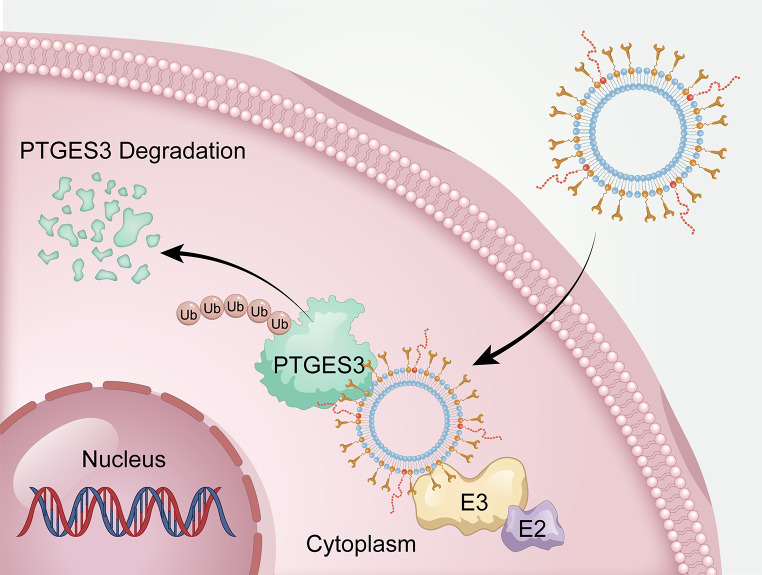
Abstract
Background
Hepatocellular carcinoma (HCC) is the leading cause of cancer-related deaths worldwide, and the lack of effective biomarkers for early detection leads to poor therapeutic outcomes. Prostaglandin E Synthase 3 (PTGES3) is a putative prognostic marker in many solid tumors; however, its expression and biological functions in HCC have not been determined. The proteolysis-targeting chimera (PROTAC) is an established technology for targeted protein degradation. Compared to the small-molecule PROTAC, the peptide PROTAC (p-PROTAC) utilizes peptides bound to target proteins to mediate the ubiquitination and degradation of undruggable proteins. This study aimed to use the PROTAC technology to develop a PTGES3 degrader liposome complex containing a PTGES3-binding peptide and the E3 ubiquitin ligase ligand pomalidomide for regulating cell function and provide a novel pathway for treating HCC.
Results
In this study, we demonstrated that PTGES3 is highly expressed in HCC at the transcriptional and protein levels; furthermore, PTGES3 was identified as a novel drug target that could potentially treat HCC. Hence, we developed PTGES3-PROTACs by adjusting the ligand ratio to optimize the efficacy of degradation agents. The results revealed that PTGES3-PROTAC effectively degraded PTGES3 protein and strongly weakened the HCC malignant phenotype in vitro and in vivo.
Conclusions
Our findings revealed that the highly selective PTGES3 proteolysis is a potential therapeutic strategy for HCC, and PTGES3 degraders PTGES3-PROTACs can be developed as safe and effective drugs for HCC treatment.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13062-024-00580-0.
Keywords: PTGES3, Peptide proteolysis-targeting chimera, Liposome, Hepatocellular Carcinoma
Background
Hepatocellular carcinoma (HCC) is one of the most prevalent and deadliest types of cancer, hindering social and economic development [1, 2]. Common risk factors for HCC include persistent hepatitis B and C viral infections and fatty liver disease [3]. Early-stage HCC can be treated with potentially curative therapeutic options such as radiofrequency ablation, transarterial therapy, and surgical resection. However, early-stage HCC symptoms are not detectable at the time of first diagnosis; therefore, many individuals with HCC often present at an intermediate or advanced stage where radical treatment is no longer effective [4]. Oral administration of Sorafenib, a kinase inhibitor, is the primary therapeutic option for advanced HCC [5]. Lenvatinib (which is as effective as Sorafenib) and regorafenib are standard treatments for advanced HCC [6]. Chemotherapy is ineffective in treating HCC. Therefore, discovering highly sensitive and specific protein biomarkers and developing effective treatment methods are crucial for HCC diagnosis and treatment. However, the clinical application of biomarkers for HCC surveillance and early diagnosis has not achieved the expected results [7]. Therefore, there is a need to identify better diagnostic biomarkers and develop therapeutic strategies for HCC.
Targeted protein degradation technology based on the precision medicine concept is widely used for HCC treatment. Therefore, the proteolysis-targeting chimera (PROTAC) is a promising technology for targeted protein degradation [8]. This small-molecule hijacking of the protein of interest (POI) brings it closer to the E3 ubiquitin ligase, promoting POI ubiquitination and protein degradation via the ubiquitin-proteasome pathway [9]. PROTAC successfully degrades several key pathogenic proteins. NRF2 is a transcription factor that regulates the cellular defense against oxidative damage by participating in oxidative stress responses [10]. Keap1 is considered an endogenous Nrf2 inhibitor [11]. A previous study developed a PROTAC named SD2267, which induces KEAP1 degradation and NRF2 activation to combat oxidative stress-related diseases [12]. Additionally, RIPK1-specific PROTACs have been developed to overcome radiation or immunotherapy resistance and enhance anticancer treatments [13]. Wang et al. [14] identified NR4A1 as an effective target for cancer immunotherapy and developed a PROTAC that targets NR4A1. Although PROTACs that target many proteins have been developed and applied, they cannot target undruggable proteins without small-molecule binders. Even among druggable targets, traditional treatment strategies that occupy active sites often require high-dose drug exposure, thereby increasing the potential threat of off-target toxicity. With advances in chemically induced proximity technologies, novel PROTAC approaches have been developed to target undruggable proteins. Unlike traditional PROTAC, peptide-PROTAC (p-PROTAC) have high specificity and low toxicity and are a promising alternative for the proteolysis of undruggable proteins by binding peptides to POI [15]. However, p-PROTACs exhibited poor in vivo efficacy, mainly owing to poor cell permeability and low accumulation in diseased tissues. Previous studies have used liposomes as PROTAC carriers because they can prevent drug degradation, target drugs, and improve cell permeability.
Perfecting p-PROTAC will enable this technology to address the limitations of traditional drug development methods and can be used to treat more diseases. PTGES3 can be targeted using this new strategy. Previous studies revealed that heat shock protein 90 (Hsp90) regulates the folding and activation of several proteins [16]. Similar to many other molecular chaperones, Hsp90 has important ATPase activity and relies on ATP hydrolysis to power a conformational circuit that aids in the folding of target proteins [17]. P23 and Aha1 are two co-chaperones that have contrasting effects on the ATPase activity of Hsp90 [18]. P23 suppresses Hsp90 ATPase and promotes the stability of client-bound Hsp90 [19]. Subsequently, p23 was renamed prostaglandin E Synthase 3 (PTGES3) because of its prostaglandin E2 (PGE2) synthetase properties [20]. Previous studies have emphasized the functional role of PTGES3 in the malignant progression of tumors. For example, in human prostate tumors, the high PTGES3 expression and its ability to promote the acquisition of a more aggressive phenotype have been confirmed, supporting the practicality of PTGES3 as a promising therapeutic target in prostate cancer [21, 22]. Elevated PTGES3 levels facilitate tumor progression in breast cancer and provide a fundamental principle for inhibiting PTGES3 during anticancer chemotherapy [23–25]. In lung adenocarcinoma, PTGES3 could be considered as a novel drug target and predictive biomarker for poor prognosis [26, 27]. Contrastingly, Chen et al. have reported that PTGES3 plays an inhibitory role in ovarian cancer invasion [28]; however, the PTGES3 expression and function in HCC need to be elucidated. Therefore, it is necessary to clarify the PTGES3 expression and biological function in HCC and to develop p-PROTAC for PTGES3 degradation.
This study aimed to use PROTAC technology to develop a system for regulating cell function and provide a novel pathway for treating HCC by creating a PTGES3 degrader liposome complex containing a PTGES3-binding peptide and the E3 ubiquitin ligase ligand pomalidomide.
Methods
Cell lines and culturing
Human HCC cell lines (Hep3B and Huh7) were purchased from Pricella Biotechnology Co., Ltd (Wuhan, China). MHCC-97 H, MHCC-97 L and human hepatic epithelial cell line THLE-2 were stored in our laboratory. HCC cells were maintained in DMEM culture medium containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified chamber with 5% CO2.
PTGES3 knockdown was achieved by transient transfection of siRNA into cells using Lipofectamine 2000 transfection reagent (11668, Invitrogen). PTGES3 siRNAs were synthesized by Hanheng Company (Shanghai, China). After transfection for 3 days, PTGES3 protein expression was detected. The target sequences of siPTGES3 were as follows: siPTGES3#1, forward, 5′-GGACUAUGUCUUCAUUGAAUUdTdT-3′, reverse, 5′-AAUUCAAUGAAGACAUAGUCCdTdT-3′; siPTGES3#2, forward, 5′- GGCUUAGUGUCGACUUCAAUAdTdT-3′, reverse, 5′- UAUUGAAGUCGACACUAAGCCdTdT-3′; siPTGES3#3, forward, 5′- CCAAAUGAUUCCAAGCAUAAAdTdT-3′, reverse, 5′- UUUAUGCUUGGAAUCAUUUGGdTdT-3′.
Hep3B and Huh7 cells were incubated with 15 µmol/L MG132. Subsequently, we conducted co-immunoprecipitation (CO-IP) and western blotting assays to test the ubiquitination level of PTGES3.
Fluorescein isothiocyanate (FITC) labeled synthetic FIP peptides and PTGES3-PROTAC were added to the culture medium, and the cells were processed for proliferation and migration tests.
HCC tissue microarray
HCC tissue microarray (HLivH180Su30) was purchased from OUTDO BIOTECH Company (Shanghai, China). Human liver cancer tissue microarray slide was subjected to standard immunohistochemistry (IHC) protocols. The PTGES3 (1:500, 67736-1-Ig, Proteintech Group) expression in human HCC tissue microarray was evaluated.
Immunohistochemistry (IHC) staining analysis
Fresh tissues were fixed in 4% paraformaldehyde for 48 h at room temperature and further embedded with paraffin. In the light of standard instructions, slices were processed to IHC staining. The results were observed under a light microscope. Anti-PTGES3 and anti-Ki67 (27309-1-AP, Proteintech Group) were used as primary antibodies.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA extraction from HCC cells (CW0584, CWBIO), cDNA generation (CW2569, CWBIO) and real-time PCR (CW0957, CWBIO) were conducted according to the manufacturer’s protocols. The expression of PTGES3 mRNA was normalized by GAPDH. The specific qRT-PCR primer sequences were displayed as follows in this study: PTGES3-forward, 5-CAGTTGTCTCGGAGGAAGTGATA-3, PTGES3-reverse, 5-GCTTTGCCCTTTCTTTTGTTA-3; GAPDH-forward, 5-GACACCCACTCCTCCACCTTT-3, GAPDH-reverse, 5-TCTCTTCCTCTTGTGCTCTTGCT-3.
Western blotting
For western blotting analysis, total protein lysate from indicated cells was obtained by ice-cold Enhanced RIPA buffer with protease inhibitor PMSF ((P0013B and ST505, Beyotime Biotechnology). Protein concentrations were determined by a BCA Protein Detection Kit (P0012S, Beyotime Biotechnology). Equal amounts of cellular protein extracts were loaded onto SDS-PAGE gel for electrophoresis, and then transferred to the polyvinylidene fluoride membranes for immunoblotting. Following blocking with 5% skim milk solution, the membranes were incubated sequentially with diluted primary antibodies overnight at 4 °C and HRP-conjugated secondary antibodies for 1.5 h at room temperature. Finally, bands were visualized using the chemiluminescence imaging system (SCG-W3000, Servicebio) and measured via ImageJ software. Primary antibodies were listed as follows in the present study: anti-PTGES3, anti-β-actin and anti-ubiquitin (1:1000, 15216-1-AP, HRP-60008 and 10201-2-AP, Proteintech Group). The second antibody (1:5000, SA00001-2) was purchased from the Proteintech Group Company.
Cell proliferation tests
Cell counting kit-8 assays (CCK-8 assay) were performed according to manufacturer’s instructions (C0037, Beyotime Biotechnology). Briefly, HCC cells were planted in 96-well plates. Samples were incubated (37 °C, 2 h) with CCK-8 solution and analyzed by spectrometer at a wavelength of 450 nm (OD450).
For the EdU staining proliferation test (C0071S, Beyotime Biotechnology), HCC cells were stained and visualized after be labeled by EdU. EdU-positive immunofluorescence was marked green; all cell nuclei were marked blue by DAPI solution.
For colony formation assay, HCC cells were seeded in 6-well plates (1000 cells/well) in triplicate. After fixation and staining, colonies were counted.
Cell migration assays
Cell culture inserts (PTEP24H48, Millipore) were used to perform migration tests. HCC cells were added to the upper chamber of the transwell. The migrated cells were observed and counted under a microscope.
For wound healing assays, HCC cells were seeded in 6-well plates and cultured until complete fusion. The fused monolayer was scraped off by a clean pipette tip. Scratch distance was recorded by microscope. Serum free media containing FIP-2 and PTGES3-PROTAC were added to six well plates. 48 h later, the migration distance of cells across the wound was measured using a microscope.
Animal models assays
The present study used a subcutaneous model involving 5-week-old male BALB/c mice for investigating tumour growth.
In order to investigate the in vivo biological function of PTGES3 on HCC, a total of 2 × 106 transfected HCC cells (Hep3B and Huh7 cells) were injected subcutaneously into the right axilla of each mouse. After planting cells for 4 weeks, the mice were euthanized by CO2. The implanted tumor was obtained and weighed. Tumor volume was measured weekly according to the formula: volume (mm3) = length × width2/2. Subsequently, subcutaneous tumors were further analyzed via IHC staining.
To verify the in vivo antitumor effect of PROTACs, we established a subcutaneous implanted tumor BALB/c mice model by injecting 2 × 106 HCC cells (Hep3B and Huh7 cells) into subcutaneous tissue. After a week, tumor bearing nude mice were injected with 20 mg/kg FIP-2 or PTGES3-PROTAC via intraperitoneal injection, every other day for 14 days. Four weeks after tumor implantation, the mice were euthanized. Main organs and subcutaneous tumors were obtained and analyzed.
In vitro phage displays
Human PTGES3 protein was coated in a 96 well plate overnight at 4 °C. 2% defatted milk powder was used to block at room temperature for 1 h. Subsequently, phage solution, diluted with 2% skim milk powder, was added to the plate coated with PTGES3 protein, and incubated at room temperature for 2 h. Bound phages were eluted by Tris-HCl (pH = 3.0) and used as input for the next round of biopanning. Three rounds of biopanning led to enrichment of specific phages. The enriched phage sheath DNA was sequenced.
Peptide and PTGES3 degrader liposomes complex production
The ChomiX Biotech Company (Nanjing, China) provided assistance in peptide synthesis. TAT (GRKKRRQRRRPPQQ) was selected as a cell penetrating peptide coupled to the C-terminus of six FIP peptides.
C(DSPE-PEG2000-MAL)-FIP-2 was generated through the addition reaction of C-FIP-2 and DSPE-PEG2000-MAL (CAS NO.: 474922-22-0); DSPE-PEG2000-pomalidomide was produced by the reaction of DSPE-PEG2000-NHS (CAS NO.: 1445723-73-8) with the amino group of pomalidomide (CAS NO.: 19171-19-8).
The raw materials for synthesizing liposome complex include: HSPC (92128-87-5, AVT (Shanghai) Pharmaceutical Tech CO. Ltd.), cholesterol (C104032-5 g, aladdin), DSPE-mPEG2000 (147867-65-0, AVT (Shanghai) Pharmaceutical Tech CO. Ltd.), Coumarin 6 (C100929-100mg, aladdin), DSPE-PEG2000-pomalidomide and C(DSPE-PEG2000-MAL)-FIP-2. We first prepared PTGES3 degrader liposomes (1:1). Briefly, DSPE-PEG2000-pomalidomide: C(DSPE-PEG2000-MAL)-FIP-2 in a 1:1 molar ratio were dissolved in 2.5 ml chloroform containing 14.2 mg HSPC, 5.6 mg cholesterol, 0.3 mg Coumarin 6 and 11.8 mg DSPE-mPEG. Chloroform was evaporated using a rotary evaporator water bath. After ultrasonicating the resultant powder, liposomes were obtained. Similarly, the PTGES3 degrader liposomes (1:5) and (5:1) were successfully prepared as per the aforementioned method.
Immunofluorescence (IF) staining
HCC cells were implanted in a 6-well plate containing coverslips and allowed to culture overnight. Six synthesized FIP peptides were added to the culture medium at a final concentration of 10 µM, and incubated for 6 h. Subsequently, the cells were fixed and permeabilized. The slide was blocked for 1 h and incubated overnight with anti-PTGES3 antibody (67736-1-Ig, Proteintech Group) at 4 °C. The next day, slide was washed three times with PBS and stained with fluorescent secondary antibody. The slide was mounted by Antifade Mounting Medium containing DAPI and photographed under a confocal microscope.
Ubiquitination analysis
Ubiquitination level of PTGES3 was tested by CO-IP with anti-PTGES3 antibody (15216-1-AP, Proteintech Group). Prior to collecting cellular proteins, proteasome inhibitor MG132 (15 µM) was administered to HCC cells. The rest of the steps were conducted according to the instructions of the rProtein A/G Magnetic IP/Co-IP Kit (AM001, Nanjing ACE Biotechnology) and further verified by SDS–PAGE for western blotting. The PVDF membranes were visualized with enhanced chemiluminescence reagent (180–506, Tanon). Rabbit IgG (A7016, Beyotime Biotechnology) was used as control group.
Serum liver and kidney indexes in nude mice
The blood sample was centrifuged at 3000–4000 rpm for 10 min, and the supernatant was taken and stored in a refrigerator at 4 °C for subsequent testing. Alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN) and creatinine (CREA) were measured according to the instructions of the kits (S03030, S03040, S03036 and S03076, Rayto).
Statistical analysis
GraphPad Prism 8.0.1 software was adopted for statistical analysis and graph generation. Data were displayed as mean ± s.d. or a representative photograph of independent experiments. Student’s t-test and ANOVA were applied for data analysis (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Results
High PTGES3 expression is significantly associated with unfavorable clinical outcomes
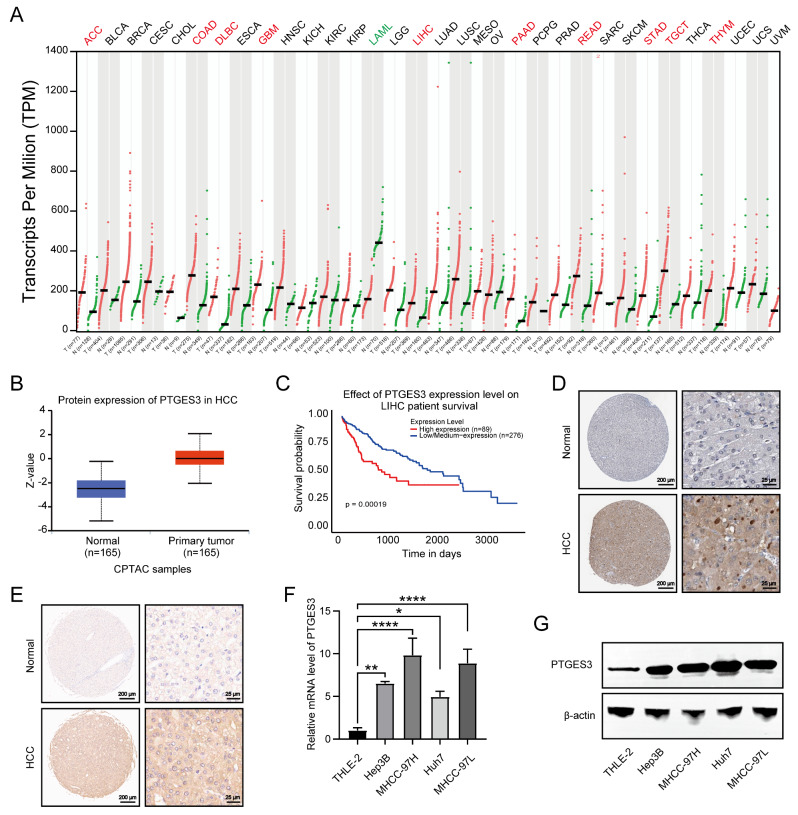
The Gene Expression Profiling Interactive Analysis server was used to explore the PTGES3 dysregulated expression in pan-cancer samples and paired normal tissues [29]. Among the 33 cancer types, PTGES3 mRNA was highly expressed in 10 (including HCC) compared with those in normal tissues (Fig. 1A). The UALCAN platform revealed that PTGES3 protein levels were higher in HCC tissues than that in the adjacent tissues (Fig. 1B). Furthermore, Kaplan–Meier survival analysis revealed that patients with HCC overexpressing PTGES3 had a poor prognosis (Fig. 1C). Additionally, publicly available IHC data from the online Human Protein Atlas (HPA) database indicates that PTGES3 protein expression is higher in HCC samples than that in paired non-cancerous hepatic tissues (Fig. 1D). Consistent with the results from the HPA database, our IHC results from the microarray of HCC tissues from patients with HCC revealed that the PTGES3 protein level was upregulated in HCC samples compared with that in the samples from the surrounding cells (Fig. 1E). We investigated the PTGES3 expression in HCC cells, and the results revealed that the PTGES3 mRNA and protein expression was elevated in both HCC cell lines (Fig. 1F and G) compared with that in THLE-2 cells. In summary, the results of this study suggest that PTGES3 is overexpressed in HCC cells and is correlated with a poor prognosis.
Fig. 1.
PTGES3 was overexpressed and predicted poor prognosis in hepatocellular carcinoma (HCC). (A) Bioinformatics analysis of PTGES3 mRNA expression in multiple tumor samples by an online website GEPIA. (B) Protein expression analysis of PTGES3 in 165 HCC tumor samples and 165 normal tissues by UALCAN. (C) Kaplan-Meier survival analysis was used to measure the correlation between PTGES3 expression and overall survival in HCC from UALCAN website. (D) Immunohistochemical (IHC) staining pictures of PTGES3 protein in HCC and paracancerous samples from the Human Protein Atlas online database (n = 3). (E) Representative IHC images of PTGES3 protein in paired HCC specimens and their normal liver tissues from the tissue microarray (n = 90). (F, G) PTGES3 mRNA and protein expressions of HCC cell lines and normal liver epithelial THLE-2 cells from qRT-PCR (F) and western blotting (G) results (n = 3)
Altered PTGES3 expression impacts HCC cell proliferation and migration in vitro
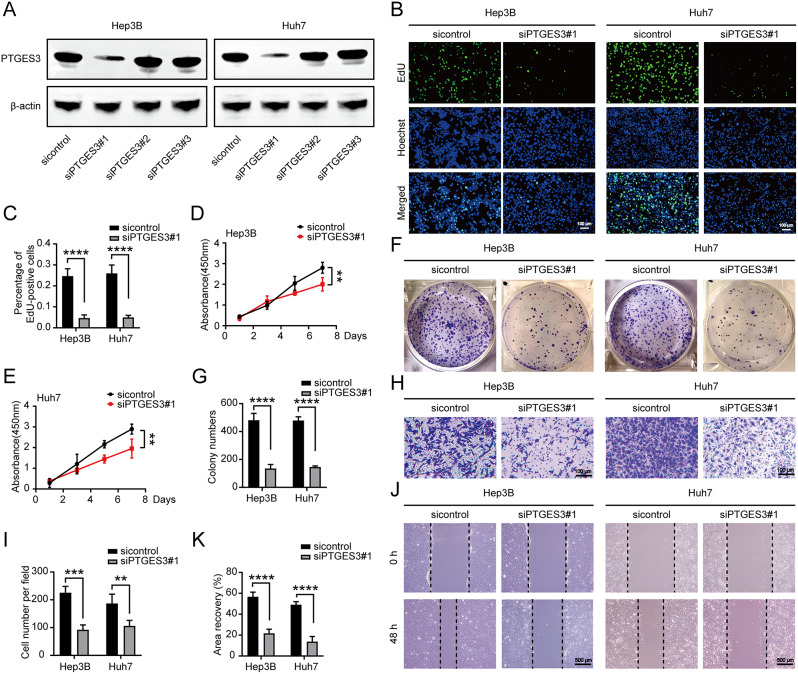
PTGES3 plays an important role in developing multiple tumors; therefore, we determined whether PTGES3 is involved in HCC progression. To determine the effect of PTGES3 on HCC cells, we first constructed three siRNAs to knockdown PTGES3 in Hep3B and Huh7 cell lines and found that siPTGES3#1 had a significant manipulation ability (Fig. 2A). EdU incorporation (Fig. 2B and C), Cell Counting Kit-8 (Fig. 2D and E), and clone formation assays (Fig. 2F and G) revealed that PTGES3 silencing suppressed HCC cell growth. Furthermore, we aimed to demonstrate the effect of PTGES3 on the regulation of tumor migration capacity using transwell and wound healing assays. Our results revealed that PTGES3 reduction considerably impaired cell migration ability (Fig. 2H–K). Overall, PTGES3 knockdown reduced the proliferation and migration of HCC cells in vitro.
Fig. 2.
PTGES3 knockdown weakened hepatocellular carcinoma (HCC) cells proliferation and migration in vitro. (A) Western blotting was conducted to detect the capacity to manipulate PTGES3 expression by siRNAs (n = 3). (B, C) EdU assays showed the growth of HCC cells (n = 3). (D, E) CCK-8 experiments revealed the proliferative ability of HCC cells (n = 3). (F, G) HCC cells were subjected to colony formation assays (n = 3). (H, I) Transwell showed that the migrated ability of HCC cells (n = 3). (J, K) Wound healing assays were performed to test the migration of HCC cells (n = 3)
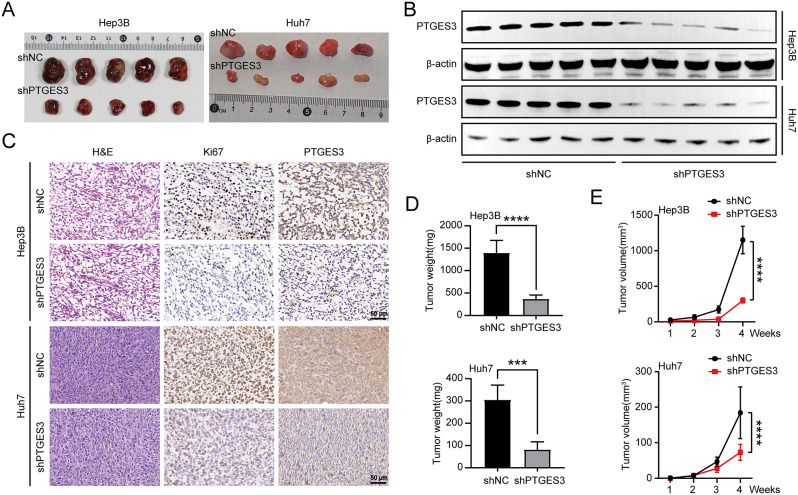
PTGES3 knockout impedes HCC cell growth in vivo
Hep3B and Huh7 cells were used to establish subcutaneous xenograft nude mice model to assess whether PTGES3 affected HCC tumorigenesis in vivo (Fig. 3A). PTGES3 expression analysis in xenograft tumors revealed that transfecting shPTGES3 lentivirus into HCC cells markedly silenced PTGES3 (Fig. 3B). The IHC results for xenograft tumors indicated that PTGES3 staining was reduced by PTGES3 silencing. IHC analysis also revealed that the proportion of Ki67-positive cells in PTGES3 depletion xenograft tumors was significantly lower than that in the control group, indicating that PTGES3 plays an important role in HCC cell proliferation in vivo (Fig. 3C). The findings of this study indicated that PTGES3 knockdown significantly impeded tumor weight and volume compared with those of the corresponding control group (Fig. 3D and E), indicating that PTGES3 knockdown inhibits the growth of HCC cells in vivo.
Fig. 3.
The ablation of PTGES3 decreased the tumor growth in vivo. (A) Images of implanted tumors from nude mice injected with Hep3B and Huh7 cells subcutaneously (n = 5). (B) Western blotting measured the PTGES3 expression in tumor samples (n = 5). (C) Immunohistochemical (IHC) staining of PTGES3 protein in implanted tumors with stable transfection by shNC or shPTGES3 (n = 5). (D) Tumor weight of nude mice (n = 5). (E) Tumor volume of nude mice (n = 5)
Peptides specifically bind to the PTGES3 protein
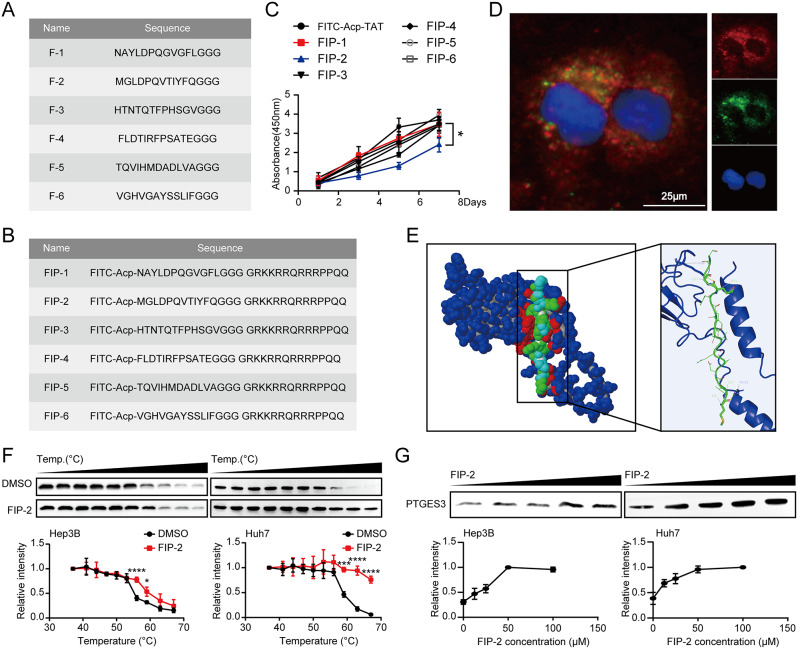
Peptides are more desirable therapeutic molecules than small-molecule drugs due to their high selectivity and fewer side effects [30]. Several new peptides have emerged as effective linkers, suggesting they can be used to develop small-molecule degraders such as p-PROTACs [31, 32]. Peptide phage display libraries and the human PTGES3 protein were used to screen peptides targeting the PTGES3 protein. The bound phages were eluted and sequenced after three rounds of screening. Based on the sequencing results, we obtained six potential peptides targeting PTGES3 (Fig. 4A). Although some peptides can enter cells without carriers, their permeability through the cell membrane is limited. Cell-penetrating peptides can cross the cell membrane and transport biological products and therapeutic agents into cells; however, some are more effective than others [33]. TAT, a typical cell-penetrating peptide, efficiently translocate across cellular membranes and transports biological products [34]. To ensure that the six candidate peptides could penetrate the cell membrane, we selected TAT as a carrier and coupled it with FITC at the C-terminal- and N-termini of the peptides. We synthesized six peptides with cell membrane-penetrating functions (Fig. 4B). Subsequently, mass spectrometry was performed (Figure S1). We confirmed that FIP-2 suppressed HCC cell growth (Fig. 4C). IF assays revealed that FITC-labeled FIP-2 could not only successfully penetrate cells, but also effectively co-localize with PTGES3 (Fig. 4D). Furthermore, molecular docking results revealed an interaction between F-2 and PTGES3 (Fig. 4E) [35, 36]. We employed complementary methods to directly analyze protein-peptide interactions in HCC cell lysates to demonstrate the binding of FIP-2 to PTGES3. Temperature- and dose-dependent cellular thermal shift measurements revealed that FIP-2 affected the PTGES3 thermal stability (Fig. 4F and G), supporting a direct interaction between FIP-2 and PTGES3. Collectively, these data indicated that FIP-2 can bind to PTGES3 and can be selected as a peptide for developing p-PROTACs.
Fig. 4.
Identification of peptides targeting PTGES3 protein. (A) Name and sequence of peptides bound to PTGES3 protein. (B) Name and sequence of synthetic peptides conjugated with FITC and TAT sequence (GRKKRRQRRRPPQQ). (C) CCK-8 showed the growth rate of Huh7 cells treated with six synthetic peptides (n = 3). (D) IF experiments showed the cellular localization of PTGES3 protein (red) and FITC labeled FIP-2 (green) in Huh7 cells (n = 3). (E) The optimized binding modes between PTGES3 protein and F-2, Blue: PTGES3 protein; Green: F-2 peptide. (F) Temperature-dependent cellular thermal shift assay displayed the elevated thermal stability of PTGES3 in HCC cell lysates treated with FIP-2 (n = 3). (G) Concentration-dependent cellular thermal shift experiment revealed the increased thermal stability of PTGES3 treated with FIP-2 in Hep3B and Huh7 cell lysates at 56 and 59°C, respectively (n = 3)
Liposome complexes are modified by pomalidomide and peptides
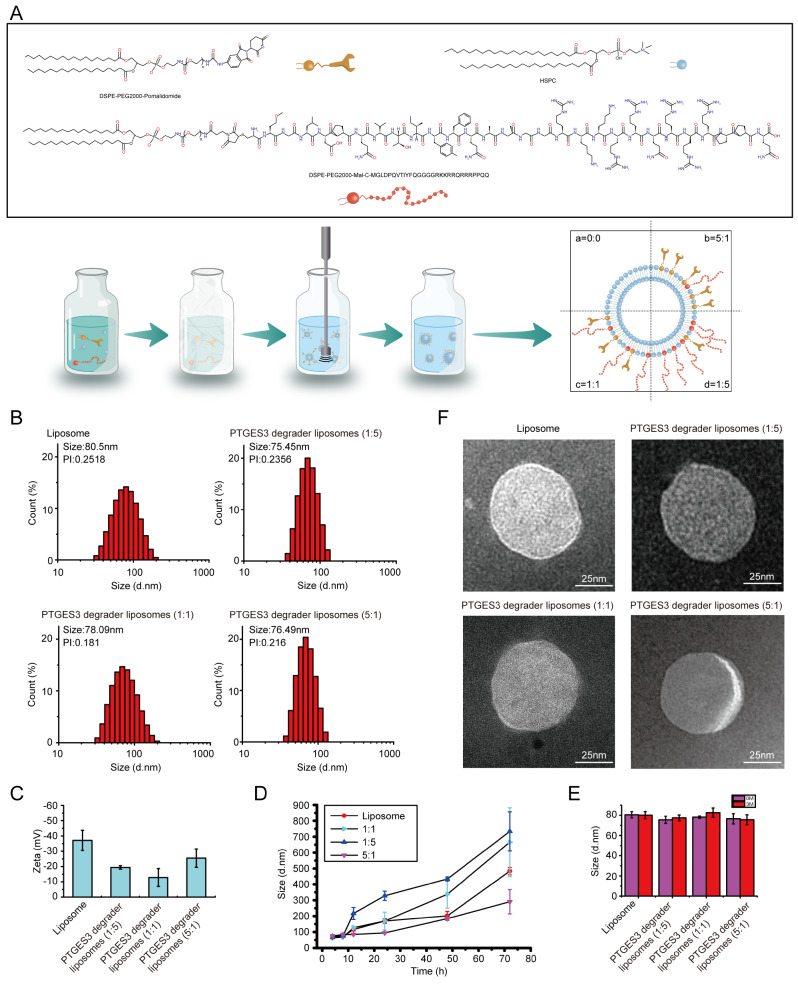
Liposomes containing phospholipids and cholesterol (without drugs) were prepared for subsequent experiments. Next, the PTGES3 degrader liposomes were successfully prepared (including DSPE-PEG2000-pomalidomide and C(DSPE-PEG2000-MAL)-FIP-2). To optimize the ligand ratios of the liposome complexes, we set the pomalidomide: FIP-2 ratios to 1: 5, 1: 1, and 5: 1 (Fig. 5A). As shown in Fig. 5B, the diameter of the empty liposome was 80.5 nm with a polydispersity index (PI) less than 0.30. The dynamic light scattering (DLS) of the three PTGES3 degrader liposomes we synthesized indicated that PTGES3 degrader liposomes were uniform and smaller in size. The zeta potential of PTGES3 degrader liposomes confirmed that the surface of liposomes was modified with FIP-2 and pomalidomide, and the ratio of FIP-2 to pomalidomide varied among the three types of PTGES3 degrader liposomes (Fig. 5C). We measured the stability of PTGES3 degrader liposomes in vitro in presence of serum and various enzymes and observed that within 24 h, those liposomes maintained extremely high stability, especially PTGES3 degrader liposomes (5:1, Fig. 5D). Additionally, after storage at 4 °C for 3 months, the particle size remained nearly unchanged, indicating the stability of the prepared particles (Fig. 5E). The transmission electron microscopy imaging revealed that the PTGES3 degrader liposomes had a uniform spherical morphology (Fig. 5F).
Fig. 5.
Preparation and characterization of PTGES3 liposome peptide-proteolysis targeting chimera (p-PROTAC). (A) Schematic diagram of the PTGES3 liposome p-PROTAC platform. (B) Particle size distribution of PTGES3 degrader liposomes collected via DLS. (C) Zeta potential of PTGES3 degrader liposomes (n = 3). (D) Stability of PTGES3 degrader liposomes in vitro in presence of serum and various enzymes (n = 3). (E) Stability of PTGES3 degrader liposomes Stored at 4 °C for 3 Months (n = 3). (F) Transmission electron microscope picture of PTGES3 degrader liposomes (n = 3)
Effect of liposomes on the PTGES3 protein degradation
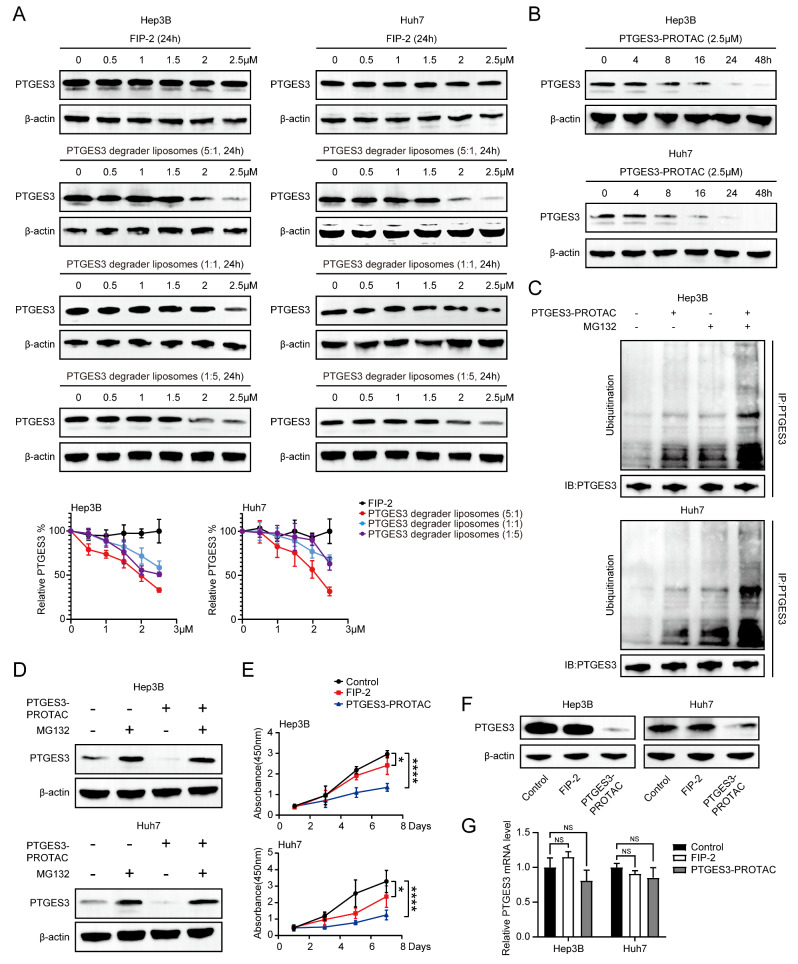
To confirm the effectiveness of the PTGES3 degrader liposomes in vitro, we performed western blotting to determine whether the PTGES3 degradation agents significantly induced PTGES3 degradation in Hep3B and Huh7 cells. When treating cells with small molecules, we observed a dose-dependent degradation of PTGES3, a significant degradation at 2.5 µM, and the 5: 1 (pomalidomide: FIP-2) had the best PTGES3 degradability. Contrastingly, the 1: 5 and 1: 1 ratio did not result in significant degradation (Fig. 6A). PTGES3 degrader liposomes (5:1) were named PTGES3-PROTACs. Similarly, western blotting assays revealed that the PTGES3 degradation in Hep3B and Huh7 cells was time-dependent and reached maximum degradation at 48 h (Fig. 6B). We characterized the uptake and distribution of the liposome system in Huh7 cells using the coumarin 6 fluorescence–labeling technology. PTGES3-PROTAC aggregated in the cytoplasm of HCC cells (Figure S2). Next, to explore the specific PTGES3 degradation mechanisms, CO-IP assays with an anti-PTGES3 antibody were performed to determine the PTGES3 ubiquitination level. The combination of the PTGES3-PROTACs and MG132 groups exhibited significantly higher PTGES3 ubiquitination levels than those of the sole PTGES3-PROTACs and MG132 groups (Fig. 6C). Thus, MG132 was used in subsequent experiments, and it effectively rescued the PTGES3 degradation by PTGES3-PROTACs in HCC cells, indicating that degradation occurred via an ubiquitin–proteasome-dependent pathway (Fig. 6D). Next, we investigated whether liposomes linked to pomalidomide and peptides exerted inhibitory effects. CCK-8 assays demonstrated that PTGES3-PROTACs had a stronger inhibitory effect on HCC cell proliferation than that of FIP-2 (Fig. 6E). We measured the PTGES3 transcriptional and translational levels to determine the specific mechanisms underlying liposomal inhibition, indicating that PTGES3-PROTACs decreased PTGES3 protein levels but not mRNA levels (Fig. 6F and G), suggesting that PROTAC inhibits HCC cell proliferation by eliminating most of the oncogenic PTGES3 protein. FIP-2 also inhibited the malignant phenotype of HCC cells by binding to PTGES3; however, there is a need for future studies to determine the specific molecular mechanisms involved.
Fig. 6.
PTGES3-PROTAC induced PTGES3 proteolysis in a transcriptional independent manner. (A) Western blotting examination showed the PTGES3 degradation in HCC cells treated with different doses of FIP-2 or PTGES3 degrader liposomes (n = 3). (B) Western blotting assays displayed the PTGES3 degradation in HCC cells treated with PTGES3-PROTAC at different time points (n = 3). (C) Western blotting analysis of the ubiquitinated levels of PTGES3 in HCC cells treated with PTGES3-PROTAC and/or MG132 (n = 3). (D) Western blotting analysis of the protein levels of PTGES3 in HCC cells treated with PTGES3-PROTAC and/or MG132 (n = 3). (E) CCK-8 assays were performed to test proliferation of HCC cells treated with FIP-2 or PTGES3-PROTAC (n = 3). (F) Western blotting assays showed the PTGES3 protein expression in HCC cells treated with FIP-2 or PTGES3-PROTAC (n = 3). (G) Detection of PTGES3 mRNA expression by qRT-PCR in HCC cells treated with FIP-2 or PTGES3-PROTAC (n = 3)
PTGES3-PROTACs inhibit the proliferation and migration of HCC cells
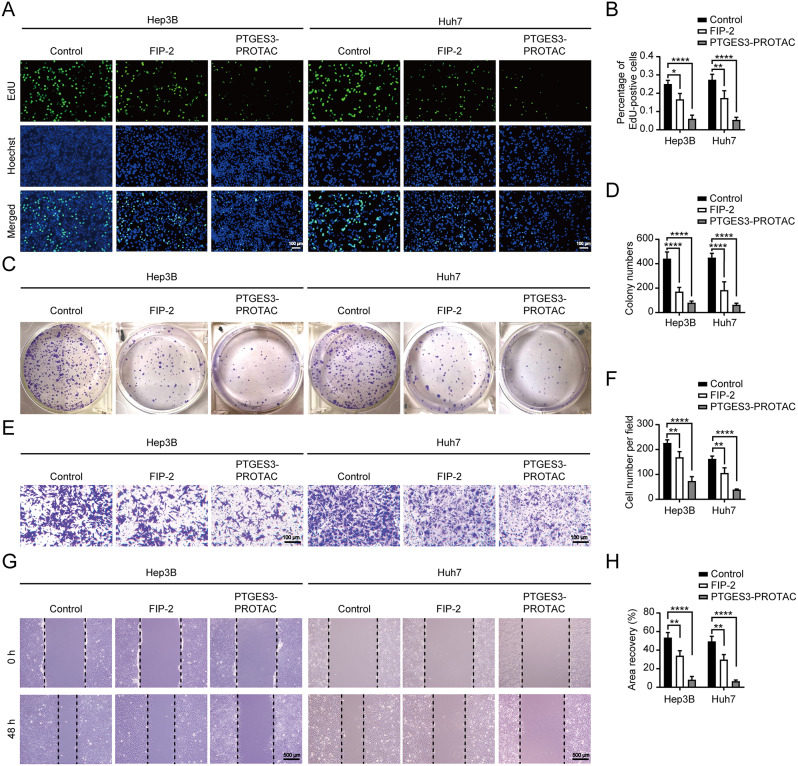
A previous study conducted a comprehensive analysis using multiple databases to determine whether PTGES3 had predictive and targeted therapeutic value in HCC [37]. Hep3B and Huh7 cells were treated with 2.5 µM PTGES3-PROTACs to determine the effect of liposomes on the malignant phenotype of HCC cells. EdU incorporation and clone formation assays indicated that PTGES3-PROTACs suppressed HCC cell proliferation (Fig. 7A–D). Transwell and scratch assays were used to asses cell migration. The migratory ability of HCC cells in the PTGES3-PROTAC-treated group was relatively weaker than that of the control group (Fig. 7E–H). Collectively, these data demonstrated that PTGES3-PROTACs suppressed tumor cell proliferation and migration, indicating their potential for the development of therapeutic drugs against tumors.
Fig. 7.
PTGES3-PROTAC inhibited the malignant phenotype of HCC cells in vitro. (A, B) Representative and quantified results of the EdU incorporation assays. HCC cells were treated with FIP-2 or PTGES3-PROTAC (n = 3). (C, D) The proliferation abilities were analyzed and quantified by colony formation assays. FIP-2 or PTGES3-PROTAC was used to treat HCC cells (n = 3). (E, F) Transwell detection of HCC cells treated with FIP-2 or PTGES3-PROTAC. Representative photographs of transwell assays were presented (E). Statistical analysis of the transwell assays (F, n = 3). (G, H) Representative images and quantified data of the wound healing assays. FIP-2 or PTGES3-PROTAC treating HCC showed the reduced migration ability (n = 3)
PTGES3-PROTACs repress tumor growth in vivo and were not toxic to normal tissues
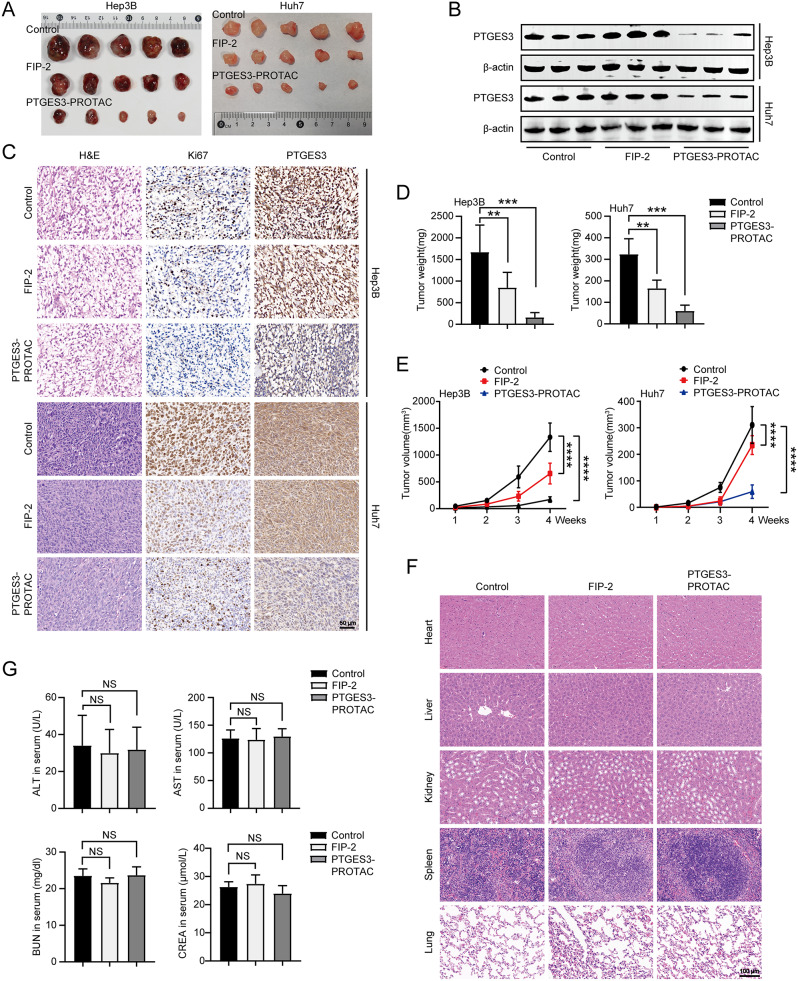
To explore the biological function of PTGES3-PROTACs in vivo, we subcutaneously injected Hep3B and Huh7 cells into nude mice transplanted with tumors. First, in vivo fluorescence imaging was performed by injecting PTGES3-PROTACs carrying DiR iodide into the tail vein to observe the delivery distribution and efficiency. PTGES3-PROTACs were enriched at tumor sites through the enhanced permeability and retention effect (Figure S3). Subsequently, the three groups were injected with sterile physiological saline, 20 mg/kg FIP-2, or 20 mg/kg PTGES3-PROTAC. On the 28th day, the mice were euthanized. The heart, liver, spleen, lungs, kidneys, and subcutaneous tissues were obtained. Xenograft tumors were collected (Fig. 8A), and western blotting and IHC assays were conducted to determine the role of PTGES3-PROTACs in vivo. PTGES3-PROTACs affected the PTGES3 protein expression level in vivo (Fig. 8B and C). Additionally, we observed a reduced proportion of Ki67-positive cells after PTGES3 knockout, indicating that PTGES3 affected tumor proliferation in vivo. The subcutaneous tumors obtained from the three groups were weighed, and the results revealed that the tumors in the PTGES3-PROTACs treatment group were significantly lighter than those in the other groups (Fig. 8D). We measured the tumor volume in each group weekly and found that the tumor volume in the PTGES3-PROTACs treatment group was lower than that in the control group (Fig. 8E). Combined, these data demonstrate that PTGES3-PROTACs significantly inhibit tumor growth in vivo.
Fig. 8.
PTGES3-PROTAC suppressed tumor growth in vivo. (A) Representative photographs of excised tumors from xenograft tumor models (n = 5). (B) The protein levels of PTGES3 expression in subcutaneous xenografts from the control, FIP-2 treatment and PTGES3-PROTAC treatment groups (n = 3). (C) Immunohistochemistry images of Ki67 and PTGES3 protein in xenograft tumors from the indicated groups (n = 3). (D) Subcutaneous xenograft masses were excised and weighed at the endpoint (n = 5). (E) Tumor growth curves of subcutaneous tumor formation in nude mice treated with FIP-2 or PTGES3-PROTAC (n = 5). (F) H&E staining of major organ tissues harvested from tumor-bearing nude mice subcutaneously injected with Hep3B cells. The results showed no systemic toxicity (n = 5). (G) Activities of serum alanine aminotransferase (ALT), aspartic acid transferase (AST), creatinine and blood urea nitrogen from nude mice subcutaneously injected with Hep3B cells (n = 5)
Hematoxylin and eosin (H&E) staining of various mouse organs were conducted to determine drug toxicity, and the results revealed that PTGES3-PROTACs were not toxic (Fig. 8F and S4A). Additionally, the serum liver and kidney indices indicated that PTGES3-PROTACs were not toxic to the liver or kidney, consistent with the H&E staining results (Fig. 8G and S4B).
Discussion
Cancer is a major global health concern. In cancer cells, signaling pathways undergo significant changes that affect various cellular activities [38]. These changes are critical because they have adverse effects, including causing resistance to conventional chemotherapeutic drugs. Therefore, there is a need to identify new effective targets based on changes in tumor cell molecular biology [39]. STAT3, at the intersection of numerous carcinogenic signaling pathways, is a promising therapeutic target for many cancers [40]. Dysregulation of the hippo signaling pathway leads to uncontrolled tumor growth and spread. Targeting the YAP and TAZ kinases has therapeutic potential [41, 42]. Additionally, many studies have revealed that AKT plays an important role in the biological functions of the PI3K/AKT signaling pathway. Synthetic and natural AKT-targeting compounds are currently being comprehensively explored [43]; however, the performance of the current treatment strategies remains poor. Identifying reliable biomarkers has become increasingly urgent and critical with the emergence of bottlenecks in screening and treating patients with HCC. Using public online websites we revealed that PTGES3 is highly expressed in HCC tissues compared to that in normal liver tissues, consistent with the results of this study. We knocked down the PTGES3 protein to determine the PTGES3 biological function in HCC, and the results revealed that the PTGES3 downregulation decreased cell proliferation and migration. Overall, PTGES3 can potentially be a novel diagnostic and therapeutic biomarker for HCC.
The p-PROTAC design can achieve specificity and effective POI degradation and expand the scope of “non-pharmaceutical” protein targeting. Notably, poor membrane permeability and low cellular uptake hinder the successful application of p-PROTAC. Cell membrane-penetrating peptides have been used to increase the intracellular concentration of small molecules to improve membrane penetration efficiency. Previous studies have revealed that the membrane-penetrating peptide TAT (GRKKRRQRRRPPQQ) can successfully carry targeted peptides into cells at high intracellular concentrations [44, 45]. Thus, TAT coupled to the FIP-2 C-terminus was selected as the cell-penetrating peptide. We found that FIP-2 carrying TAT could not only enter cells, but also effectively co-localize with the PTGES3 protein. Previous studies have attempted to address the challenge of precisely targeting lesion areas using PROTAC in vivo because restricting its protein degradation activity to the lesion site can ensure precise treatment and prevent side effects. Encapsulating PROTAC in different drug delivery carriers can help overcome the physicochemical challenges and ultimately deliver it to the desired target site [46]. Liposomes are safe and universal delivery systems with high payload transportation capabilities [47], which increase drug solubility and bioavailability. They improve the in vivo therapeutic efficacy of drugs by stabilizing compounds, increasing cellular and tissue uptake rates, and enhancing the biological distribution of drugs in diseased tissues while minimizing systemic toxicity [48]. Liposomes are widely used as drug delivery systems in the PROTAC design and synthesis. For example, a previous study revealed that novel PROTAC liposomes with ligands of traditional PROTAC recruited multi-target POIs and E3 ubiquitin ligases simultaneously and induced rapid and sustained degradation of POIs [49]. Additionally, a previous study prepared co-delivery liposomes to promote the effective encapsulation of PROTACs and siRNAs, thereby improving protein clearance and tumor treatment efficacy [50]. To increase cancer stem cells-targeted delivery, Wang et al. [51] coated the carrier liposomes of PROTAC with cyclic Arg-Gly-Asp. A previous study reported the anti-cancer activity of BRD4-targeted PROTACs ARV-825 and its nano-formulation development for cancer treatment, providing a new therapeutic strategy for cancer [52]. Subsequently, this study focused on the development of asialoglyco-protein receptor-directed nanoliposomes and successfully synthesized a targeted PROTAC-based nanotherapeutic compound for hepatocellular carcinoma by combining liposomes with ARV-825 [53]. Additionally, the same team reported that ARV-825 and vinofibinib-loaded lipid nanocomplexes exhibited promising in vivo antitumor activity and prolonged survival [54]. The liposomal PTGES3-degrading p-PROTAC that we designed and synthesized can easily enter cells and exhibited tumor-specific accumulation in vivo, indicating that using liposomes as carriers can overcome the p-PROTAC low cell permeability and poor target tissue distribution.
PROTAC targeting PTGES3 has not been reported. Therefore, we designed and synthesized a p-PROTAC liposome system that includes peptides targeting POI and ligands that bind to E3 enzymes. These molecules exhibited sustainable and significant PTGES3 degradation in HCC cells at low concentrations. PTGES3-PROTAC attenuates the HCC malignant phenotype in vitro and in vivo. Given the advantages of using 3D multicellular tumor spheroids (MCTSs) to test anti-cancer nano-formulations, there is a need to determine the PTGES3-PROTAC anti-cancer efficacy using MCTSs in future studies [55, 56]. Our PROTAC exhibited several advantages, including low toxicity, high drug efficacy, modulation of ligand ratios, and high stability. Furthermore, the FIP-2 peptide can significantly inhibit cell proliferation and migration by binding to PTGES3, although the underlying mechanism remains unclear. PTGES3-PROTAC demonstrated strong therapeutic effects by combining the inhibitory function of FIP-2 peptide inhibitors with the ability of the PROTAC to degrade carcinogenic proteins.
Conclusions
In this study, PTGES3 knockdown weakened the malignant phenotype in vitro and in vivo, indicating that targeting PTGES3 is a potential therapeutic approach for HCC. Using the PROTAC technology, we developed a PTGES3 degrader liposome complex containing a PTGES3-binding peptide and the E3 ubiquitin ligase ligand pomalidomide. This innovative technology may represent the next paradigm for regulating cell function and may provide a novel pathway for treating HCC. Overall, we believe that the system developed in this study provides a platform for adjusting the degradation efficiency of POI targets for clinical treatment and can be applied to degrade pathogenic protein targets of various diseases, including HCC.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Figure S1: MS detection of six FITC labeled synthetic peptides.
Supplementary Figure S2: The in vitro cellular uptake and distribution of PTGES3-PROTAC (n = 3).
Supplementary Figure S3: The in vivo distribution and efficiency of PTGES3-PROTAC by fluorescence imaging (n = 3).
Supplementary Figure S4: (A) H&E staining of major organ tissues harvested from tumor-bearing nude mice subcutaneously injected with Huh7 cells (n = 5). (B) Activities of serum alanine aminotransferase (ALT), aspartic acid transferase (AST), creatinine and blood urea nitrogen from nude mice subcutaneously injected with Huh7 cells (n = 5)
Acknowledgements
We are grateful for the technical assistance supported by the Mr. Ling Huang of Nanjing Yasmin Biotech Co. Ltd and the employees of the laboratory at Xuzhou Central Hospital.
Abbreviations
- HCC
Hepatocellular carcinoma
- PROTAC
Proteolysis-targeting chimera
- p-PROTAC
Peptide PROTAC
- Hsp90
Heat shock protein 90
- PGE2
Prostaglandin E2
- PTGES3
Prostaglandin E Synthase 3
- FITC
Fluorescein Isothiocyanate
- IHC
Immunohistochemistry
- IF
Immunofluorescence
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- BUN
Blood urea nitrogen
- CREA
Creatinine
- PI
Polydispersity index
- DLS
Dynamic light scattering
- H&E
Hematoxylin and eosin
- MCTs
3D multicellular tumor spheroids
Author contributions
All authors made substantial and intellectual contributions to this research. Under the direction of the corresponding authors, S.L., F.Y. and H.D. performed the experiments and wrote this article, and J.Z. and X. M. performed the bioinformatics analysis and molecular docking. All authors read and approved the final manuscript.
Funding
This study was supported by: project from the Xuzhou Health Commission (2018TD005), project from the Xuzhou Science and Technology Council (KC23174), and Xuzhou medical key talent project (XWRCHT20220058).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethical approval
We followed the ethical guidelines of Animal Care & Welfare Committee at Southeast University for all animal experiments (approval numbers 20240301009).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shiwei Liu, Fukang Yuan and Hui Dong contributed equally to this work.
Contributor Information
Yangsui Liu, Email: yangsui-liu@163.com.
Huansong Li, Email: xzhuansong@sina.com.
References
- 1.Flores A, Marrero JA. Emerging trends in hepatocellular carcinoma: focus on diagnosis and therapeutics. Clin Med Insights Oncol. 2014;8:71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400(10360):1345–62. [DOI] [PubMed] [Google Scholar]
- 3.Sayiner M, Golabi P, Younossi ZM. Disease Burden of Hepatocellular Carcinoma: A Global Perspective. Dig Dis Sci. 2019;64(4):910–7. [DOI] [PubMed] [Google Scholar]
- 4.He PH, Wan HF, Wan J, Jiang HY, Yang Y, Xie KL et al. Systemic therapies in hepatocellular carcinoma: Existing and emerging biomarkers for treatment response. Front Oncol. 2022;12. [DOI] [PMC free article] [PubMed]
- 5.Raza A, Sood GK. Hepatocellular carcinoma review: Current treatment, and evidence-based medicine. World J Gastroenterol. 2014;20(15):4115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–14. [DOI] [PubMed] [Google Scholar]
- 7.Beudeker BJB, Boonstra A. Circulating biomarkers for early detection of hepatocellular carcinoma. Therapeutic Adv Gastroenterol. 2020;13. [DOI] [PMC free article] [PubMed]
- 8.Li X, Song Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J Hematol Oncol. 2020;13(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson LM, Glennie L, Brewer A, Zhao JF, Crooks J, Shpiro N, et al. Target protein localization and its impact on PROTAC-mediated degradation. Cell Chem biology. 2022;29(10):1482–e5047. [DOI] [PubMed] [Google Scholar]
- 10.He F, Ru X, Wen T. NRF2, a Transcription Factor for Stress Response and Beyond. Int J Mol Sci. 2020;21(13). [DOI] [PMC free article] [PubMed]
- 11.Bellezza I, Giambanco I, Minelli A, Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim et Biophys acta Mol cell Res. 2018;1865(5):721–33. [DOI] [PubMed] [Google Scholar]
- 12.Park SY, Gurung R, Hwang JH, Kang JH, Jung HJ, Zeb A, et al. Development of KEAP1-targeting PROTAC and its antioxidant properties: In vitro and in vivo. Redox Biol. 2023;64:102783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannion J, Gifford V, Bellenie B, Fernando W, Ramos Garcia L, Wilson R, et al. A RIPK1-specific PROTAC degrader achieves potent antitumor activity by enhancing immunogenic cell death. Immunity. 2024;57(7):1514–e3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Xiao Y, Luo Y, Master RP, Mo J, Kim MC et al. PROTAC-mediated NR4A1 degradation as a novel strategy for cancer immunotherapy. J Exp Med. 2024;221(3). [DOI] [PMC free article] [PubMed]
- 15.Jin J, Wu Y, Chen J, Shen Y, Zhang L, Zhang H, et al. The peptide PROTAC modality: a novel strategy for targeted protein ubiquitination. Theranostics. 2020;10(22):10141–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson SE. Hsp90: structure and function. Top Curr Chem. 2013;328:155–240. [DOI] [PubMed] [Google Scholar]
- 17.Obermann WMJ. A motif in HSP90 and P23 that links molecular chaperones to efficient estrogen receptor α methylation by the lysine methyltransferase SMYD2. J Biol Chem. 2018;293(42):16479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehn AB, Buchner J. p23 and Aha1. Sub-cellular biochemistry. 2015;78:113 – 31. [DOI] [PubMed]
- 19.Biebl MM, Buchner J. p23 and Aha1: Distinct Functions Promote Client Maturation. Subcell Biochem. 2023;101:159–87. [DOI] [PubMed] [Google Scholar]
- 20.Tanioka T, Nakatani Y, Semmyo N, Murakami M, Kudo I. Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with cyclooxygenase-1 in immediate prostaglandin E2 biosynthesis. J Biol Chem. 2000;275(42):32775–82. [DOI] [PubMed] [Google Scholar]
- 21.Cano LQ, Lavery DN, Sin S, Spanjaard E, Brooke GN, Tilman JD, et al. The co-chaperone p23 promotes prostate cancer motility and metastasis. Mol Oncol. 2015;9(1):295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reebye V, Querol Cano L, Lavery DN, Brooke GN, Powell SM, Chotai D, et al. Role of the HSP90-associated cochaperone p23 in enhancing activity of the androgen receptor and significance for prostate cancer. Molecular endocrinology (Baltimore. Md). 2012;26(10):1694–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson NE, Lambert WM, Watkins R, Giashuddin S, Huang SJ, Oxelmark E, et al. High levels of Hsp90 cochaperone p23 promote tumor progression and poor prognosis in breast cancer by increasing lymph node metastases and drug resistance. Cancer Res. 2010;70(21):8446–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adekeye A, Agarwal D, Nayak A, Tchou J. PTGES3 is a Putative Prognostic Marker in Breast Cancer. J Surg Res. 2022;271:154–62. [DOI] [PubMed] [Google Scholar]
- 25.Yin Q, Ma H, Dong Y, Zhang S, Wang J, Liang J, et al. The integration of multidisciplinary approaches revealed PTGES3 as a novel drug target for breast cancer treatment. J translational Med. 2024;22(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang W, Wei Q, Xie H, Wu D, He H, Lv X. Effect of PTGES3 on the prognosis and immune regulation in lung adenocarcinoma. Anal Cell Pathol (Amst). 2023;2023:4522045. [DOI] [PMC free article] [PubMed]
- 27.Gao P, Zou K, Xiao L, Zhou H, Xu X, Zeng Z, et al. High expression of PTGES3 is an independent predictive poor prognostic biomarker and correlates with immune infiltrates in lung adenocarcinoma. Int Immunopharmacol. 2022;110:108954. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Wu Y, Gao Y, Wu C, Wang Y, Hou C, et al. Allosterically inhibited PFKL via prostaglandin E2 withholds glucose metabolism and ovarian cancer invasiveness. Cell Rep. 2023;42(10):113246. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49(W1):W242–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Q, Chen Z, Paul PK, Lu Y, Wu W, Qi J. Oral delivery of proteins and peptides: Challenges, status quo and future perspectives. Acta Pharm Sinica B. 2021;11(8):2416–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi YY, Dong DR, Fan G, Dai MY, Liu M. A cyclic peptide-based PROTAC induces intracellular degradation of palmitoyltransferase and potently decreases PD-L1 expression in human cervical cancer cells. Front Immunol. 2023;14:1237964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Chen M, Zhang X, Xie S, Qin J, Li J. Peptide-based PROTACs: Current Challenges and Future Perspectives. Curr Med Chem. 2024;31(2):208–22. [DOI] [PubMed] [Google Scholar]
- 33.Ramsey JD, Flynn NH. Cell-penetrating peptides transport therapeutics into cells. Pharmacol Ther. 2015;154:78–86. [DOI] [PubMed] [Google Scholar]
- 34.Vale N, Duarte D, Silva S, Correia AS, Costa B, Gouveia MJ, et al. Cell-penetrating peptides in oncologic pharmacotherapy: A review. Pharmacol Res. 2020;162:105231. [DOI] [PubMed] [Google Scholar]
- 35.Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, et al. How good is automated protein docking? Proteins. 2013;81(12):2159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12(2):255–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Sun P, Yao R, Zhang W, Zhou X, Yao J, et al. Comprehensive pan-cancer analysis of PTGES3 and its prognostic role in hepatocellular carcinoma. Front Oncol. 2023;13:1158490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaghari-Tabari M, Ferns GA, Qujeq D, Andevari AN, Sabahi Z, Moein S. Signaling, metabolism, and cancer: An important relationship for therapeutic intervention. J Cell Physiol. 2021;236(8):5512–32. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Herrero E, Fernández-Medarde A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur J Pharm biopharmaceutics: official J Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik eV. 2015;93:52–79. [DOI] [PubMed] [Google Scholar]
- 40.Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in Cancer Immunotherapy. Mol Cancer. 2020;19(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andl T, Zhou L, Yang K, Kadekaro AL, Zhang Y. YAP and WWTR1: New targets for skin cancer treatment. Cancer Lett. 2017;396:30–41. [DOI] [PubMed] [Google Scholar]
- 42.Cunningham R, Hansen CG. The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. Clinical science (London, England: 1979). 2022;136(3):197–222. [DOI] [PMC free article] [PubMed]
- 43.Song M, Bode AM, Dong Z, Lee MH. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019;79(6):1019–31. [DOI] [PubMed] [Google Scholar]
- 44.Wang K, Dai X, Yu A, Feng C, Liu K, Huang L. Peptide-based PROTAC degrader of FOXM1 suppresses cancer and decreases GLUT1 and PD-L1 expression. J experimental Clin cancer research: CR. 2022;41(1):289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S, Lv Q, Mao X, Dong H, Xu W, Du X, et al. O-GlcNAcylated RALY Contributes to Hepatocellular Carcinoma Cells Proliferation by Regulating USP22 mRNA Nuclear Export. Int J Biol Sci. 2024;20(9):3675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saraswat AL, Vartak R, Hegazy R, Patel A, Patel K. Drug delivery challenges and formulation aspects of proteolysis targeting chimera (PROTACs). Drug Discov Today. 2023;28(1):103387. [DOI] [PubMed] [Google Scholar]
- 47.Tanner P, Baumann P, Enea R, Onaca O, Palivan C, Meier W. Polymeric vesicles: from drug carriers to nanoreactors and artificial organelles. Acc Chem Res. 2011;44(10):1039–49. [DOI] [PubMed] [Google Scholar]
- 48.Guimarães D, Cavaco-Paulo A, Nogueira E. Design of liposomes as drug delivery system for therapeutic applications. Int J Pharm. 2021;601:120571. [DOI] [PubMed] [Google Scholar]
- 49.Song C, Jiao Z, Hou Z, Wang R, Lian C, Xing Y, et al. Selective Protein of Interest Degradation through the Split-and-Mix Liposome Proteolysis Targeting Chimera Approach. J Am Chem Soc. 2023;145(40):21860–70. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Jin Y, Wang J, Gu M, Wang Y, Zhang X, et al. Co-delivery of PROTAC and siRNA via novel liposomes for the treatment of malignant tumors. J Colloid Interface Sci. 2025;678(Pt A):896–907. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Zhao Y, Li X, Zhang Q, He J, Liu Y et al. Liposomal STAT3-Degrading PROTAC Prodrugs Promote Anti-Hepatocellular Carcinoma Immunity via Chemically Reprogramming Cancer Stem Cells. Nano Lett. 2024 Apr 10. [DOI] [PubMed]
- 52.Saraswat A, Patki M, Fu Y, Barot S, Dukhande VV, Patel K. Nanoformulation of PROteolysis TArgeting Chimera targeting ‘undruggable’ c-Myc for the treatment of pancreatic cancer. Nanomed (London England). 2020;15(18):1761–77. [DOI] [PubMed] [Google Scholar]
- 53.Saraswat A, Vemana HP, Dukhande VV, Patel K. Galactose-decorated liver tumor-specific nanoliposomes incorporating selective BRD4-targeted PROTAC for hepatocellular carcinoma therapy. Heliyon. 2022;8(1):e08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saraswat A, Vartak R, Hegazy R, Fu Y, Rao TJR, Billack B, et al. Oral lipid nanocomplex of BRD4 PROteolysis TArgeting Chimera and vemurafenib for drug-resistant malignant melanoma. Biomed Pharmacother. 2023;168:115754. [DOI] [PubMed] [Google Scholar]
- 55.Saraswat A, Vemana HP, Dukhande V, Patel K. Novel gene therapy for drug-resistant melanoma: Synergistic combination of PTEN plasmid and BRD4 PROTAC-loaded lipid nanocarriers. Mol therapy Nucleic acids. 2024;35(3):102292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel A, Saraswat A, Patel H, Chen ZS, Patel K. Palmitoyl Carnitine-Anchored Nanoliposomes for Neovasculature-Specific Delivery of Gemcitabine Elaidate to Treat Pancreatic Cancer. Cancers (Basel). 2022;15(1). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: MS detection of six FITC labeled synthetic peptides.
Supplementary Figure S2: The in vitro cellular uptake and distribution of PTGES3-PROTAC (n = 3).
Supplementary Figure S3: The in vivo distribution and efficiency of PTGES3-PROTAC by fluorescence imaging (n = 3).
Supplementary Figure S4: (A) H&E staining of major organ tissues harvested from tumor-bearing nude mice subcutaneously injected with Huh7 cells (n = 5). (B) Activities of serum alanine aminotransferase (ALT), aspartic acid transferase (AST), creatinine and blood urea nitrogen from nude mice subcutaneously injected with Huh7 cells (n = 5)
Data Availability Statement
No datasets were generated or analysed during the current study.