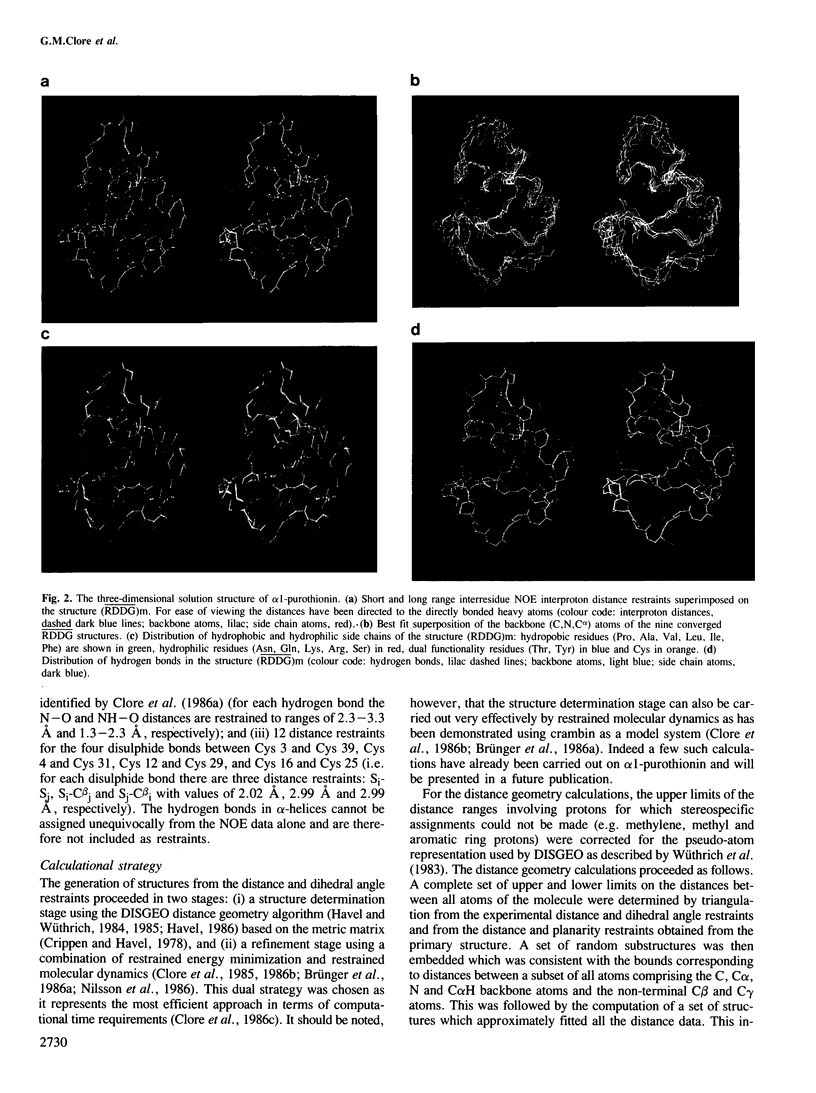
Abstract
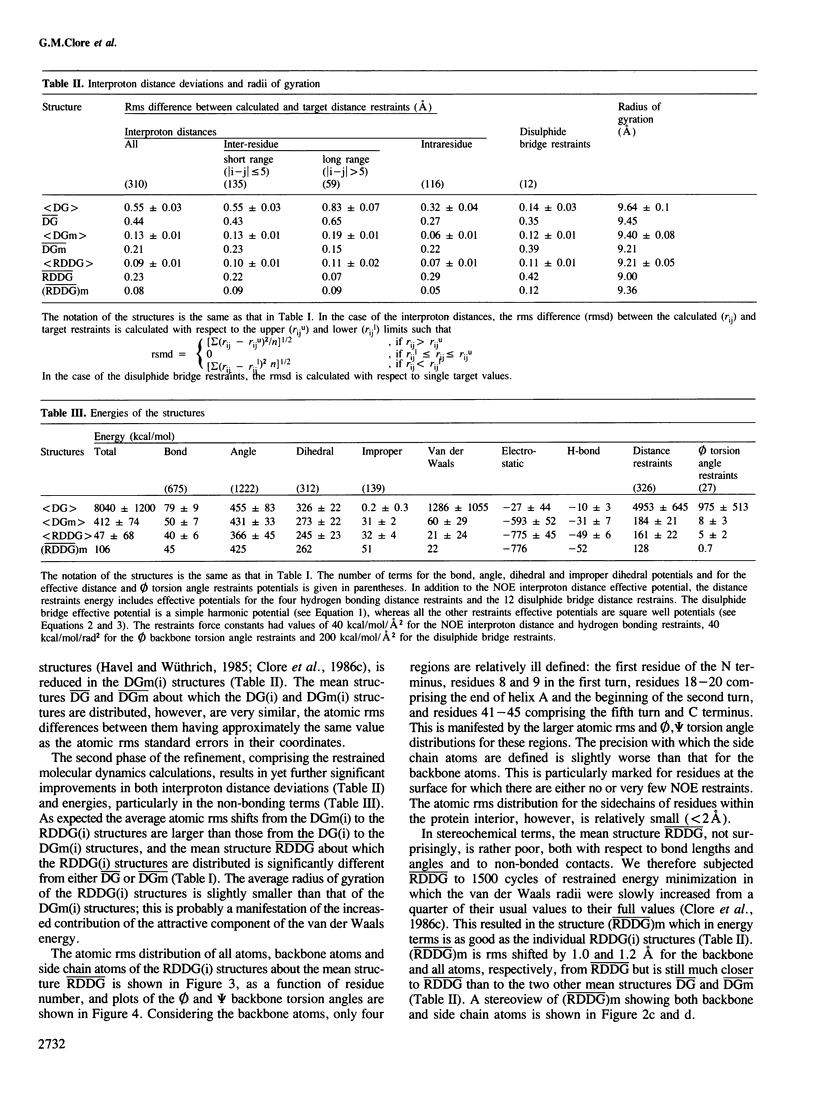
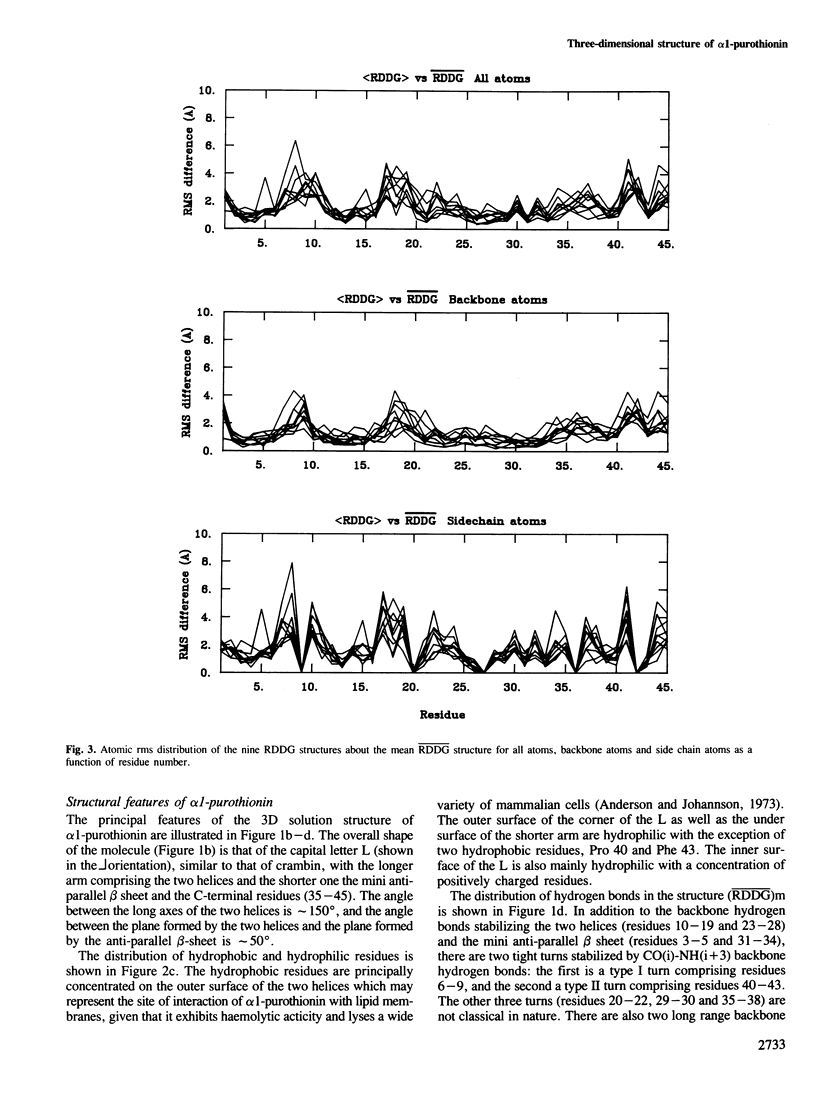
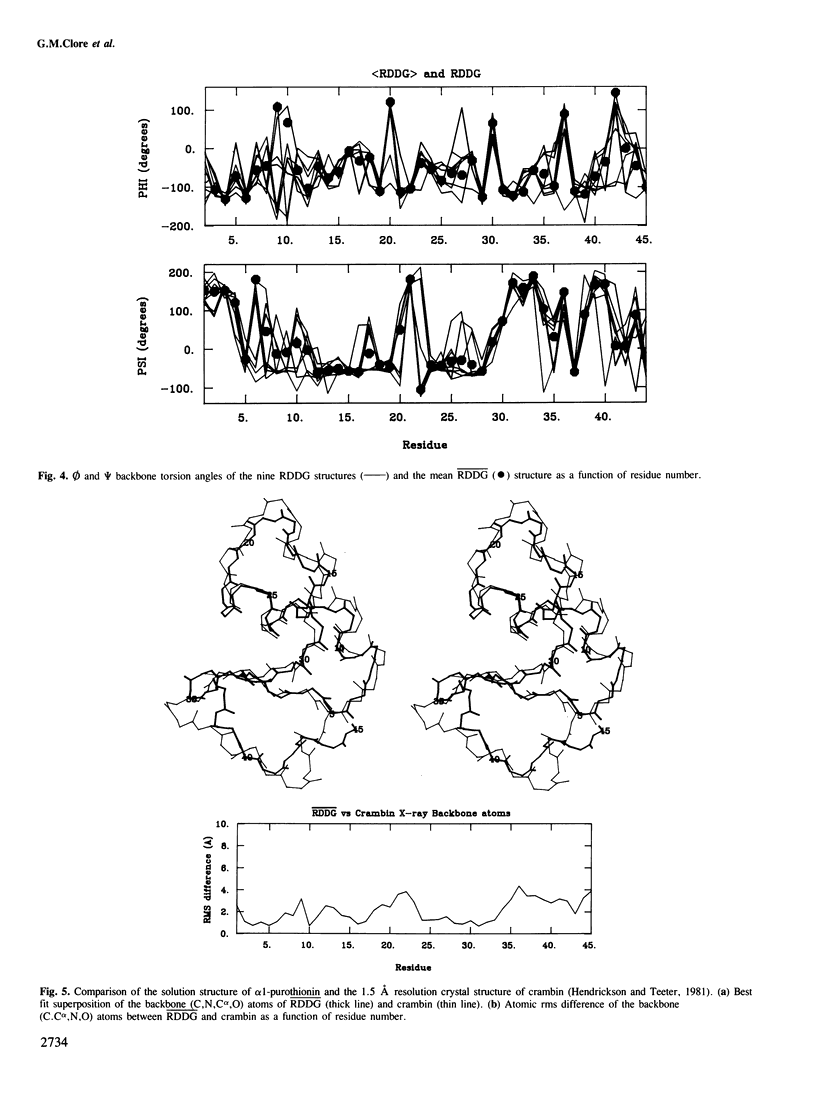
The determination of the three-dimensional solution structure of α1-purothionin using a combination of metric matrix distance geometry and restrained molecular dynamics calculations based on n.m.r. data is presented. The experimental data comprise complete sequence-specific proton resonance assignments, a set of 310 approximate interproton distance restraints derived from nuclear Overhauser effects, 27 Ø backbone torsion angle restraints derived from vicinal coupling constants, 4 distance restraints from hydrogen bonds and 12 distance restraints from disulphide bridges. The average atomic rms difference between the final nine converged structures and the mean structure obtained by averaging their coordinates is 1.5 ± 0.1 å for the backbone atoms and 2.0 ± 0.1 å for all atoms. The overall shape of α1-purothionin is that of the capital letter L, similar to that of crambin, with the longer arm comprising two approximately parallel α-helices and the shorter arm a strand and a mini anti-parallel β sheet.
Keywords: α1-purothionin, 3D structure, nuclear Overhauser effect, interproton distances, distance geometry, restrained molecular dynamics.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brünger A. T., Clore G. M., Gronenborn A. M., Karplus M. Three-dimensional structure of proteins determined by molecular dynamics with interproton distance restraints: application to crambin. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3801–3805. doi: 10.1073/pnas.83.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Solution conformation of a heptadecapeptide comprising the DNA binding helix F of the cyclic AMP receptor protein of Escherichia coli. Combined use of 1H nuclear magnetic resonance and restrained molecular dynamics. J Mol Biol. 1985 Nov 20;186(2):435–455. doi: 10.1016/0022-2836(85)90116-0. [DOI] [PubMed] [Google Scholar]
- Havel T. F., Wüthrich K. An evaluation of the combined use of nuclear magnetic resonance and distance geometry for the determination of protein conformations in solution. J Mol Biol. 1985 Mar 20;182(2):281–294. doi: 10.1016/0022-2836(85)90346-8. [DOI] [PubMed] [Google Scholar]
- Jones R. R. C-reactive protein estimation in lupus erythematosus. J R Soc Med. 1980 Feb;73(2):143–144. doi: 10.1177/014107688007300214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein R., Zuiderweg E. R., Scheek R. M., Boelens R., van Gunsteren W. F. A protein structure from nuclear magnetic resonance data. lac repressor headpiece. J Mol Biol. 1985 Mar 5;182(1):179–182. doi: 10.1016/0022-2836(85)90036-1. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Kline A. D., Braun W., Wüthrich K. Studies by 1H nuclear magnetic resonance and distance geometry of the solution conformation of the alpha-amylase inhibitor tendamistat. J Mol Biol. 1986 May 20;189(2):377–382. doi: 10.1016/0022-2836(86)90519-x. [DOI] [PubMed] [Google Scholar]
- Lattman E. Diffraction methods for biological macromolecules. Use of the rotation and translation functions. Methods Enzymol. 1985;115:55–77. doi: 10.1016/0076-6879(85)15007-x. [DOI] [PubMed] [Google Scholar]
- Mak A. S., Jones B. L. Separation and characterisation of chymotryptic peptides from alpha- and beta-purothionins of wheat. J Sci Food Agric. 1976 Mar;27(3):205–213. doi: 10.1002/jsfa.2740270302. [DOI] [PubMed] [Google Scholar]
- Mak A. S., Jones B. L. The amino acid sequence of wheat beta-purothionin. Can J Biochem. 1976 Oct;54(10):835–842. doi: 10.1139/o76-120. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Structure refinement of oligonucleotides by molecular dynamics with nuclear Overhauser effect interproton distance restraints: application to 5' d(C-G-T-A-C-G)2. J Mol Biol. 1986 Apr 5;188(3):455–475. doi: 10.1016/0022-2836(86)90168-3. [DOI] [PubMed] [Google Scholar]
- Pardi A., Billeter M., Wüthrich K. Calibration of the angular dependence of the amide proton-C alpha proton coupling constants, 3JHN alpha, in a globular protein. Use of 3JHN alpha for identification of helical secondary structure. J Mol Biol. 1984 Dec 15;180(3):741–751. doi: 10.1016/0022-2836(84)90035-4. [DOI] [PubMed] [Google Scholar]
- Sippl M. J., Scheraga H. A. Cayley-Menger coordinates. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2283–2287. doi: 10.1073/pnas.83.8.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter M. M., Mazer J. A., L'Italien J. J. Primary structure of the hydrophobic plant protein crambin. Biochemistry. 1981 Sep 15;20(19):5437–5443. doi: 10.1021/bi00522a013. [DOI] [PubMed] [Google Scholar]
- Whitlow M., Teeter M. M. Energy minimization for tertiary structure prediction of homologous proteins: alpha 1-purothionin and viscotoxin A3 models from crambin. J Biomol Struct Dyn. 1985 Feb;2(4):831–848. doi: 10.1080/07391102.1985.10506327. [DOI] [PubMed] [Google Scholar]
- Williams R. W., Teeter M. M. Raman spectroscopy of homologous plant toxins: crambin and alpha 1- and beta-purothionin secondary structures, disulfide conformation, and tyrosine environment. Biochemistry. 1984 Dec 18;23(26):6796–6802. doi: 10.1021/bi00321a080. [DOI] [PubMed] [Google Scholar]
- Williamson M. P., Havel T. F., Wüthrich K. Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol. 1985 Mar 20;182(2):295–315. doi: 10.1016/0022-2836(85)90347-x. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Billeter M., Braun W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J Mol Biol. 1983 Oct 5;169(4):949–961. doi: 10.1016/s0022-2836(83)80144-2. [DOI] [PubMed] [Google Scholar]