Abstract
Aldosterone-producing adenoma (APA) is a leading cause of primary aldosteronism (PA), a condition marked by excessive aldosterone secretion. CYP11B2, the aldosterone synthase, plays a critical role in aldosterone biosynthesis and the development of APA. Despite its significance, encoding regulatory mechanisms governing CYP11B2, particularly its degradation, remain poorly understood. In this study, we sought to uncover novel regulators of CYP11B2 stability by conducting a siRNA screen targeting E3 ubiquitin ligases. Our results identified TRIM2 as a key negative regulator of CYP11B2, where its overexpression led to a significant reduction in CYP11B2 protein levels and a concomitant decrease in aldosterone production in adrenal tumor cells. Mechanistically, we demonstrated that TRIM2 interacts with CYP11B2 via its RBCC domain, promoting K29/48-linked polyubiquitination and destabilization of CYP11B2. Further results revealed that TRIM2 is downregulated in APA tissues, showing differential expression between the zona glomerulosa (ZG) and zona fasciculata (ZF) of normal adrenal tissue. These findings highlight TRIM2 as a novel modulator of aldosterone synthesis through CYP11B2 degradation, offering a potential therapeutic target for APA.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05545-0.
Keywords: Adrenal cortex, Protein degradation, Hormone biosynthesis regulation, E3 ligases in endocrine tumors
Introduction
Aldosterone (ALD) is a mineralocorticoid hormone synthesized by the adrenal glands, contributing to the regulation of electrolyte balance and blood volume. Primary aldosteronism is a state of extracellular fluid volume expansion, low renin condition, excessive and non-suppressible aldosterone production [1–3]. Primary aldosteronism stands as one of the most prevalent causes of endocrine hypertension, affecting approximately 6–10% of hypertensive patients [4–6]. Aldosterone-producing adenoma (APA) is a common functional benign tumor of the adrenal cortex and a prevalent subtype of primary aldosteronism [7].
Cytochrome P450 11B2 (CYP11B2), also referred to as aldosterone synthase, is a constituent of the cytochrome P450 (CYP) superfamily and assumes a pivotal role in catalyzing three successive oxygenation reactions during the synthesis of aldosterone [8]. Moreover, it is increasingly acknowledged as a promising therapeutic target for cardiovascular conditions associated with primary aldosteronism. As commonly understood, CYP11B2 catalyzes the conversion of deoxycorticosterone to corticosterone, corticosterone to 18OH-corticosterone, and ultimately 18OH-corticosterone to aldosterone [9]. Elevated CYP11B2 expression is the key factor leading to excessive production and secretion of aldosterone in APAs.
Recent investigations have employed diverse methodologies, unveiling numerous novel and pivotal insights into the molecular pathogenesis of aldosterone-producing adenomas (APAs), notably the upregulation of aldosterone synthase (CYP11B2) in APAs [10–13]. The transcriptional regulation of CYP11B2 is governed by the renin-angiotensin system (RAS). Within the zona glomerulosa of the adrenal cortex, angiotensin II (Ang II) binds to Ang II type 1 receptors (AT1R), members of the G protein-coupled receptor family. Upon ligand activation, AT1R initiates various signaling cascades including inositol triphosphate (IP3), Ca2+/calmodulin (CaM), and MAPK signaling pathways [14, 15]. Transcription factors such as NR4A and CREB/ATF1 families are known to be activated via these signaling routes, thereby promoting the transcriptional activity of CYP11B2 [16, 17]. Additionally, somatic mutations, such as potassium channel KCNJ5, ATPase function-related genes ATP1A1 and ATP2B3, as well as mutations influencing calcium channel activity in CACNA1D and CACNA1H, along with mutations in CLCN2 affecting [18] chloride ion channels, can all promote the expression of CYP11B2 by influencing its gene transcription activity [13, 19–21]. However, there is limited reported research concerning the protein-level regulatory mechanisms of functional CYP11B2, necessitating further exploration.
Various modifications, including phosphorylation, glycosylation, nitration, and ubiquitination, have been documented for P450s [22–26]. Protein stability of P450s is primarily regulated by phosphorylation and ubiquitination mechanisms [22, 27]. Proteins serve as the fundamental building blocks of life processes, with ubiquitination standing as the second most prevalent form of post-translational modification among proteins. Ubiquitin ligases (UBEs) are responsible for attaching ubiquitin molecules to specific protein substrates, countering modifications catalyzed by deubiquitinating enzymes (DUBs). However, the specific ubiquitination and stabilization functions of the ubiquitination enzyme (UBE) responsible for CYP11B2 remain largely unexplored. In this study, we demonstrate that a UBE-TRIM2-regulates aldosterone production in adrenal tumor cells by regulating the expression of CYP11B2. Mechanistically, TRIM2 ubiquitinates and destabilizes CYP11B2 in a catalytic activity-dependent manner. Furthermore, TRIM2 expression was downregulated in APA samples, suggesting that it may be involved in the pathogenesis of APA and may be a target for therapeutic intervention in APA.
Materials and methods
Cell culture and reagents
The human adrenocortical carcinoma cell line (H295R) was obtained from the American Type Culture Collection (CRL-2128; ATCC) and grown in H295R cell culture medium (CM-0399; Procell, Wuhan, China) at 37 °C in 5% CO2. 293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS (Gibco, China). Cells were maintained in a humidified incubator at 37 °C with 5% CO2. Antibodies against TRIM2 (20356-1-AP, Proteintech), CYP11B2 (MABS1251, Millipore), CYP11B1 (MABS502, Millipore), β-Actin (sc-47778, Santa Cruz), MYC (18583, CST), GFP (ab290, Abcam), HA (TA180128, OriGene), and Ub (3933, CST) were purchased from specified commercial sources.
siRNA library screening
We procured the ON-TARGETplus siRNA library, which includes a pool of E3 ligase siRNAs, from Dharmacon (GE Healthcare, catalog G-105635-01). H295R cells were cultured in 96-well plates at a density of 4 × 103 cells per well. The transfection reagent (Lipofectamine 3000 Transfection Reagent) was diluted using Opti-MEM and then added to each well containing the siRNA pool. After a 15-minute incubation period, the siRNA (50 nM)/Opti-MEM/Lipo3000 mixture was transfected into the H295R cells. 48 h post-transfection, cells were harvested, and lysates were prepared for immunoblotting analysis to assess endogenous CYP11B2 expression. Detailed siRNA sequences for E3 ligases are provided in Table 1.
Table 1.
The List of siRNA Sequences
| Gene Name | Gene ID | Sequence |
|---|---|---|
| PCGF3 | 10,336 | GCACUUAGAUUCCCAUCGG |
| PCGF3 | 10,336 | GUACAUCGGUCAUGACAGA |
| PCGF3 | 10,336 | GGAGUUCUAUCACAAAUUG |
| PCGF3 | 10,336 | GCGACCGUCUUGCAUCUGA |
| VPS41 | 27,072 | GAACGGUCUUGGAUGAACA |
| VPS41 | 27,072 | AGUCAUAGUUCAAGCAGUU |
| VPS41 | 27,072 | UGACAUAGCAGCACGCAAA |
| VPS41 | 27,072 | CAGCAUGUGUAUUUGCAUA |
| TRIML1 | 339,976 | UCAGAGAGCCUGUGUGUAA |
| TRIML1 | 339,976 | CAGCAAAUCAGAAGCCUAA |
| TRIML1 | 339,976 | ACACAACUAUUAAGACGAU |
| TRIML1 | 339,976 | GGACCUGUUCUCACUAAUA |
| PHF13 | 148,479 | GGGGAGAGGUGUUCGGUUU |
| PHF13 | 148,479 | CCGUGGUAGCUGUGCGUUU |
| PHF13 | 148,479 | ACUGAAGGCAAACGGACUA |
| PHF13 | 148,479 | CAAACAGCCAGGACGCAUU |
| TRIM22 | 10,346 | GUAGAUGUGUCUGGAAAGA |
| TRIM22 | 10,346 | AGUGAAAGCUGGACAUUGA |
| TRIM22 | 10,346 | UAAACGAGGUGGUCAAGGA |
| TRIM22 | 10,346 | GUACGCACCUGCACAUUUA |
| MARCH6 | 10,299 | ACGGAAAUCUGGCAAACAA |
| MARCH6 | 10,299 | CUAAGGAAUUUGAAUGAUC |
| MARCH6 | 10,299 | CAUACAAUGUCAUGCUCUA |
| MARCH6 | 10,299 | UCAUAGAUCUCGUCGCUUA |
| TRIM46 | 80,128 | UGACAAAGAGCCUGACAUA |
| TRIM46 | 80,128 | GCGAAUACAGUGAAGAUGU |
| TRIM46 | 80,128 | GCCAACGCCUGGUAUGUCA |
| TRIM46 | 80,128 | GCUCAGGUCUGGUGGGCUA |
| TRIM45 | 80,263 | GCACCGAGGAGUCUACUUA |
| TRIM45 | 80,263 | GGACAUACUACAUUUCCUA |
| TRIM45 | 80,263 | GCUCAGAUCCACAUAAUAA |
| TRIM45 | 80,263 | GUGCAGGGCUCGCCAUUCA |
| RNF4 | 6047 | GAAUGGACGUCUCAUCGUU |
| RNF4 | 6047 | GCAAUAAAUUCUAGACAAG |
| RNF4 | 6047 | GACAGAGACGUAUAUGUGA |
| RNF4 | 6047 | GCUAAUACUUGCCCAACUU |
| IBRDC1 | 154,214 | CAUAGCGGUUGUAAUCGUU |
| IBRDC1 | 154,214 | GUUGUUAAGUGAUAUGCUA |
| IBRDC1 | 154,214 | CAUAUGACCUGCUCACAAU |
| IBRDC1 | 154,214 | GAACUUGGCCGUAUUGAUU |
| RFP2 | 10,206 | GAAGGGAGUGUGCGGAAUU |
| RFP2 | 10,206 | GACACUGGCACAUUCAUUA |
| RFP2 | 10,206 | UAAAACAGCCGAUUUCAUA |
| RFP2 | 10,206 | GAGACCAGCUCCAUUCAAG |
| RNF190 | 162,333 | UCAUUUAGGUUCCGAGAUG |
| RNF190 | 162,333 | CCAAACAGAGUUCUAGUGA |
| RNF190 | 162,333 | GAGCAGAGGUUUGCAGAAC |
| RNF190 | 162,333 | GAGAGAGGUUGUCAAGAAA |
| TRIM72 | 493,829 | CAUGCAUGCGUAAGGAGAA |
| TRIM72 | 493,829 | GAGCAGACAUUGCGGCACU |
| TRIM72 | 493,829 | GGGAGGUGGAUGUUGGCGA |
| TRIM72 | 493,829 | CCAGAAUACUGACAAGCGU |
| LNX1 | 84,708 | GGAAGAAUUACUCUAACUA |
| LNX1 | 84,708 | GCACGGCCCUUUGAGAGAU |
| LNX1 | 84,708 | CGAUAGUACUCAAAGCUUU |
| LNX1 | 84,708 | GGAGAAUGACCGUGUGUUA |
| MYLIP | 29,116 | GGUGAAAGUUUAUGGCUAA |
| MYLIP | 29,116 | CCAGAACACUGCCAAGUAU |
| MYLIP | 29,116 | GACUUUAGCCCAAUUAAUA |
| MYLIP | 29,116 | UAACAGAGACGCACGCAUU |
| PJA2 | 9867 | GCAGGAGGGUAUCAGACAA |
| PJA2 | 9867 | GAAGCACCCUAAACCUUGA |
| PJA2 | 9867 | GUUAGAUUCUGUACCAUUA |
| PJA2 | 9867 | AGACUGCUCUGGCCCAUUU |
| MDM4 | 4194 | CCACGAGACGGGAACAUUA |
| MDM4 | 4194 | CGUCAGAGCUUCUCCGUAA |
| MDM4 | 4194 | CCUAAAGAUGCGUAUAUAA |
| MDM4 | 4194 | AAGCAUGGGAGAACAGUUA |
| TRIM47 | 91,107 | CCAAAGGUGUCAAGAGGGU |
| TRIM47 | 91,107 | GUACGGGACGGCAAGAUGA |
| TRIM47 | 91,107 | CCCAAGACCUCGAGAGUAC |
| TRIM47 | 91,107 | CAUCAAGAGUGCAGCCGUA |
| TRIM49 | 57,093 | CAAGAUGUACCCUAUUUCA |
| TRIM49 | 57,093 | CAUAUUACUCUGCAUCAUG |
| TRIM49 | 57,093 | GUUAAUCAAAGCUCCCUAA |
| TRIM49 | 57,093 | GAAGAUAGAUGGAAAGGCG |
| ARIH1 | 25,820 | CGAGAUAUUUCCCAAGAUU |
| ARIH1 | 25,820 | GAGAGUCGACGAAGGGUUU |
| ARIH1 | 25,820 | CCAAAUGCCAUGUCACAAU |
| ARIH1 | 25,820 | GGAUAUGCCUUGUCAGAUC |
| ZNRF1 | 84,937 | ACGAUGAUGUGCUGACUAA |
| ZNRF1 | 84,937 | GAACAGAUCUUGUCCGGAA |
| ZNRF1 | 84,937 | GGAAAUGCACUUUAUAAUG |
| ZNRF1 | 84,937 | GCAUAGUGGUUUCAAGUGC |
| RNF103 | 7844 | GUUCAUGUGCCAAUAAAUA |
| RNF103 | 7844 | UUACCAAUGUGGCGAUUUA |
| RNF103 | 7844 | GCCAUUGUGUGGUAUGAAA |
| RNF103 | 7844 | GAGCUUGGUUCUAGUUAAU |
| PHF16 | 9767 | AGGAACAGAUCUUCGGUUU |
| PHF16 | 9767 | CUGCUGAGGUAUUCCGGAA |
| PHF16 | 9767 | GGAUGGAACCGAUCACGAA |
| PHF16 | 9767 | CAGUGACAGUUCAGACGAA |
| LRSAM1 | 90,678 | CAGAUCAGGAGCCAGAUUA |
| LRSAM1 | 90,678 | GAACGAUUCCAGCAGAUUC |
| LRSAM1 | 90,678 | UGACGGAGUUAGAAGCCAA |
| LRSAM1 | 90,678 | GCAGAUGACAUUCUCGACA |
| RNF149 | 284,996 | GCUAGAAGCCGGCAGGAGU |
| RNF149 | 284,996 | GAGUCUAGCUUUACCAGAU |
| RNF149 | 284,996 | UAUACUGGCUCUCAGAUUG |
| RNF149 | 284,996 | CAUGAUGAUUAUCUCGUUA |
| PHF10 | 55,274 | GCGCAGUGAUGAAGUGAUU |
| PHF10 | 55,274 | ACUUAAACCGGGAACGCAU |
| PHF10 | 55,274 | AAAGAACGUCAACGAAUUA |
| PHF10 | 55,274 | AAUAAAGGCACUUCGGACA |
| SH3RF2 | 153,769 | GCGGCCAGCUCCCUCAUUA |
| SH3RF2 | 153,769 | GGACAGGUCAGCACUUAUC |
| SH3RF2 | 153,769 | GCAAAGGCCUUAUGCAACU |
| SH3RF2 | 153,769 | GGGCGGAGCAGCAUGAGAA |
| PHF23 | 79,142 | GCAAGAAGCGGAAGUUAAA |
| PHF23 | 79,142 | AAGAAGAGAUGGCAACAGU |
| PHF23 | 79,142 | CGACAGUGCUACCUUGCUU |
| PHF23 | 79,142 | AAACGGCGGAGAACAAUUG |
| ZNF330 | 27,309 | CCUAAAUGUGGGCAUGAAA |
| ZNF330 | 27,309 | GAUCGAAGCAAUUGAAGUA |
| ZNF330 | 27,309 | GCAAUAGGGUGUAGCGUUU |
| ZNF330 | 27,309 | GCUGAGAACCGCCGAGAAC |
| RFPL2 | 10,739 | GAGAAUCUGUUCACCGCAA |
| RFPL2 | 10,739 | GAGCGUAUCUGCUGAGGAG |
| RFPL2 | 10,739 | GAAGUGGGCGCAUCAGACA |
| RFPL2 | 10,739 | CUUCGUAGACCGCAAGUUA |
| RAD18 | 56,852 | GCUCUCUGAUCGUGAUUUA |
| RAD18 | 56,852 | GAAAUGAGUGGUUCUACAU |
| RAD18 | 56,852 | GGGAGCAGGUUAAUGGAUA |
| RAD18 | 56,852 | CCAAGAAACAAGCGUAAUA |
| MLL3 | 58,508 | CCAGGUCAAUCAACAGUUA |
| MLL3 | 58,508 | CCAAAGCAUUUCAUCAGUA |
| MLL3 | 58,508 | GCAGUUACCAGAUACUUUA |
| MLL3 | 58,508 | GCAAUGGUCUUUCUGGAUA |
| TRIM2 | 23,321 | GGUCAACUAUGGCCUCAAA |
| TRIM2 | 23,321 | GCAAGAGUGUGCUGCUUAU |
| TRIM2 | 23,321 | GUAUAUGCCUGGAACGGUA |
| TRIM2 | 23,321 | CAACCAAUGUGUGCAGAUA |
| RFFL | 117,584 | CAUGACAUCUCUACCGAAA |
| RFFL | 117,584 | GAGGAGAACCUGUGUAAGA |
| RFFL | 117,584 | CCUGAGAGCUUGCAUCGGU |
| RFFL | 117,584 | GGACUCACCCAUUGACUGU |
| RNF150 | 57,484 | UCGCAUGGCUCGUCUUUUA |
| RNF150 | 57,484 | GACGUCAUCUUUACUACUA |
| RNF150 | 57,484 | AAGCAGUGAUUCUGACAUU |
| RNF150 | 57,484 | CCAAUGACGUUGUCCGGAU |
| RNF25 | 64,320 | AGGCUGAGCGAAACCGAUA |
| RNF25 | 64,320 | GGUCAAAUCAGCAAAGGUU |
| RNF25 | 64,320 | GACCAGGAUUCACAGUAUG |
| RNF25 | 64,320 | UGAGUCAGCUGUAGAUGUC |
| RNF148 | 378,925 | GAUCACAUCGGAAUUAGGA |
| RNF148 | 378,925 | CUACUUAGCUUUCCUGACU |
| RNF148 | 378,925 | GAGGCGAAGUCAAAUAAAG |
| RNF148 | 378,925 | GCAUAGUUCUGUUUCAUCU |
| MKRN1 | 23,608 | UCAAGUCUCUCAUCGAUAG |
| MKRN1 | 23,608 | GAGUGGGACUUGUUUCAUG |
| MKRN1 | 23,608 | GCAAGUGGAGGAGUGCUAA |
| MKRN1 | 23,608 | UGACUUGGAUCUAUAGCAA |
| TRIM61 | 391,712 | GGAGAGAGUGGAACUAAUU |
| TRIM61 | 391,712 | UCAGAAAGACCUAGAGCUU |
| TRIM61 | 391,712 | CUGGGUAGUUUGACUGAAA |
| TRIM61 | 391,712 | GGAUCUACAUGAUAGUUUC |
Plasmids and siRNAs
We subcloned HA-tagged TRIM2 (amino acids 1-744) and its truncations, including TRIM2-R (amino acids 1-111), TRIM2-BCC (amino acids 108–321), TRIM2-fn (amino acids 318–472), and TRIM2-NHL (amino acids 469–744), into the pcDNA 3.1 vector. Transfection of the plasmids into cells was performed using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions, in serum-free OptiMEM medium (Gibco). The DNA construct sequences were thoroughly validated through DNA sequencing. For TRIM2 knockdown, the following siRNAs were utilized: TRIM2-si1: 5’-GGACGAUCUUAACCACCAA-3’, TRIM2-si2: 5’-GGGUGUAGCAGUGGAUUUCAA-3’, and siNC: 5’-UUCUCCGAACGGUGUCACGUTT-3’, all procured from Shanghai, China Gene Pharma Ltd. Opti-MEM medium, Lipofectamine® 3000, and P3000TM (Invitrogen) were employed for transfecting cells with siRNA and plasmids according to manufacturer’s instructions after the H295R cells reached 80–90% confluent. The transfection efficiency was detected by quantitative polymerase chain reaction (PCR) at 24 h after transfection.
Measurement of aldosterone concentration
When H295R cells were grown to confluence in 24-multiwell plates, they were transfected with HA-TRIM2 or empty vector for 48 h. Angiotensin II (ANGII) was then added to the media at a concentration of 100 nmol/L, and the cells were incubated for 6 h. Before treating the cells with ANGII, they were incubated in serum-free medium for 24 h. Aldosterone concentrations of the media were thereafter measured by Aldosterone EIA kit (Cayman Chemical, Ann Arbor, MI, USA) according to the manufacturer’s instructions.
Total RNA isolation and qRT-PCR
We isolated total RNA from APA cells using the RNeasy Mini Kit (Cat. #74101, Qiagen, Germany) according to the provided protocol. Subsequently, cDNA synthesis was performed using the ReverTrace qRT-PCR Kit (Toyobo, China) for reverse transcription reactions. Each qRT-PCR reaction consisted of 3 μl primer, 4.5 μl cDNA, and 7.5 μl iQTM SYBR® Green Supermix (Bio-Rad, USA). The primer sequences for qRT-PCR are provided in Table 2. The cycle threshold (Ct) was determined relative to GAPDH.
Table 2.
The specific primer sequences
| ID | Primer | Primer sequences (5’to3’) | Base count |
|---|---|---|---|
| 1 | TRIM2-F | TGCGCCAGATTGACAAGCA | 19 |
| 2 | TRIM2-R | GCACCTCTCGCAGAAAGTG | 19 |
| 3 | CYP11B2-F | GCACCTGCACCTGGAGATG | 19 |
| 4 | CYP11B2-R | CACACACCATGCGTGGTCC | 19 |
| 5 | CYP11B1-F | ACTAGGGCCCATTTTCAGGT | 20 |
| 6 | CYP11B1-R | GGCAGCATCACACACACC | 18 |
| 7 | GAPDH-F | TGCACCACCAACTGCTTAG | 19 |
| 8 | GAPDH-R | GATGCAGGGATGATGTTC | 18 |
Proliferation assays
The MTT test involved seeding 3000 BLCA cells per well in a 96-well plate. Next, 20 μL of MTT solution was added to every well, and the plate was subjected to a 4-hour incubation period. A microplate reader (SpectraMax M2, USA) was used to measure the absorption values at 540 nm after the precipitate was dissolved in DMSO. A colony formation assay was performed for 10 days on a 6-well dish containing 1000 cells per well. Then, 4% paraformaldehyde was used to fix the colonies, and 0.1% crystal violet was used to stain them.
Western blotting
The process of total protein isolation and western blotting commenced with the centrifugation and ice-based lysis of cells for a duration of thirty minutes, employing RIPA buffer supplemented with protease inhibitor and phosphatase inhibitor (Sigma-Aldrich, USA). Subsequently, the cells underwent treatment using an ultrasonic crusher for 10 s followed by centrifugation at 14,000 × g for 10 min. Protein separation and detection procedures were executed in accordance with the methods previously outlined [28, 29]. Finally, immune response bands were visualized using the electromagnetic interference XRS imaging system (Bio-Rad, USA).
Co-immunoprecipitation
Following transfection for 48 h, cells were harvested and subjected to lysis using a cell lysis buffer containing a proteasome inhibitor. The lysis process was conducted on ice for 30 min, followed by centrifugation at 14,000 × g at 4 °C for 10 min. A small aliquot of the resulting supernatant was utilized as Input for western blot analysis. Magnetic beads, bound to the corresponding antibodies, were then added to the remaining supernatant and allowed to incubate at 4 °C with gentle shaking overnight to facilitate the conjugation of antigen-antibody magnetic beads. After the immunoprecipitation reaction, the magnetic bead coupling complex was washed with buffer. Following this, 15 μl of 1 × SDS buffer was added and the mixture was heated at 100 °C for 5 min. The proteins were then separated via SDS-PAGE electrophoresis, followed by western blotting to identify the interacting proteins.
Immunofluorescence
H295R cells underwent a series of procedures: they were initially washed with PBS and subsequently fixed with 4% PFA for 20 min at room temperature. Following fixation, the cells were again washed three times with PBS and then subjected to overnight incubation at 4 °C with the desired antibodies. After another three washes with PBS, the cells were incubated with fluorescein-labeled secondary antibodies for 40 min at 37 °C. Following an additional three washes with PBS, the cells were incubated with DAPI (D8417, Sigma-Aldrich, Germany) at room temperature for 30 min and mounted in mounting medium. The slides were subsequently analyzed using a laser scanning confocal microscope (LSM880, Zeiss, Germany).
Human adrenal tissues and immunohistochemistry
The human adrenal tissue samples used in this study were obtained from primary aldosteronism (PA) patients who underwent lateral adrenalectomy at Zhongnan Hospital of Wuhan University. The diagnosis of PA was determined according to the clinical practice guidelines of the American Endocrine Society [30]. Normal adrenal tissues were obtained from the adjacent tissue within 1 cm of the tumor margins. All adrenal tumors and normal adjacent tissues were pathologically confirmed. Patients provided informed consent for participation in the study. The protein expressions of CYP11B2 and TRIM2 in human aldosterone-producing adenomas (APAs) and normal adrenal tissues were detected using immunohistochemistry (IHC). Initially, fresh tumors were fixed in 4% paraformaldehyde (PFA) for 24 h. Subsequently, they were embedded in paraffin and sectioned into 5 μm slices. The sections were then subjected to IHC staining using the CYP11B2 antibody (MABS1251, Millipore, dilution 1:200) and the TRIM2 antibody (20356-1-AP, Proteintech, dilution 1:200). The DAB chromogen was applied for incubation, followed by counterstaining with hematoxylin. Finally, the IHC-detected images were scanned and analyzed using a molecular microscope (Olympus BX53). In this study, tissue microarrays (TMAs) and IHC analysis were also utilized to assess protein expression and distribution, which have been widely applied in previous studies [28]. The IHC staining scores were evaluated using the following criteria: 0 indicated negative staining, 1 indicated weak positive staining, 2 indicated moderate positive staining, and 3 indicated strong positive staining. The positivity rate was assessed based on the percentage of cells showing positive staining, with the scoring as follows: 0 indicated negative staining, 1–25% scored 1, 26–50% scored 2, 51–75% scored 3, and 76–100% scored 4. To calculate the total staining score, the staining intensity score was multiplied by the staining positivity rate score. The immunohistochemical grading was performed in a blinded manner by two independent pathologists.
Mass spectrometry analysis
After transfecting plasmids into H295R cells, the cells were incubated for 48 h and IP was performed. After the immunoprecipitation reaction, the magnetic bead coupling complex was washed with cold PBS buffer. Mass spectrometry analysis was conducted using timsTOF Pro (Bruker, USA). The mass spectrometry data were processed using MaxQuant (V1.6.6) software and the Andromeda database search algorithm. The Proteome Reference Database for Humans in UniProt was used for the search.
Molecular docking
Molecular docking is utilized to validate the binding activity between proteins or between proteins and small molecules. The HDOCK online platform (http://hdock.phys.hust.edu.cn/) was employed for molecular docking in this study. HDOCK can analyze different conformations of protein-protein docking, binding activities under different conformations, and amino acid residues within 5Å of interaction distance. Proteins with UniProt IDs P19099 and Q9C040, corresponding to CYP11B2 and TRIM2, were downloaded from the UniProt database and subjected to docking analysis. Ligplus software can be used to analyze the two-dimensional interactions between the two proteins. PyMOL (version 4.3.0) software is employed for visualizing the amino acid residues involved in the interaction between the two proteins.
Statistical analyses
The data are presented as the mean ± standard deviation (SD) of three independent experiments. The two-tailed t-test was used to analyze normally distributed data, while non-parametric Kruskal-Wallis test was used for non-normally distributed data. Data with more than two groups using one-way ANOVA. Spearman’s test was employed to investigate the correlations between variables. All statistical analyses were performed using GraphPad Prism 9.00 software. Image analysis and quantification were conducted using ImageJ v1.45 software. In statistical analysis, p-values less than 0.05 were considered statistically significant.
Results
TRIM2 inhibits CYP11B2 protein level and aldosterone production
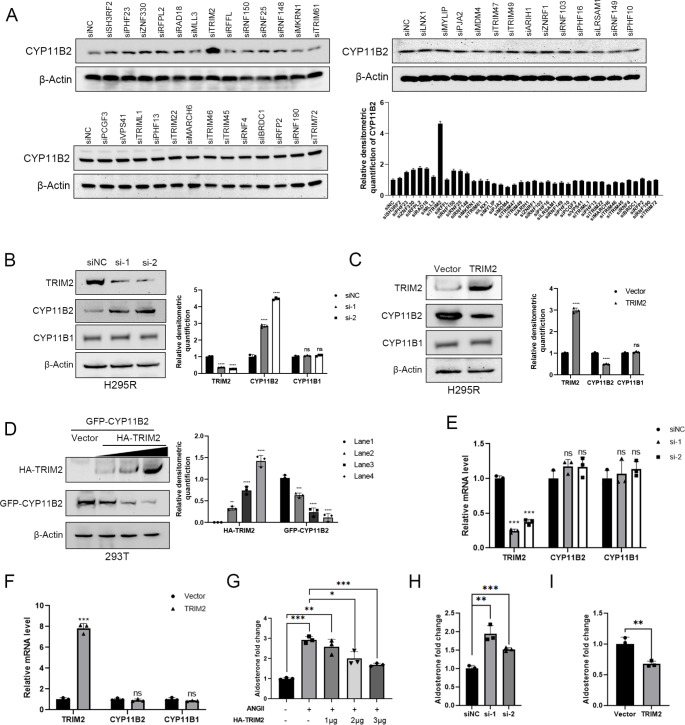
To systematically identify the E3 ligases regulating CYP11B2, we randomly selected 39 E3 ligases from an siRNA library and conducted an unbiased siRNA screen to monitor the levels of CYP11B2. Among these E3 ligases, we observed that TRIM2 could potentially serve as a candidate for ubiquitinating and destabilizing CYP11B2, as siRNA-mediated depletion of TRIM2 significantly increased the abundance of CYP11B2 (Fig. 1A). We then used two non-overlapping siRNAs to confirm that knockdown of TRIM2 increased the protein level of CYP11B2, and this effect was specific to CYP11B2 without affecting CYP11B1 expression(Fig. 1B). Conversely, TRIM2 overexpression reduced CYP11B2 protein expression, while having no impact on CYP11B1 expression (Fig. 1C). Similarly, overexpression of wild-type TRIM2 significantly decreased the expression of CYP11B2 in a dose-dependent manner (Fig. 1D). However, RT-PCR analysis revealed that TRIM2 does not alter the mRNA abundance of CYP11B2 (Fig. 1E-F). Furthermore, we found that overexpression of TRIM2 could inhibit aldosterone production induced by ANGII in H295R cells (Fig. 1G). Additionally, we examined both TRIM2 knockdown and overexpression in H295R cells under basal conditions. These results show that TRIM2 knockdown increases aldosterone levels (Fig. 1H), while overexpression significantly reduces aldosterone production (Fig. 1I). We also confirmed that TRIM2 overexpression had no effect on H295R cell proliferation, ruling out potential toxicity effects (Figure S1A-C).
Fig. 1.
TRIM2 depletion increases CYP11B2 protein expression in adrenal tumor cells. A Specific E3 ligases were knocked down by siRNA transfection, followed by protein immunoblot analysis of CYP11B2 in H295R cells. Cells were seeded into a 96-well plate, and then ON-TARGET plus siRNA library (final siRNA concentration of 50 nM) mixed with Lipofectamine 3000 and Opti-MEM was added to each well. B TRIM2 depletion increased the expression of CYP11B2 but had no effect on CYP11B1 protein expression using two different siRNA oligos in H295R cells. C TRIM2 overexpression decreased CYP11B2 protein expression in H295R cells. D In 293T cells, after transfecting with different doses of GFP-CYP11B2 and HA-TRIM2 plasmids, cells were cultured for 48 h. Western blotting was performed using anti-GFP antibody to determine the levels of exogenous CYP11B2. E-F TRIM2 depletion or overexpression have no effects on the expression of CYP11B2 and CYP11B1 mRNA level using two different siRNAs. G Aldosterone inhibition by TRIM2 overexpression in the presence of 100 nmol/L Angiotensin II (ANGII, an aldosterone activator). H Aldosterone were detected after TRIM2 knckdown using two distinct siRNAs without ANGII treatment. I Aldosterone were detected after TRIM2 overexpression without ANGII treatment. Aldosterone levels were described as fold changes from control group. Statistical significance was assessed using two-tailed t-tests. Error bars represent the standard deviations of three independent experiments. *P value < 0.05; **P value < 0.01; ***P value < 0.001; ****P value < 0.0001
TRIM2 regulates CYP11B2 stability in a ubiquitin-proteasome-dependent manner
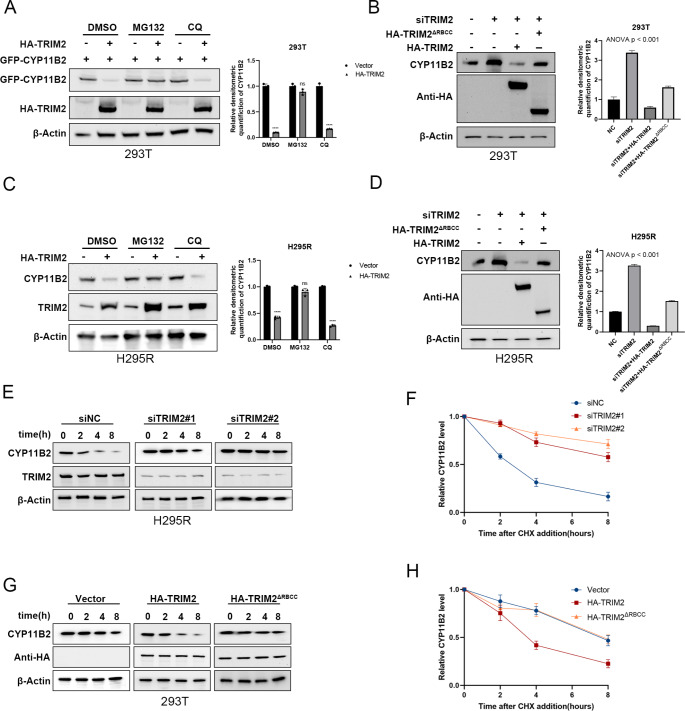
We hypothesize that TRIM2 may control the turnover of CYP11B2 through the ubiquitin-proteasome pathway since overexpression of TRIM2 reduces the protein levels without affecting the mRNA levels. We transfected 293T cells with GFP-tagged CYP11B2 plasmids and HA-tagged TRIM2 plasmids. We observed that the proteasome inhibitor MG132 partially inhibited TRIM2-mediated degradation of CYP11B2, while the autophagy inhibitor chloroquine (CQ) had no inhibitory effect (Fig. 2A). The catalytically inactive mutant ΔRBCC (TRIM2ΔRBCC) did not reduce CYP11B2 protein levels, indicating that TRIM2 decreases CYP11B2 protein levels through its E3 ligase activity (Fig. 2B). Furthermore, in H295R cells treated with the proteasome inhibitor MG132, CYP11B2 protein levels were not influenced by TRIM2, confirming that TRIM2 regulates CYP11B2 degradation through the ubiquitin-proteasome pathway (Fig. 2C). Similarly, overexpression of wild-type (WT) TRIM2, but not its catalytically inactive mutant (TRIM2ΔRBCC), could reverse the increase in CYP11B2 caused by TRIM2 knockdown (Fig. 2B and D). To further demonstrate that TRIM2 affects the stability of CYP11B2, H295R and 293T cells were treated with cycloheximide (CHX) for 0, 2, 4, and 8 h. In cells lacking TRIM2, the half-life of CYP11B2 was prolonged. Overexpression of TRIM2-WT, but not TRIM2ΔRBCC, reduced the stability of CYP11B2 (Fig. 2E-F). These results indicate that TRIM2 destabilizes CYP11B2 through the ubiquitin-proteasome pathway.
Fig. 2.
TRIM2 regulates CYP11B2 stability in a ubiquitin-proteasome-dependent manner. A GFP-CYP11B2 and HA-TRIM2 were transfected into 293T cells for 24 h, then the cells were treated with dimethyl sulphoxide (DMSO), 10 μM MG132 (#S2619, Selleck) or 20 μM chloroquine (CQ, #S8808, Selleck) for 8 h. The cells were collected and anti-GFP immunoreactivity were detected by western blot. B 293T cells were transfected with TRIM2 (wild type or TRIM2ΔRBCC) together with TRIM2 siRNA. The CYP11B2 level were measured. C HA-TRIM2 were transfected into H295R cells for 24 h, then the cells were treated with dimethyl sulphoxide (DMSO), 10 μM MG132 (#S2619, Selleck) or 20 μM chloroquine (CQ, #S8808, Selleck) for 8 h. The cells were collected and anti-GFP immunoreactivity were detected by western blot. D H295R cells were transfected with TRIM2 (wild type or TRIM2ΔRBCC) together with TRIM2 siRNA. The CYP11B2 level was measured. E-F HA-TRIM2 plasmid was transfected into H295R cells, and then 100 μg/ml cycloheximide (CHX, #S7418, Selleck) was respectively added at the specified time points for 0 h, 4 h, 8 h, and 12 h. Then, the cells were harvested, and western blot determined the half-life of CYP11B2 protein. G-H HA-TRIM2 plasmid was transfected into 293T cells, and then 100 μg/ml cycloheximide (CHX, #S7418, Selleck) was respectively added at the specified time points for 0 h, 2 h, 4 h, and 8 h. Western blot detected GFP antibodies to determine the half-life of GFP-CYP11B2 protein. Statistical significance was assessed using two-tailed t-tests. Error bars represent the standard deviations of three independent experiments. *P value < 0.05; **P value < 0.01; ***P value < 0.001; ****P value < 0.0001
TRIM2 interacts with CYP11B2
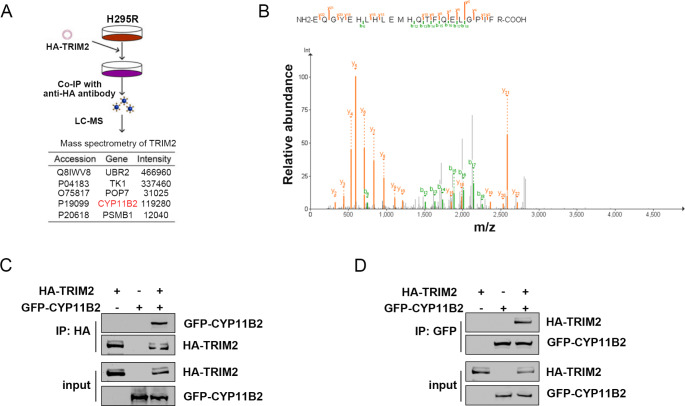
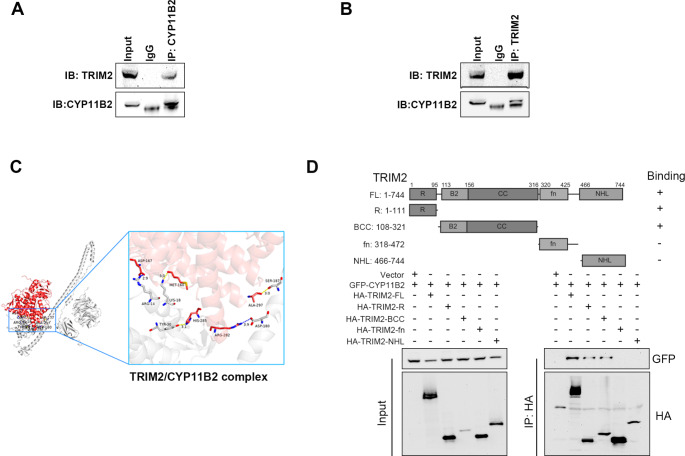
Firstly, we conducted mass spectrometry analysis of HA-TRIM2-overexpressing proteins in H295R cells to elucidate the precise molecular mechanism by which TRIM2 regulates the ubiquitination of CYP11B2. We found that CYP11B2 was included in the list of proteins interacting with TRIM2 (Fig. 3A-B). Upon overexpression of these two proteins in 293T cells, the interaction between HA-TRIM2 and GFP-CYP11B2 was confirmed (Fig. 3C-D). Subsequently, TRIM2 and CYP11B2 were localized by immunofluorescence analysis. Moreover, endogenous TRIM2 interacted with CYP11B2 in H295R cells (Fig. 4A-B). In immunostained adrenocortical tumor cells, CYP11B2 and TRIM2 were co-localized (Figure S2A). By constructing a series of truncation plasmids, we were able to determine the specific binding region between TRIM2 and CYP11B2. It has been reported that TRIM proteins share a common structural feature, the TRIM or RBCC motif, which contains a conserved RING domain responsible for ubiquitin transfer. The protein molecular docking model indicates that the RBCC domain of TRIM2 can interact with CYP11B2 (Fig. 4C). To identify the key structural domains involved in this interaction, we constructed HA-tagged truncations of TRIM2 and investigated the interaction of each truncation with CYP11B2 (Fig. 4D). In this study, we found that the RBCC domain, which includes the RING-type domain, is crucial for maintaining this interaction.
Fig. 3.
TRIM2 interacts with CYP11B2. A Flow chart analyzing the proteins interacting with TRIM2 in H295R cells by mass spectrometry. B Liquid chromatography-tandem mass spectrometry analysis of CYP11B2-associated peptides corresponding to HA-TRIM2. C-D 293T cells were transfected with the described plasmids for 48 h, and co-IP was performed with anti-GFP or anti-HA antibodies
Fig. 4.
RBCC domain of TRIM2 interacts with CYP11B2. A-B Co-IP assay showing that endogenous CYP11B2 interacts with TRIM2 in H295R cells. C Docking model of the CYP11B2/TRIM2 complex. D 293T cells were transfected with the described plasmids for 48 h, and co-IP was performed with anti-HA antibody
TRIM2 ubiquitylates CYP11B2
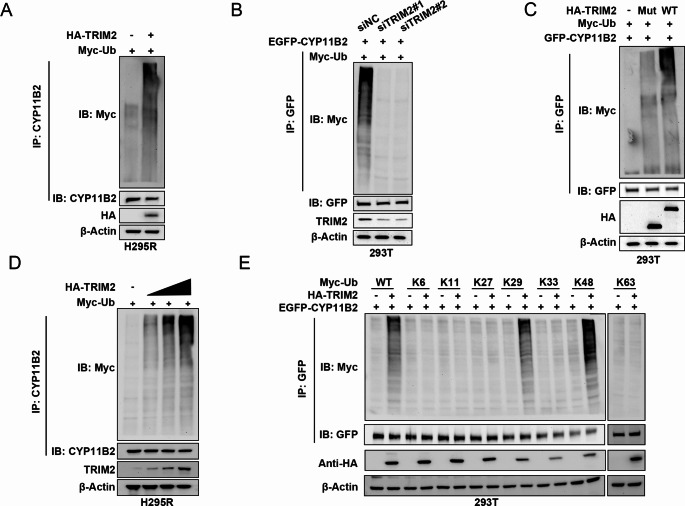
As TRIM2 is a ubiquitin ligase, we further investigated whether CYP11B2 is a substrate of TRIM2. As shown in Fig. 5A, overexpression of TRIM2 significantly increased the ubiquitination levels of CYP11B2. Conversely, knockdown of TRIM2 markedly reduced the ubiquitination levels of CYP11B2 (Fig. 5B). Moreover, ectopic expression of TRIM2-WT (but not TRIM2ΔRBCC) significantly increased the ubiquitination of CYP11B2 in cells (Fig. 5C). In vivo ubiquitination assays indicated that TRIM2 dose-dependently increased the ubiquitination levels of CYP11B2 (Fig. 5D). To further investigate which type of ubiquitin chain TRIM2 adds from CYP11B2, we conducted ubiquitination assays using different types of mutant ubiquitins (K6, K11, K27, K29, K33, K48, and K63). The results indicated that TRIM2 can efficiently add K29 and K48-linked ubiquitin chains to CYP11B2 protein (Fig. 5E). These findings suggest that TRIM2 may act as a CYP11B2-directed E3 ligase, leading to ubiquitination and destabilization of CYP11B2.
Fig. 5.
TRIM2 polyubiquitylates CYP11B2. A H295R cells transfected with the empty vector and HA-TRIM2 for 48 h before collection. CYP11B2 was immunoprecipitated with anti-CYP11B2 and immunoblotted with anti-Myc. B Immunoblotting was used to detect the ubiquitination of CYP11B2 in 293T cells co-transfected with EGFP-CYP11B2, Myc-Ub, and siTRIM2. C Immunoblotting to detect the ubiquitination of CYP11B2 in HEK293 cells co-transfected with GFP-CYP11B2, Myc-Ub and HA-TRIM2 (wild type or RBCC domain deletion). D TRIM2 promoted the ubiquitin chain of CYP11B2 in a dose-dependent manner. E Myc-WT, K6, K11, K27, K29, K33, K48 or K63 Ub were co-transfected with EGFP-CYP11B2 and HA-TRIM2 into 293T cells. After 48 h, cell lysates were subjected to ubiquitination assay and the ubiquitination level of CYP11B2 detected by Myc antibody
TRIM2 and CYP11B2 expression are negatively correlated in APA samples
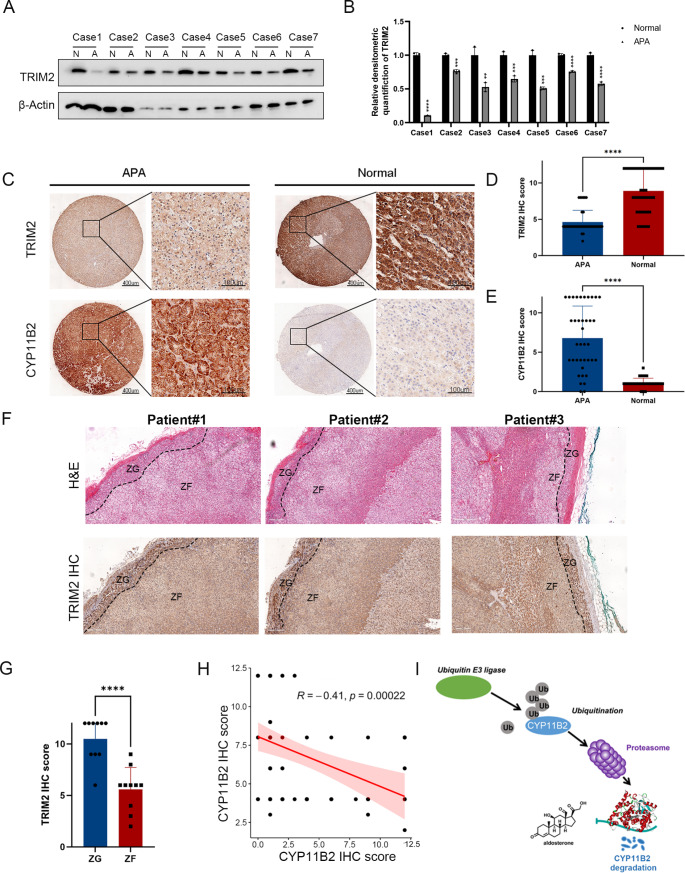
To clarify the expression of TRIM2 in APA tissues, we obtained 7 pairs of human APA tissues and paired normal tissues adjacent to the tumor. We performed western blot and found that compared with paired normal tissues, the expression of TRIM2 in APA tissues was reduced (Fig. 6A-B). To further investigating the relationship between TRIM2 and CYP11B2 in APA samples, we determined the expression levels of these two proteins in APA tissues and normal tissues. In APA samples, protein levels of TRIM2 were significantly lower than those in normal tissues as determined by immunohistochemistry (Fig. 6C-E). We performed immunohistochemistry staining of normal tissues adjacent to the tumor, specifically examining TRIM2 expression in both the zona glomerulosa (ZG) and zona fasciculata (ZF). Interestingly, our new results reveal that TRIM2 expression is notably higher in the ZG compared to the ZF (Fig. 6F-G), which aligns well with its proposed role in regulating aldosterone synthesis. Furthermore, there was a significant negative correlation between the protein levels of TRIM2 and CYP11B2 in APA tissues (Fig. 6H). Our results also demonstrate that CYP11B2 ubiquitination levels are higher in normal adjacent tissues compared to APA tissues (Figure S2B). These results support the notion that TRIM2 and CYP11B2 may be associated with regulatory relationships.
Fig. 6.
TRIM2 correlates with CYP11B2 protein levels in human APA samples. A-B Total protein was extracted from APA tissues (APA) and paired normal tissues adjacent to the tumor (Normal), and the expression of TRIM2 was detected by western blot. β-Actin was used as an internal reference. C-E 77 samples were used for IHC analysis, including normal tissues adjacent to the tumor (n = 39) and APA tissues (n = 38). Specific primary antibodies against CYP11B2 (MABS1251, Millipore), TRIM2 (Proteintech, 20356-1-AP) were used for IHC. Statistical graphs were used to show the total staining scores of TRIM2 and CYP11B2 in APA and normal tissues. The staining scoring method can be found in Materials and Methods. F 10 normal samples were used for H&E and IHC analysis. Specific primary antibodies against TRIM2 (Proteintech, 20356-1-AP) were used for IHC. G Statistical graphs were used to show the total staining scores of TRIM2 in zona glomerulosa (ZG) and zona fasciculata (ZF). H Correlation analysis showed that the expression of CYP11B2 negatively correlated with TRIM2 in human APA samples. I The mechanism diagram of the study. TRIM2 is a E3 ubiquitin ligase that destabilizes CYP11B2 and inhibits the production of aldosterone though its ubiquitylation activity. The n number represents n biologically independent experiments in each group. The data are presented as the mean ± SD (bar plots). *P value < 0.05; **P value < 0.01; ***P value < 0.001
Discussion
Adrenocortical adenomas (APAs) that produce aldosterone are the primary cause of clinically diagnosed primary aldosteronism. However, the mechanisms underlying the occurrence of APAs remain largely unknown. Post-translational modifications (PTMs) play a pivotal role in regulating protein expression by modulating their function, stability, and interactions within the cell, making them essential components of cellular biology regulation [31]. Among PTMs, ubiquitination stands out as an important mechanism contributing significantly to cellular homeostasis and protein degradation [32]. However, there is currently limited literature on the degradation mechanism of CYP11B2. Our study revealed that TRIM2 interacts with and ubiquitinates CYP11B2, leading to its degradation by the proteasome. Knockdown of TRIM2 increased the level of CYP11B2 and aldosterone secretion in adrenal cells, while overexpression of TRIM2 has the opposite effect. These findings suggest that TRIM2 may critically regulate aldosterone synthesis by targeting CYP11B2 degradation.
While our findings highlight TRIM2’s significant role in CYP11B2 regulation, it is important to acknowledge that protein homeostasis typically involves multiple regulatory mechanisms [33]. Our siRNA library screen, while identifying TRIM2 as a key regulator, also suggested the potential involvement of other E3 ligases in CYP11B2 regulation. Furthermore, other post-translational modifications such as phosphorylation [34] and SUMOylation [35] might work in concert with ubiquitination to fine-tune CYP11B2 levels. This multi-layered regulation could explain why TRIM2 manipulation alone does not completely abolish aldosterone production, suggesting the existence of compensatory mechanisms.
Significantly, our study revealed that TRIM2 displays differential expression between the zona glomerulosa (ZG) and zona fasciculata (ZF) of normal tissues adjacent to adrenal tumor. The enrichment of TRIM2 in the ZG, where CYP11B2 is primarily expressed, suggests an evolved regulatory mechanism to maintain appropriate aldosterone production under normal conditions. This spatial organization might serve as a crucial checkpoint preventing excessive aldosterone synthesis [36]. The downregulation of TRIM2 in APAs disrupts this homeostatic mechanism, potentially contributing to autonomous aldosterone production.
The therapeutic implications of our findings are particularly noteworthy in the context of primary aldosteronism treatment. Current treatment options are limited, primarily consisting of mineralocorticoid receptor antagonists or surgical intervention for unilateral disease [1]. The identification of TRIM2 as a negative regulator of aldosterone production opens new therapeutic possibilities. Developing TRIM2-specific activators could provide a novel approach for medical treatment of primary aldosteronism, particularly beneficial for patients who are poor surgical candidates or those with bilateral disease.
Recent advances in therapeutic approaches targeting protein degradation pathways offer promising directions for translating our findings [37]. The success of proteolysis-targeting chimeras (PROTACs) in cancer treatment demonstrates the feasibility of therapeutically manipulating protein degradation pathways [38]. Similar strategies could be explored for enhancing TRIM2-mediated degradation of CYP11B2. However, several challenges need to be addressed, including achieving tissue specificity to avoid off-target effects, as TRIM2 is expressed in multiple tissues and has other known substrates. Additionally, the development of small molecule activators of E3 ligases remains technically challenging [39], although recent advances in this field are encouraging.
Future research directions should focus on several key areas. First, comprehensive proteomics studies could identify additional post-translational modifications of CYP11B2 and their potential interplay with TRIM2-mediated regulation. Second, structural studies of the TRIM2-CYP11B2 interaction could facilitate the design of specific therapeutic modulators. Third, investigation of potential resistance mechanisms to TRIM2-mediated regulation could help predict and overcome therapeutic limitations. Finally, development of biomarkers based on TRIM2 expression or activity might help in patient stratification and treatment selection.
In conclusion, we demonstrated that TRIM2 functions as a crucial E3 ubiquitin ligase for CYP11B2, destabilizing CYP11B2 and inhibiting aldosterone synthesis through its ubiquitination activity. This finding not only provides new insights into the regulatory mechanisms of aldosterone production but also reveals TRIM2’s potential as a therapeutic target for primary aldosteronism treatment. The zone-specific expression pattern of TRIM2 and its dysregulation in APAs suggest a fundamental role in maintaining aldosterone homeostasis, while the possibility of therapeutic targeting opens new avenues for treating primary aldosteronism.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank Yayun Fang, Dany Shan, and others for their excellent technical help. This study was supported by the Youth Interdisciplinary Special Fund of Zhongnan Hospital of Wuhan University (Grant No. ZNQNJC2023012), Excellent Doctoral Program of Zhongnan Hospital of Wuhan University (ZNYB2020006), Science and Technology Innovation Cultivation Fund of Zhongnan Hospital of Wuhan University (CXYP2022011).
Data availability
All the data and supporting materials are available within the article and Supplemental information.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Zhongnan Hospital of Wuhan University (approval number: 2020014), and All patients had signed the informed consents before the study.
Consent for publication
All the authors agree with the publication of the manuscript.
Conflict of interest
The authors declare that there is no conflict of interests
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Liang Chen and Xuan Hu contributed equally to this work.
Contributor Information
Yong Li, Email: liyongstudent@126.com.
Wan Xiang, Email: xiangwan@whu.edu.cn.
Zhe Meng, Email: mengzhe@whu.edu.cn.
References
- 1.Reincke M, Bancos I, Mulatero P, Scholl UI, Stowasser M, Williams TA (2021) Diagnosis and treatment of primary aldosteronism. Lancet Diabetes Endocrinol 9(12):876–892. 10.1016/s2213-8587(21)00210-2 [DOI] [PubMed] [Google Scholar]
- 2.Schrier RW (2006) Water and sodium retention in edematous disorders: role of vasopressin and aldosterone. Am J Med 119(7 Suppl 1):S47–53. 10.1016/j.amjmed.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 3.Yang T, He M, Hu C (2018) Regulation of aldosterone production by ion channels: from basal secretion to primary aldosteronism. Biochim Biophys Acta Mol Basis Dis 1864(3):871–881. 10.1016/j.bbadis.2017.12.034 [DOI] [PubMed] [Google Scholar]
- 4.Turcu AF, Yang J, Vaidya A (2022) Primary aldosteronism - a multidimensional syndrome. Nat Rev Endocrinol 18(11):665–682. 10.1038/s41574-022-00730-2 [DOI] [PubMed] [Google Scholar]
- 5.Hellman DE, Kartchner M, Komar N, Mayes D, Pitt M (1980) Hyperaldosteronism, hyperparathyroidism, medullary sponge kidneys, and hypertension. JAMA 244(12):1351–1353 [PubMed] [Google Scholar]
- 6.Vaidya A, Mulatero P, Baudrand R, Adler GK (2018) The expanding spectrum of primary aldosteronism: implications for diagnosis, pathogenesis, and treatment. Endocr Rev 39(6):1057–1088. 10.1210/er.2018-00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milliez P, Girerd X, Plouin PF, Blacher J, Safar ME, Mourad JJ (2005) Evidence for an increased rate of cardiovascular events in patients with primary aldosteronism. J Am Coll Cardiol 45(8):1243–1248. 10.1016/j.jacc.2005.01.015 [DOI] [PubMed] [Google Scholar]
- 8.Rainey WE (1999) Adrenal zonation: clues from 11beta-hydroxylase and aldosterone synthase. Mol Cell Endocrinol 151(1–2):151–160. 10.1016/s0303-7207(99)00051-9 [DOI] [PubMed] [Google Scholar]
- 9.Curnow KM, Tusie-Luna MT, Pascoe L, Natarajan R, Gu JL, Nadler JL et al (1991) The product of the CYP11B2 gene is required for aldosterone biosynthesis in the human adrenal cortex. Mol Endocrinol 5(10):1513–1522. 10.1210/mend-5-10-1513 [DOI] [PubMed] [Google Scholar]
- 10.Oki K, Plonczynski MW, Luis Lam M, Gomez-Sanchez EP, Gomez-Sanchez CE (2012) Potassium channel mutant KCNJ5 T158A expression in HAC-15 cells increases aldosterone synthesis. Endocrinology 153(4):1774–1782. 10.1210/en.2011-1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beuschlein F, Boulkroun S, Osswald A, Wieland T, Nielsen HN, Lichtenauer UD et al (2013) Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension. Nat Genet 45(4):440. 10.1038/ng.2550 [DOI] [PubMed] [Google Scholar]
- 12.Scholl UI, Goh G, Stölting G, de Oliveira RC, Choi M, Overton JD et al (2013) Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat Genet 45(9):1050–1054. 10.1038/ng.2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattangady NG, Karashima S, Yuan L, Ponce-Balbuena D, Jalife J, Gomez-Sanchez CE et al (2016) Mutated KCNJ5 activates the acute and chronic regulatory steps in aldosterone production. J Mol Endocrinol 57(1):1–11. 10.1530/jme-15-0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Te Riet L, van Esch JH, Roks AJ, van den Meiracker AH, Danser AH (2015) Hypertension: renin-angiotensin-aldosterone system alterations. Circ Res 116(6):960–975. 10.1161/circresaha.116.303587 [DOI] [PubMed] [Google Scholar]
- 15.Burrello J, Buffolo F, Domenig O, Tetti M, Pecori A, Monticone S et al (2020) Renin-angiotensin-aldosterone system triple-a analysis for the screening of primary aldosteronism. Hypertension 75(1):163–172. 10.1161/hypertensionaha.119.13772 [DOI] [PubMed] [Google Scholar]
- 16.Demura M, Wang F, Yoneda T, Karashima S, Mori S, Oe M et al (2011) Multiple noncoding exons 1 of nuclear receptors NR4A family (nerve growth factor-induced clone B, Nur-related factor 1 and neuron-derived orphan receptor 1) and NR5A1 (steroidogenic factor 1) in human cardiovascular and adrenal tissues. J Hypertens 29(6):1185–1195. 10.1097/HJH.0b013e32834626bb [DOI] [PubMed] [Google Scholar]
- 17.Zhai YS, Li J, Peng L, Lu G, Gao X (2022) Phosphorylation of CaMK and CREB-mediated cardiac aldosterone synthesis induced by arginine vasopressin in rats with myocardial infarction. Int J Mol Sci 23(23). 10.3390/ijms232315061 [DOI] [PMC free article] [PubMed]
- 18.Seidel E, Schewe J, Scholl UI (2019) Genetic causes of primary aldosteronism. Exp Mol Med 51(11):1–12. 10.1038/s12276-019-0337-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams TA, Monticone S, Schack VR, Stindl J, Burrello J, Buffolo F et al (2014) Somatic ATP1A1, ATP2B3, and KCNJ5 mutations in aldosterone-producing adenomas. Hypertension 63(1):188–195. 10.1161/hypertensionaha.113.01733 [DOI] [PubMed] [Google Scholar]
- 20.Tan GC, Negro G, Pinggera A, Tizen Laim NMS, Mohamed Rose I, Ceral J et al (2017) Aldosterone-producing adenomas: histopathology-genotype correlation and identification of a novel CACNA1D mutation. Hypertension 70(1):129–136. 10.1161/hypertensionaha.117.09057 [DOI] [PubMed] [Google Scholar]
- 21.Reimer EN, Walenda G, Seidel E, Scholl UI (2016) CACNA1H(M1549V) mutant calcium channel causes autonomous aldosterone production in HAC15 cells and is inhibited by mibefradil. Endocrinology 157(8):3016–3022. 10.1210/en.2016-1170 [DOI] [PubMed] [Google Scholar]
- 22.Aguiar M, Masse R, Gibbs BF (2005) Regulation of cytochrome P450 by posttranslational modification. Drug Metab Rev 37(2):379–404. 10.1081/dmr-46136 [DOI] [PubMed] [Google Scholar]
- 23.He BB, Liu J, Cheng Z, Liu R, Zhong Z, Gao Y et al (2023) Bacterial cytochrome P450 catalyzed post-translational macrocyclization of ribosomal peptides. Angew Chem Int Ed Engl 62(46):e202311533. 10.1002/anie.202311533 [DOI] [PubMed] [Google Scholar]
- 24.Gomez A, Nekvindova J, Travica S, Lee MY, Johansson I, Edler D et al (2010) Colorectal cancer-specific cytochrome P450 2W1: intracellular localization, glycosylation, and catalytic activity. Mol Pharmacol 78(6):1004–1011. 10.1124/mol.110.067652 [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Lin X, Jiang Y, Qin X, Ma N, Yao F et al (2023) Engineering cytochrome P450BM3 enzymes for direct nitration of unsaturated hydrocarbons. Angew Chem Int Ed Engl 62(13):e202217678. 10.1002/anie.202217678 [DOI] [PubMed] [Google Scholar]
- 26.Correia MA, Wang Y, Kim SM, Guan S (2014) Hepatic cytochrome P450 ubiquitination: conformational phosphodegrons for E2/E3 recognition? IUBMB Life 66(2):78–88. 10.1002/iub.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T, Harada N (2014) Post-translational dual regulation of cytochrome P450 aromatase at the catalytic and protein levels by phosphorylation/dephosphorylation. Febs j 281(21):4830–4840. 10.1111/febs.13021 [DOI] [PubMed] [Google Scholar]
- 28.Liu T, Xiang W, Chen Z, Wang G, Cao R, Zhou F et al (2023) Hypoxia-induced PLOD2 promotes clear cell renal cell carcinoma progression via modulating EGFR-dependent AKT pathway activation. Cell Death Dis 14(11):774. 10.1038/s41419-023-06298-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Chen L, Zhou F, He Z, Wang G, Luo Y (2023) NRP1 promotes prostate cancer progression via modulating EGFR-dependent AKT pathway activation. Cell Death Dis 14(2):159. 10.1038/s41419-023-05696-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaidya A, Carey RM (2020) Evolution of the primary aldosteronism syndrome: updating the approach. J Clin Endocrinol Metab. 105(12):3771-83. 10.1210/clinem/dgaa606 [DOI] [PMC free article] [PubMed]
- 31.Lee JM, Hammarén HM, Savitski MM, Baek SH (2023) Control of protein stability by post-translational modifications. Nat Commun 14(1):201. 10.1038/s41467-023-35795-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swatek KN, Komander D (2016) Ubiquitin modifications. Cell Res 26(4):399–422. 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koga H, Kaushik S, Cuervo AMJA (2011) Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res Rev 10(2):205–215. 10.1016/j.arr.2010.02.001 [DOI] [PMC free article] [PubMed]
- 34.Oesch-Bartlomowicz B, Oesch FJB, communications br (2005) Phosphorylation of cytochromes P450: first discovery of a posttranslational modification of a drug-metabolizing enzyme. Biochem Biophys Res Commun 338(1):446–449. 10.1016/j.bbrc.2005.08.092 [DOI] [PubMed]
- 35.Tomasi ML, Ramani K, Ryoo M, Cossu C, Floris A, Murray BJ et al (2018) SUMOylation regulates cytochrome P450 2E1 expression and activity in alcoholic liver disease. Febs j 32(6):3278. 10.1111/febs.13021 [DOI] [PMC free article] [PubMed]
- 36.Pawlonka J, Buchalska B, Buczma K, Borzuta H, Kamińska K, Cudnoch-Jędrzejewska A (2024) Targeting the renin-angiotensin-aldosterone system (RAAS) for cardiovascular protection and enhanced oncological outcomes: review. Curr Treat Options Oncol 25(11):1406–1427. 10.1007/s11864-024-01270-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nalawansha DA, Crews CM (2020) PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem biology 27(8):998–1014. 10.1016/j.chembiol.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alabi SB, Crews CM (2021) Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J Biol Chem 296:100647. 10.1016/j.jbc.2021.100647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Słabicki M, Yoon H, Koeppel J, Nitsch L, Roy Burman SS, Di Genua C et al (2020) Small-molecule-induced polymerization triggers degradation of BCL6. Nature 588(7836):164–168. 10.1038/s41586-020-2925-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data and supporting materials are available within the article and Supplemental information.