Abstract
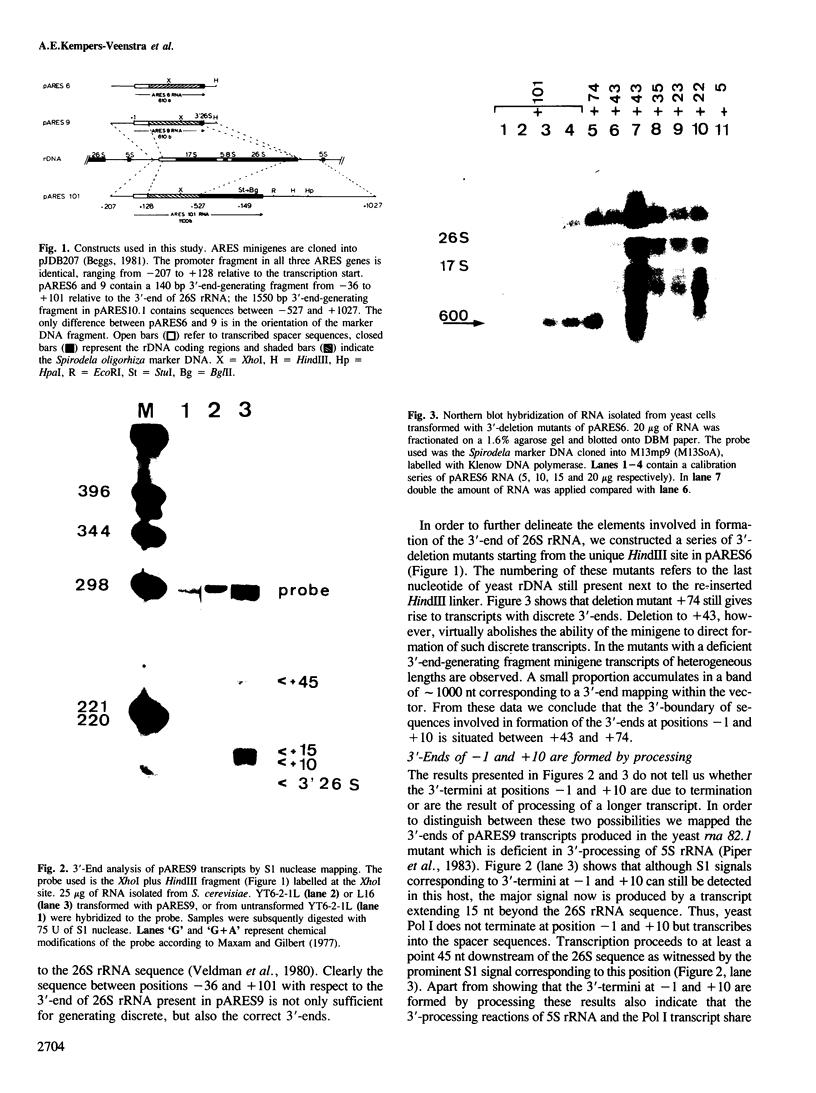
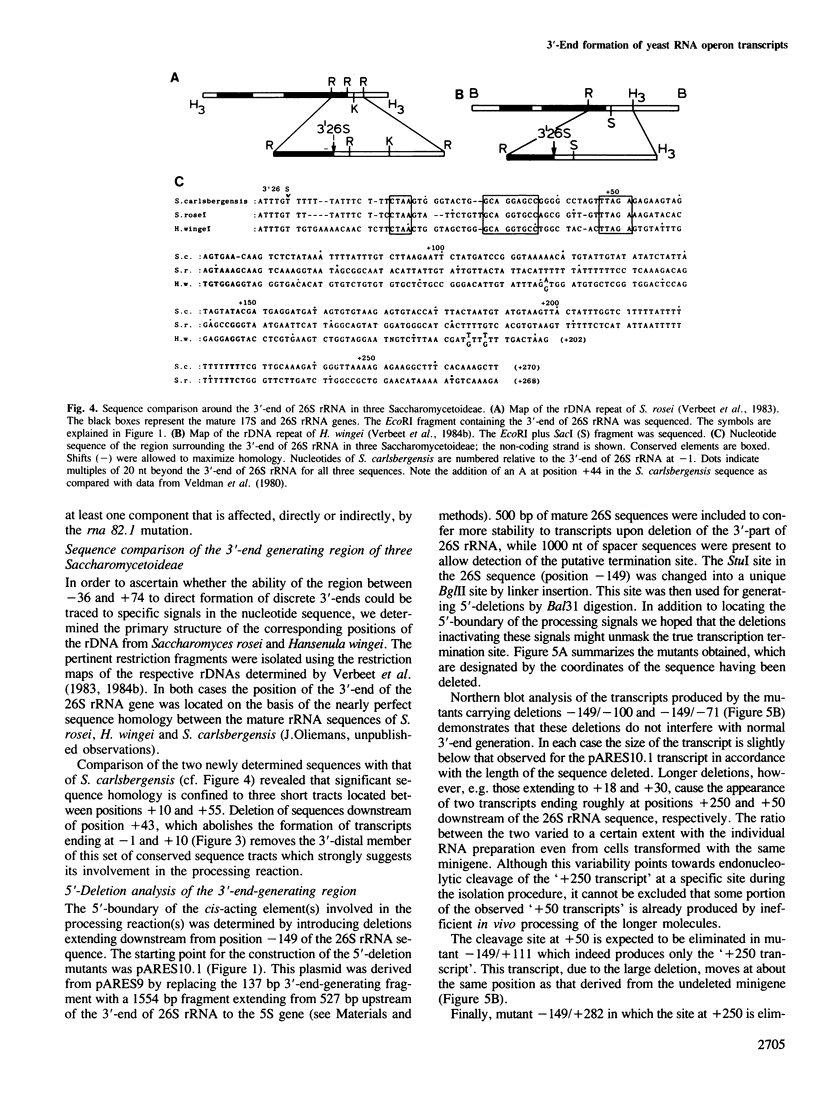
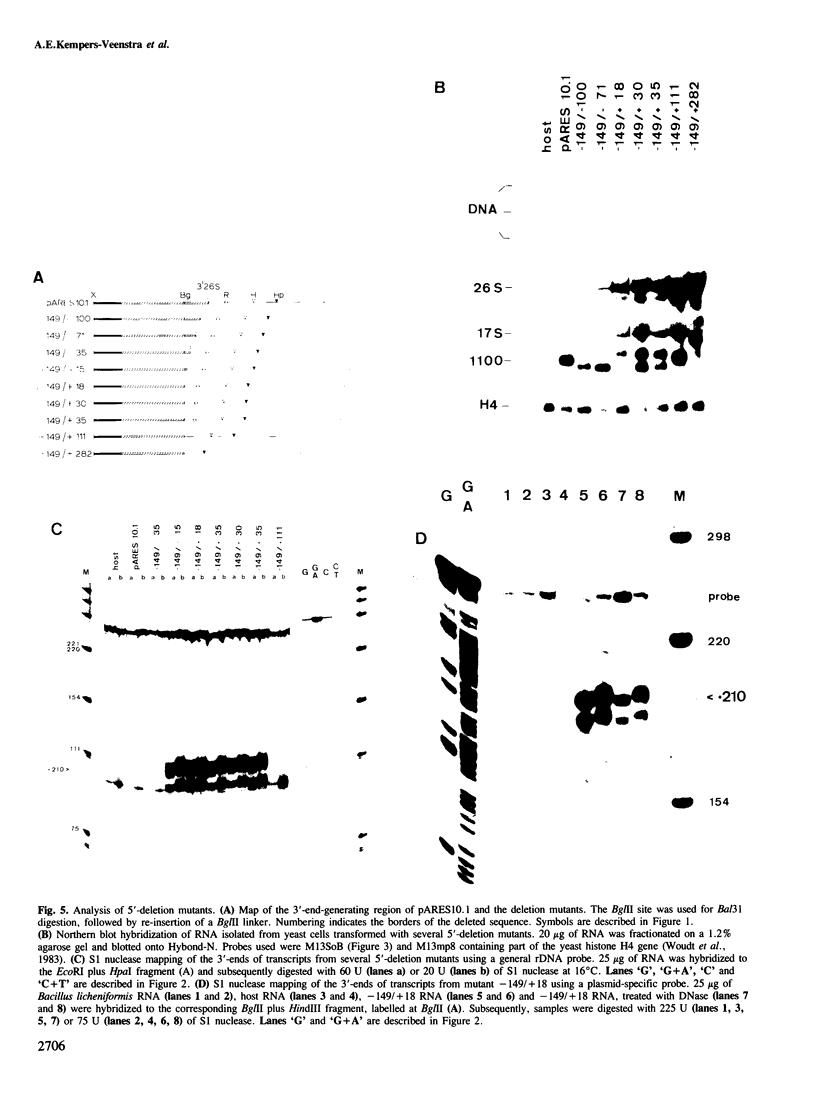
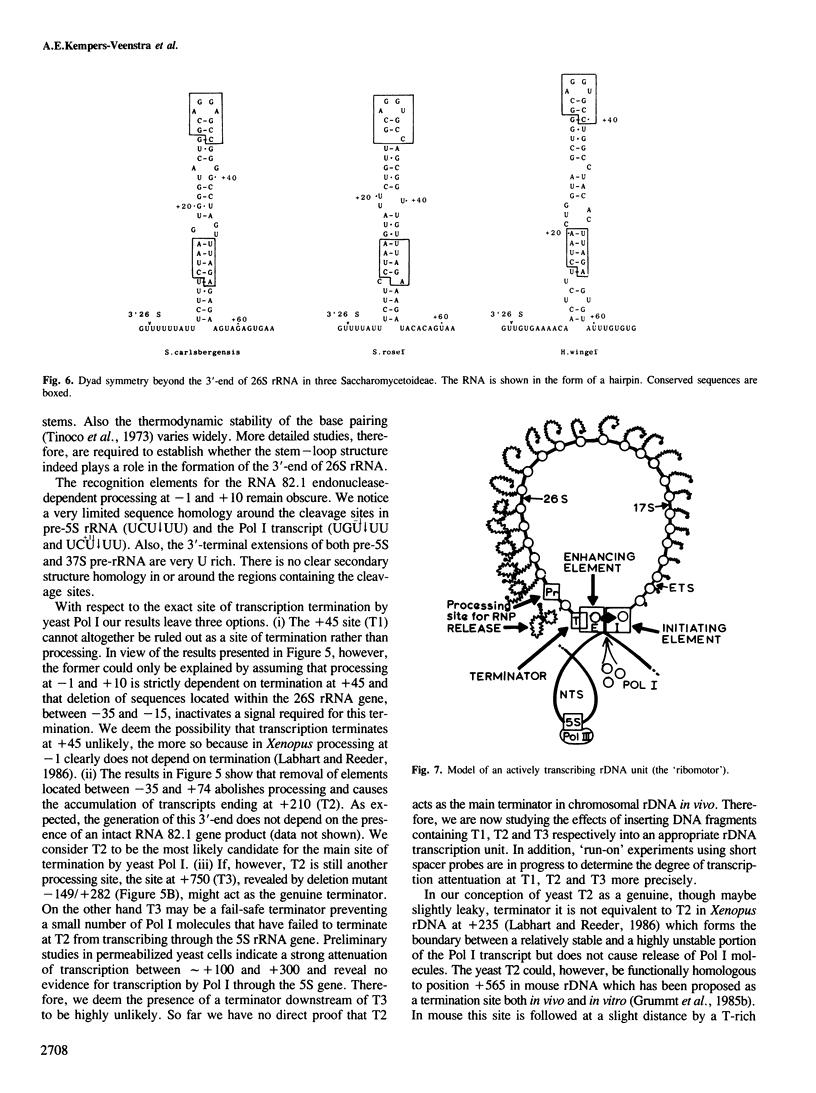
Deletion analysis of artificial rRNA minigenes transformed into Saccharomyces cerevisiae revealed that a 110 bp long fragment corresponding to positions -36 to +74 relative to the 3'-end of the 26S rRNA gene, is both necessary and sufficient for obtaining transcripts whose 3'-termini are identical to those of 26S and 37S (pre-)rRNA. These termini are produced via processing of longer transcripts because in an rna 82.1 mutant the majority of the minigene transcripts extend further downstream. Since the rna 82.1 mutation inactivates an endonuclease involved in the 3'-processing of 5S pre-rRNA it is concluded that the maturation of 37S- and that of 5S pre-rRNA requires a common factor. Comparison of the spacer sequences between Saccharomyces carlsbergensis, Saccharomyces rosei and Hansenula wingei revealed several conserved sequence blocks within the region between +10 and +55. These conserved sequence tracts, which are part of a longer region showing dyad symmetry, are supposed to be involved in the interaction with the processing component(s). Deletion of the sequences required for the formation of the 3'-ends of 26S rRNA and 37S pre-rRNA revealed a putative terminator for transcription by RNA polymerase I situated at position +210. This site maps within a DNA fragment that also contains the enhancing element for rDNA transcription by RNA polymerase I.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N., Engberg J., Gall J. G. The nucleotide sequence at the transcription termination site of the ribosomal RNA gene in Tetrahymena thermophila. Nucleic Acids Res. 1982 Mar 11;10(5):1503–1513. doi: 10.1093/nar/10.5.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Sorbaz H., Hofmann A., Roth E. Spacer sequences downstream of the 28S RNA coding region are part of the mouse rDNA transcription unit. Nucleic Acids Res. 1985 Apr 11;13(7):2293–2304. doi: 10.1093/nar/13.7.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Musters W., Dekker A. F., Klootwijk J., Planta R. J. Deletion mapping of the yeast Pol I promoter. Curr Genet. 1985;10(4):253–260. doi: 10.1007/BF00365621. [DOI] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., van Heerikhuizen H., Musters W., Klootwijk J., Planta R. J. Transcription of an artificial ribosomal RNA gene in yeast. EMBO J. 1984 Jun;3(6):1377–1382. doi: 10.1002/j.1460-2075.1984.tb01980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kief D. R., Warner J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Nov;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami R., Mishima Y., Urano Y., Sakai M., Muramatsu M. Cloning and determination of the transcription termination site of ribosomal RNA gene of the mouse. Nucleic Acids Res. 1982 Mar 25;10(6):1963–1979. doi: 10.1093/nar/10.6.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Xenopus ribosomal gene enhancers function when inserted inside the gene they enhance. Nucleic Acids Res. 1985 Dec 20;13(24):8999–9009. doi: 10.1093/nar/13.24.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R. K., Dawid I. B. The nucleotide sequence at the transcription termination site of ribosomal RNA in Drosophila melanogaster. Nucleic Acids Res. 1981 Apr 24;9(8):1801–1811. doi: 10.1093/nar/9.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R. K. The organization and transcription of eukaryotic ribosomal RNA genes. Prog Nucleic Acid Res Mol Biol. 1984;31:115–160. doi: 10.1016/s0079-6603(08)60376-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Mitchelson K., de Winter R. The promotion of ribosomal transcription in eukaryotes. Oxf Surv Eukaryot Genes. 1985;2:207–250. [PubMed] [Google Scholar]
- Piper P. W., Bellatin J. A., Lockheart A. Altered maturation of sequences at the 3' terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J. 1983;2(3):353–359. doi: 10.1002/j.1460-2075.1983.tb01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeet M. P., Klootwijk J., van Heerikhuizen H., Fontijn R. D., Vreugdenhil E., Planta R. J. A conserved sequence element is present around the transcription initiation site for RNA polymerase A in Saccharomycetoideae. Nucleic Acids Res. 1984 Jan 25;12(2):1137–1148. doi: 10.1093/nar/12.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeet M. P., Klootwijk J., van Heerikhuizen H., Fontijn R., Vreugdenhil E., Planta R. J. Molecular cloning of the rDNA of Saccharomyces rosei and comparison of its transcription initiation region with that of Saccharomyces carlsbergensis. Gene. 1983 Jul;23(1):53–63. doi: 10.1016/0378-1119(83)90216-0. [DOI] [PubMed] [Google Scholar]
- Verbeet M. P., van Heerikhuizen H., Klootwijk J., Fontijn R. D., Planta R. J. Evolution of yeast ribosomal DNA: molecular cloning of the rDNA units of Kluyveromyces lactis and Hansenula wingei and their comparison with the rDNA units of other Saccharomycetoideae. Mol Gen Genet. 1984;195(1-2):116–125. doi: 10.1007/BF00332733. [DOI] [PubMed] [Google Scholar]
- Woudt L. P., Pastink A., Kempers-Veenstra A. E., Jansen A. E., Mager W. H., Planta R. J. The genes coding for histone H3 and H4 in Neurospora crassa are unique and contain intervening sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5347–5360. doi: 10.1093/nar/11.16.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]