Abstract
Purpose of Review
Heart failure (HF) is often accompanied by a constellation of comorbidities, leading to diverse patient presentations and clinical trajectories. While traditional methods have provided valuable insights into our understanding of HF, network medicine approaches seek to leverage these complex relationships by analyzing disease at a systems level. This review introduces the concepts of network medicine and explores the use of comorbidity networks to study HF and heart disease.
Recent Findings
Comorbidity networks are used to understand disease trajectories, predict outcomes, and uncover potential molecular mechanisms through identification of genes and pathways relevant to comorbidity. These networks have shown the importance of non-cardiovascular comorbidities to the clinical journey of patients with HF. However, the community should be aware of important limitations in developing and implementing these methods.
Summary
Network approaches hold promise for unraveling the impact of comorbidities in the complex presentation and genetics of HF. Methods that consider comorbidity presence and timing have the potential to help optimize management strategies and identify pathophysiological mechanisms.
Keywords: Networks, Comorbidity, Heart failure
Introduction
The emergence of big data in the cardiology field has presented an unprecedented opportunity to unravel the complexities underlying cardiovascular diseases [1]. Heart failure (HF), characterized by its high prevalence, heterogeneous nature, and diverse clinical presentations, represents an ideal candidate for the implementation of new big data-derived approaches [2–4]. While traditional reductionist methods have provided valuable insights into our understanding of the disease so far, they often struggle to fully capture the systemic nature of HF and its associated comorbidities [5]. Network medicine offers a complementary analytical framework that aims to leverage this inherent heterogeneity [6]. By analyzing relationships between multiple diseases/conditions simultaneously, these methods can reveal patterns that may not be immediately apparent in conventional epidemiological studies [7]. This network-based view aligns with the growing understanding of HF as a systemic disorder rather than a purely cardiac condition [8, 9]. In this review, we examine the current state of disease and comorbidity networks studying HF, discussing the methodological foundations, key findings, and potential clinical implications of the literature. We also critically assess the limitations of these approaches and consider future directions that may enhance their utility in research and clinical practice.
Comorbidities in Heart Failure
Comorbidity is the presence of additional complicating disorders in patients with a primary disease of interest [10]. In heart failure (HF) and its subphenotypes, comorbidity is common in patient populations across the world [11]. In some studies, more than 85% of patients reported having at least two conditions in addition to HF [12, 13]. Despite this, there is no agreed-upon definition of comorbidity in terms of causal relationship to the primary disease of interest [14], and studies use different definitions, which place varying degrees of importance on the index disease [15], causality, and presence of additional conditions [16]. When multiple diseases are present in addition to the condition of interest, the term multimorbidity is frequently used [17].
Patients with HF can experience a wide variety of comorbid conditions. However, most investigations have focused on a small number of the most common comorbidities. Commonly assessed comorbidities include coronary artery disease, hypertension, diabetes, atrial fibrillation, chronic obstructive pulmonary disorder, kidney disease, and obesity, amongst others [18–22]. In patients with HF, comorbidity is associated with poorer quality of life [23], increased complications, and higher rates of readmission and death [18, 24–26]. Noncardiovascular comorbidities especially have been more strongly associated with risk of death and hospitalization [27, 28].
Comorbidities have been particularly important in the context of subtypes of HF.Patients with HFare commonly grouped by left ventricular ejection fraction, with patients with an LVEF ≤ 40 identified as HF with reduced ejection fraction (HFrEF) and those with an LVEF ≥ 50 categorized as HF with preserved ejection fraction (HFpEF) [29]. The differences in comorbidity in these two groups have been posited as impacting changes in the heart before disease [30], and guidelines for managing specific comorbidities, such as hypertension or diabetes, play a particularly crucial role in tailoring treatment strategies for patients with HFpEF, as these comorbidities often contribute significantly to the pathophysiology and symptom burden in this subtype of heart failure [31, 32]. Some literature has found that HFpEF patients have higher rates of comorbidity [28], and especially non-cardiac comorbidities, than their HFrEF counterparts [33–35], while others have found prevalence rates across EF-based subtypes to be fairly consistent [36]. Comorbidities common in HFrEF have been described as more likely to be cardiac diseases [37] than those in HFpEF patients. There is also some evidence that the relationship between comorbidity and negative outcomes such as mortality [38] differs between EF-based subtypes [36, 39, 40]. The relationship between comorbidities and HF subtype can be complicated by age and sex [41]. For example, anemia is a more frequent comorbidity in HFpEF compared to HFrEF, and is more common in women with HF regardless of subtype than men [42].
Leveraging Network Medicine to Understanding Comorbidities in Disease
The desire to understand the complex relationships between comorbidity and HF, and the availability of big data resources [43] has resulted in embracing methods that can leverage this information. Network medicine can be defined as the application of network methodologies to approach the study of human diseases [6]. First applied to study physical interactions at the molecular level within the cell [44, 45], investigators soon adapted these methods to explore human disease relationships by hypothesizing that diseases sharing molecular characteristics might also display phenotypic similarities [46]. Network medicine relies on the hypothesis that if two diseases are related, changes in the network that cause one disease will likely affect the manifestation of other diseases as well [47].
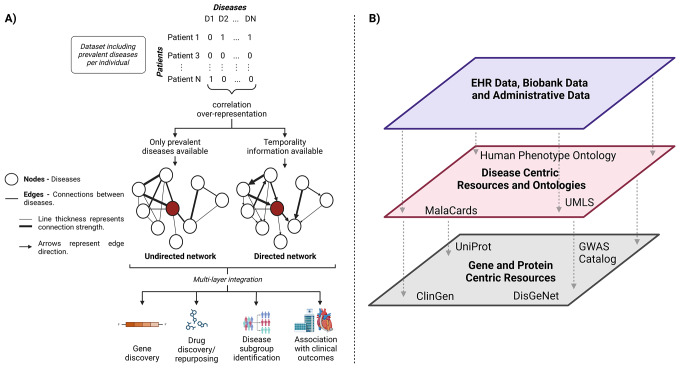
Conceptually, a network is a collection of nodes connected by edges [47]. Networks can be undirected, directed, and/or weighted, with edges providing information about the directionality and strength of the relationship between the nodes [48] (Fig. 1A). In the case of comorbidity networks, the network nodes represent diseases and the edges depict connections between two nodes, often representing a correlation between or an observed-to-expected ratio of two diseases [46, 49, 50]. Several types of measures including degree characterize networks, measuring the number of connections in a node, betweenness, a measure of the number of nodes a node can influence, closeness, measuring the average from a node to the rest of the nodes in the network [48]. A list of network-related vocabulary can be found in the included Glossary of Terms.
Fig. 1.
Overview of methods of network construction. Networks can be constructed (A) from data where information on each disease is present for each patient. In these networks, diseases are represented by nodes and the connections between diseases by edges. After network construction they can be used for a variety of study types or can be (B) integrated with data or prior knowledge resources that allow the linking of different levels of information about medicine and biology
To construct a comorbidity network, the definition of nodes and edges must align with the research question. In comorbidity networks, the nodes are typically based on a disease ontology. Since many ontologies exist (e.g. MESH, HPO, ICD, PheWAS, DO), each developed for different purposes and with different levels of resolution, selecting the appropriate one requires careful consideration of the research question. Sensitivity analyses have shown that ontology choice significantly influences network topology [51]. Then, statistical tests are often applied to assess whether two diseases co-occur more frequently than expected based on their prevalence [49]. These models can be enhanced in various ways including incorporating temporal relationships [52, 53] or multivariate analyses to control for confounders [54, 55]. From these models, edges are filtered, usually by selecting cutoffs based on effect sizes or p-values globally (on network level) or locally (e.g. on node level [56]). In traditional comorbidity networks, diseases serve as the nodes, and statistical associations between them form the edges. However, alternative approaches, such as patient networks, can be used for patient stratification or classification, where the nodes represent patients and the edges reflect the similarity [57, 58]of their comorbidity profiles [59]. A central challenge in both approaches is designing a network that accurately captures the relationships under study, particularly in defining meaningful edges that reflect comorbid associations.
Once built, networks can be layered with other types of network data to create “multi-layer” networks [60](Fig. 1B). These can be homogeneous if only one node type is used, or heterogeneous when different types of networks are combined. Heterogeneous multi-layer networks have been suggested to integrate clinical information, omics, and more to identify genes and proteins involved in pathological processes [57, 58]. Prior knowledge is one important source of information that links phenotypic data to biological knowledge [61]. Resources such as DISGENET [62], Malacards [63], UniProtKB [64], and ClinVar [65], amongst others, can provide important information about linking phenotypic information to disease level knowledge, and disease ontologies [66, 67] and language systems [68] can help combine knowledge from different data sources with different nomenclatures.
Comorbidity networks often use large numbers of comorbidities, and therefore, most analyses are performed in datasets from electronic health records or administrative data. These data sources allow the capture of a broad spectrum of comorbid conditions in populations who have actively sought care. However, some network analyses only analyze a small spectrum of comorbidity, creating a phenotype algorithm for each one [69, 70]. At the core, though, these methods focus on the interconnectedness of comorbidities, aiming to provide insights into the complex web of comorbid relationships and potential pathways between different diseases [6, 71] rather than make patient-level statements.
Other common computational methods in the study of the heterogeneity of HF, such as clustering [72, 73], latent class analyses [74, 75], and factor analyses [13], have been used as alternatives to, or in combination with, comorbidity network analysis as they can provide easily interpretable insights about patient sub-populations. These methods focus on patients to group individuals based on their shared characteristics without necessarily exploring the intricate relationships between the variables themselves [76]. We will only loosely touch on these methods as they intersect with using networks to study HF comorbidities. We recommend these reviews for readers interested in the methodology behind clustering in HF [77–80].
Comorbidity Networks of Heart Failure and Heart Disease
Networks have been used to investigate different aspects of HF. In the following sections, we discuss networks developed in HF and one of its most frequent underlying etiologies, ischemic heart disease (IHD). We have loosely grouped literature by the underlying purpose for which the network was developed.
Networks have Evaluated Comorbidity Relationships in HF and IHD
An important use of disease networks is to explore the connections between comorbidities in disease subpopulations. Carmona-Perez et al. evaluated the comorbidity patterns in HF and COPD patients, specifically comparing men and women [81] with one or both diseases. Some diseases, such as kidney disease and respiratory disorders, were amongst the 10 most highly connected nodes in the HF network of both males and females. Others were more sex specific, with arthritis one of the 10 nodes with the highest links in the HF network in women only and peripheral vascular disorder only in the male-only HF network.
Networks developed within age and sex-based subgroups have shown that comorbidity relationships may be different in independent populations. In an IHD network, males had a more complex network than females, and the connections between comorbidities were also different [82]. For example, conditions relating to cerebrovascular disease were more common in men, while kidney failure and related alterations of metabolism were more comorbid in females. Across age-stratified networks, they found overall consistency of prevalent comorbidities, but that comorbidity relationships changed, becoming more complicated in older patients. A study by Martins et al. used clustering as the primary method to determine groups of associated comorbidity, but leveraged network methods to visualize the results of these clusters [83]. The network visualization revealed differences in comorbidity prevalence and complexity between clusters and allowed important disease pairs in the clusters to be identified. The relationship between anemia and hypertension and anemia and CKD was important in one cluster, while in another the connections of AF, to obesity and hypertension were important.
This was also evident in a study from the Colombian heart failure registry [84]. In this study, sex- and LVEF-stratified networks revealed significant differences in the relationships between comorbidities, highlighting significant correlations between valvular disease and atrial fibrillation, as well as valvular disease and coronary heart disease exclusive to women, while the correlation between valvular disease and T2DM was exclusive to men. Moreover, a significant correlation between chronic kidney disease and valvular disease was observed only in HFrEF patients, while the strength of the correlation between T2DM and chronic kidney disease in HFpEF almost doubled the one in HFrEF.
Finally, at least one example has shown that in populations of patients with HF grouped over chronological time, the relationship between comorbidities in HF has changed over time, but the disease communities have remained stable [85].
Communities in Comorbidity Networks have been Associated with Outcomes
Comorbidity networks have also been leveraged to provide information about the risk of clinically relevant outcomes. A large study of the Swedish Heart Failure Registry used graphical models to assess the relationship between comorbidities and metrics of patient health including patient-reported outcomes. It found non-cardiovascular comorbidities were a major driver of patient health and that for some patients might be driving the burden of symptoms normally assessed in HF [86]. Networks have also been used to study the usage of clinical services by patients with HF, demonstrating a diffuse flow of patients between clinical resources, but one that changed depending on whether a patient had been admitted [87].
Regarding “hard” clinical outcomes, Zheng et al. used latent class analysis to subgroup patients with HFpEF and HFrEF by comorbidities. Then, they used network methods to examine the relationships between the comorbid conditions in each cluster. They found that different connections between common comorbidities were important in each cluster, but the relevant comorbidities were highly overlapping. Comorbidity-based populations were associated with differential outcomes rates, including all-cause mortality and readmission, despite otherwise similar clinical characteristics. They used this to argue for hierarchical comorbidity management in chronic heart failure patients [88].
Similarly, a study in IHD used network analyses and machine learning to predict which disease clusters would put patients at risk of progression to heart failure. They built a disease co-occurrence network for IHD and personal networks for individual patients. They used network measures as features in machine learning models, concluding that network measures were more important as features than demographic information such as age and sex [89]. Finally, at least one study used networks to identify diagnoses and procedures associated with increased hospital costs for affected patients [90].
Networks to Connect HF to Genes
While the number of studies that have analyzed ejection fraction-based subtypes of HF using clustering methods is quite large, few studies were primarily network-based. In our previous work, we built a comorbidity network from patients with HF visiting a German university hospital [91]. Ejection fraction data was then used to subphenotype HFpEF and HFrEF patients and learn discriminant comorbidity profiles. Statistical methods were employed to contrast disease communities more representative of the HFpEF and HFrEF populations. We found that the characteristics of HFpEF patients were overrepresented in the endocrine and pulmonary disease clusters. Moreover, using existing data about the relationship of diseases to genes, we developed a heterogeneous multi-layer network and used a random walk to predict which genes were closer in the network context to the comorbidity profiles discriminant for HFpEF or HFrEF. As a result, we identified genes involved in fibrosis, hypertrophy, oxidative stress, and endoplasmic reticulum stress, which were significantly overrepresented in a murine transcriptomic disease signature of HFpEF, providing additional support for their relevance in this context. This work serves as a proof of concept, suggesting that not only can comorbid diseases share molecular profiles, but multi-organ syndromes like HFpEF may be linked to recurrent molecular patterns across organs. However, studying systemic diseases remains challenging, and experimental strategies are particularly needed to evaluate organism-wide changes in response to disease.
On the other hand, Cruz-Ávila et al. built comorbidity networks for a CVD population and divided the population into age brackets as determined by 10-year increments [92]. In their network, they found that arrhythmias, heart failure, and kidney disease had the greatest number of connections. They noted that many possible links between comorbidities were absent and that comorbidity relationships were heterogeneous. Additionally, they reported that congenital disorders had a prominent place in the network in children, while in adults, complex diseases became the central nodes. To identify comorbidity-associated genes, they used ClinVar to link diseases to genetics and then performed a pathway analysis. HF was well connected in age-stratified networks from age 11 on.
Heart Failure in Networks Focused on Other Diseases
Although not designed to assess HF specifically, networks focused on other diseases or generally on multimorbid patients without a specific index disease have also captured HF and its subtypes. This may be important for highlighting comorbidities before HF [93] and understanding HF in patients with complex healthcare trajectories. Querying these networks may provide valuable insights for identifying at-risk patients or interventional opportunities before the onset of heart failure.
Morbinet, while focused on type 2 diabetes, captured heart failure, heart disease, and ischemic heart disease as important nodes [94]. Heart failure was most strongly linked with diabetes and pulmonary heart disease, though it was also less strongly linked to non-cardiovascular comorbidities, including gout and liver disease. A second network built for type 2 diabetes also identified HF as an important node in a cardiovascular disease-heavy cluster and found that diseases in the cluster were less common in the networks of non-diabetics [95]. This cluster contained comorbidities such as anemia, lymphoma, and rheumatoid arthritis in addition to circulatory and vascular disorders. These networks demonstrate the complicated relationship between diabetes and HF and the breadth of comorbidities associated with both. In contrast to the importance of HF in diabetes networks, in a study of lung cancer patients, the authors found that while the patients with HF all had multiple other comorbidities, HF itself did not play a strong role in the development of the comorbidity pattern an individual had [96]. This demonstrates that networks of other HF-relevant phenotypes will also recapitulate knowledge relevant to HF. Similarly, a network focused on hypothyroidism identified HF as a well-connected node that directionally led to hypothyroidism in the directed network [97].
General multimorbidity networks have also assessed mortality and hospitalization due to HF. A multimorbidity network in Veterans found differential rates of deceased persons 8 years after HF in different subclusters of the HF temporal network [98]. This demonstrates that comorbidity networks may be a valid way to identify high-risk populations. Other studies found differences in the number and length of stay in patients who develop HF depending on the order of CVD development [99]. However, another temporal network study found that hypertensive heart disease and heart failure were important nodes for predicting future heart failure, a result potentially at odds with some known developmental pathways for HF [100]. Data assessing multimorbidity in multiple racial/ethnic populations from the UK found that HF was associated with high multimorbidity coefficients in all populations [101]. They also found that many of the most common comorbidities in HF populations were exclusion criteria for clinical trials, providing an important assessment of the real-world applicability of trial data.
Limitations of Network Analyses
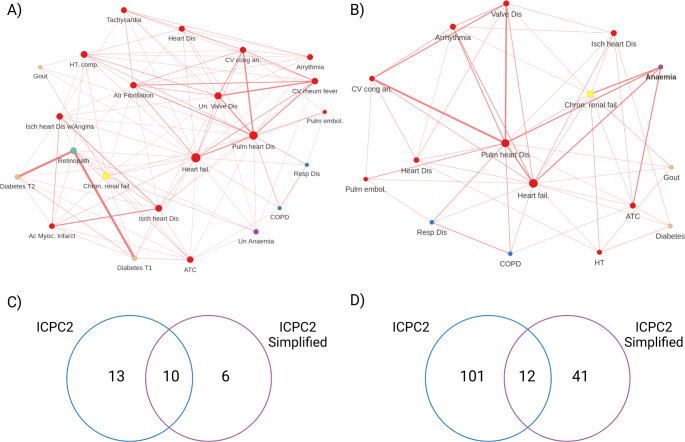
Network analyses can offer valuable insights into the complex landscape of comorbidities in HF; however, they also suffer from important limitations that researchers and clinicians alike must consider [6]. At their core, these approaches focus on disease-level relationships rather than individual patient characteristics, which can limit direct clinical applicability [102]. Moreover, the heavy reliance on diagnostic codes from electronic health records or administrative databases introduces potential biases and may not capture the full clinical picture, particularly for rare or underdiagnosed conditions [103]. The type of code and data preprocessing may also impact the network results (Fig. 2).
Fig. 2.
Comparison of two networks built using the Morbinet shiny browser [104] demonstrates how small processing changes can affect networks. The network built using an odds ratio for association of at least 1.8 with (A) International Classification of Primary Care, 2nd edition (ICPC2) codes contains more nodes and connections than that built with (B) simplified ICPC2 codes. Comparison of (C) number of shared nodes and (D) edges are shown in Venn Diagrams with ICPC2 data in blue and simplified ICPC2 data in purple
A significant challenge lies in the temporal and causal aspects of disease relationships. Many network analyses provide a static view, failing to account for the order of disease onset or the causal links between conditions. This limitation can hinder our understanding of disease progression and the development of targeted interventions [105]. Furthermore, the generalizability of these networks to diverse populations or healthcare settings may be limited, as they are often built using data from specific systems or regions [6].
Notably, the methodological aspects of network analyses also present challenges. The structure and insights derived from these networks can be highly sensitive to the specific metrics and algorithms used in their creation [102]. An example of this is provided in Fig. 2. This sensitivity makes comparisons across studies difficult and raises questions about the reproducibility of findings [105]. Unfortunately, external validation of network-derived insights in independent cohorts or through prospective studies is often lacking, limiting confidence in the generalizability of findings. Additionally, the focus on pairwise disease relationships in many analyses may oversimplify the complex interactions between multiple conditions [51], potentially missing important higher-order relationships [106].
Interpreting the clinical relevance of identified relationships remains a critical challenge [107]. Not all statistically significant connections in a network may be meaningful in a clinical context, and distinguishing between spurious associations and truly important relationships requires careful interpretation and validation. As networks become more complex, incorporating more diseases and relationships, they can become increasingly difficult to interpret and visualize effectively, potentially obscuring key insights [105, 108].
Finally, while theoretically possible, integrating diverse data types remains practically challenging. Combining clinical, genetic, and molecular data in a meaningful way is an ongoing area of research that has yet to be fully realized in many network studies [109]. This is compounded by a lack of consensus about terminology, which may limit the ability to incorporate datasets and compare results [110].
Conclusion
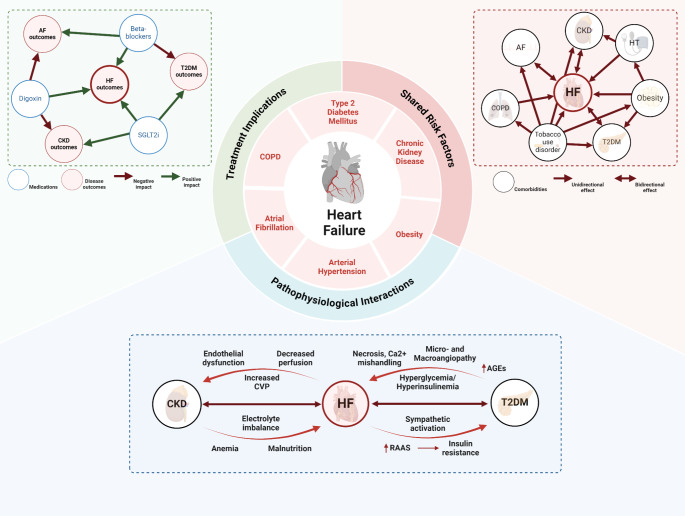
Comorbidity networks can be leveraged to elucidate relationships between diseases that might otherwise go unrecognized. By harnessing disease heterogeneity at the individual level [58], network approaches have been important for advancing our understanding of the intricate connections between related diseases [111]. To fully exploit the potential of comorbidity networks, we must move beyond merely describing subpopulations and strive to make and validate claims about the epidemiology, pathophysiology, and potential avenues for treatment (Fig. 3). Linking networks to genomic data sources will be critical to understanding how the genetic heterogeneity in HF populations contributes to the variety of clinical trajectories patients experience and potentially identify treatment opportunities. This has been demonstrated even in patient populations with heart diseases that are often genetic but lack a single genetic driver [112]. Networks that delve into the biological underpinnings of disease subpopulations could pave the way for future innovations.
Fig. 3.
Schematic overview of potential of comorbidity network representations to explore the complexity and implications of comorbidities in heart failure. The central panel highlights the complex interplay between heart failure and its most common comorbidities, beyond the mere coexistence of diseases to clinical impact. The upper right panel highlights the commonality of risk associations derived from epidemiological studies between the different conditions surrounding HF. The network in the upper left panel represents the complex interplay between therapeutic classes and different outcomes across HF and related comorbidities, highlighting the differential impact of each therapy concerning the assessed outcome. Finally, the lower panel summarizes the pathophysiological interplay between HF, chronic kidney disease (CKD), and type 2 diabetes mellitus from a molecular perspective, highlighting bidirectional effects related to multisystemic pathways
Disease networks also represent interactions with healthcare systems by representing comorbidity from clinical databases. Networks that consider the order and timing of diseases can provide information about clinical trajectories in patients with HF, even well before it manifests. By providing a realistic representation of comorbidity relationships and how patients experience treatment for these conditions, these networks can inform the development of clinical trials, ensuring that populations experiencing comorbidities can benefit from innovative therapeutic approaches. This shift in focus from description to practical information is crucial to unlocking the true potential of comorbidity networks in HF research.
Glossary of Terms
- Network
A collection of nodes (entities) connected by edges (relationships)
- Node
In comorbidity networks, a node typically represents a disease or condition
- Edge
A connection between nodes, often representing a statistical relationship (e.g., correlation, co-occurrence) between diseases
- Degree
The number of connections a node has to other nodes in the network
- Betweenness
A measure of a node’s centrality, indicating how often it lies on the shortest path between other nodes
- Closeness
A measure of how close a node is to all other nodes in the network
- Hub
A node with a high degree of connectivity, often representing a disease with many comorbid relationships
- Module
A group of nodes that are more densely connected to each other than to the rest of the network
- Cluster
A group of similar nodes, often identified through algorithmic analysis of the network structure
- Network motif
Recurring patterns of interconnections in a network
- Phenotype
Observable characteristics of an organism resulting from the interaction of its genotype with the environment
- Multi-layer network
A network with multiple types of relationships or data integrated into a single structure
- Directed network
A network where edges have a direction, indicating a causal or temporal relationship
- Weighted network
A network where edges have associated values indicating the strength of relationships
- Random walk
A mathematical method used to explore network structures and identify important nodes or relationships
Author Contributions
SAGO, JDL, and RTL wrote the manuscript text. SAGO and RTL prepared the figures. All authors reviewed the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
No funding was received.
Data Availability
No datasets were generated or analysed during the current study.
Declarations
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rhodes CJ, Sweatt AJ, Maron BA. Harnessing Big Data to Advance Treatment and understanding of Pulmonary Hypertension. Circ Res. 2022;130:1423–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanzer JD, Leuschner F, Kramann R, Levinson RT, Saez-Rodriguez J. Big Data approaches in heart failure research. Curr Heart Fail Rep. 2020;17:213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savarese G, Becher PM, Lund LH, Seferovic P, Rosano GMC, Coats AJS. Global burden of heart failure: a comprehensive and updated review of epidemiology. Cardiovasc Res. 2023;118:3272–87. [DOI] [PubMed] [Google Scholar]
- 4.Emmons-Bell S, Johnson C, Roth G. Prevalence, incidence and survival of heart failure: a systematic review. Heart. 2022;108:1351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comte B, Baumbach J, Benis A, et al. Network and Systems Medicine: position paper of the European collaboration on Science and Technology Action on Open Multiscale Systems Medicine. Netw Syst Med. 2020;3:67–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Yuan Z, Ji J, Li H, Xue F. Network or regression-based methods for disease discrimination: a comparison study. BMC Med Res Methodol. 2016;16:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou R, Xia Y-Y, Li Z, Wu L-D, Shi Y, Ling Z-Y, Zhang J-X. HFpEF as systemic disease, insight from a diagnostic prediction model reminiscent of systemic inflammation and organ interaction in HFpEF patients. Sci Rep. 2024;14:5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellumkonda L, Tyrrell D, Hummel SL, Goldstein DR. Pathophysiology of heart failure and frailty: a common inflammatory origin? Aging Cell. 2017;16:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinstein AR. THE PRE-THERAPEUTIC CLASSIFICATION OF CO-MORBIDITY IN CHRONIC DISEASE. J Chronic Dis. 1970;23:455–68. [DOI] [PubMed] [Google Scholar]
- 11.Gerhardt T, Gerhardt LMS, Ouwerkerk W, et al. Multimorbidity in patients with acute heart failure across world regions and country income levels (REPORT-HF): a prospective, multicentre, global cohort study. Lancet Glob Health. 2023;11:e1874–84. [DOI] [PubMed] [Google Scholar]
- 12.Heidenreich PA, Bozkurt B, Aguilar D, et al. 2022 AHA/ACC/HFSA Guideline for the management of Heart failure: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice guidelines. Circulation. 2022. 10.1161/CIR.0000000000001063.
- 13.Gimeno-Miguel A, Gracia Gutiérrez A, Poblador-Plou B, Coscollar-Santaliestra C, Pérez-Calvo JI, Divo MJ, Calderón-Larrañaga A, Prados-Torres A, Ruiz-Laiglesia FJ. Multimorbidity patterns in patients with heart failure: an observational Spanish study based on electronic health records. BMJ Open. 2019;9:e033174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortin M, Soubhi H, Hudon C, Bayliss EA, van den Akker M. Multimorbidity’s many challenges. BMJ. 2007;334:1016–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison C, Fortin M, van den Akker M, Mair F, Calderon-Larranaga A, Boland F, Wallace E, Jani B, Smith S. Comorbidity versus multimorbidity: why it matters. J Multimorb Comorb. 2021;11:2633556521993993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valderas JM, Starfield B, Sibbald B, Salisbury C, Roland M. Defining comorbidity: implications for understanding health and health services. Ann Fam Med. 2009;7:357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skou ST, Mair FS, Fortin M, Guthrie B, Nunes BP, Miranda JJ, Boyd CM, Pati S, Mtenga S, Smith SM. Multimorbidity. Nat Rev Dis Primers. 2022;8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Screever EM, van der Wal MHL, van Veldhuisen DJ, et al. Comorbidities complicating heart failure: changes over the last 15 years. Clin Res Cardiol. 2023;112:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loosen SH, Roderburg C, Curth O, Gaensbacher J, Joerdens M, Luedde T, Konrad M, Kostev K, Luedde M. The spectrum of comorbidities at the initial diagnosis of heart failure a case control study. Sci Rep. 2022;12:2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan MS, Samman Tahhan A, Vaduganathan M, Greene SJ, Alrohaibani A, Anker SD, Vardeny O, Fonarow GC, Butler J. Trends in prevalence of comorbidities in heart failure clinical trials. Eur J Heart Fail. 2020;22:1032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Triposkiadis F, Giamouzis G, Parissis J, Starling RC, Boudoulas H, Skoularigis J, Butler J, Filippatos G. Reframing the association and significance of co-morbidities in heart failure. Eur J Heart Fail. 2016;18:744–58. [DOI] [PubMed] [Google Scholar]
- 22.Bavishi A, Patel RB. Addressing comorbidities in Heart failure: hypertension, Atrial Fibrillation, and diabetes. Heart Fail Clin. 2020;16:441–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cascino TM, Kittleson MM, Lala A, et al. Comorbid conditions and Health-Related Quality of Life in Ambulatory Heart failure patients. Circ Heart Fail. 2020;13:e006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Wel MC, Jansen RWMM, Bakx JC, Bor HHJ, Olderikkert MGM, van Weel C. Non-cardiovascular co-morbidity in elderly patients with heart failure outnumbers cardiovascular co-morbidity. Eur J Heart Fail. 2007;9:709–15. [DOI] [PubMed] [Google Scholar]
- 25.Lee KS, Park D-I, Lee J, Oh O, Kim N, Nam G. Relationship between comorbidity and health outcomes in patients with heart failure: a systematic review and meta-analysis. BMC Cardiovasc Disord. 2023;23:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haregu TN, Nanayakkara S, Carrington M, Kaye D. Multimorbidity and multiple causes of death in heart failure. J Public Health. 2021;29:1181–7. [Google Scholar]
- 27.Manemann SM, Chamberlain AM, Boyd CM, Gerber Y, Dunlay SM, Weston SA, Jiang R, Roger VL. Multimorbidity in Heart failure: Effect on outcomes. J Am Geriatr Soc. 2016;64:1469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomasoni D, Vitale C, Guidetti F, et al. The role of multimorbidity in patients with heart failure across the left ventricular ejection fraction spectrum: data from the Swedish Heart failure Registry. Eur J Heart Fail. 2024;26:854–68. [DOI] [PubMed] [Google Scholar]
- 29.Kittleson Michelle M, Panjrath Gurusher S, Kaushik A, Davis Leslie L, Anita D, Dixon Dave L, Januzzi James L, Clyde Y W. 2023 ACC Expert Consensus decision pathway on management of heart failure with preserved ejection fraction. J Am Coll Cardiol. 2023;81:1835–78. [DOI] [PubMed] [Google Scholar]
- 30.Simmonds SJ, Cuijpers I, Heymans S, Jones EAV. Cellular and Molecular differences between HFpEF and HFrEF: a step ahead in an Improved Pathological understanding. Cells. 2020. 10.3390/cells9010242. [DOI] [PMC free article] [PubMed]
- 31.Deichl A, Wachter R, Edelmann F. Comorbidities in heart failure with preserved ejection fraction. Herz. 2022;47:301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Authors/Task Force Members:, McDonagh TA, Metra M, et al. 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail. 2022;24:4–131. [DOI] [PubMed] [Google Scholar]
- 33.Pandey A, Vaduganathan M, Arora S, Qamar A, Mentz RJ, Shah SJ, Chang PP, Russell SD, Rosamond WD, Caughey MC. Temporal trends in Prevalence and Prognostic implications of comorbidities among patients with Acute Decompensated Heart failure: the ARIC Study Community Surveillance. Circulation. 2020;142:230–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ather S, Chan W, Bozkurt B, Aguilar D, Ramasubbu K, Zachariah AA, Wehrens XHT, Deswal A. Impact of noncardiac comorbidities on morbidity and mortality in a predominantly male population with heart failure and preserved versus reduced ejection fraction. J Am Coll Cardiol. 2012;59:998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ergatoudes C, Schaufelberger M, Andersson B, Pivodic A, Dahlström U, Fu M. Non-cardiac comorbidities and mortality in patients with heart failure with reduced vs. preserved ejection fraction: a study using the Swedish Heart failure Registry. Clin Res Cardiol. 2019;108:1025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savarese G, Settergren C, Schrage B, et al. Comorbidities and cause-specific outcomes in heart failure across the ejection fraction spectrum: a blueprint for clinical trial design. Int J Cardiol. 2020;313:76–82. [DOI] [PubMed] [Google Scholar]
- 37.Levinson RT, Vaitinidin NS, Farber-Eger E, Roden DM, Lasko TA, Wells QS, Mosley JD. Heart failure clinical care analysis uncovers risk reduction opportunities for preserved ejection fraction subtype. Sci Rep. 2021;11:18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riedel O, Ohlmeier C, Enders D, Elsässer A, Vizcaya D, Michel A, Eberhard S, Schlothauer N, Berg J, Garbe E. The contribution of comorbidities to mortality in hospitalized patients with heart failure. Clin Res Cardiol. 2018;107:487–97. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Savarese G, Dahlström U, Lund LH, Fu M. Age-dependent differences in clinical phenotype and prognosis in heart failure with mid-range ejection compared with heart failure with reduced or preserved ejection fraction. Clin Res Cardiol. 2019;108:1394–405. [DOI] [PubMed] [Google Scholar]
- 40.Kroshian G, Joseph J, Kinlay S, Peralta AO, Hoffmeister PS, Singh JP, Yuyun MF. Atrial fibrillation and risk of adverse outcomes in heart failure with reduced, mildly reduced, and preserved ejection fraction: a systematic review and meta-analysis. J Cardiovasc Electrophysiol. 2024;35:715–26. [DOI] [PubMed] [Google Scholar]
- 41.Jensen AB, Moseley PL, Oprea TI, Ellesøe SG, Eriksson R, Schmock H, Jensen PB, Jensen LJ, Brunak S. Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat Commun. 2014;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mentz RJ, Kelly JP, von Lueder TG, et al. Noncardiac comorbidities in heart failure with reduced versus preserved ejection fraction. J Am Coll Cardiol. 2014;64:2281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weintraub WS. Role of Big Data in Cardiovascular Research. J Am Heart Assoc. 2019;8:e012791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barabási AL. Network medicine — from obesity to the Diseasome. N Engl J Med 2007;357:404–7. 10.1056/NEJMe078114. [DOI] [PubMed]
- 45.Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118:4947–57. [DOI] [PubMed] [Google Scholar]
- 46.Hidalgo CA, Blumm N, Barabási A-L, Christakis NA. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol. 2009;5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabási A-L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz Amores G, Martínez-Antonio A. Basics on network theory to analyze biological systems: a hands-on outlook. Funct Integr Genomics. 2022;22:1433–48. [DOI] [PubMed] [Google Scholar]
- 49.Fotouhi B, Momeni N, Riolo MA, Buckeridge DL. Statistical methods for constructing disease comorbidity networks from longitudinal inpatient data. Appl Netw Sci. 2018;3:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park J, Lee D-S, Christakis NA, Barabási A-L. The impact of cellular networks on disease comorbidity. Mol Syst Biol. 2009;5:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunson JC, Agresta TP, Laubenbacher RC. Sensitivity of comorbidity network analysis. JAMIA Open. 2020;3:94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang Y, Guo C, Li H. Comorbidity progression analysis: patient stratification and comorbidity prediction using temporal comorbidity network. Health Inform Sci Syst. 2024;12:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y, Chen S, Miao Z, Delen D, Gin A. Clustering temporal disease networks to assist clinical decision support systems in visual analytics of comorbidity progression. Decis Support Syst. 2021;148:113583. [Google Scholar]
- 54.Epskamp S. Psychometric network models from time-series and panel data. Psychometrika. 2020;85:206–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Epskamp S, Kruis J, Marsman M. Estimating psychopathological networks: be careful what you wish for. PLoS ONE. 2017;12:e0179891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serrano MÁ, Boguñá M, Vespignani A. Extracting the multiscale backbone of complex weighted networks. Proc Natl Acad Sci. 2009;106:6483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Infante T, Del Viscovo L, De Rimini ML, Padula S, Caso P, Napoli C. Network medicine: a clinical approach for precision medicine and personalized therapy in coronary heart disease. J Atheroscler Thromb. 2020;27:279–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R-S, Maron BA, Loscalzo J. Multiomics Network Medicine Approaches To Precision Medicine and therapeutics in Cardiovascular diseases. Arterioscler Thromb Vasc Biol. 2023;43:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pai S, Hui S, Isserlin R, Shah MA, Kaka H, Bader GD. netDx: interpretable patient classification using integrated patient similarity networks. Mol Syst Biol. 2019. 10.15252/msb.20188497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, Li D, Havlin S. From a single network to a network of networks. Natl Sci Rev. 2014;1:346–56. [Google Scholar]
- 61.Ideker T, Dutkowski J, Hood L. Boosting signal-to-noise in complex biology: prior knowledge is power. Cell. 2011;144:860–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019;48:D845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rappaport N, Nativ N, Stelzer G et al. (2013) MalaCards: an integrated compendium for diseases and their annotation. Database 2013:bat018. [DOI] [PMC free article] [PubMed]
- 64.UniProt Consortium. UniProt: the Universal protein knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gargano MA, Matentzoglu N, Coleman B, et al. The human phenotype ontology in 2024: phenotypes around the world. Nucleic Acids Res. 2023;52:D1333–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baron JA, Johnson CS-B, Schor MA, et al. The DO-KB knowledgebase: a 20-year journey developing the disease open science ecosystem. Nucleic Acids Res. 2024;52:D1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falsetti L, Viticchi G, Zaccone V, et al. Clusters of comorbidities in the short-term prognosis of Acute Heart failure among Elderly patients: a retrospective cohort study. Medicina. 2022. 10.3390/medicina58101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uszko-Lencer NHMK, Janssen DJA, Gaffron S, et al. Clustering based on comorbidities in patients with chronic heart failure: an illustration of clinical diversity. ESC Heart Fail. 2022;9:614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sonawane AR, Weiss ST, Glass K, Sharma A. Network Medicine in the age of Biomedical Big Data. Front Genet. 2019;10:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meijs C, Brugts JJ, Lund LH, et al. Identifying distinct clinical clusters in heart failure with mildly reduced ejection fraction. Int J Cardiol. 2023;386:83–90. [DOI] [PubMed] [Google Scholar]
- 73.Bisson A, Fawzy M, Romiti A, Proietti GF, Angoulvant M, El-Bouri D, Lip WYH, Fauchier G L. Phenotypes and outcomes in non-anticoagulated patients with atrial fibrillation: an unsupervised cluster analysis. Arch Cardiovasc Dis. 2023;116:342–51. [DOI] [PubMed] [Google Scholar]
- 74.Gulea C, Zakeri R, Quint JK. Model-based comorbidity clusters in patients with heart failure: association with clinical outcomes and healthcare utilization. BMC Med. 2021;19:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tromp J, Tay WT, Ouwerkerk W, et al. Multimorbidity in patients with heart failure from 11 Asian regions: a prospective cohort study using the ASIAN-HF registry. PLoS Med. 2018;15:e1002541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiwie C, Baumbach J, Röttger R. Comparing the performance of biomedical clustering methods. Nat Methods. 2015;12:1033–8. [DOI] [PubMed] [Google Scholar]
- 77.Meijs C, Handoko ML, Savarese G, Vernooij RWM, Vaartjes I, Banerjee A, Koudstaal S, Brugts JJ, Asselbergs FW, Uijl A. Discovering distinct phenotypical clusters in heart failure across the ejection Fraction Spectrum: a systematic review. Curr Heart Fail Rep. 2023;20:333–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van de Veerdonk MC, Savarese G, Handoko ML, Beulens JWJ, Asselbergs F, Uijl A. Multimorbidity in Heart failure: leveraging cluster analysis to guide tailored treatment strategies. Curr Heart Fail Rep. 2023;20:461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qiu C, Yu DS-F, Song D, Wang X. The prognostic impact of symptom clusters in patients with heart failure: a systematic review and meta-analysis. J Adv Nurs. 2022;78:2713–30. [DOI] [PubMed] [Google Scholar]
- 80.Palazzuoli A, Ruocco G, Gronda E. Noncardiac comorbidity clustering in heart failure: an overlooked aspect with potential therapeutic door. Heart Fail Rev. 2022;27:767–78. [DOI] [PubMed] [Google Scholar]
- 81.Carmona-Pírez J, Poblador-Plou B, Díez-Manglano J, Morillo-Jiménez MJ, Marín Trigo JM, Ioakeim-Skoufa I, Gimeno-Miguel A, Prados-Torres A. Multimorbidity networks of chronic obstructive pulmonary disease and heart failure in men and women: evidence from the EpiChron Cohort. Mech Ageing Dev. 2021;193:111392. [DOI] [PubMed] [Google Scholar]
- 82.Zhou D, Wang L, Ding S, Shen M, Qiu H. Phenotypic Disease Network Analysis To Identify Comorbidity Patterns in hospitalized patients with ischemic heart Disease using large-Scale Administrative Data. Healthc (Basel). 2022. 10.3390/healthcare10010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martins C, Neves B, Teixeira AS, Froes M, Sarmento P, Machado J, Magalhães CA, Silva NA, Silva MJ, Leite F. Identifying subgroups in heart failure patients with multimorbidity by clustering and network analysis. BMC Med Inf Decis Mak. 2024;24:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell-Quintero S, Echeverría LE, Gómez-Mesa JE, et al. Comorbidity profile and outcomes in patients with chronic heart failure in a latin American country: insights from the Colombian heart failure registry (RECOLFACA). Int J Cardiol. 2023;378:123–9. [DOI] [PubMed] [Google Scholar]
- 85.Ieva F, Bitonti D. Network analysis of comorbidity patterns in heart failure patients using administrative data. Epidemiol Biostat Public Health. 2022. 10.2427/12779. [Google Scholar]
- 86.Lawson CA, Solis-Trapala I, Dahlstrom U, Mamas M, Jaarsma T, Kadam UT, Stromberg A. Comorbidity health pathways in heart failure patients: a sequences-of-regressions analysis using cross-sectional data from 10,575 patients in the Swedish Heart failure Registry. PLoS Med. 2018;15:e1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Merrill JA, Sheehan BM, Carley KM, Stetson PD. Transition networks in a cohort of patients with congestive heart failure: a novel application of informatics methods to inform care coordination. Appl Clin Inf. 2015;6:548–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng C, Han L, Tian J, et al. Hierarchical management of chronic heart failure: a perspective based on the latent structure of comorbidities. ESC Heart Fail. 2022;9:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou D, Qiu H, Wang L, Shen M. Risk prediction of heart failure in patients with ischemic heart disease using network analytics and stacking ensemble learning. BMC Med Inf Decis Mak. 2023;23:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gong K, Xue Y, Kong L, Xie X. Cost prediction for ischemic heart disease hospitalization: interpretable feature extraction using network analysis. J Biomed Inf. 2024;154:104652. [DOI] [PubMed] [Google Scholar]
- 91.Lanzer JD, Valdeolivas A, Pepin M, Hund H, Backs J, Frey N, Friederich H-C, Schultz J-H, Saez-Rodriguez J, Levinson RT. A network medicine approach to study comorbidities in heart failure with preserved ejection fraction. BMC Med. 2023;21:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cruz-Ávila HA, Vallejo M, Martínez-García M, Hernández-Lemus E. Comorbidity Networks in Cardiovascular diseases. Front Physiol. 2020;11:1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Christiansen MN, Køber L, Torp-Pedersen C, Gislason GH, Schou M, Smith JG, Vasan RS, Andersson C. Preheart failure comorbidities and impact on prognosis in heart failure patients: a nationwide study. J Intern Med. 2020;287:698–710. [DOI] [PubMed] [Google Scholar]
- 94.Aguado A, Moratalla-Navarro F, López-Simarro F, Moreno V. MorbiNet: multimorbidity networks in adult general population. Analysis of type 2 diabetes mellitus comorbidity. Sci Rep. 2020;10:2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khan A, Uddin S, Srinivasan U. Comorbidity network for chronic disease: a novel approach to understand type 2 diabetes progression. Int J Med Inf. 2018;115:1–9. [DOI] [PubMed] [Google Scholar]
- 96.Feng J, Mu X-M, Ma L-L, Wang W. Comorbidity patterns of older Lung Cancer patients in Northeast China: An Association Rules Analysis Based on Electronic Medical Records. Int J Environ Res Public Health. 2020. 10.3390/ijerph17239119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moratalla-Navarro F, Moreno V, López-Simarro F, Aguado A. MorbiNet Study: Hypothyroidism Comorbidity Networks in the Adult General Population. J Clin Endocrinol Metab. 2021;106:e1179–90. [DOI] [PubMed] [Google Scholar]
- 98.do Valle IF, Ferolito B, Gerlovin H, et al. Network-medicine framework for studying disease trajectories in U.S. veterans. Sci Rep. 2022;12:12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dervić E, Sorger J, Yang L, Leutner M, Kautzky A, Thurner S, Kautzky-Willer A, Klimek P. Unraveling cradle-to-grave disease trajectories from multilayer comorbidity networks. NPJ Digit Med. 2024;7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo M, Yu Y, Wen T, Zhang X, Liu B, Zhang J, Zhang R, Zhang Y, Zhou X. Analysis of disease comorbidity patterns in a large-scale China population. BMC Med Genomics. 2019;12:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuan V, Denaxas S, Patalay P, et al. Identifying and visualising multimorbidity and comorbidity patterns in patients in the English National Health Service: a population-based study. Lancet Digit Health. 2023;5:e16–27. [DOI] [PubMed] [Google Scholar]
- 102.Silverman EK, Schmidt HHHW, Anastasiadou E, et al. Molecular networks in Network Medicine: Development and applications. Wiley Interdiscip Rev Syst Biol Med. 2020;12:e1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Benincasa G, Marfella R, Della Mura N, Schiano C, Napoli C. Strengths and opportunities of Network Medicine in Cardiovascular diseases. Circ J. 2020;84:144–52. [DOI] [PubMed] [Google Scholar]
- 104.Morbinet. https://morbinet.org/shiny. Accessed 28 Jun 2024.
- 105.Jordan DG, Winer ES, Salem T. The current status of temporal network analysis for clinical science: considerations as the paradigm shifts? J Clin Psychol. 2020;76:1591–612. [DOI] [PubMed] [Google Scholar]
- 106.Hanauer DA, Ramakrishnan N. Modeling temporal relationships in large scale clinical associations. J Am Med Inf Assoc. 2013;20:332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arrell DK, Terzic A. Interpreting networks in systems biology. Clin Pharmacol Ther. 2013;93:389–92. [DOI] [PubMed] [Google Scholar]
- 108.Conte F, Fiscon G, Licursi V, Bizzarri D, D’Antò T, Farina L, Paci P. A paradigm shift in medicine: a comprehensive review of network-based approaches. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194416. [DOI] [PubMed] [Google Scholar]
- 109.Bodein A, Scott-Boyer M-P, Perin O, Lê Cao K-A, Droit A. Interpretation of network-based integration from multi-omics longitudinal data. Nucleic Acids Res. 2022;50:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sadegh S, Skelton J, Anastasi E, et al. Lacking mechanistic disease definitions and corresponding association data hamper progress in network medicine and beyond. Nat Commun. 2023;14:1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Halu A, Liu S, Baek SH, Hobbs BD, Hunninghake GM, Cho MH, Silverman EK, Sharma A. Exploring the cross-phenotype network region of disease modules reveals concordant and discordant pathways between chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Hum Mol Genet. 2019;28:2352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maron BJ, Maron MS, Maron BA, Loscalzo J. Moving beyond the Sarcomere to explain heterogeneity in hypertrophic cardiomyopathy: JACC Review topic of the Week. J Am Coll Cardiol. 2019;73:1978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.