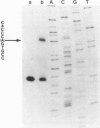
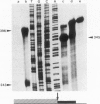
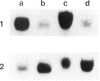
Abstract
We describe the structure and transcriptional activity of the 5' portion of the alpha-cardiac actin gene of BALB/c mice. Southern blotting and DNA sequencing reveal that the promoter and first three exons of the gene are present as perfect repeats in a direct duplication of 9.5 kbp situated immediately upstream of the gene. Both promoters are active in adult cardiac tissue. Transcripts from the partial gene duplication give rise to novel RNAs that are spliced correctly in the actin region and polyadenylated. The level of mature alpha-cardiac actin mRNA is only 16.5% that found in mice that do not possess the duplication. This is due, at least in part, to interference at the transcriptional level. Transcripts from the alpha-skeletal actin gene accumulate to abnormally high levels in the hearts of such mutant mice. This result suggests tight regulatory coupling for this actin gene pair.
Full text
PDF

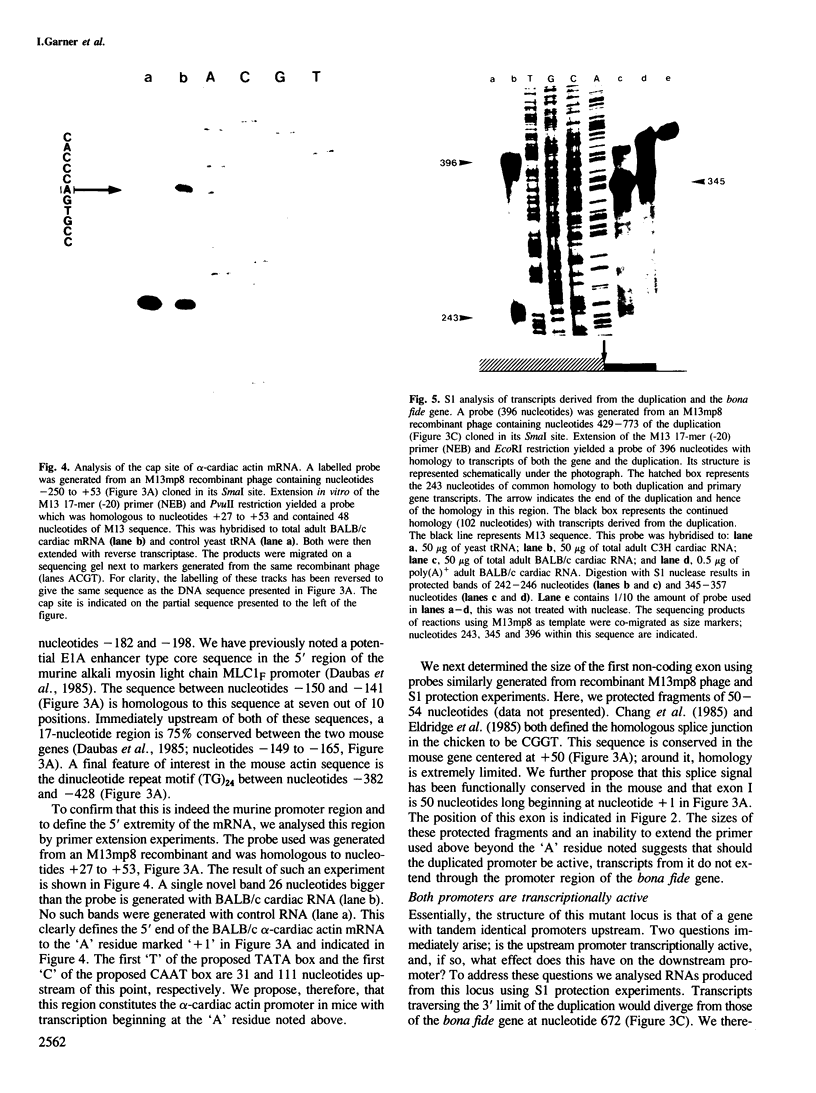





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso S., Minty A., Bourlet Y., Buckingham M. Comparison of three actin-coding sequences in the mouse; evolutionary relationships between the actin genes of warm-blooded vertebrates. J Mol Evol. 1986;23(1):11–22. doi: 10.1007/BF02100994. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Bains W., Ponte P., Blau H., Kedes L. Cardiac actin is the major actin gene product in skeletal muscle cell differentiation in vitro. Mol Cell Biol. 1984 Aug;4(8):1449–1453. doi: 10.1128/mcb.4.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau H. M., Chiu C. P., Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983 Apr;32(4):1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Blau H. M., Pavlath G. K., Hardeman E. C., Chiu C. P., Silberstein L., Webster S. G., Miller S. C., Webster C. Plasticity of the differentiated state. Science. 1985 Nov 15;230(4727):758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- Bodary S., Grossi G., Hagenbüchle O., Wellauer P. K. Members of the Amy-2 alpha-amylase gene family of mouse strain CE/J contain duplicated 5' termini. J Mol Biol. 1985 Mar 5;182(1):1–10. doi: 10.1016/0022-2836(85)90022-1. [DOI] [PubMed] [Google Scholar]
- Buckingham M. E. Actin and myosin multigene families: their expression during the formation of skeletal muscle. Essays Biochem. 1985;20:77–109. [PubMed] [Google Scholar]
- Caravatti M., Minty A., Robert B., Montarras D., Weydert A., Cohen A., Daubas P., Buckingham M. Regulation of muscle gene expression. The accumulation of messenger RNAs coding for muscle-specific proteins during myogenesis in a mouse cell line. J Mol Biol. 1982 Sep;160(1):59–76. doi: 10.1016/0022-2836(82)90131-0. [DOI] [PubMed] [Google Scholar]
- Carmon Y., Czosnek H., Nudel U., Shani M., Yaffe D. DNAase I sensitivity of genes expressed during myogenesis. Nucleic Acids Res. 1982 May 25;10(10):3085–3098. doi: 10.1093/nar/10.10.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. S., Rothblum K. N., Schwartz R. J. The complete sequence of the chicken alpha-cardiac actin gene: a highly conserved vertebrate gene. Nucleic Acids Res. 1985 Feb 25;13(4):1223–1237. doi: 10.1093/nar/13.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. A gene chimaera of SV40 and mouse beta-globin is transcribed and properly spliced. Nature. 1981 Jan 29;289(5796):378–382. doi: 10.1038/289378a0. [DOI] [PubMed] [Google Scholar]
- Cullen B. R., Lomedico P. T., Ju G. Transcriptional interference in avian retroviruses--implications for the promoter insertion model of leukaemogenesis. Nature. 1984 Jan 19;307(5948):241–245. doi: 10.1038/307241a0. [DOI] [PubMed] [Google Scholar]
- Czosnek H., Nudel U., Mayer Y., Barker P. E., Pravtcheva D. D., Ruddle F. H., Yaffe D. The genes coding for the cardiac muscle actin, the skeletal muscle actin and the cytoplasmic beta-actin are located on three different mouse chromosomes. EMBO J. 1983;2(11):1977–1979. doi: 10.1002/j.1460-2075.1983.tb01687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czosnek H., Nudel U., Shani M., Barker P. E., Pravtcheva D. D., Ruddle F. H., Yaffe D. The genes coding for the muscle contractile proteins, myosin heavy chain, myosin light chain 2, and skeletal muscle actin are located on three different mouse chromosomes. EMBO J. 1982;1(11):1299–1305. doi: 10.1002/j.1460-2075.1982.tb01314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubas P., Robert B., Garner I., Buckingham M. A comparison between mammalian and avian fast skeletal muscle alkali myosin light chain genes: regulatory implications. Nucleic Acids Res. 1985 Jul 11;13(13):4623–4643. doi: 10.1093/nar/13.13.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge J., Zehner Z., Paterson B. M. Nucleotide sequence of the chicken cardiac alpha actin gene: absence of strong homologies in the promoter and 3'-untranslated regions with the skeletal alpha actin sequence. Gene. 1985;36(1-2):55–63. doi: 10.1016/0378-1119(85)90069-1. [DOI] [PubMed] [Google Scholar]
- Emerman M., Temin H. M. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984 Dec;39(3 Pt 2):449–467. [PubMed] [Google Scholar]
- Felsani A., Maione R., Ricci L., Amati P. Coordinate expression of myogenic functions and polyoma virus replication. Cold Spring Harb Symp Quant Biol. 1985;50:753–757. doi: 10.1101/sqb.1985.050.01.093. [DOI] [PubMed] [Google Scholar]
- Flanagan J. G., Lefranc M. P., Rabbitts T. H. Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell. 1984 Mar;36(3):681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Gunning P., Ponte P., Blau H., Kedes L. alpha-skeletal and alpha-cardiac actin genes are coexpressed in adult human skeletal muscle and heart. Mol Cell Biol. 1983 Nov;3(11):1985–1995. doi: 10.1128/mcb.3.11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P., Ponte P., Kedes L., Eddy R., Shows T. Chromosomal location of the co-expressed human skeletal and cardiac actin genes. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1813–1817. doi: 10.1073/pnas.81.6.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982 Jul 22;298(5872):396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. Molecular structure and evolutionary origin of human cardiac muscle actin gene. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5901–5905. doi: 10.1073/pnas.79.19.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hen R., Borrelli E., Sassone-Corsi P., Chambon P. An enhancer element is located 340 base pairs upstream from the adenovirus-2 E1A capsite. Nucleic Acids Res. 1983 Dec 20;11(24):8747–8760. doi: 10.1093/nar/11.24.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D. Three-dimensional structure of the complex of actin and DNase I at 4.5 A resolution. EMBO J. 1985 Aug;4(8):2113–2118. doi: 10.1002/j.1460-2075.1985.tb03900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlik C. C., Coutu M. D., Fyrberg E. A. A nonsense mutation within the act88F actin gene disrupts myofibril formation in Drosophila indirect flight muscles. Cell. 1984 Oct;38(3):711–719. doi: 10.1016/0092-8674(84)90266-6. [DOI] [PubMed] [Google Scholar]
- Karlik C. C., Fyrberg E. A. An insertion within a variably spliced Drosophila tropomyosin gene blocks accumulation of only one encoded isoform. Cell. 1985 May;41(1):57–66. doi: 10.1016/0092-8674(85)90061-3. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Angelides K. J., Holloman W. K. Left-handed DNA and the synaptic pairing reaction promoted by Ustilago rec1 protein. Cell. 1985 Jan;40(1):139–145. doi: 10.1016/0092-8674(85)90317-4. [DOI] [PubMed] [Google Scholar]
- Konieczny S. F., Emerson C. P., Jr Differentiation, not determination, regulates muscle gene activation: transfection of troponin I genes into multipotential and muscle lineages of 10T1/2 cells. Mol Cell Biol. 1985 Sep;5(9):2423–2432. doi: 10.1128/mcb.5.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt J., Gunning P., Porreca P., Ng S. Y., Lin C. S., Kedes L. Molecular cloning and characterization of mutant and wild-type human beta-actin genes. Mol Cell Biol. 1984 Oct;4(10):1961–1969. doi: 10.1128/mcb.4.10.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. R., Karn J., Brenner S. Molecular analysis of the unc-54 myosin heavy-chain gene of Caenorhabditis elegans. Nature. 1981 Jun 4;291(5814):386–390. doi: 10.1038/291386a0. [DOI] [PubMed] [Google Scholar]
- Mayer Y., Czosnek H., Zeelon P. E., Yaffe D., Nudel U. Expression of the genes coding for the skeletal muscle and cardiac actions in the heart. Nucleic Acids Res. 1984 Jan 25;12(2):1087–1100. doi: 10.1093/nar/12.2.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloul D., Aloni B., Calvo J., Yaffe D., Nudel U. Developmentally regulated expression of chimeric genes containing muscle actin DNA sequences in transfected myogenic cells. EMBO J. 1984 May;3(5):983–990. doi: 10.1002/j.1460-2075.1984.tb01917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty A. J., Alonso S., Caravatti M., Buckingham M. E. A fetal skeletal muscle actin mRNA in the mouse and its identity with cardiac actin mRNA. Cell. 1982 Aug;30(1):185–192. doi: 10.1016/0092-8674(82)90024-1. [DOI] [PubMed] [Google Scholar]
- Minty A. J., Alonso S., Guénet J. L., Buckingham M. E. Number and organization of actin-related sequences in the mouse genome. J Mol Biol. 1983 Jun 15;167(1):77–101. doi: 10.1016/s0022-2836(83)80035-7. [DOI] [PubMed] [Google Scholar]
- Minty A. J., Caravatti M., Robert B., Cohen A., Daubas P., Weydert A., Gros F., Buckingham M. E. Mouse actin messenger RNAs. Construction and characterization of a recombinant plasmid molecule containing a complementary DNA transcript of mouse alpha-actin mRNA. J Biol Chem. 1981 Jan 25;256(2):1008–1014. [PubMed] [Google Scholar]
- Minty A., Kedes L. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol Cell Biol. 1986 Jun;6(6):2125–2136. doi: 10.1128/mcb.6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogami K., O'Donnell P. T., Bernstein S. I., Wright T. R., Emerson C. P., Jr Mutations of the Drosophila myosin heavy-chain gene: effects on transcription, myosin accumulation, and muscle function. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1393–1397. doi: 10.1073/pnas.83.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Rich A. Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature. 1983 Jun 23;303(5919):674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte P., Gunning P., Blau H., Kedes L. Human actin genes are single copy for alpha-skeletal and alpha-cardiac actin but multicopy for beta- and gamma-cytoskeletal genes: 3' untranslated regions are isotype specific but are conserved in evolution. Mol Cell Biol. 1983 Oct;3(10):1783–1791. doi: 10.1128/mcb.3.10.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackwitz H. R., Zehetner G., Frischauf A. M., Lehrach H. Rapid restriction mapping of DNA cloned in lambda phage vectors. Gene. 1984 Oct;30(1-3):195–200. doi: 10.1016/0378-1119(84)90120-3. [DOI] [PubMed] [Google Scholar]
- Robert B., Barton P., Minty A., Daubas P., Weydert A., Bonhomme F., Catalan J., Chazottes D., Guénet J. L., Buckingham M. Investigation of genetic linkage between myosin and actin genes using an interspecific mouse back-cross. Nature. 1985 Mar 14;314(6007):181–183. doi: 10.1038/314181a0. [DOI] [PubMed] [Google Scholar]
- Robert B., Daubas P., Akimenko M. A., Cohen A., Garner I., Guenet J. L., Buckingham M. A single locus in the mouse encodes both myosin light chains 1 and 3, a second locus corresponds to a related pseudogene. Cell. 1984 Nov;39(1):129–140. doi: 10.1016/0092-8674(84)90198-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Yamada Y., de Crombrugghe B. DNA sequence comparison of the regulatory signals at the 5' end of the mouse and chick alpha 2 type I collagen genes. J Biol Chem. 1984 Jun 25;259(12):7411–7415. [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Sutoh K. Identification of myosin-binding sites on the actin sequence. Biochemistry. 1982 Jul 20;21(15):3654–3661. doi: 10.1021/bi00258a020. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Bugaisky G., Buckingham M. Simultaneous expression of skeletal muscle and heart actin proteins in various striated muscle tissues and cells. A quantitative determination of the two actin isoforms. J Biol Chem. 1986 Feb 5;261(4):1838–1843. [PubMed] [Google Scholar]
- Vandekerckhove J., Weber K. Actin typing on total cellular extracts: a highly sensitive protein-chemical procedure able to distinguish different actins. Eur J Biochem. 1981 Jan;113(3):595–603. doi: 10.1111/j.1432-1033.1981.tb05104.x. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Weber K. Chordate muscle actins differ distinctly from invertebrate muscle actins. The evolution of the different vertebrate muscle actins. J Mol Biol. 1984 Nov 5;179(3):391–413. doi: 10.1016/0022-2836(84)90072-x. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Weber K. The complete amino acid sequence of actins from bovine aorta, bovine heart, bovine fast skeletal muscle, and rabbit slow skeletal muscle. A protein-chemical analysis of muscle actin differentiation. Differentiation. 1979;14(3):123–133. doi: 10.1111/j.1432-0436.1979.tb01021.x. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wright W. E. Synthesis of rat myosin light chains in heterokaryons formed between undifferentiated rat myoblasts and chick skeletal myocytes. J Cell Biol. 1981 Oct;91(1):11–16. doi: 10.1083/jcb.91.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]