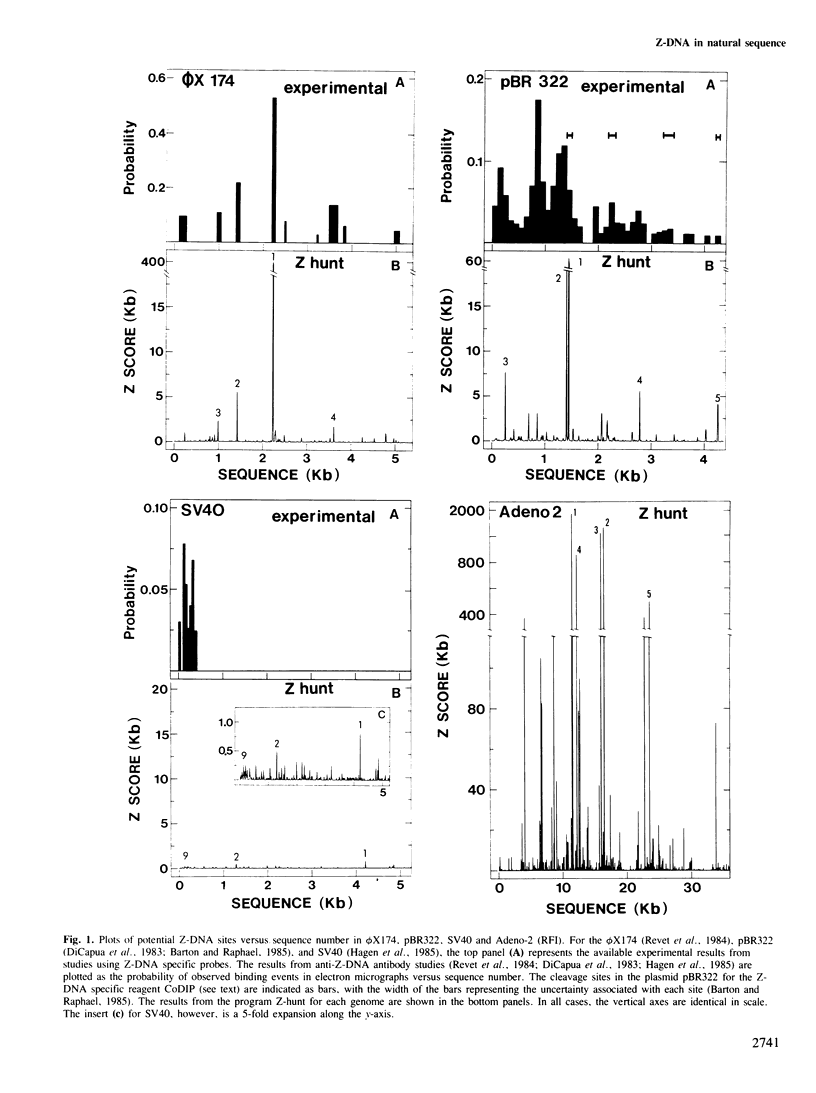
Abstract
The ease with which a particular DNA segment adopts the left-handed Z-conformation depends largely on the sequence and on the degree of negative supercoiling to which it is subjected. We describe a computer program (Z-hunt) that is designed to search long sequences of naturally occurring DNA and retrieve those nucleotide combinations of up to 24 bp in length which show a strong propensity for Z-DNA formation. Incorporated into Z-hunt is a statistical mechanical model based on empirically determined energetic parameters for the B to Z transition accumulated to date. The Z-forming potential of a sequence is assessed by ranking its behavior as a function of negative superhelicity relative to the behavior of similar sized randomly generated nucleotide sequences assembled from over 80,000 combinations. The program makes it possible to compare directly the Z-forming potential of sequences with different base compositions and different sequence lengths. Using Z-hunt, we have analyzed the DNA sequences of the bacteriophage phi X174, plasmid pBR322, the animal virus SV40 and the replicative form of the eukaryotic adenovirus-2. The results are compared with those previously obtained by others from experiments designed to locate Z-DNA forming regions in these sequences using probes which show specificity for the left-handed DNA conformation.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton J. K., Raphael A. L. Site-specific cleavage of left-handed DNA in pBR322 by lambda-tris(diphenylphenanthroline)cobalt(III). Proc Natl Acad Sci U S A. 1985 Oct;82(19):6460–6464. doi: 10.1073/pnas.82.19.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Capua E., Stasiak A., Koller T., Brahms S., Thomae R., Pohl F. M. Torsional stress induces left-handed helical stretches in DNA of natural base sequence: circular dichroism and antibody binding. EMBO J. 1983;2(9):1531–1535. doi: 10.1002/j.1460-2075.1983.tb01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Ellison M. J., Feigon J., Kelleher R. J., 3rd, Wang A. H., Habener J. F., Rich A. An assessment of the Z-DNA forming potential of alternating dA-dT stretches in supercoiled plasmids. Biochemistry. 1986 Jun 17;25(12):3648–3655. doi: 10.1021/bi00360a026. [DOI] [PubMed] [Google Scholar]
- Ellison M. J., Kelleher R. J., 3rd, Wang A. H., Habener J. F., Rich A. Sequence-dependent energetics of the B-Z transition in supercoiled DNA containing nonalternating purine-pyrimidine sequences. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8320–8324. doi: 10.1073/pnas.82.24.8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigon J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Z-DNA forms without an alternating purine-pyrimidine sequence in solution. Science. 1985 Oct 4;230(4721):82–84. doi: 10.1126/science.4035359. [DOI] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Hagen F. K., Zarling D. A., Jovin T. M. Electron microscopy of SV40 DNA cross-linked by anti-Z DNA IgG. EMBO J. 1985 Mar;4(3):837–844. doi: 10.1002/j.1460-2075.1985.tb03706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Transition of a cloned d(AT)n-d(AT)n tract to a cruciform in vivo. Nucleic Acids Res. 1985 Jun 25;13(12):4343–4363. doi: 10.1093/nar/13.12.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschemeyer A. E., Rich A. Nucleoside conformations: an analysis of steric barriers to rotation about the glycosidic bond. J Mol Biol. 1967 Jul 28;27(2):369–384. doi: 10.1016/0022-2836(67)90026-5. [DOI] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., McIntosh L. P., Arndt-Jovin D. J., Zarling D. A., Robert-Nicoud M., van de Sande J. H., Jorgenson K. F., Eckstein F. Left-handed DNA: from synthetic polymers to chromosomes. J Biomol Struct Dyn. 1983 Oct;1(1):21–57. doi: 10.1080/07391102.1983.10507425. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Angelides K. J., Holloman W. K. Left-handed DNA and the synaptic pairing reaction promoted by Ustilago rec1 protein. Cell. 1985 Jan;40(1):139–145. doi: 10.1016/0092-8674(85)90317-4. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986 Feb 28;44(4):545–554. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Konopka A. K., Reiter J., Jung M., Zarling D. A., Jovin T. M. Concordance of experimentally mapped or predicted Z-DNA sites with positions of selected alternating purine-pyrimidine tracts. Nucleic Acids Res. 1985 Mar 11;13(5):1683–1701. doi: 10.1093/nar/13.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer E. M., Sousa R., Rich A. Anti-Z-DNA antibody binding can stabilize Z-DNA in relaxed and linear plasmids under physiological conditions. EMBO J. 1985 Dec 30;4(13B):3655–3660. doi: 10.1002/j.1460-2075.1985.tb04131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller F. D., Jorgenson K. F., Winkfein R. J., van de Sande J. H., Zarling D. A., Stockton J., Rattner J. B. Natural occurrence of left-handed (Z) regions in PM2 DNA. J Biomol Struct Dyn. 1983 Dec;1(3):611–620. doi: 10.1080/07391102.1983.10507468. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Lafer E. M., Peck L. J., Wang J. C., Stollar B. D., Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982 Dec;31(2 Pt 1):309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature. 1983 Jun 23;303(5919):674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- Panyutin I., Lyamichev V., Mirkin S. A structural transition in d(AT)n.d(AT)n inserts within superhelical DNA. J Biomol Struct Dyn. 1985 Jun;2(6):1221–1234. doi: 10.1080/07391102.1985.10507634. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revet B., Zarling D. A., Jovin T. M., Delain E. Different Z DNA forming sequences are revealed in phi X174 RFI by high resolution darkfield immuno-electron microscopy. EMBO J. 1984 Dec 20;3(13):3353–3358. doi: 10.1002/j.1460-2075.1984.tb02303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Stockton J. F., Miller F. D., Jorgenson K. F., Zarling D. A., Morgan A. R., Rattner J. B., van de Sande J. H. Left-handed Z-DNA regions are present in negatively supercoiled bacteriophage PM2 DNA. EMBO J. 1983;2(12):2123–2128. doi: 10.1002/j.1460-2075.1983.tb01712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii A. V., Frank-Kamenetskii M. D. Left-handed Z form in superhelical DNA: a theoretical study. J Biomol Struct Dyn. 1984 Jun;1(6):1325–1333. doi: 10.1080/07391102.1984.10507523. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Gessner R. V., van der Marel G. A., van Boom J. H., Rich A. Crystal structure of Z-DNA without an alternating purine-pyrimidine sequence. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3611–3615. doi: 10.1073/pnas.82.11.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang A. J., Quigley G. J., Kolpak F. J., van der Marel G., van Boom J. H., Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981 Jan 9;211(4478):171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- Zarling D. A., Arndt-Jovin D. J., Robert-Nicoud M., McIntosh L. P., Thomae R., Jovin T. M. Immunoglobulin recognition of synthetic and natural left-handed Z DNA conformations and sequences. J Mol Biol. 1984 Jul 5;176(3):369–415. doi: 10.1016/0022-2836(84)90495-9. [DOI] [PubMed] [Google Scholar]


