemmli (1970) [23]. Proteins were transferred to nitrocellulose membranes using the Transblot SD Semi Dry Transfer Cell (BioRad) at 46 mA for 1 h. Membranes were blocked with 5% low-fat milk in TBS-T for 1 h, washed with TBS-T, and incubated overnight with the primary polyclonal antibody CalS12/PMR4/Callose synthase specific to peptide derived from Arabidopsis thaliana CALS12 protein sequence (Agrisera) (1:1000). After washing, membranes were incubated with HRP-conjugated anti-rabbit secondary antibody (Agrisera) (1:20,000 for 2 h), washed again, and chemiluminescence was visualized with the Super Bright reagent (Agrisera). Blots were scanned using an Epson Perfection V700 Photo Scanner. Three biological replicates were used for each treatment, with two technical replicates each, resulting in six measurements per treatment.
Expression of BnC05.CalS.a, BnC09.CalS.a and BnA10.CalS.b genes in the roots: RNA isolation, cDNA synthesis and real-time PCR
The accumulation of the BnC05.CalS.a, BnC09.CalS.a and BnA10.CalS.b transcripts were measured using RT-q PCR amplification. The analyses were carried out using a QuantStudio™ 6 Pro Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). Approximately 0.05 g of the plant tissue (central part of the root - roots were selected from root hair zone and lateral root zone which were not woody) was used to extract RNA using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The quality and quantity of the RNA were checked using a UV-Vis Spectrophotometer Q5000 (Quawell, San Jose, CA, USA). Approximately 800 ng of RNA template was used for reverse transcription reaction combined with genomic DNA enzymatic elimination using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). For the Real-Time PCR assay, cDNA was diluted 1:10 with dH2O. Primer sequences were prepared based on the publication by Liu et al. (2018) [11] (Tab. ST1., supplementary material). They were prepared according to the manufacturer’s information (Genomed) and then diluted in a 1:10 ratio for the reaction. The GAPDH and Actin coding genes were selected as the reference genes. Primer sequences for GAPDH were prepared based on the publication by Liu et al. (2018) [11] and for Actin on the publication by Rys et al. (2020) [24] (Tab. ST1., supplementary material). For each sample of the separate gene, we used: 12.5 µL of SYBR™ Green PCR Master Mix, 2.5 µL of each primer (10 µM), 6.5 µL of sterile, DNAse-free water, and 1 µL of cDNA. For each treatment, a pooled sample was run in four technical replicates along with negative control and 0, 10, 100, and 1000 times diluted one of the samples to create a melting curve. The amplification programs were set at the following conditions: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 60 s.
Immunohistological detection of arabinogalactans, pectic and extensin epitopes
Immunofluorescence staining was done according to Godel-Jędrychowska et al. (2023) [25]. Briefly, sections were submerged in a blocking buffer consisting of 2% (v/v) fetal calf serum (FCS) and 2% (w/v) bovine serum albumin (BSA) in PBS for 30 min. Sections were treated with the primary monoclonal antibodies (Plant Probes, Leeds, UK; diluted 1:20 in blocking buffer) and kept overnight at 4 °C. The antibodies used in the present studies are shown in Tab. ST2, see online supplementary material. The secondary antibody (Alexa Fluor 488 goat anti-rat IgG, Jackson ImmunoResearch Laboratories; diluted 1:100 in the blocking buffer) was applied for 2 h. To visualize the cellulose, the sections were counterstained with 0.01% (w/v) Fluorescent Brightener 28 (Sigma-Aldrich) in PBS for 10 min and mounted in a Fluoromount (Sigma-Aldrich) anti-fading medium. To confirm the specificity of a secondary antibody, negative controls were made in which the primary antibody step was omitted, and the blocking buffer was applied instead. Each negative control section exhibited no fluorescence signal (not shown). Sections were analyzed using a Nikon Eclipse Ni-U microscope equipped with a Nikon Digital DS-Fi1-U3 camera with the corresponding software (Nikon, Tokyo, Japan) at a maximum excitation wavelength of 490 nm (AlexaFluor 488) or 330 nm (Fluorescent Brightener 28). The epifluorescence microscopy images were prepared as figures using Corel Draw x12 and Corel Photo-Paint software (the brightness and contrast were adjusted).
Statistical analysis
Statistical analysis and graphic presentation of the results were performed using Statistica 13.1 (StatSoft, Tulsa, OK, USA) and PAST 4.06 [26]. The main effects of the treatments on the number of cells with callose deposition and the accumulation of CalS12 protein were determined with one or two-factor analysis of variance (ANOVA). The significance of differences among the treatment means was calculated with Duncan’s multiple range test at 0.05 probability level. The figures include mean values ± standard deviation (SD). Relative quantification of BnC05.CalS.a, BnC09.CalS.a and BnC10.CalS.b transcript levels was performed based on the modified standard curve method proposed by Pffafl (2001) [27]. The amplification efficiency of each examined gene was calculated based on the slope of the standard curve. The expression level of each target gene normalized according to the geometric means of endogenous reference genes – GAPDH and Actin. Values of gene expression were shown as the log2 fold change.
In immunohistological measurements, after the assessment of the distribution of epitopes in 13 different locations of various tissues in roots, the frequency scale was converted into values from 0 to 3, where +++ very strong signal in all cell walls (= 3; +++ epitope present in walls of all cells of a given tissue), ++ epitope present in the most of walls of cells (= 2; ++ epitope present in the walls of at least 75% of the cells of given tissue), + epitope present in the part of walls of cells (= 1; + epitope present in the walls of less than 50% of the cells of a given tissue), - epitope not detected (= 0; Table S3-S6). Then, the significance of differences in terms of the average frequency of particular pectic, APG, and extensin epitopes occurrence in plant roots between experimental groups was tested with Kruskal–Wallis tests (p < 0.05) followed by Dunn’s post-hoc tests. Multiple correspondence analysis (MCA) was used to visualize the occurrence of particular epitopes in different tissues across experimental groups to investigate the associations between the location of a given epitope (tissue) and the experimental group. The frequency of individual epitopes in tissues constitutes the mean value from three observations. The tissues in which a certain epitope was not found were excluded from the analysis. The analysis was performed separately for pectin and AGP epitopes.
Results
Histological analysis of the roots structure
Control roots growing in well-watered conditions were characterized by a lack of fibres in the cortex (Fig. 2A). Fibres were detected in the roots of the remaining variants of the experiment (Fig. 2B-F) in the form of two-cell to several-cell clusters in drought treated roots (Fig. 2B). They were most abundant in roots treated with silicon in well-watered conditions, where they formed an almost continuous cylinder around the root circumference (Fig. 2C). In roots treated with silicon in drought, fibres formed clusters with several cells (Fig. 2D). They were least numerous in roots treated with silicon complex in well-watered conditions (Fig. 2E), but drought resulted in the formation of multi-cellular clusters of fibres around the root periphery.
Fig. 2.
Presence of fibres on cross sections in roots growing under different conditions. A, B – control roots, A - well-watered, B – under drought; C, D – roots treated with silicic acid, C - well-watered, D – under drought; E, F – roots treated with silicon complex, E - well-watered, F – under drought (arrows point to fibres; x- xylem; c – cambium; ph – phloem; cor – cortex; the analysis was carried out using a fluorescence microscope and the ability of the lignified walls to autofluorescence; scale bars = 50 μm)
Callose deposition and ultrastructural analysis of roots
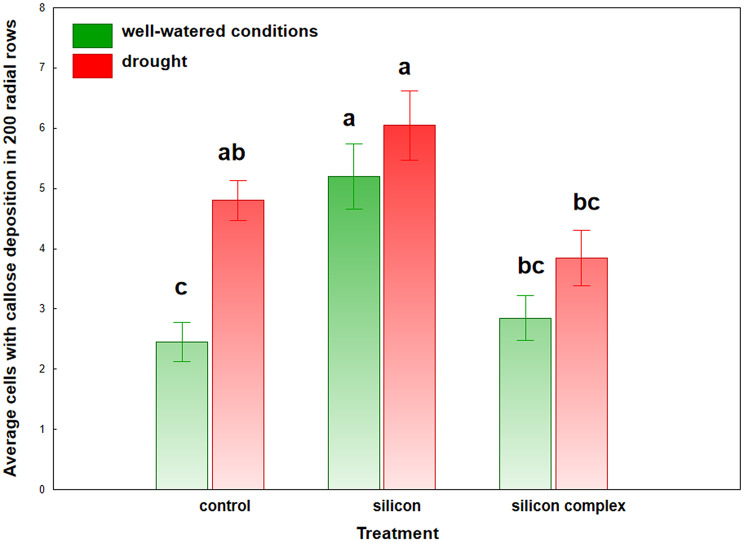
The lowest number of cells with callose was found in control roots and those treated with the silicon complex, which grew in well-watered conditions (Fig. 3).
Fig. 3.
Number of cells with callose deposition. Values which do not differ significantly are marked with the same letter (Duncan’s multiple range test at 0.05 probability level)
The highest number of cells with callose was found in control roots treated with drought stress and in roots treated with silicon, regardless of whether they were subjected to drought stress or grew in well-watered conditions. The differences between these roots and the control roots growing in well-watered conditions were statistically significant. Statistically significant differences in the number of cells with callose were also found between roots treated with silicon and those treated with the silicon complex, regardless of the growth conditions (Fig. 3).
Aniline blue staining indicated that callose occurred in walls of cells on the phloem side in roots from all experimental variants (Figs. 4A and F, 5A and F and 6A and F).
Fig. 4.
Histology and ultrastructure of callose deposition in control roots. A, B, C, D, E– well-watered; F, G, H, I – under drought; (A, F – cross-sections; B, C, D, E, G, H, I – TEM images). A – E Control roots growing in well-watered conditions. A – Callose deposition in root cross-section (aniline blue staining). B – Organization of phloem cells. Two sieve elements are linked by a sieve plate and associated with companion cell and phloem parenchyma cell. C – Developing sieve plate with callose deposits. D – Branched plasmodesma (arrows) between a differentiating sieve element and a companion cell. The callose is visible on the sieve element side. E – Accumulation of callose in the cell wall between the sieve element and the phloem parenchyma cell. F – I Control roots growing in drought. F– Callose deposition in root cross-section (aniline blue staining). G – Mature sieve plate with electron-lucent callose deposits. The sieve plate pores are plugged with P-protein. H – Callose deposition at the plasmodesma. I – Aggregates of callose in the cell wall of phloem parenchyma cell and sieve plate. Abbreviations: white arrows – callose deposits, c – cambium, Cal – callose, CC – companion cell, cor – cortex, cw – cell wall, Mi – middle lamella, Pd – plasmodesma, ph – phloem, Pp – P-protein filaments, Po – sieve pore, PC – phloem parenchyma cell, Pw – primary wall, SE – sieve element, Sp – sieve plate, x – xylem; the color of the walls of xylem cells and fibres is their autofluorescence; scale bars = 50 μm (A, F), 2 μm (B), 1 μm (C, G, I), 0.4 μm (D, E, H)
Fig. 5.
Histology and ultrastructure of callose deposition in roots treated with silicic acid. A, B, C, D, E – well-watered; F, G, H, I, J – under drought; (A, F cross-sections; B, C, D, E, G, H, I, J – TEM images). A – E Silicon treated roots growing in well-watered conditions. A – Callose deposition in root cross-section (aniline blue staining). B – Organization of phloem cells. Two sieve elements are connected by a sieve plate and associated with companion cells and phloem parenchyma cell. C – Callose deposits in the developing sieve plate. D – Callose is associated with the branched plasmodesma (arrows) on the sieve element side. E – Callose deposits in the cell wall between the parenchyma cell and sieve element. F – J Silicon treated roots growing in drought. G – Mature sieve plate with extensive electron-lucent deposits of callose around the pores and network of P-protein in the sieve elements lumen. H – High magnification of sieve plate with callose and the accumulation of dispersive P-proteins. The sieve plate pores are plugged with P-protein. I – Plasmodesma between a differentiating sieve element and a companion cell. The callose is visible on the sieve element side. J – Callose accumulation in the cell wall between the parenchyma cell and sieve element. Abbreviations: white arrows – callose deposits; c – cambium, Cal – callose, CC – companion cell, cor – cortex, cw– cell wall, Pd – plasmodesma, ph – phloem, Pp – P-protein filaments, PC– phloem parenchyma cell, SE – sieve element, Sp – sieve plate, x – xylem; the color of the walls of xylem cells and fibres is their autofluorescence; scale bars = 50 μm (A, F), 1 μm (B, G, J), 0.4 μm (C, D, E, H, I)
Fig. 6.
Histology and ultrastructure of callose deposition in roots treated with silicon complex. A, B, C, D, E – well-watered; F, G, H, I, J – under drought; (A, F cross-sections; B, C, D, E, G, H, I, J – TEM images). A– E Silicon complex treated roots growing in drought. A – Callose deposition in root cross-section (aniline blue staining). B – Organization of phloem cells. Sieve elements are linked by a sieve plate and associated with companion cells. C – Developing a sieve plate with callose deposits around the plasmodesmata. D – Branched plasmodesma (arrows) with callose deposit on the sieve element side. E – Abundant electron-lucent aggregates of callose in the cell wall thickenings of phloem parenchyma cells. F–J Silicon complex treated roots growing in drought. F – Callose deposition in root cross-section (aniline blue staining). G – Organization of phloem cells. H – Developing a sieve plate with callose deposits around the plasmodesmata. I – Callose deposits at the branched plasmodesmata (arrows) between companion cell and sieve element. J – Callose accumulation in the cell wall between the parenchyma cell and sieve element. Abbreviations: white arrows – callose deposits; Cal – callose, CC – companion cell, cw– cell wall, Pd – plasmodesma, PC– phloem parenchyma cell, SE – sieve element, Sp – sieve plate, x – xylem; the color of the walls of xylem cells and fibres is their autofluorescence; scale bars = 50 μm (A, F), 2 μm (B), 1 μm (E, G), 0.4 μm (C, D, H, I, J)
TEM analyses were also performed to localize callose (Cal) in the phloem cells. Electronograms are representative of the obtained results. Our observations particularly focused on the plasmodesmata (Pd) among phloem sieve elements (SEs), companion cells (CCs), and phloem parenchyma cells (PCs). In control roots in well-watered conditions, electron-lucent deposits of callose were seen on the cell walls of developing sieve plates (Fig. 4B, C). The SE and CC were interconnected by branched Pd. Callose deposits were observed at Pd on the SE side (Fig. 4D). Moreover, the callose aggregates were localized in the cell wall thickenings of PC cells not only around plasmodesmata but also in areas of the walls lacking Pd (Fig. 4E). In roots under drought, in mature SE, callose was present as a distinctive electron-lucent layer that surrounds the sieve plate cell wall. Masses of P-protein fibrils surged to the sieve plates and into the pores. The sieve plate pores were plugged with P-protein (Fig. 4G). During the differentiation of the SEs and CC, callose deposits were noticed at Pd on the SE side (Fig. 4H). Moreover, callose accumulation between the cell walls of SEs and PCs was seen to merge and form large masses (Fig. 4I). In silicon-treated roots in well-watered conditions at the beginning of phloem perforation, callose was deposited surrounding the Pd of sieve plates (Fig. 5B, C). The branched plasmodesmata between the SEs and CCs contained callose deposits on the sieve element side (Fig. 5D). In addition, the callose aggregates were observed in the cell wall thickenings of PCs not only around the Pd but also in those areas of the walls where no Pd was detected (Fig. 5E). In the case of roots treated with silicon in drought, mature sieve plates showed an accumulation of high levels of callose collars around the sieve pores. The sieve elements lumen contained a dispersed network of P-protein (Fig. 5G, H). On the SE side, callose deposits were observed at Pd (Fig. 5I). Callose deposits were also visible in the thickened cell walls between SE/PC (Fig. 5J). In silicon complex treated roots in well-watered conditions during the early development of sieve plates, callose was deposited around the Pd (Fig. 6B, C) but also in the branched Pd between the SE and CC (Fig. 6D). Abundant callose aggregates were also noted in the cell wall thickenings of PC cells not only around the Pd but also in those areas of the walls lacking Pd similar to the previous experiments (Fig. 6E). In the case of silicon complex treated roots in drought, in the cell walls of developing sieve plates, callose deposits were detected around simple Pd (Fig. 6G, H), branched plasmodesmata between CC/SE (Fig. 6I), or in the thickened cell wall between the SE/PC around the plasmodesmata (Fig. 6J).
Effect of silicon supplementation on the accumulation of CalS12 protein and expression of BnC09.CalS.a, BnC05.CalS.a and BnA10.CalS.b genes in the roots of oilseed rape plants growing under well-watered conditions and drought
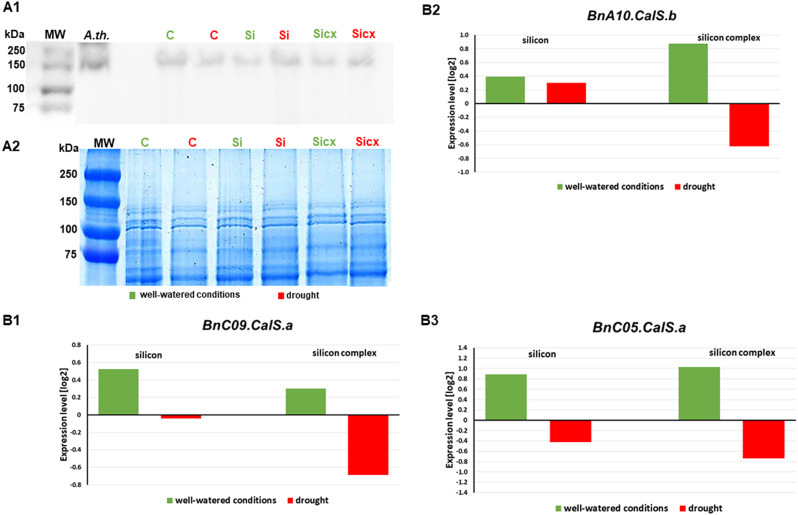
The results of CalS12 protein level analyses indicated the differences between the samples concerning both the plant growth conditions and the type of silicon treatment (Fig. 7 A1-2).
Fig. 7.
The effect of silicon supplementation (silicon – Si or silicon complex – Si cx) and watered with water (control – C) on accumulation level of CalS12 protein (A1). Immunoblot analysis of the CalS12 protein in the roots of oilseed rape plants growing in different conditions using CALS12/PMR4/Callose synthase polyclonal antibody; MW – molecular weight marker (Thermo Scientific PageRuler Prestained Protein Ladder), A. th. – Arabidopsis thaliana. Loading control (A2), gel stained with Coomassie Brilliant Blue solution after electrophoretic separation of proteins isolated from studied plant material (5.5 µg total protein isolated from investigated samples was loaded on each lane). The expression of genes involved in callose synthase synthesis (B1-B3). The expression profiles of BnC09.CalS.a, BnC10.CalS.b and BnC05.CalS.a genes in roots of oilseed rape under well-watered conditions (green) and drought (red). The expression level of each examined gene was relative to non-treated plants in well-watered (green bars) and drought (red bars) conditions. Genes transcript accumulation is rendered as the log2 fold change in the expression of a specific gene in a particular sample in contrast to the endogenous reference genes – Actin and GAPDH
In the roots of plants growing in well-watered conditions, a higher level of this protein was observed in the control than in plants supplemented with silicon independently on the silicon source, while the opposite was true in drought. Transcriptional analysis of BnC09.CalS.a gene encoding CalS12 showed that after silicon supplementation in well-watered conditions, the expression level of this gene increased, while in drought it decreased relative to the control (Fig. 7 B1), which was inversely proportional to the content of this protein (Fig. 7 A1). Transcriptional analysis of two other callose synthase (CalS) genes—BnA10.CalS.b and BnC05.CalS.a (Fig. 7 B2-3), showed that in well-watered conditions silicon supplementation, similar to the BnC09.CalS.a gene, causes a significant increase in its expression relative to controls. In drought, in the case of the BnA10.CalS.b gene, a slight increase was observed after silicon treatment and a decrease after silicon complex treatment of the plants (Fig. 7 B2), and in the case of the BnA10.CalS.b gene (Fig. 7 B3), a decrease in its transcriptional activity was observed in both types of silicon treatment.
Effect of silicon on the presence of pectic, arabinogalactan proteins (AGP) and extensin epitopes in the roots of oilseed rape plants growing under well-watered conditions and drought
Representative photographs showing the locations of the analyzed epitopes are in Figs. SF1-SF6 and Tabs. ST3-ST6 (see online supplementary material).
Control roots growing in well-watered conditions
The pectic epitopes recognised by LM19 antibody were not detected in walls of pith and xylem vessels and xylem parenchyma. The LM20 epitope was detected in walls of cells from each analysed root tissues. Pectic epitope recognised by LM5 antibody was not detected in vessels from pith and xylem parenchyma. Pectic epitope recognised by LM6 antibody was present in cambium, phloem, cortex and pith parenchyma. The AGP detected by JIM13 antibody was observed in all tissues apart from phelloderm. AGP detected by LM2 antibody was abundantly present in all tissues apart from cork cells. AGP recognised by JIM16 antibody was present in paratracheal parenchyma, cambium and phloem. AGP recognised by JIM11 antibody was observed in pith paratracheal parenchyma, cambium, phloem and cortex (Fig. SF1, ST3-ST6).
Control roots growing in drought
Pectic epitope recognised by LM19 antibody was not detected only in walls of pith and xylem vessels and fibres. The pectic epitope detected by LM20 was not present only in pith. The pectic epitope recognised by LM5 antibody was detected in parenchyma cells of pith, cambium, phloem, cortex and fibres. The LM6 antibody was not present in walls of pith, xylem vessels and parenchyma, fibres and periderm. AGP epitope recognised by JIM13 antibody was not detected in pith parenchyma, cambium, phloem, cortex, phelloderm and cork cells. Epitope recognised by LM2 antibody was detected in walls of pith parenchyma, xylem paratracheal parenchyma, cambium and phloem. The JIM16 signal was not detected in any tissue. Signal generated by JIM11 antibody was present in pith parenchyma, cambium, phloem and cortex (Fig. SF2, ST3-ST6).
Silicon treated roots growing in well-watered conditions
Signal generated by the LM19 pectic epitope was detected only in cambium, phloem and fibres. The signal generated by LM20 antibody was detected in cells of each root tissues. The LM5 epitope was not detected in pith an xylem vessels, fibres and phellem and cork cells. The LM6 epitope was present in pith and xylem parenchyma, cambium, phloem, cortex and fibres. The signal generated by the JIM13 antibody was detected in pith, xylem, phellogen and cork cells. The LM2 antibody was detected in pith paratracheal parenchyma, xylem parenchyma, cambium, phloem, cortex, phelloderm and phellogen. The signal generated by JIM16 antibody was present only in cambium and phloem. In case of JIM11 antibody signal was not detected in xylem and cork cells (Fig. SF3, ST3-ST6).
Silicon treated roots growing in drought
The LM19 and LM20 epitopes were present in all analysed tissues. The LM5 antibody was not detected in pith an xylem vessels, xylem parenchyma and fibres. In the case of LM6 antibody, this epitope was not detected only in pith vessels, phellogen and cork cells. Signal generated by JIM13 antibody was weak and detected in vessel and paratracheal parenchyma of pith, xylem parenchyma, fibres, phelloderm and phellogen. The LM2 antibody was present in pith and xylem parenchyma, cambium and phloem. The JIM16 was detected in pith parenchyma, xylem, cambium and phloem. The JIM11 antibody were found only pith paratracheal parenchyma and cambium (Fig. SF4, ST3-ST6).
Silicon complex treated roots in well-watered conditions
The LM19 was detected in each analysed root tissues, however in xylem was present only in differentiating part, similarly to LM20 antibody. Moreover the LM20 was not detected in pith vessels, cambium and cork cells. The LM5 antibody was detected in pith and xylem parenchyma, cambium, phloem and cortex. Signal generated by LM6 antibody was not detected in vessels from pith, fibres and cork cells. The JIM13 AGP epitope was detected only in xylem paratracheal parenchyma, fibres and phellogen. The LM2 epitope was not present in pith vessels, xylem, cortex, fibres phelloderm and cork cells. The JIM16 and JIM11 epitopes were not detected in most cells (Fig. SF5, ST3-ST6).
Silicon complex treated roots growing in drought
The LM19 was not detected only in pith vessels and xylem. In the case of LM20 antibody signal was absent in periderm. The LM5 antibody was not detected in pith and xylem vessels, xylem parenchyma and periderm and the LM6 epitopes was absent in walls of pith vessels, xylem parenchyma and periderm. The JIM13 epitope was not detected only in pith vessels, pith parenchyma, cortex and cork cells. The LM2 antibody was detected in pith paratracheal parenchyma, xylem parenchyma, cambium, phloem and phellogen. The JIM16 antibody was not detected and the JIM11 antibody was detected only in pith parenchyma and phelloderm (Fig. SF6, ST3-ST6) .
The JIM13 epitope had significantly higher frequency in silicon optimal irrigation compared to silicon complex optimal irrigation group (Table 1). As regards JIM16 epitope, significantly lower frequency of this epitope was found in the roots from well-watered control group than in roots from silicon optimal irrigation group in which the highest frequency was recorded. In the case of LM19 pectic epitope, the highest frequency was recorded in roots from silicon optimal irrigation group, which differ significantly from the remaining groups. Control roots growing under drought was characterized by the lowest frequency of LM20 epitope and this group differ significantly from the remaining groups with the exception of silicon complex well-watered group.
Table 1.
Mean frequency (± SE; n = 13) of the AGP and pectic epitopes in different locations of the root tissues in particular experimental groups along with the results of Kruskal-Wallis tests (H values; p < 0.05). The different letters next to means indicate statistically significant differences (p < 0.05) according to Dunn’s post-hoc tests.
| Control, well-watered conditions | Control, drought | Silicon, well-watered conditions | Silicon, drought | Silicon complex, well-watered conditions | Silicon complex, drought | H | p | |
|---|---|---|---|---|---|---|---|---|
| JIM11 | 0.23 ± 0.17 | 0.15 ± 0.10 | 0.54 ± 0.18 | 0.23 ± 0.17 | 0.23 ± 0.17 | 0.23 ± 0.17 | 5.51 | 0.357 |
| JIM13 | 0.85 ± 0.27ab | 1.31 ± 0.26ab | 1.54 ± 0.29b | 0.46 ± 0.18ab | 0.23 ± 0.12a | 0.85 ± 0.19ab | 17.73 | 0.003 |
| JIM16 | 0.23 ± 0.17ab | 0.00 ± 0.00a | 0.54 ± 0.14b | 0.23 ± 0.17ab | 0.31 ± 0.17ab | 0.08 ± 0.08ab | 13.17 | 0.022 |
| LM2 | 0.62 ± 0.27 | 0.54 ± 0.22 | 0.85 ± 0.22 | 1.08 ± 0.37 | 0.92 ± 0.29 | 0.62 ± 0.21 | 2.31 | 0.804 |
| LM5 | 1.46 ± 0.39 | 0.85 ± 0.30 | 1.15 ± 0.36 | 1.46 ± 0.33 | 1.00 ± 0.36 | 1.08 ± 0.31 | 2.75 | 0.738 |
| LM6 | 0.23 ± 0.17 | 1.00 ± 0.30 | 1.85 ± 0.32 | 1.08 ± 0.35 | 1.46 ± 0.33 | 1.08 ± 0.31 | 5.70 | 0.336 |
| LM19 | 2.00 ± 0.34b | 1.92 ± 0.26b | 0.38 ± 0.14a | 1.92 ± 0.33b | 2.08 ± 0.21b | 2.08 ± 0.31b | 21.70 | 0.001 |
| LM20 | 1.85 ± 0.27b | 0.54 ± 0.18a | 1.69 ± 0.24b | 1.69 ± 0.29b | 0.77 ± 0.20ab | 1.77 ± 0.28b | 21.66 | 0.001 |
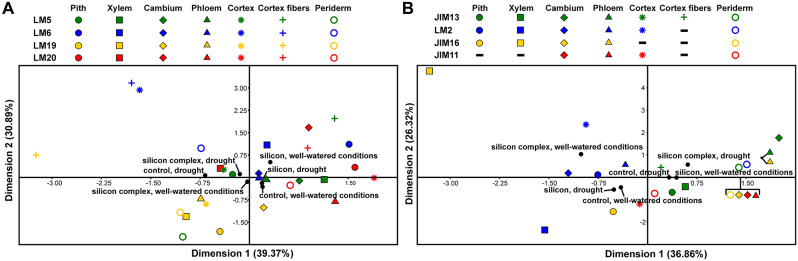
The CA diagram of pectic epitopes in different root tissues shows a gradient along the first axis, with control roots in drought on the left and silicon-treated roots in well-watered conditions on the right (Fig. 8; Figs. SF1-SF6). The groups on the right (silicon-treated roots in well-watered and drought conditions, and control roots in well-watered conditions) had higher frequencies of LM5, LM20, and LM6 epitopes compared to other groups, especially the drought-stressed control roots on the left. Drought-stressed control and silicon complex-treated roots on the left had the highest LM19 epitope frequencies in cortex fibres. Silicon-treated roots under well-watered conditions were farthest from LM19 epitopes, indicating the lowest frequency. Notably, silicon-treated roots in drought and control roots under well-watered conditions were closely positioned, indicating similar pectic epitope frequency patterns.
Fig. 8.
Scatter diagram presenting the results of multiple correspondence analysis (MCA) for particular experimental groups concerning pectic epitopes (A) and for particular experimental groups concerning AGP and extensin epitopes in different root tissues (B). Colors indicate epitopes and shapes indicate plant tissues. Experimental groups are marked with black circles
Discussion
Silicon supplementation of plants under drought conditions exhibits a protective role by improving plant hydration and increasing water use efficiency [28]. According to our studies, this effect concerns not only high silicon accumulators but also species like oilseed rape which is known to poorly accumulate silicon [4–6]. Research on the role of silicon in plants primarily concerns the physiological, biochemical, and molecular levels, while its influence on plant histology and cytology has not been commonly studied even though it is also significant. Changes in the structure of organs and tissues, as well as the remodeling of cell walls, are inherent results of the action of various types of stresses constituting a defense mechanism. This leads to a whole range of other adaptations at deeper levels of plant functioning. In this study, we have shown for the first time that silicon supplementation influences deposition of low- and highly-esterified pectins and AGP recognized by JIM13 and JIM16 antibody which could mean that reconstruction of cell wall in B. napus roots is taking place, probably to prevent the effects of drought. Our finding of cortical cell differentiation into fibres under the influence of drought and Si supplementation is consistent with the commonly recognized response of cells to stress [29]. The observed changes in the differentiation of cortical cells can be interpreted as a manifestation of tissue strengthening, which can counteract changes in turgor, especially under drought-stress conditions [30]. Moreover, the presence of thick cell wall of fibres and middle lamella may constitute a reservoir of water protecting from water loss in drought conditions. A similar role of fibres was observed in the organs of non-woody xerophytic and hydrophytic species where in addition to mechanical support, G-fibres were involved in water storage [31]. According to Bessadok et al. (2007) [32] water enters in the fibres and breaks the secondary bonds between the macromolecules of cellulose and water molecules could link to the network via hydrogen bonds resulting in a swelling of the material [33].
Callose deposition on plant cells can be considered a marker of silicon-influenced tissue remodeling, as callose polysaccharides provide suitable microenvironments for silicic acid condensation [8]. The increased abundance of cells with callose under various stress conditions is well-documented [17, 18, 34]. We also observed an increase in the number of cells with callose in silicon-treated roots under water deficit conditions, supporting the hypothesis that silicon induces a protective mechanism involving callose synthesis in roots of oilseed rape growing under drought stress.
Callose deposition in plant cells involves the activity of specific enzymes known as callose synthases (CalS), which belong to the group of glucan synthase-like enzymes. These enzymes catalyze the synthesis of callose using UDP-glucose as the initial substrate [35]. In plants, callose is synthesized on the cell membrane from UDP-glucose, which binds directly to the catalytic subunit of callose synthase and is degraded by β-1,3-glucanases [36]. All CalS are encoded by genes belonging to the callose synthase gene family, and they are conserved across plant species [37]. Members of the callose synthase gene family are differentially expressed in various tissues and they might be specifically activated in the response to various biotic and abiotic stressors [15]. In Brassica napus, 12 BnCalS genes out of 32, showing the highest similarity with 12 AtCalS genes, were characterized, and their tissue-specific expression patterns were analyzed by Liu et al. 2018 [11]. Until now, most research on the change in expression of CalS genes in plants is related to their involvement in processes such as pollen development, cell plate formation during cytokinesis, and defense against pathogens [11, 15]. Similarly the reactions of these genes in B. napus tissues in response to a fungal pathogen attack were studied [11]. Recent studies have uncovered a novel function of callose in silicon accumulation. In Arabidopsis thaliana, impaired callose synthesis led to reduced silicon content in leaves [17]. Similarly, in rice overexpression of a callose hydrolase altered silicon distribution by degrading callose [38]. These findings suggest that callose plays a crucial role in silicon deposition within plant cell walls. Among the genes studied by Liu et al. (2018) [11] in Brassica napus, several showed root-specific expression and shared similarity with corresponding genes in Arabidopsis thaliana. Based on this, we selected three genes, BnC05.CalS.a, BnC09.CalS.a and BnA10.CalS.b, to investigate for the first time the changes in their expression in Brassica napus var. napus growing under different hydration conditions and after silicon supplementation. We found that under well-watered conditions, silicon supplementation increased the expression of all tested genes in the roots. However, under drought conditions, silicon supplementation led to decrease in these genes activity. Silicon complex had stronger inhibitory effect than orthosilicic acid. In our previous studies we focused also on the changes in the level of CalS12 protein encoded in A. thaliana by AtCalS12 – a homolog of BnC09.CalS.a gene, investigated in our studies. In A. thaliana roots it was expressed in the stele and quiescent cells and controls root development by regulating the symplast transport via plasmodesmata [11, 39]. Our results did not indicate a positive correlation between BnC09.CalS.a gene expression and the level of CalS12 protein. Specifically, silicon increased BnC09.CalS.a gene expression and reduced CalS12 protein accumulation under well-watered conditions, but this correlation was reversed under drought conditions. This finding may be explained by post-translational regulation of callose synthase activity through phosphorylation [40]. However, it must be underlined that the complex interaction between genes expression and protein level in (B) napus might result from genes redundancy. B. napus ( 2n = 38) is an allotetraploid species that was formed by hybridization between ancestors of Brassica oleracea (2n = 18) and Brassica rapa (2n = 20), followed by allopolyploidy [41]. In allopolyploid species epigenetic modifications and transcriptional regulation can play a crucial role in regulating gene expression. It is also important to consider that, under drought stress, other silicon-induced defense mechanisms, besides callose synthesis, might play a more prominent role in stress protection. Silicon’s ability to form covalent bonds with cell wall components suggests that it can actively remodel the cell wall, leading to structural changes that benefit the plant cell under stress conditions [42].
Our study aimed to deepen understanding of the molecular components of Brassica napus cell walls, focusing on how silicon supplementation influences drought responses. We utilized cytochemical methods and monoclonal antibodies, particularly targeting AGP, extensin, and pectic epitopes known to affect wall extensibility and mechanical strength [43–45].
Silicon accumulation in plants is proposed to involve covalent bonding with cell wall components like pectin, potentially influencing wall structure and remodeling [46]. Recent research has revealed that silicon can interact with plant cell walls in intricate ways. Beyond non-covalent associations with amorphous silica, Si can form covalent bonds with polysaccharides like hemicellulose and pectin, as well as lignin. These covalent bonds, likely formed through Si-O-C linkages between monosilicic acid and cis-diol groups in cell wall components, highlight a novel role for Si in shaping plant cell wall structure and function [43]. Covalently bound organosilicon, distinct from amorphous inorganic silica, may significantly contribute to plant growth and stress tolerance. Si-induced cell wall remodelling has been shown to benefit plasma membrane integrity, osmotic and salt stress tolerance, and nutrient homeostasis. Additionally, it is possible that these cell wall modifications trigger cell wall integrity signalling pathways, further enhancing plant growth under stress conditions [47, 48]. This study revealed changes in the presence of unmethyl-esterified homogalacturonans in root cell walls under silicon treatment and drought stress, suggesting that wall stiffening is possibly linked to drought tolerance and turgor maintenance [49]. This response parallels known changes in cell wall chemistry under stress conditions as described in existing literature [45, 50–52].
The results of our analysis reveal varying distributions of epitopes across different root tissues. Pectic epitope differences, especially in walls of xylem cells under silicon treatment and drought stress, were mainly associated with galactans. Galactans were absent in xylem of most of drought-stressed roots. Arabinans hydrate faster than galactans, suggesting their role as pectic plasticizers, maintaining cell wall flexibility under stress. LM6 antibody recognized more pectic epitopes in roots responding to drought, indicating increased synthesis and localization of arabinans in the cell wall. Galactan sidechains of rhamnogalacturonan I (RG-I) contribute to cell stiffness. Reduced presence of LM5 antibody-recognized pectic epitopes in drought-treated roots suggests decreased cell stiffness, potentially explaining the adverse effects of drought on root growth [52, 53].
The conducted studies showed that roots grown under well-watered conditions and treated with silicon exhibited a significant reduction in low-esterified pectins compared to roots from other experimental variants. On the other hand, the level of highly esterified pectins increased compared to control roots growing under drought stress. In normal growth, HG is highly methyl-esterified when exported into cell walls and then within the wall are de-esterified by pectin methylesterases (PMEs) [54]. This suggests that silicon leads to the presence of esterified pectins in well-watered roots, possibly due to PME functional disorders [55]. While both organosilicon and inorganic SiO2 coexist in plants, differentiating their specific contributions to plant growth and stress responses remains a challenge. A multidisciplinary approach combining genetics, omics, imaging, and biochemical techniques will be crucial to unravel the functional roles of these two Si forms and their underlying molecular mechanisms.
Our study unveils how silicon and drought stress impact AGPs and extensins in root cell walls. In silicon-treated, drought-stressed roots, the absence of AGP epitope recognized by LM2 antibody contrasts with control roots across various treatments. Th obtained results indicate changes in the presence of AGP and extension depending on the treatment. There seems to be an interaction between drought stress, silicone form and changes in the chemical composition of the cell walls. While literature data suggest a link between AGP synthesis and stress, the specific response to drought and silicon remains unclear. Overall, our cytohistochemical studies enhance our understanding of plant responses to stress, elucidating the intricate changes in cell wall structure and tissue differentiation essential for adaptation to changing environmental conditions.
Conclusions
Silicon supplementation under drought stress can induce anatomical changes in root cortex, including the formation of fibres.
Callose deposition increased significantly in response to drought stress, both in control and silicon-treated roots. This suggests that callose may play a protective role against drought-induced cellular damage. Silicon supplementation can differentially modulate the expression of callose synthase genes in Brassica napus roots depending on plant growth conditions, suggesting a complex interplay between silicon signalling and cell wall defense mechanisms against drought.
Silicon supplementation altered the cell wall composition, affecting the abundance of low- and high-esterified pectins, arabinans, galactans, and AGPs. These changes may contribute to enhanced drought tolerance.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are deeply grateful to dr Monika Kula-Maximenko from the Franciszek Górski Institute of Plant Physiology Polish Academy of Sciences and Mateusz Malisz from Jagiellonian University for help in biochemical analyses.
Abbreviations
- AGP
Arabinogalactan protein
- c
Cambium
- Cal
Callose
- CC
Companion cell
- cor
Cortex
- cw
Cell wall
- HG
Homogalacturonan
- HRP
Horseradish peroxidase
- PC
phloem parenchyma cell
- Pd
Plasmodesmata
- ph
Phloem
- PMEs
Pectin methylesterases
- Po
Sieve pore
- Pp
P-protein filaments
- RG-I
Rhamnogalacturonan I
- SE
Sieve element
- Si
Silicon
- Sp
Sieve plate
- Ml
Middle lamelle
- Pw
Primary wall
- x
Xylem
Author contributions
Conception of the project, experimental design, plant cultivation and treatment, samples collection: D.S-G. Histological analysis: K.G-J., E.K., M.T.; immunohistological analysis: K.G-J., E.K.; ultrastructural analysis; M. K-K.; accumulation of CalS12 protein: D.S-G., M.L-K; genes expression analysis: D.S-G, M.R, E.G., K.U.; statistical analysis: K.S, D. S-G., M.R. Formal analysis, project administration and funding: D.S-G. Visualization: D.S-G., K.G-J., E.K., M. K-K., M.L-K, M.R, K.S. Writing original draft manuscript: D.S-G, K.G-J., E.K., M.K-K, M.L-K. Validation and writing - review & editing: D.S-G, K.G-J., E.K., M.K-K, M.T., E.G., K.S., M.R., K.U., M.K., M.L-K. Supervision - D.S-G, M.L-K. All authors critically revised and approved the final manuscript.
Funding
Project “Imaging changes in the physicochemical properties of oilseed rape tissues under the influence of silicon in optimal hydratation and drought conditions” - National Science Center Miniatura 2022/06/X/NZ3/00555.
Data availability
Availability of data and materialsMaterials described in the manuscript, including all relevant raw data, will be freely available to any scientist, after the sending such asking to corresponding author. All raw date are stored on the PC computer of corresponding author. Moreover they are collected on the separate external drive stored by corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Saja-Garbarz Diana, Email: d.saja@ifr-pan.edu.pl.
Godel-Jędrychowska Kamila, Email: kamila.godel@gmail.com.
References
- 1.Coskun D, Britto DT, Huynh WQ, Kronzucker HJ. The role of silicon in higher plants under salinity and drought stress. Front Plant Sci. 2016;7:1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guntzer F, Keller C, Meunier JD. Benefits of plant silicon for crops: a review. Agron Sustain Dev. 2012;32:201–13. [Google Scholar]
- 3.Chen DQ, Wang SW, Yin L, Deng X. How does silicon mediate plant water uptake and loss under water deficiency? Front Plant Sci. 2018;9:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saja-Garbarz D, Ostrowska A, Kaczanowska K, Janowiak F. Accumulation of silicon and changes in water balance under drought stress in Brassica napus var. napus L. Plants-Basel. 2021;10:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saja-Garbarz D, Libik-Konieczny M, Fellner M, Jurczyk B, Janowiak F. Silicon-induced alterations in the expression of aquaporins and antioxidant system activity in well-watered and drought-stressed oilseed rape. Plant Physiol Biochem. 2022;174:73–86. [DOI] [PubMed] [Google Scholar]
- 6.Saja-Garbarz D, Libik-Konieczny M, Janowiak F. Silicon improves root functioning and water management as well as alleviates oxidative stress in oilseed rape under drought conditions. Front Plant Sci. 2024;15:1359747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coskun D, Deshmukh R, Sonah H, Menzies JG, Reynolds O, Ma JF, Kronzucker HJ, Bélanger RR. The controversies of silicon’s role in plant biology. New Phytol. 2019;221:67–85. [DOI] [PubMed] [Google Scholar]
- 8.Mandlik R, Thakral V, Raturi G, Shine S, Nikolić M, Tripathi DK, Sonah H, Deshmukh R. Significance of silicon uptake, transport, and deposition in plants. J Exp Bot. 2020;71:6703–18. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Adiram-Filiba N, Blum S, Sanchez-Lopez JA, Tzfadia O, Omid A, Volpin H. et. al. Siliplant 1 protein precipitates silica in sorghum silica cells. J Exp Bot. 2020;71:6830–43. [DOI] [PubMed] [Google Scholar]
- 10.He C, Ma J, Wang L. A hemicellulose-bound form of silicon with potential to improve the mechanical properties and regeneration of the cell wall of rice. New Phytol. 2015;206:1051–62. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Zou Z, Fernando WGD. Characterization of Callose Deposition and Analysis of the Callose Synthase Gene Family of Brassica napus in Response to Leptosphaeria maculans. Int J Mol Sci. 2018;19:3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzezicka E, Kozieradzka-Kiszkurno M. Callose deposition analysis with special emphasis on plasmodesmata ultrastructure during megasporogenesis in Sedum. (Crassulaceae) Protoplasma. 2024;261:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen XY, Kim JY. Callose synthesis in higher plants. Plant Signal Behav. 2009;4(6):489–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zavaliev R, Ueki S, Epel BL, Citovsky V. Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma. 2011;248:117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ning L, Zeng L, Peiyao Y, Peiyao Y, Yanling Z, Shenxiu D, Li-Jun H. The multifarious role of callose and callose synthase in plant development and environment interactions. Front Plant Sci. 2023;14. 10.3389/fpls.2023.1183402. [DOI] [PMC free article] [PubMed]
- 16.Feng J, Chen Y, Xiao X, Qu Y, Li P, Lu Q, Huang J. Genome-wide analysis of the CalS gene family in cotton reveals their potential roles in fiber development and responses to stress. PeerJ. 2021;9: e12557. [DOI] [PMC free article] [PubMed]
- 17.Li N, Lin Z, Yu P, Zeng Y, Du S, Huang L-J. The multifarious role of callose and callose synthase in plant development and environment interactions. Front Plant Sci. 2023;14. 10.3389/fpls.2023.1183402. [DOI] [PMC free article] [PubMed]
- 18.Brugiére T, Exley C. Callose-associated silica deposition in Arabidopsis. J Trace Elem Med Biol. 2017;39:86–90. [DOI] [PubMed] [Google Scholar]
- 19.Sala K, Karcz J, Rypień A, Kurczyńska EU. Unmethyl-esterified homogalacturonan and extensins seal Arabidopsis graft union. BMC Plant Biol. 2019;19:151. 10.1186/s12870-019-1748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G. et. al. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell. 2015;33:216–30. [DOI] [PubMed] [Google Scholar]
- 21.Brzezicka E, Kozieradzka-Kiszkurno M. Developmental, ultrastructural and cytochemical investigations of the female gametophyte in Sedum rupestre L. (Crassulaceae). Protoplasma. 2021;258:529–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford MM. A rapid and sensitive method for the quantitation of microgramquantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. [DOI] [PubMed] [Google Scholar]
- 24.Rys M, Pociecha E, Oliwa J, Ostrowska A, Jurczyk B, Saja D. Janeczko A Deacclimation of winter oilseed rape: Insight into physiological changes. Agronomy. 2020;10:1565. [Google Scholar]
- 25.Godel-Jędrychowska K, Milewska-Hendel A, Sala K, Barański R, Kurczyńska E. The impact of gold nanoparticles on somatic embryogenesis using the example of Arabidopsis thaliana. Int J Mol Sci. 2023;24:10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9. [Google Scholar]
- 27.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y-X, Xu X-B, Hu Y-H, Han W-H, Yin J-L, Li H-L. Gong H-J. Silicon improves salt tolerance by increasing root water uptake in Cucumis sativus L. Plant Cell Rep. 2015;34:1629–46. [DOI] [PubMed] [Google Scholar]
- 29.Esmaeili N, Cai Y, Tang F, Zhu X, Smith J, Mishra N, Hequet E, Ritchie G, Jones D, et al. Towards doubling fibre yield for cotton in the semiarid agricultural area by increasing tolerance to drought, heat and salinity simultaneously. Plant Biotechnol J. 2021;19(3):462–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luyckx M, Hausman JF, Lutts S, Guerriero G. Impact of Silicon in Plant Biomass Production: Focus on Bast Fibres, Hypotheses, and Perspectives. Plants-Basel. 2017;96(3):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piva TC, Machado SR, Edna S-D. Anatomical and ultrastructural studies on gelatinous fibers in the organs of non-woody xerophytic and hydrophytic species. Botany. 2019;97(10):529–36. [Google Scholar]
- 32.Bessadok A, Marais S, Gouanvé F, Colasse L, Zimmerlin I, Roudesli S, Metayer M. Effect of chemical treatments of Alfa (Stipatenacissima) fibres on water-sorption properties. Compos Sci Technol. 2007;67:685–97. [Google Scholar]
- 33.Pejic BM, Kostic MM, Skundric PD, Praskalo JZ. The effects of hemicelluloses and lignin removal on water uptake behavior of hemp fibers. Bioresour Technol. 2008;99:7152–9. [DOI] [PubMed] [Google Scholar]
- 34.Guerriero G, Stokes I, Exley C. Is callose required for silicification in plants? Biol Lett. 2018;14:20180338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Cheng W, Wang J, Cheng T, Lin X, Zhang Q, Li C. Genome-Wide Identification of Callose Synthase Family Genes and Their Expression Analysis in Floral Bud Development and Hormonal Responses in Prunus mume. Plants. 2023;12:4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong Z, Zhang Z, Olson JM, Verma DP. A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell. 2001;3(4):769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X, Han X, Lu TG. Callose synthesis during reproductive development in monocotyledonous and dicotyledonous plants. Plant Signal Behav. 2016;11(2):e1062196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kido N, Yokoyama R, Yamamoto T, Furukawa J, Iwai H, Satoh S, Nishitani K. The matrix polysaccharide (1;3,1;4)-b-D-Glucan is involved in silicon-dependent strengthening of rice cell wall. Plant Cell Physiol. 2015;56:1679. [DOI] [PubMed] [Google Scholar]
- 39.Yan D, Yadav SR, Paterlini A, Nicolas WJ, Petit JD, Brocard L, Belevich I, Grison MS, Vaten A, Karami L, et al. Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat Plants. 2019;5:604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA. 2010;107(36):15986–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H. et. al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–3. [DOI] [PubMed] [Google Scholar]
- 42.He C, Ma J, Wang L. A hemicellulose-bound form of silicon with potential to improve the mechanical properties and regeneration of the cell wall of rice. New Phytol. 2015;206(3):1051–62. [DOI] [PubMed] [Google Scholar]
- 43.Sheng H, Chen S. Plant silicon-cell wall complexes: identification, model of covalent bond formation and biofunction. Plant Physiol Biochem. 2020;155:13–9. [DOI] [PubMed] [Google Scholar]
- 44.Willats WG, Marcus SE, Knox JP. Generation of a monoclonal antibody specific to (1→5)-α-l-arabinan. Carbohydr Res. 1998;308:149–52. [DOI] [PubMed] [Google Scholar]
- 45.Emam MM, Khattab HE, Helal NM, Deraz AE. Effect of selenium and silicon on yield quality of rice plant grown under drought stress. AJCS. 2014;8(4):596–605. [Google Scholar]
- 46.Cui J, Li F. 2023. Silica nanoparticles inhibit arsenic uptake into rice suspension cells via improving pectin synthesis and the mechanical force of the cell wall. In: van der Wal, Ahmad, Petrusevski, Weijma, Savic, van der Wens, Beerendonk, Bhattacharya, Bundschuh & Naidu, editors. Arsenic in the Environment: Bridging Science to Practice for Sustainable Development. 2023;ISBN 978-1-032-32928-4.
- 47.Sheng H, Ma J, Pu J, Wang L. Cell wall-bound silicon optimizes ammonium uptake and metabolism in rice cells. Ann Bot. 2018;122:303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelsdorf T, Gigli-Bisceglia N, Veerabagu M, McKenna JF, Vaahtera L, Augstein F, van der Does D. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci Signal. 2018;11. 10.1126/scisignal.aao3070. [DOI] [PubMed]
- 49.Nardini A. 2022. Hard and tough: the coordination between leaf mechanical resistance and drought tolerance. Flora. 2022;288:152023.
- 50.Sujkowska-Rybkowska M, Borucki W. Pectins esterification in the apoplast of aluminum-treated pea root nodules. J Plant Physiol. 2015;20(184):1–7. [DOI] [PubMed] [Google Scholar]
- 51.Ganie SA, Ahammed GJ. Dynamics of cell wall structure and related genomic resources for drought tolerance in rice. Plant Cell Rep. 2021;40(3):437–59. [DOI] [PubMed] [Google Scholar]
- 52.Tenhaken R. Cell wall remodeling under abiotic stress. Front Plant Sci. 2015;7(5):771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaskowiak J, Kwasniewska J, Milewska-Hendel A, Kurczyńska EU, Szurman-Zubrzycka M, Szarejko I. Aluminum Alters the Histology and Pectin Cell Wall Composition of Barley Roots. Int J Mol Sci. 2019;20:3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf S, Mouille G, Pelloux J. Homogalacturonan methyl-esterification and plant development. Mol Plant. 2009;2(5):851–60. [DOI] [PubMed] [Google Scholar]
- 55.Pelloux J, Rusterucci C, Mellerowicz EJ. New insights into pectin methyl-esterase structure and function. Trends Plant Sci. 2007;12:267–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Availability of data and materialsMaterials described in the manuscript, including all relevant raw data, will be freely available to any scientist, after the sending such asking to corresponding author. All raw date are stored on the PC computer of corresponding author. Moreover they are collected on the separate external drive stored by corresponding author.