Abstract
Self-incompatibility (SI) is a genetic mechanism to prevent self-fertilization and thereby promote outcrossing in hermaphroditic plant species through discrimination of self and nonself-pollen by pistils. In many SI systems, recognition between pollen and pistils is controlled by a single multiallelic locus (called the S-locus), in which multiple alleles (called S-alleles) are segregating. Because of the extreme level of polymorphism of the S-locus, identification of S-alleles has been a major issue in many SI studies for decades. Here, we report an RNA-seq–based method to explore allelic diversity of the S-locus by employing the long-read sequencing technology of the Oxford Nanopore MinION and applied it for the gametophytic SI system of Petunia (Solanaceae), in which the female determinant is a secreted ribonuclease called S-RNase that inhibits the elongation of self-pollen tubes by degrading RNA. We developed a method to identify S-alleles by the search of S-RNase sequences, using the previously reported sequences as queries, and found in total 62 types of S-RNase including 45 novel types. We validated this method through Sanger sequencing and crossing experiments, confirming the sequencing accuracy and SI phenotypes corresponding to genotypes. Then, using the obtained sequence data together with polymerase chain reaction–based genotyping in a larger sample set of 187 plants, we investigated the diversity, frequency, and the level of shared polymorphism of S-alleles across populations and species. The method and the dataset obtained in Petunia will be an important basis for further studying the evolution of S-RNase–based gametophytic SI systems in natural populations.
Keywords: self-incompatibility, S-RNase, Petunia, nonself-recognition, trans-specific polymorphism
Significance.
Flowering plants have evolved molecular mechanisms called self-incompatibility (SI) for discriminating self- and nonself-pollen at pistils to prevent self-fertilization, which is often deleterious due to inbreeding depression. The specificity of SI is usually determined by multiple highly divergent alleles (called S-alleles) segregating at a single locus, and identification of S-alleles has been a major issue in many SI systems. Here, we report a new method to identify S-alleles by employing a long-read sequencing technology and applied it for the gametophytic SI system of Petunia, identifying 62 types of S-alleles including 45 novel types. The method and the dataset obtained in this study will be an important basis for the research of SI evolution.
Introduction
Self-incompatibility (SI) is a genetic mechanism to prevent self-fertilization and thereby promote outcrossing in hermaphroditic plant species through discrimination of self- and nonself-pollen by pistils (De Nettancourt 2001; Takayama and Isogai 2005). SI systems are reported in more than 100 plant families and occur in about 40% of species (Igic et al. 2008). In many SI systems, recognition between pollen and pistils is controlled by a single multiallelic locus (called the S-locus), in which multiple alleles (called S-alleles) are segregating (Takayama and Isogai 2005; Fujii et al. 2016). At the S-locus, a female specificity gene and a male specificity gene are tightly linked, and pollen is rejected when the same specificity of the S-allele is expressed by both pollen and pistils. There are two major distinct forms of SI mechanisms, the sporophytic SI (SSI) and the gametophytic SI (GSI), which differ by the genetic determination of the pollen specificity phenotype (Takayama and Isogai 2005).
In most of the SI systems reported, the S-locus shows an extreme level of polymorphism (Schierup and Vekemans 2008). Multiple S-alleles are segregating in a population, and S-allele sequences are highly divergent from each other (Castric and Vekemans 2004). Such high allelic and sequence variation at the S-locus reflects negative frequency–dependent selection, where pollen of rare S-alleles is rejected by pistils at lower rates than those of common S-alleles (Wright 1939; Vekemans and Slatkin 1994; Castric and Vekemans 2004; Schierup and Vekemans 2008). Identification of S-alleles has been a major issue in many studies of SI systems for decades, because they are a fundamental basis for studying the genetic and evolutionary features of SI, including the number of segregating S-alleles in populations, associations between S-alleles and SI phenotypes, trans-specific sharing of polymorphism, detecting natural selection on the S-locus genes, and S-allele distribution across populations (Richman et al. 1995; Castric and Vekemans 2004; Igic et al. 2007; Kato et al. 2007; Raspé and Kohn 2007; Kim et al. 2009; Dzidzienyo et al. 2016; Ma et al. 2017; De Franceschi et al. 2018; Durand et al. 2020).
However, the identification and genotyping of S-alleles have been challenging because of their extreme level of polymorphism. Since male and female specificity genes were identified in multiple SI systems (Takayama and Isogai 2005; Fujii et al. 2016), molecular cloning and Sanger sequencing of those genes have been major approaches for identification of S-alleles. These methods require polymerase chain reaction (PCR) and general primers designed in conserved regions intended to amplify specificity-determining genes irrespective of S-alleles (Janssens et al. 1995; Richman et al. 1995; Verdoodt et al. 1998; Charlesworth et al. 2000; Robbins et al. 2000; Broothaerts 2003; Mable et al. 2003; Matsumoto et al. 2006; Kubo et al. 2010, 2015; Dzidzienyo et al. 2016; Sheick et al. 2020). One of the major obstacles to these approaches is that a set of general primers is rarely perfect, often failing to amplify some of the S-alleles, particularly highly divergent ones. It has also been suggested that PCR would amplify closely related sequences that are not linked to the S-locus (Mable et al. 2003).
More recently, next-generation sequencing technologies have been applied for the identification and genotyping of S-alleles. Jørgensen et al. (2012) and Mable et al. (2018, 2017) adopted a barcoded amplicon-based method and successfully identified S-alleles in multiple Arabidopsis species, although these methods were sensitive to biases of PCR amplification. Tsuchimatsu et al. (2017) exploited short-read–based whole-genome resequencing data from 1,083 natural accessions of self-compatible Arabidopsis thaliana and revealed allelic variation of the S-locus. This method was based on mapping to reference sequences available for all three S-alleles segregating in A. thaliana, thus would not directly be applicable to the identification of many novel S-alleles in self-incompatible species. More recently, Genete et al. (2020) developed an integrated pipeline to predict S-alleles from short-read data by combining mapping-based and de novo assembly approaches, successfully identifying previously reported S-alleles and novel ones in A. halleri. These methods using next-generation sequencing data were so far mostly designed and applied for SSI systems such as those of Arabidopsis, rarely for GSI systems. While the study by De Franceschi et al. (2018) was an example of proposing a resequence-based approach for the GSI system of Rosaceae, their method was intended to gain full-length coding sequences rather than obtaining novel S-allele sequences from large-scale samples.
Here, we report an RNA-seq–based method to explore allelic diversity of the S-locus by employing the long-read sequencing technology of the Oxford Nanopore MinION and applied it to the GSI system of Petunia (Solanaceae). In this system, the female determinant is a secreted ribonuclease called S-RNase, inhibiting the elongation of self-pollen tubes by degrading RNA (Takayama and Isogai 2005). The male determinant is a set of F-box proteins, called S-locus F-box (SLF), functioning as a component of the Skp1–Cullin1–F-box (SCF)-type E3 ubiquitin ligase that generally mediates ubiquitination of target proteins for degradation by the 26S proteasome (Takayama and Isogai 2005). This system has been considered a collaborative nonself-recognition system, in which the product of each SLF interacts with a subset of nonself-S-RNases, and the products of multiple SLF types are required for the entire suite of nonself-S-RNases to be recognized (Kubo et al. 2010, 2015; Fujii et al. 2016). In this study, we first developed a pipeline to identify S-alleles based on the sequences of S-RNase, using the previously reported sequences as queries (Fig. 1; supplementary table S1, Supplementary Material online). MinION-based RNA-seq enabled us to directly obtain full-length transcripts of S-RNase, avoiding the possibility of misassembly that could occur with the de novo assembly of short-read sequences. With this method, we particularly aimed to identify novel S-alleles, because it has been difficult with conventional methods such as PCR-based ones. Next, we validated this method through crossing experiments and Sanger sequencing using primers designed for the newly obtained sequences. Then, using the obtained sequence data together with PCR-based genotyping in a larger sample set, we investigated the level of diversity including selection signals in the S-RNase gene, frequency, and distributions of S-alleles across populations and species. The method and the dataset obtained in Petunia will be an important basis for further studying the evolution of nonself-recognition systems in natural populations.
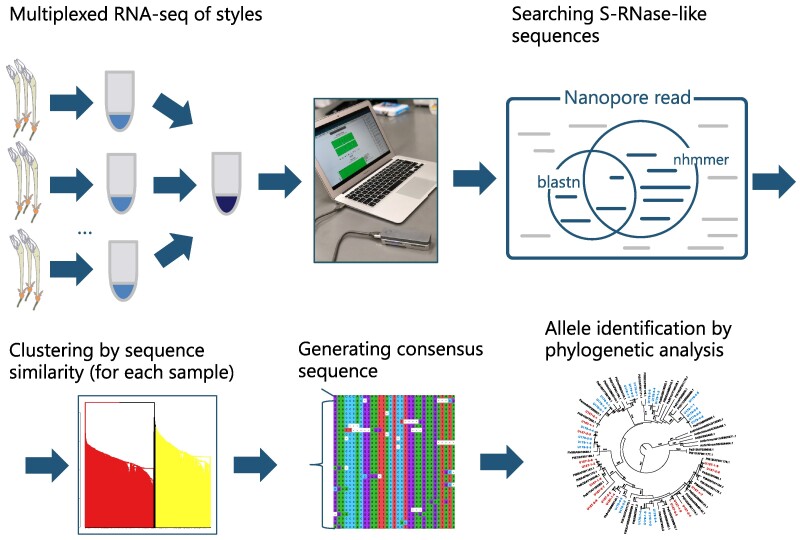
Fig. 1.
Overview of the developed pipeline to identify S-RNase sequences. We performed individually barcoded multiplexed RNA-seq of styles using the Nanopore MinION sequencer and searched for S-RNase–like sequences using previously reported S-RNase sequences as queries. Then, two heterozygous S-RNase sequences were separated by clustering analysis based on sequence similarity, and consensus sequences were generated for each S-allele. Finally, we generated a phylogenetic tree and assigned S-alleles based on the monophyly.
Results
Identification of S-RNase by Nanopore Sequencing
Our developed pipeline is summarized in Fig. 1. In short, we performed individually barcoded multiplexed RNA-seq of styles and searched for S-RNase–like sequences using previously reported S-RNase sequences as queries. Then, two heterozygous S-RNase sequences were separated by clustering analysis, and consensus sequences were generated for each S-allele. Finally, we generated a phylogenetic tree and assigned S-alleles based on the monophyly.
We first performed RNA-seq of styles using the Oxford Nanopore MinION sequencer for 86 individuals in total (supplementary table S2, Supplementary Material online). After trimming adaptor sequences, mean number of reads and total base pairs per sample were 219,078 and 85,739,454 bp, respectively (supplementary table S2, Supplementary Material online). Using BLAST (v.2.2.31; Altschul et al. 1990) and HMMER (v.3.3.2; Finn et al. 2011), we searched for S-RNase–like sequences using publicly available 37 S-RNase sequences as queries and found on average 654 S-RNase–like reads per sample, ranging from 13 to 5,994 (supplementary table S3, Supplementary Material online). In each individual, we separated those S-RNase–like sequences by clustering analysis based on sequence similarity and obtained a consensus sequence for each cluster. Except two reported S-RNase–like sequences unlinked to SI (Panon-S and PiRNX2 [Lee et al. 1992]), we in total obtained 172 sequences and generated a phylogenetic tree together with previously reported S-RNase sequences of Petunia and those of Antirrhinum as outgroups (Fig. 2a). We then assigned S-alleles based on the monophyly of this tree, identifying 62 alleles including 45 novel S-alleles. It is important to note that, except for the alleles unlinked to SI (Panon-S and PiRNX2), we found two S-alleles copies from most of the individuals surveyed, suggesting the validity of our approach (supplementary table S3, Supplementary Material online). We note that, in GSI systems, the S-locus should be heterozygous and that two S-alleles should be identified from a single individual.
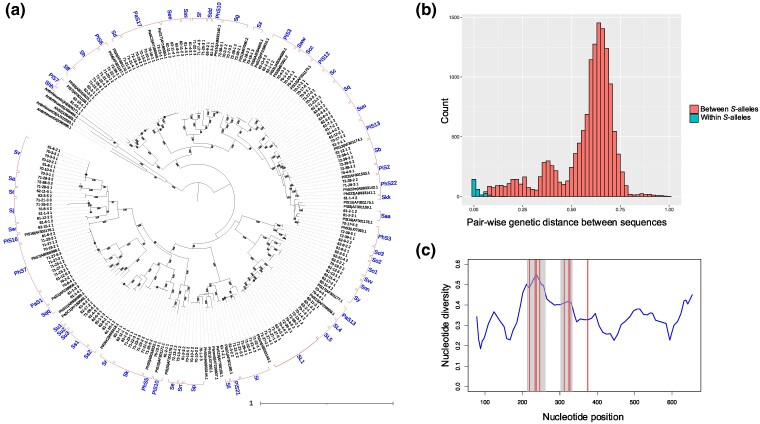
Fig. 2.
Identification of S-RNase sequences in Petunia. a) A maximum likelihood phylogenetic tree of the Petunia S-RNase sequences generated by Nanopore sequencing with five Antirrhinum ones as outgroups. The percentage of 1,000 bootstrap replicates are shown on the branches when ≥90%. Assigned names of S-alleles are also shown. b) Pair-wise genetic distance between S-RNase sequences. Distances between different and the same S-allele pairs are indicated by red and blue, respectively. The distributions of genetic distances for within and between S-alleles were significantly different (P < 2.2 × 10−16, Wilcoxon rank-sum test). c) Nucleotide diversity along the coding region of the S-RNase genes (blue line) and positively selected sites (red vertical lines). Gray shaded areas indicate hypervariable regions I and II.
Figure 2b shows the distribution of pair-wise genetic distances between all the S-RNase sequences in the tree with the information of our S-allele assignments. While our S-allele assignments are not strictly based on the genetic distance between sequences but on the monophyly in the tree, this histogram shows that the two S-alleles were largely assigned as the same ones if their genetic distance is in the left-most peak. The distributions of genetic distances for within and between S-alleles were significantly different (P < 2.2 × 10−16, Wilcoxon rank-sum test), suggesting that our S-allele assignments have consistency with the genetic distance between S-alleles.
To evaluate the sequencing accuracy of Nanopore-based genotyping, we performed Sanger sequencing for in total 69 S-alleles and compared them with Nanopore-based sequences (supplementary table S4, Supplementary Material online). Primers were designed for each S-allele based on the Nanopore-based sequences. We found that the sequencing accuracy was 99.1% (median value), suggesting that the Nanopore-based sequencing is mostly accurate and reliable.
Using these sequences obtained by the Sanger sequencing method, we evaluated the genetic diversity along the coding region of the S-RNase gene by sliding window analysis (Fig. 2c). Nucleotide diversity of the gene was particularly elevated in the hypervariable regions I and II compared with its average (mean π = 0.341), as previously reported (Ioerger et al. 1991). Since high genetic diversity is indicative of positive selection, we formally detected sites under positive selection in S-RNase using the PAML4 software with the Bayes empirical Bayes (BEB) method (Yang 2007). The likelihood of the model M2a (positive selection) was significantly higher than the model M1a (nearly neutral; likelihood ratio test, P = 1.78 × 10−24), indicating that several nucleotide positions are under positive selection. Nine positive selection sites with a posterior probability >0.95 were identified, and eight of them were located in the hypervariable regions I or II (Fig. 2c).
Frequency and Distribution of S-Alleles
To investigate the frequency and distribution of S-alleles in a larger sample set, we performed PCR-based genotyping using primers designed based on the Nanopore-based sequences. We first tested the validity of the PCR-based genotyping by Sanger sequencing of several individuals that were assigned as the same S-alleles (Sv and Sg alleles; supplementary fig. S1, Supplementary Material online). Both trees show that individuals assigned to harbor the same S-alleles by PCR-based genotyping had almost identical sequences, confirming the validity of PCR-based genotyping.
We performed genotyping of in total 187 individuals from 3 populations of P. axillaris subsp. axillaris and 2 populations of P. inflata (supplementary tables S5 and S6, Supplementary Material online). Note that our sample set of 187 individuals was comprised of half-sibs of 40 families. Among the total 374 copies of 187 individuals, we could not identify 20 copies (5.3%), suggesting the presence of unknown alleles.
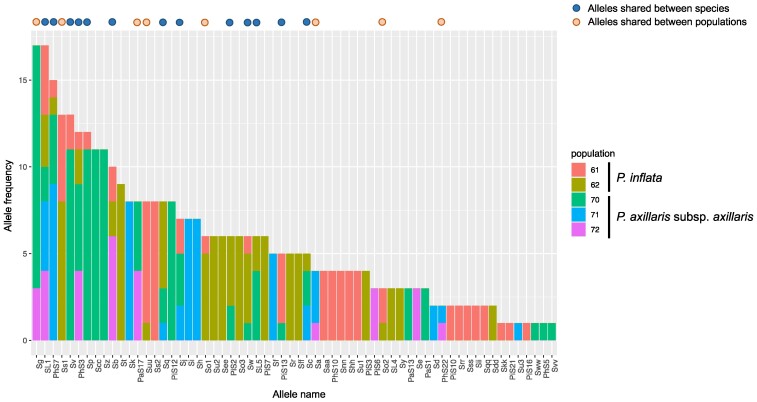
We then investigated the frequency and distribution of S-alleles across populations and species (Fig. 3). The frequency of S-alleles varied greatly, ranging from 0.28% (singleton) to 4.8%, although isoplethy is theoretically expected in GSI systems (Wright 1939). The number of S-alleles per population ranged from 9 to 27. Among the 62 S-alleles identified, 21 S-alleles (33.9%) were found in more than one population and 14 S-alleles (22.6%) were found in 2 species, suggesting the pervasive shared polymorphism between populations and species.
Fig. 3.
Frequency and distribution of S-alleles. Frequencies of each population are shown for each S-allele, based on the PCR-based genotyping of in total 187 individuals from three populations of P. axillaris subsp. axillaris and two populations of P. inflata. S-alleles shared between species or between populations are indicated on the top of the histogram.
The total number of S-alleles was estimated for each population using two different methods: the E2 estimator (O’Donnell and Lawrence 1984) and a curve fitting with a two-parameter Michaelis–Menten model (Busch et al. 2010). The former ranged from 11 to 31, and the latter from 12 to 41, both of which were higher than the actual numbers of S-alleles observed (Table 1). This result suggests that we could not exhaustively identify all the S-alleles segregating in each population, consistent with the presence of unknown S-alleles in our PCR-based search of 187 individuals (supplementary table S5, Supplementary Material online).
Table 1.
Observed and estimated numbers of S-alleles in five populations
| Species | Population code | Number of copies investigateda | Observed number of S-allelesa | Estimated number of S-alleles (O'Donnell and Lawrence 1984) | Estimated number of S-alleles (Busch et al. 2010) |
|---|---|---|---|---|---|
| P. axillaris subsp. axillaris | 70 | 105 | 22 | 25.56 | 27.74 |
| P. axillaris subsp. axillaris | 71 | 52 | 13 | 14.68 | 16.74 |
| P. axillaris subsp. axillaris | 72 | 29 | 9 | 11.82 | 12.15 |
| P. inflata | 61 | 74 | 27 | 31.60 | 41.39 |
| P. inflata | 62 | 94 | 24 | 27.97 | 31.09 |
aBased on PCR-based genotyping.
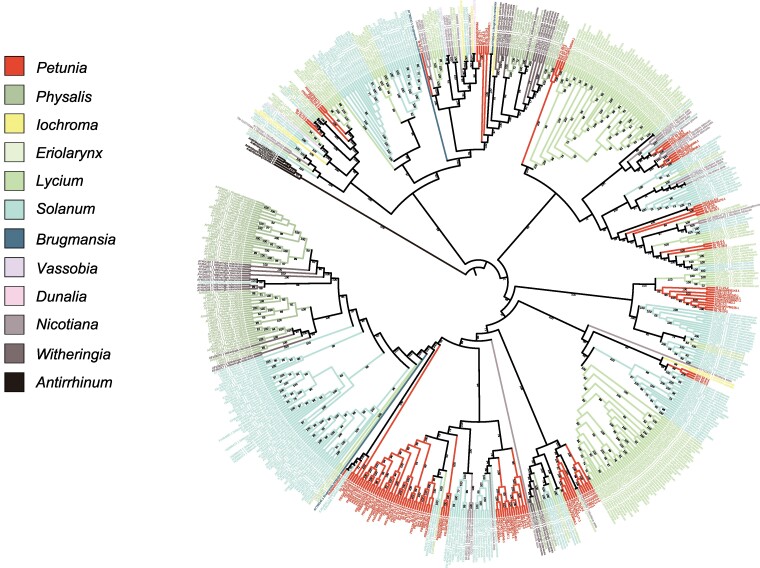
To understand how S-alleles are shared across genera in Solanaceae, we constructed a phylogenetic tree by using S-RNase sequences of Petunia and other genera, Physalis, Iochroma, Eriolarynx, Lycium, Solanum, Brugmansia, Vassobia, Dunalia, Nicotiana, and Witheringia, in total 646 sequences, including Antirrhinum as outgroups. We found that the tree was not strictly clustered by genera, but many S-alleles were shared between genera (Fig. 4). Except Dunalia from which only one sequence was reported, all the genera had multiple genus-specific clades ranging from 2 (Vassobia) to 36 (Solanum), suggesting that multiple S-alleles segregated before the origins of these clades followed by genus-specific expansions.
Fig. 4.
A maximum likelihood phylogenetic tree of S-RNase using 646 sequences from Petunia and other genera of Solanaceae, Physalis, Iochroma, Eriolarynx, Lycium, Solanum, Brugmansia, Vassobia, Dunalia, Nicotiana, and Witheringia, in total 646 sequences, including Antirrhinum as outgroups. The values on the branches indicate the percentage of 1,000 bootstrap replicates.
Genotypes Correspond to Phenotypes
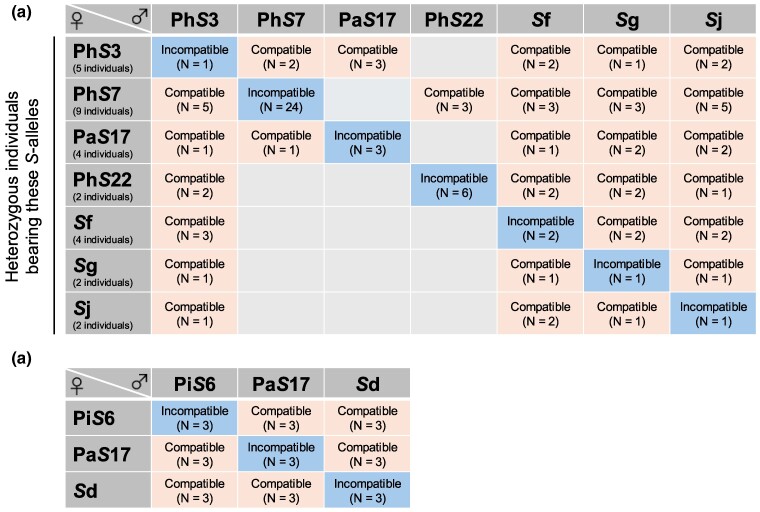
Our S-allele assignments based on sequence similarity may not necessarily correspond to SI specificities, and thus it is important to show the link between genotypes and the SI phenotypes. We therefore performed crossing experiments using P. axillaris subsp. axillaris individuals and observed SI phenotypes using total nine S-alleles (Fig. 5; supplementary table S7, Supplementary Material online). We first performed crosses between seven S-alleles (PhS7, PhS22, PaS17, PhS3, Sg, Sf, and Sj) including three novel ones (Sg, Sf, and Sj) and overall observed the incompatibility phenotype in crosses between individuals that have the same S-alleles but observed the compatible phenotype in crosses between individuals that have different S-alleles (Fig. 5a; supplementary table S7, Supplementary Material online). We also performed crosses between closely related S-alleles (PiS6, PaS17, and Sd) and consistently observed compatible reactions in all outcrosses (Fig. 5b), suggesting that these closely related S-alleles have different SI specificities. It is also suggested that our assignment of S-alleles based on sequence similarity would largely reflect the SI specificities, demonstrating the validity of our method.
Fig. 5.
Crossing experiment to examine whether SI phenotypes are associated with S-RNase genotypes. Individuals of P. axillaris subsp. axillaris were used for experiments. We observed pollen tubes by aniline staining and <20 pollen tubes reaching the bottom tip of styles were considered as a criterion of incompatible crosses. a) Seven homozygous lines generated by forced selfing were used as pollen donors (PhS7, PhS22, PaS17, PhS3, Sg, Sf, and Sj), and heterozygous individuals that have at least one of these S-alleles were used as pistil donors (see supplementary table S7, Supplementary Material online for details). b) Homozygous lines of PiS6, PaS17, and Sd alleles were used as pollen and pistil donors. Note that these three S-alleles (PiS6, PaS17, and Sd) had closely related S-RNase sequences.
Discussion
A MinION-Based Method to Detect S-RNase Alleles
In this study, we developed an RNA-seq–based method to identify and genotype S-RNase genes exploiting the long-read sequencer, MinION. We identified 62 alleles including 45 novel S-alleles through the sequencing of 86 individuals (172 copies). Based on the obtained sequence information, we designed allele-specific primers and performed PCR-based genotyping for 187 individuals to investigate the S-allele distributions and frequencies in a larger sample set.
The validity of our approach was supported by multiple lines of evidence. First, in the individuals used for MinION-based RNA-seq, we identified two S-alleles per diploid individual, when previously reported nonspecific allele copies (Panon-S and PiRNX2) were excluded. The PCR genotyping based on these MinION sequences also identified two S-alleles per individual (supplementary table S5, Supplementary Material online). These results suggest that our method could overall identify true S-allele sequences properly rather than merely list up S-RNase–like sequences. Second, crossing experiments based on the genotypes of S-RNase confirmed that the SI phenotypes were largely correlated with genotypes, although the number of examined S-alleles and replicates was relatively small (Fig. 5; supplementary table S7, Supplementary Material online).
Several methods have been used to genotype the S-RNase gene in GSI systems, such as 2D gel electrophoresis (Sassa et al. 1994), restriction enzyme–based genotyping (cleaved amplified polymorphic sequences and restriction fragment length polymorphism; Takasaki et al. 2004; Kim et al. 2009), and PCR with allele-specific or general primers (Janssens et al. 1995; Richman et al. 1995; Verdoodt et al. 1998; Robbins et al. 2000; Broothaerts 2003; Matsumoto et al. 2006, 1999; Kubo et al. 2010, 2015; Dzidzienyo et al. 2016; Sheick et al. 2020), but false positives, false negatives, and the throughput of the experiments have been major obstacles. We demonstrate that our MinION-based method can identify novel S-alleles and detect known S-alleles in Petunia, which would be applicable to other S-RNase–based GSI systems. We emphasize that our MinION-based method is particularly useful to find novel S-alleles. Once these novel S-alleles are identified, allele-specific primers can be designed and applied to a larger sample set, as indeed demonstrated in this study.
Despite the validity of our method to detect S-RNase alleles, here we note a few caveats. First, although we confirmed MinION-generated sequences through Sanger sequencing, there were a few mismatches in multiple S-alleles. Therefore, while our method has enough power to discover novel S-alleles and genotype them, it would be desirable to confirm by Sanger sequencing for the complete determination of the nucleotide sequences. Second, this method relies on the relatively high expression levels of the S-RNase gene in pistils, which provide hundreds of reads and help reduce sequence errors. Therefore, this method may not be applicable when the expression level of S-RNase is low. Third, our method benefited from the relatively abundant available S-RNase sequences of Petunia as queries, thus it may not work as efficiently as this study in taxa with little a priori information of S-RNase.
Allele Number, Distribution, and Frequency
We estimated the total number of S-alleles for each studied population from two species, P. axillaris and P. inflata, using two methods (O’Donnell and Lawrence 1984; Busch et al. 2010) The former ranged from 11 to 29 and the latter from 12 to 41, both of which were higher than the actual numbers of S-alleles observed (Table 1). We note that these should still be underestimates because our sample set consists of half-sib families. They are more likely to share the same S-alleles between individuals than random sampling, which the applied models assume.
In this study, among the 62 S-alleles identified, 21 S-alleles (33.9%) were found from more than one population. Richman et al. (1995) investigated S-alleles in two wild populations of Solanum carolinense and found 11 and 12 S-alleles for each, 10 of them were shared between populations. A study using Prunus lannesiana var. speciosa investigated S-alleles in seven populations, finding that S-alleles are shared 41% to 83% (Kato et al. 2007). These differences in the fraction of shared S-alleles may be due to the sample size, the genetic divergence between populations, and the classification of S-alleles. If our assignment of S-alleles based on the sequence similarity does not correspond to the SI specificity in some cases, the fraction of shares S-alleles between populations may be overestimated or underestimated. For example, a few high-frequency S-alleles found in multiple populations (e.g. SL1) showed population-specific clades (Fig. 2a); thus, it might be possible that S-allele specificities are different between populations. In such a case, the fraction of shares S-alleles is overestimated.
Toward an Understanding of S-Allele Evolution
There have been extensive studies of the S-RNase–based SI system in Petunia, as the collaborative nonself-recognition system was first discovered in this genus. Despite the wealth of knowledge of the functional aspect of the S-RNase–based SI system, the comprehensive information on S-locus diversity in wild populations of Petunia remained largely unexplored. Indeed, while about 40 S-RNase partial or full-length sequences have been reported so far, many of them were obtained from cultivated species Petunia × hybrida or a relatively limited number of strains in wild populations (Clark et al. 1990; Ai et al. 1992, 1990; Coleman and Kao 1992; Clark and Sims 1994; Entani et al. 1999; Wang et al. 2001, 2003; Tsukamoto et al. 2005, 2003; Sassa and Hirano 2006; Kubo et al. 2010, 2015). Among the S-RNase–based GSI systems, the survey of S-allele diversity in wild populations is still limited to a handful number of species, including S. carolinense (Richman et al. 1995), S. chilense (Igic et al. 2007), Sorbus aucuparia (Raspé and Kohn 2007), Prunus lannesiana var. speciosa (Kato et al. 2007), and Malus sieversii (Ma et al. 2017). One of the major questions in this field would be to demonstrate how a new specificity of SI evolves in the nonself-recognition system. Although there are several theoretical investigations on the origin of new S-alleles (Kubo et al. 2015; Fujii et al. 2016; Bod’ová et al. 2018; Harkness and Brandvain 2021), the information in wild populations is essential, including the number of S-alleles and variation in SLF repertories between S-alleles. Here, we laid the foundation for the studies of the evolution of SI specificities in the nonself-recognition system by surveying S-alleles in wild Petunia populations. In particular, the recently diverged S-RNase sequence pairs would serve as a good system to study the origin of new SI specificities, as demonstrated in the SSI system (Chantreau et al. 2019).
Materials and Methods
Plant Materials
We used a total of 187 samples from 3 populations of P. axillaris subsp. axillaris (populations 70 [34°28′00′′S, 57°49′48′′W], 71 [34°30′58′′S, 54°20′44′′W] and 72 [34° 53′ 17′′ S, 56° 15′ 35′′W]) and 2 populations of P. inflata (populations 61 [28°25′07′′S, 58°57′26′′W] and 62 [29°00′30′′S, 59°08′26′′W]) (Ando et al. 1995; Ando et al. 1998). We used multiple seeds from each individual plant collected in those five populations; thus, samples from a single individual are half-sib families. Our sample set consisted of a total of 40 half-sib families. See supplementary tables S2 and S5, Supplementary Material online for details. Plants were grown in chambers (16L8D, 22 °C) or a greenhouse at the Kashiwanoha Campus, Chiba University.
Library Preparation and Sequencing
We collected styles from flower buds and stored them in RNA later at −80 °C (De Wit et al. 2012). We extracted total RNA using RNeasy Mini Kit (QIAGEN) and TRI Reagent (Molecular Research Center, Inc.) following the manufacturing protocols. We performed multiplexed sequencing using the PCR-cDNA Barcoding Kit (SQK-PCB109; Oxford Nanopore Technologies). Libraries were prepared according to the manufacturing protocol (PCB_9092_v109_revD_10Oct2019), and 12 barcodes were used per library. We used the MinION device and flow cells R9.4.1 for sequencing. Three libraries were sequenced per flow cell, by washing the flow cell after each run with the Flow Cell Wash Kit (EXP-WSH003 or EXP-WSH004).
Identification of S-RNase by Long-Read Sequencing
For the obtained long-read sequences, we first trimmed barcode and adaptor sequences using Porechop (https://github.com/rrwick/Porechop; 2024 May 3). We then filtered low-quality reads (Q-value ≤6) and trimmed the first 50 bp of 5′ ends using Nanofilt (De Coster et al. 2018). Basecalling and demultiplexing were performed by the library Guppy (v4.2.2 and v6.1.7). Versions used for each analysis are provided in supplementary table S2, Supplementary Material online. We searched for S-RNase–like sequences among those filtered sequences as follows. First, we converted filtered fastq files to fasta files by seqkit (Shen et al. 2016). Then, we used publicly available 37 S-RNase sequences as queries (supplementary table S1, Supplementary Material online). Using these queries, we searched for S-RNase–like sequences using blastn (v.2.2.31; Altschul et al. 1990) and HMMER (v.3.3.2; Finn et al. 2011) with default parameters. We used sequences that were detected in both blastn and HMMER for the following analyses.
We separated heterozygous S-RNase alleles by clustering analysis based on sequence similarity for each individual. For each individual, S-RNase–like sequences were aligned, and distance matrix between sequences was calculated by mafft (Katoh et al. 2002). Using the obtained matrix, we performed hierarchical clustering using the R library dendextend (Galili 2015). We classified sequence clusters based on the threshold value of 0.7 and omitted clusters to which <10 sequences belonged. We realigned sequences within each cluster and omitted low-frequency indels (<0.05) as they are likely to be sequence errors by a custom R script. Finally, we obtained a consensus sequence for each cluster using EMBOSS cons (Rice et al. 2000).
We then combined all the obtained S-RNase–like sequences and previously reported S-RNase sequences of Petunia and generated an alignment using mafft (v.7.310, parameters: –adjustdirectionaccurately). We also included five Antirrhinum sequences as outgroups. After excluding two S-RNase–like sequences that are unlinked to SI (Panon-S and PiRNX2 [Lee et al. 1992]) and sequence clusters that have <50 reads or 500 bp in total length, a maximum likelihood tree was generated using the software IQ-TREE (v.2.1.4-beta, parameters: -m MFP -AIC -B 1000; Nguyen et al. 2015). Note that we omitted plant samples that had only one S-RNase–like sequences other than Panon-S and PiRNX2 satisfying the criteria (number of reads per cluster and sequence length). We classified S-RNase based on the monophyly in the tree and assigned S-alleles for each individual. We note that it is not trivial to determine the threshold for delimiting S-alleles. Although we confirmed that the tree-based determination was mostly concordant with the pair-wise genetic distance between sequences (Fig. 2b) and that genotypes of S-alleles were completely linked with SI phenotypes as long as investigated in this study (Fig. 5), there might be a few discordances between assigned genotypes and SI specificities.
Sanger Sequencing and PCR-Based Genotyping
To evaluate the sequencing accuracy of Nanopore-based genotyping, we performed Sanger sequencing for each S-allele and compared them with Nanopore-based sequences. We newly designed primers to amplify S-RNases based on the obtained Nanopore sequences (supplementary table S8, Supplementary Material online) and performed PCR using Tks Gflex DNA Polymerase (Takara) according to the manufacturer’s protocol.
To estimate allele frequency in a larger sample set, we also performed PCR-based genotyping for a total of 187 individuals including those not used for Nanopore sequencing (supplementary table S5, Supplementary Material online). To test the validity of PCR-based genotyping, we performed Sanger sequencing of multiple copies from the same S-alleles and generated phylogenetic trees for those S-alleles by the maximum likelihood method using IQ-TREE (v.2.1.4-beta, parameters: -m MFP -AIC -B 1000; Nguyen et al. 2015). While PCR-based genotyping detected two S-alleles per diploid heterozygous individual in most cases, we observed amplifications of more than two S-alleles in a few individuals (detailed in supplementary tables S5, S6, and S8, Supplementary Material online). For those amplifications, we validated PCR fragments by Sanger sequencing, confirming that they are misamplification of other S-alleles detected in the same individuals (supplementary tables S5, S6, and S8, Supplementary Material online).
Estimation of the Number of S-Alleles in Each Population
The total numbers of S-alleles were estimated by using two methods: the E2 estimator based on the sample size, allele frequencies, and the number of S-alleles identified (O’Donnell and Lawrence 1984) and a curve fitting method using a two-parameter Michaelis–Menten model (Busch et al. 2010). We employed these methods because both of them do not assume equal frequencies of S-alleles. For the method of Busch et al. (2010), the estimates were based on the average number of unique S-alleles inferred from 1,000 times of resampling without replacement. For the curve fitting, we used the drc package (Ritz and Streibig 2005) in R (R Core Team 2023). In the Michaelis–Menten model, f (x) = Smax/(1 +va K/x), Smax corresponds to the total number of alleles expected in the population, which was reported for each population.
Test of Association With SI Phenotypes
To evaluate our method to detect S-alleles based on the sequence similarity of the S-RNase, we examined whether SI phenotypes are associated with S-RNase genotypes by crossing experiments. We observed the elongation of pollen tubes in crosses within the same S-alleles and between the S-alleles. For crossing experiments, we used individuals of P. axillaris subsp. axillaris.
We performed two sets of crossing experiments. For the first one, seven homozygous lines generated by bud pollination were used as pollen donors (PhS7, PhS22, PaS17, PhS3, Sg, Sf, and Sj), and heterozygous individuals that have at least one of these S-alleles were used as pistil donors (supplementary table S7, Supplementary Material online). For the second ones, we used homozygous lines of PiS6, PaS17, and Sd alleles as pollen and pistil donors (reciprocal crosses). These three S-alleles (PiS6, PaS17, and Sd) had closely related S-RNase sequences, as they constitute a monophyletic clade together with PiS9 (Fig. 2a).
We observed pollen tubes by aniline staining as follows. We first removed anthers from flower buds and closed them by staplers to avoid contamination. Styles were pollinated 24 h after emasculation and harvested 48 h after pollination. Harvested styles were fixed in a 3:1 mixture of ethanol and acetic acid, softened for 8 h in 1 M NaOH at 65 °C and stained with aniline blue in a 2% K3PO4 solution overnight at room temperature. Styles were mounted on slides to examine the pollen tubes using epifluorescence microscopy (Olympus BX53). We counted the number of pollen tubes that reached the bottom tip of styles. Less than 20 pollen tubes were considered as a criterion of incompatible crosses.
Phylogenetic Analysis With S-RNase of Other Genera
To understand the phylogenetic relationship between Petunia S-RNase obtained in this study and previously reported S-RNase including those from other genera of Solanaceae, we constructed a phylogenetic tree by generating a large sequence alignment. The list of sequences used for the tree is available in supplementary table S1, Supplementary Material online. A phylogenetic tree was generated by the maximum likelihood method using IQ-TREE (v.2.1.4-beta, parameters: -m MFP -AIC -B 1000; Nguyen et al. 2015).
Detection of Positively Selected Sites and Sliding Window Analysis
To detect amino acid positions under selection in the S-RNase gene from Petunia, we estimated nonsynonymous/synonymous substitution ratio for each site by using the PAML4 software (Yang 2007). The codeml package of PAML4 was used to calculate posterior probabilities of codon sites under positive selection with the BEB method. The likelihood of two models M1a (nearly neutral) and M2a (positive selection) were compared by a likelihood ratio test. For this analysis, we used a total of 79 sequences of Petunia obtained by Sanger sequencing or by previous studies (supplementary table S1, Supplementary Material online). A phylogenetic tree used as an input for PAML4 was generated by a maximum likelihood method with the General Time Reversible model using MEGA11 (Tamura et al. 2021).
To evaluate the genetic diversity along the S-RNase gene from Petunia, we performed sliding window analysis using DnaSP v. 6.12.03 (Rozas et al. 2017). The window size was 50, and the step size was 10. Used sequences were mostly the same as those used for PAML4, but we omitted a few relatively short sequences due to designed primer positions (supplementary tables S1 and S8, Supplementary Material online).
Supplementary Material
Acknowledgments
This work was supported by the FOREST program of the Japan Science and Technology Agency (JPMJFR2046 to T.T.), Grants-in-Aid for Scientific Research (18H04813, 19H03271, 20H04856, 22K21352, and 23H02537 to T.T., 21K06079 to K.K., and 17H04605 to K.U.), Toray Science and Technology Grant, and Inamori Research Grant to T.T.
Contributor Information
Taiga Maenosono, Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku 113-0033, Tokyo, Japan; Graduate School of Science and Technology, Chiba University, Chiba 263-8522, Japan.
Kazuho Isono, Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku 113-0033, Tokyo, Japan.
Takanori Kuronuma, Center for Environment, Health and Field Sciences, Chiba University, Kashiwa 277-0882, Japan.
Miho Hatai, Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku 113-0033, Tokyo, Japan.
Kaori Chimura, Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku 113-0033, Tokyo, Japan.
Ken-ichi Kubo, Department of Frontier Bioscience, Nagahama Institute of Bio-Science and Technology, Nagahama, Japan.
Hisashi Kokubun, Graduate School of Horticulture, Chiba University, Matsudo 271-8510, Japan.
Julián Alejandro Greppi, Instituto de Floricultura, CNIA, INTA, Buenos Aires, Argentina.
Hitoshi Watanabe, Center for Environment, Health and Field Sciences, Chiba University, Kashiwa 277-0882, Japan.
Koichi Uehara, College of Liberal Arts and Sciences, Chiba University, Chiba 263-8522, Japan.
Takashi Tsuchimatsu, Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku 113-0033, Tokyo, Japan.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Data Availability
Sequence data have been deposited in DDBJ under accession numbers LC819163 to LC819242.
Literature Cited
- Ai Y, Singh A, Coleman CE, Ioerger TR, Kheyr-Pour A, Kao T. Self-incompatibility in Petunia inflata: isolation and characterization of cDNAs encoding three S-allele-associated proteins. Sex Plant Reprod. 1990:3(2):130–138. 10.1007/BF00198857. [DOI] [Google Scholar]
- Ai Y, Tsai D-S, Kao T. Cloning and sequencing of cDNAs encoding two SSS. Plant Mol Biol. 1992:19(3):523–528. 10.1007/BF00023404. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990:215(3):403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ando T, Iida S, Kokubun H, Ueda Y, Marchesi E. Distribution of infraspecific taxa of Petunia axillaris (Solanaceae) in Uruguay as revealed by discriminant analyses. Acta Phytotaxon Geobot. 1995:45:95–109. 10.18942/bunruichiri.KJ00001079048. [DOI] [Google Scholar]
- Ando T, Tsukamoto T, Akiba N, Kokubun H, Watanabe H, Ueda Y, Marchesi E. Differentiation in the degree of self-incompatibility in Petunia axillaris (Solanaceae) occurring in Uruguay. Acta Phytotaxon Geobot. 1998:49:37–47. 10.18942/bunruichiri.KJ00001077367. [DOI] [Google Scholar]
- Bod’ová K, Priklopil T, Field DL, Barton NH, Pickup M. Evolutionary pathways for the generation of new self-incompatibility haplotypes in a nonself-recognition system. Genetics. 2018:209(3):861–883. 10.1534/genetics.118.300748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broothaerts W. New findings in apple S-genotype analysis resolve previous confusion and request the re-numbering of some S-alleles. Theor Appl Genet. 2003:106(4):703–714. 10.1007/s00122-002-1120-0. [DOI] [PubMed] [Google Scholar]
- Busch JW, Joly S, Schoen DJ. Does mate limitation in self-incompatible species promote the evolution of selfing? The case of Leavenworthia alabamica. Evolution. 2010:64(6):1657–1670. 10.1111/j.1558-5646.2009.00925.x. [DOI] [PubMed] [Google Scholar]
- Castric V, Vekemans X. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Mol Ecol. 2004:13(10):2873–2889. 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- Chantreau M, Poux C, Lensink MF, Brysbaert G, Vekemans X, Castric V. Asymmetrical diversification of the receptor-ligand interaction controlling self-incompatibility in Arabidopsis. Elife. 2019:8:e50253. 10.7554/eLife.50253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Awadalla P, Mable BK, Schierup MH. Population-level studies of multiallelic self-incompatibility loci, with particular reference to Brassicaceae. Ann Bot. 2000:85:227–239. 10.1006/anbo.1999.1015. [DOI] [Google Scholar]
- Clark KR, Okuley JJ, Collins PD, Sims TL. Sequence variability and developmental expression of S-alleles in self-incompatible and pseudo-self-compatible petunia. Plant Cell. 1990:2(8):815–826. 10.1105/tpc.2.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KR, Sims TL. The S-ribonuclease gene of Petunia hybrida is expressed in nonstylar tissue, including immature anthers. Plant Physiol. 1994:106(1):25–36. 10.1104/pp.106.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Kao T. The flanking regions of two Petunia inflata S alleles are heterogeneous and contain repetitive sequences. Plant Mol Biol. 1992:18(4):725–737. 10.1007/BF00020014. [DOI] [PubMed] [Google Scholar]
- De Coster W, D’hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018:34(15):2666–2669. 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franceschi P, Bianco L, Cestaro A, Dondini L, Velasco R. Characterization of 25 full-length S-RNase alleles, including flanking regions, from a pool of resequenced apple cultivars. Plant Mol Biol. 2018:97(3):279–296. 10.1007/s11103-018-0741-x. [DOI] [PubMed] [Google Scholar]
- De Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Springer Berlin; 2001. [Google Scholar]
- De Wit P, Pespeni MH, Ladner JT, Barshis DJ, Seneca F, Jaris H, Therkildsen NO, Morikawa M, Palumbi SR. The simple fool’s guide to population genomics via RNA-seq: an introduction to high-throughput sequencing data analysis. Mol Ecol Resour. 2012:12(6):1058–1067. 10.1111/1755-0998.12003. [DOI] [PubMed] [Google Scholar]
- Durand E, Chantreau M, Le Veve A, Stetsenko R, Dubin M, Genete M, Llaurens V, Poux C, Roux C, Billiard S, et al. Evolution of self-incompatibility in the Brassicaceae: lessons from a textbook example of natural selection. Evol Appl. 2020:13(6):1279–1297. 10.1111/eva.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzidzienyo DK, Bryan GJ, Wilde G, Robbins TP. Allelic diversity of S-RNase alleles in diploid potato species. Theor Appl Genet. 2016:129(10):1985–2001. 10.1007/s00122-016-2754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entani T, Takayama S, Iwano M, Shiba H, Che F-S, Isogai A. Relationship between polyploidy and pollen self-incompatibility phenotype in Petunia hybrida Vilm. Biosci Biotechnol Biochem. 1999:63(11):1882–1888. 10.1271/bbb.63.1882. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011:39(Web server issue):W29–W37. 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016:2(9):16130. 10.1038/nplants.2016.130. [DOI] [PubMed] [Google Scholar]
- Galili T. Dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015:31(22):3718–3720. 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genete M, Castric V, Vekemans X. Genotyping and de novo discovery of allelic variants at the Brassicaceae self-incompatibility locus from short-read sequencing data. Mol Biol Evol. 2020:37(4):1193–1201. 10.1093/molbev/msz258. [DOI] [PubMed] [Google Scholar]
- Harkness A, Brandvain Y. Non-self recognition-based self-incompatibility can alternatively promote or prevent introgression. New Phytol. 2021:231(4):1630–1643. 10.1111/nph.17249. [DOI] [PubMed] [Google Scholar]
- Igic B, Lande R, Kohn JR. Loss of self-incompatibility and its evolutionary consequences. Int J Plant Sci. 2008:169(1):93–104. 10.1086/523362. [DOI] [Google Scholar]
- Igic B, Smith WA, Robertson KA, Schaal BA, Kohn JR. Studies of self-incompatibility in wild tomatoes: I. S-allele diversity in Solanum chilense (Dun.) Reiche (Solanaceae). Heredity (Edinb). 2007:99(5):553–561. 10.1038/sj.hdy.6801035. [DOI] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-H. Primary structural features of the self-incompatibility protein in Solanaceae. Sexual Plant Reprod. 1991:4(2):81–87. 10.1007/BF00196492. [DOI] [Google Scholar]
- Janssens GA, Goderis IJ, Broekaert WF, Broothaerts W. A molecular method for S-allele identification in apple based on allele-specific PCR. Theor Appl Genet. 1995:91(4):691–698. 10.1007/BF00223298. [DOI] [PubMed] [Google Scholar]
- Jørgensen MH, Lagesen K, Mable BK, Brysting AK. Using high-throughput sequencing to investigate the evolution of self-incompatibility genes in the Brassicaceae: strategies and challenges. Plant Ecol Divers. 2012:5(4):473–484. 10.1080/17550874.2012.748098. [DOI] [Google Scholar]
- Kato S, Iwata H, Tsumura Y, Mukai Y. Distribution of S-alleles in island populations of flowering cherry, Prunus lannesiana var. speciosa. Genes Genet Syst. 2007:82(1):65–75. 10.1266/ggs.82.65. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002:30(14):3059–3066. 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kakui H, Kotoda N, Hirata Y, Koba T, Sassa H. Determination of partial genomic sequences and development of a CAPS system of the S-RNase gene for the identification of 22 S haplotypes of apple (Malus × domestica Borkh.). Mol Breed. 2009:23(3):463–472. 10.1007/s11032-008-9249-4. [DOI] [Google Scholar]
- Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, et al. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science. 2010:330(6005):796–799. 10.1126/science.1195243. [DOI] [PubMed] [Google Scholar]
- Kubo K-I, Paape T, Hatakeyama M, Entani T, Takara A, Kajihara K, Tsukahara M, Shimizu-Inatsugi R, Shimizu KK, Takayama S. Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nat Plants. 2015:1(1):14005. 10.1038/nplants.2014.5. [DOI] [PubMed] [Google Scholar]
- Lee HS, Singh A, Kao T. RNase X2, a pistil-specific ribonuclease from Petunia inflata, shares sequence similarity with solanaceous S proteins. Plant Mol Biol. 1992:20(6):1131–1141. 10.1007/BF00028899. [DOI] [PubMed] [Google Scholar]
- Ma X, Cai Z, Liu W, Ge S, Tang L. Identification, genealogical structure and population genetics of S-alleles in Malus sieversii, the wild ancestor of domesticated apple. Heredity (Edinb). 2017:119(3):185–196. 10.1038/hdy.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK, Brysting AK, Jørgensen MH, Carbonell AKZ, Kiefer C, Ruiz-Duarte P, Lagesen K, Koch MA. Adding complexity to complexity: gene family evolution in polyploids. Front Ecol Evol. 2018:6. 10.3389/fevo.2018.00114. [DOI] [Google Scholar]
- Mable BK, Hagmann J, Kim S-T, Adam A, Kilbride E, Weigel D, Stift M. What causes mating system shifts in plants? Arabidopsis lyrata as a case study. Heredity (Edinb). 2017:118(1):52–63. 10.1038/hdy.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK, Schierup MH, Charlesworth D. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity (Edinb). 2003:90(6):422–431. 10.1038/sj.hdy.6800261. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Kitahara K, Komatsu H, Abe K. Cross-compatibility of apple cultivars possessing S-RNase alleles of similar sequence. J Hortic Sci Biotechnol. 2006:81(6):934–936. 10.1080/14620316.2006.11512178. [DOI] [Google Scholar]
- Matsumoto S, Kitahara K, Komori S, Soejima J. A new S-allele in apple, ‘Sg’, and its similarity to the ‘Sf’ allele from ‘Fuji’. HortScience. 1999:34(4):708–710. 10.21273/HORTSCI.34.4.708. [DOI] [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell S, Lawrence MJ. The population genetics of the self-incompatibility polymorphism in Papaver rhoeas. IV. The estimation of the number of alleles in a population. Heredity (Edinb). 1984:53(3):495–507. 10.1038/hdy.1984.111. [DOI] [Google Scholar]
- Raspé O, Kohn JR. Population structure at the S-locus of Sorbus aucuparia L. (Rosaceae: Maloideae). Mol Ecol. 2007:16(6):1315–1325. 10.1111/j.1365-294X.2007.03233.x. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Core Team; 2023. https://www.R-project.org/. [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000:16(6):276–277. 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richman AD, Kao T-H, Schaeffer SW, Uyenoyama MK. S-allele sequence diversity in natural populations of Solanum carolinense (Horsenettle). Heredity (Edinb). 1995:75(4):405–415. 10.1038/hdy.1995.153. [DOI] [PubMed] [Google Scholar]
- Ritz C, Streibig JC. Bioassay analysis using R. J Stat Softw. 2005:12(5):1–22. 10.18637/jss.v012.i05. [DOI] [Google Scholar]
- Robbins TP, Harbord RM, Sonneveld T, Clarke K. The molecular genetics of self-incompatibility in Petunia hybrida. Ann Bot. 2000:85:105–112. 10.1006/anbo.1999.1062. [DOI] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017:34(12):3299–3302. 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Sassa H, Hirano H. Identification of a new class of pistil-specific proteins of Petunia inflata that is structurally similar to, but functionally distinct from, the self-incompatibility factor HT. Mol Genet Genomics. 2006:275(1):97–104. 10.1007/s00438-005-0067-7. [DOI] [PubMed] [Google Scholar]
- Sassa H, Mase N, Hirano H, Ikehashi H. Identification of self-incompatibility-related glycoproteins in styles of apple (Malus x domestica). Theor Appl Genet. 1994:89(2-3):201–205. 10.1007/BF00225142. [DOI] [PubMed] [Google Scholar]
- Schierup MH, Vekemans X. Genomic consequences of selection on self-incompatibility genes. Curr Opin Plant Biol. 2008:11(2):116–122. 10.1016/j.pbi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Sheick R, Serra S, Tillman J, Luby J, Evans K, Musacchi S. Characterization of a novel S-RNase allele and genotyping of new apple cultivars. Sci Hortic. 2020:273:109630. 10.1016/j.scienta.2020.109630. [DOI] [Google Scholar]
- Shen W, Le S, Li Y, Hu F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 2016:11(10):e0163962. 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T, Okada K, Castillo C, Moriya Y, Saito T, Sawamura Y, Norioka N, Norioka S, Nakanishi T. Sequence of the S 9-RNase cDNA and PCR-RFLP system for discriminating S 1-to S 9-allele in Japanese pear. Euphytica. 2004:135(2):157–167. 10.1023/B:EUPH.0000014907.50575.d0. [DOI] [Google Scholar]
- Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005:56(1):467–489. 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021:38(7):3022–3027. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Goubet PM, Gallina S, Holl A-C, Fobis-Loisy I, Bergès H, Marande W, Prat E, Meng D, Long Q, et al. Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes. Mol Biol Evol. 2017:34(8):1878–1889. 10.1093/molbev/msx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Ando T, Takahashi K, Omori T, Watanabe H, Kokubun H, Marchesi E, Kao T. Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by loss of pollen function. Plant Physiol. 2003:131(4):1903–1912. 10.1104/pp.102.018069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Ando T, Watanabe H, Marchesi E, Kao T. Duplication of the S-locus F-box gene is associated with breakdown of pollen function in an S-haplotype identified in a natural population of self-incompatible Petunia axillaris. Plant Mol Biol. 2005:57(1):141–153. 10.1007/s11103-004-6852-6. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Slatkin M. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics. 1994:137(4):1157–1165. 10.1093/genetics/137.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoodt L, Van Haute A, Goderis IJ, De Witte K, Keulemans J, Broothaerts W. Use of the multi-allelic self-incompatibtility gene in apple to assess homozygocity in shoots obtained through haploid induction. Theor Appl Genet. 1998:96(2):294–300. 10.1007/s001220050739. [DOI] [Google Scholar]
- Wang X, Hughes AL, Tsukamoto T, Ando T, Kao T-H. Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol. 2001:125(2):1012–1022. 10.1104/pp.125.2.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, McCubbin AG, Kao T. Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Mol Biol. 2003:53(4):565–580. 10.1023/B:PLAN.0000019068.00034.09. [DOI] [PubMed] [Google Scholar]
- Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939:24(4):538–552. 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007:24(8):1586–1591. 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in DDBJ under accession numbers LC819163 to LC819242.