Abstract
Background:
Haemonchosis is a major parasitic infestation in ruminant livestock, causing significant economic losses annually. The causative organisms are helminths of the genus Haemonchus spp. Detection of the causative agent is important for effective management and control of the disease. Molecular detection and characterization of parasites is a very dependable approach for parasite identification, especially where morphological characterization is unreliable.
Methods:
To detect and characterize Haemonchus species in cases of haemonchosis at a Municipal abattoir in Ibadan, Nigeria; abomasal samples were collected from cattle at the abattoir. Polymerase chain reaction (PCR) was used to detect and amplify 320 bp internal transcribed spacer-2 (ITS-2) and 400 bp external transcribed spacer (ETS) genes of the adult worms in the samples. Multiple sequence alignment and phylogenetic tree reconstruction were carried out to further confirm the presence of the worms.
Results:
PCR, multiple sequence alignment, and phylogenetic reconstruction confirmed the presence of H. placei in the abomasal samples and further confirmed the species as a distinct species of bovine worms at the abattoir. Multiple sequence alignment also revealed genetic sites that can be employed to distinguish H. placei from H. contortus and H. similis.
Conclusion:
Molecular techniques; PCR and sequence analysis are very important and reliable in the diagnosis of parasitic diseases. This will help to formulate effective control measures for eradication of the parasite.
Keywords: Cattle, Haemonchus placei, Multiple sequence alignment, Nigeria
Introduction
Members of the genus Haemonchus are recognized as having the most economic impact on ruminant livestock worldwide (1). With a cosmopolitan distribution, members of the genus include H. contortus (2), H. placei (3), and H. similis (4,5). H. placei is a cattle parasite, while H. contortus is usually found in sheep. However, both are found in other domestic and wild ruminants as well as in other hosts (6). Accurate identification of Haemonchus spp. is essential for effective parasite control strategies, as many reports of H. contortus in cattle might actually represent H. placei (7). Morphological traits of Haemonchus worm populations of cattle and sheep origin often differ. However, they do show some phenotypic traits that are similar, making measurements crucial for the classification of individual worms (6,8). Therefore, proper identification and understanding of the epidemiology of H. placei are crucial for implementing sustainable control measures and minimizing the economic impact of the disease in Africa (9).
Numerous epidemiological studies have documented the prevalence and economic importance of bovine haemonchosis in Nigeria (10), as an economically important parasite in Africa, particularly in Nigeria. It is one of the major gastrointestinal nematodes affecting cattle in the country (11), causing adverse effects on animal health and decreasing production and productivity (10). The disease is prevalent throughout Nigeria, with high distribution reported during the rainy season.
In livestock production, there is a heavy dependence on anthelmintic prophylaxis, which accounts for reports of anthelmintic resistance spanning several worm species, including Haemonchus spp. (12,13). A precise identification of worm species, coupled with accurate data collection on the epidemiology of these parasitic worms, is therefore crucial to the development of an effective approach to control helminth infestation in livestock. Owing to Morphological similarities, it is a challenging task to differentiate between H. placei and H. contortus. Consequently, many purported claims of H. contortus, specifically in Bovine species may actually have been H. placei (14).
Based on various criteria of identification (15,16), H. placei was not recognized as a separate species, a lot of research had neglected the diversity of Haemonchus spp. Conversely, others who acknowledge the diversity simply call Haemonchus worms of small ruminant’s origin H. contortus, and those of cattle origin H. placei. However, subsequent studies established the identity of H. placei as a separate species from H. contortus (14). Both species can co-infest a single host, both cattle and small ruminants (5, 17, 18). Hybrids of the two species have been produced experimentally, but there is total or partial sterility of the F1 or F2 off-spring depending on the reciprocal cross used (19), Since both species occupy the same niche, the gene flow pattern and extent between the two species is not apparent, and there are still those who debate their status as separate species (16,20). It can be assumed that if the two species are genetically distinct, then they may have different responses to anthelmintic drugs. If there is significant gene flow between the two species, then there is a high probability of the exchange of genes that confer anthelmintic resistance.
Therefore, we aimed to detect and characterize Haemonchus spp. responsible for bovine haemonchosis at a Municipal abattoir in Ibadan, Nigeria molecularly, and to differentiate it from H. contortus.
Materials and Methods
Sample Collection and DNA Extraction
Haemonchus samples were obtained from each abomasum content of 322 infected cattle at random for two months during regular visits to the municipal abattoir in 2023. The cattle originated from the derived savannah, Sahel, and sub-arid climatic regions of Nigeria as well as from neighboring countries including the Niger Republic, Chad, and as far as Mali. The breeds, ages, sex, and sources of origin were documented. The age estimation was done by the rostral dentition technique (21). Nematodes were examined using a microscope and identified up to the genus level (22). The adult worms were collected in normal saline, not exposed to any preservative solution, and preserved at −20 °C until DNA extraction. The DNA genome was extracted using the NucleoSpin® DNA Tissue extraction kit (Thermo SCIENTIFIC®, Pittsburgh, PA) by following manufacturer instructions. DNA was eluted in 50 μl nuclease-free water and stored at −20 °C until further use.
Compliance with Ethical Standards
Haemonchus samples were obtained from the abomasum content of already slaughtered animals at the government abattoirs.
PCR and sequence analysis
A pair of primers NC1 (5′-ACG TCT GGT TCA GGG TTG TT-3′) and NC2 (5′-TTA GTT TCT TTT) (23) were used to amplify a 320 bp fragment of the ITS-2 gene; whereas, a pair of primers HaemExtTransSpacFor (5′-ACC ACA GGG ATA ACT GGC TTG TGGC-3′) and HaemExtTransSpacRev (5′-AGC TCC AGA ATT ACC GCA GTT-3′) (3) were used to amplify a 400 bp fragment of the ETS gene. The PCR was carried out in 50 μl volume containing 5 μl of total DNA, of 0.2 μM of each primer, 25 μl of PCR master mix (10 mM Tris-HCl (pH 8.6), 50 mM KCl, 1.5 mM MgCl2, 50 units/ml of Taq DNA polymerase, 0.2 mM each dNTP, and 18 μl of nuclease-free water. The GeneAmp PCR system 9700 thermal cyclers (Applied Biosystems, Foster City, CA) was used for amplification under the following conditions: initial denaturation at 95 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 20 sec, annealing at a 55 °C (ITS2) and 62 °C (ETS) for 30 sec; extension at 72 °C for 1.5 min and a final extension at 72 °C for 7 min. Five of the amplified DNA fragments of each gene from the samples were purified using GeneJET PCR Purification Kit (ThermoSCIENTIFIC®, Pittsburgh, PA). Automated nucleotide sequencing was performed on an ABI 3130XL. The sequences were designated NGA-Ib1-NGA-Ib5 for each gene. The sequences have been deposited at GenBank with the accession numbers OR702552 - OR702556. These nucleotide sequences of the ITS and ETS genes of the Nigerian Haemonchus spp. sequences were analyzed for homology with other Haemonchus spp. nucleotide sequences in the NCBI (National Center of Biotechnology Information) online repository using the BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST/) (24). Multiple sequence alignment of the partial Haemonchus spp. ITS and ETS gene sequences from this study, other Haemonchus spp. ITS-2 and ETS sequences retrieved from the GenBank and F. gigantica ITS-2 as the out-group was carried out. This was done using the Clustal W algorithm in the CLC Main Workbench computer application (Qiagen, Valencia, CA). Phylogenetic analysis was computed using maximum likelihood and Kimura 2-parameter model, and bootstrap analysis at 1000 replicates on the MEGA computer application, version 7.0 (25).
Results
Polymerase Chain Reaction and Sequence Analysis
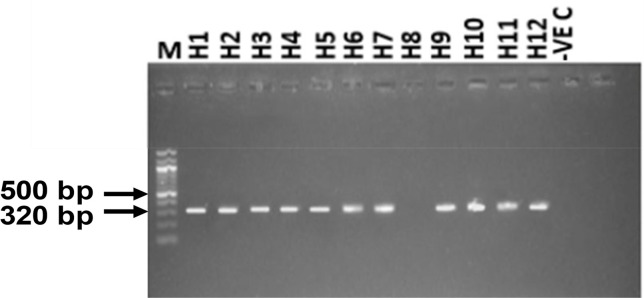
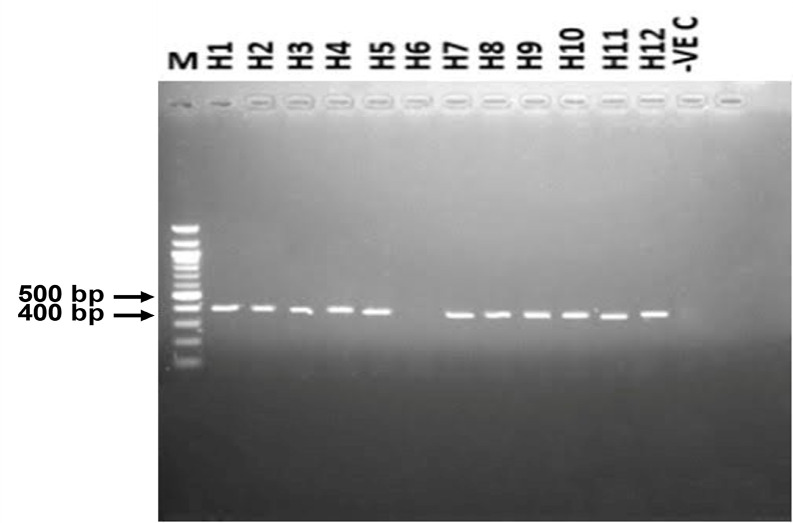
PCR amplification of H. placei partial ITS-2 and ETS genes generated 320 bp (Fig. 1) and 400 bp (Fig. 2) products, respectively.
Fig. 1:
Agarose gel electrophoresis of H. placei using ITS-2 primer in some Nigerian breeds of cattle. Lane M=100-bp molecular electrophoresis marker. White Fulani (H1 and H2), Sokoto Gudali (H3 and H4), Red Bororo (H5 and H6), Kuri (H7 and H8), cross between White Fulani and Sokoto Gudali (H9 and H10), and a cross between White Fulani and Red Bororo breeds of cattle (H11 and H12). Lane −veC served as the negative control. 5 μl of each PCR amplicon was used for agarose gel for electrophoresis. The 320 bp PCR amplicons are shown
Fig. 2:
Agarose gel electrophoresis of Haemonchus using ETS primer in some Nigerian breeds of cattle. Lane M=100-bp molecular marker. White Fulani (H1 and H2), Sokoto Gudali (H3 and H4), Red Bororo (H5 and H6), Kuri (H7 and H8), cross between White Fulani and Sokoto Gudali (H9 and H10), and a cross between White Fulani and Red Bororo breeds of cattle (H11 and H12). Lane −VE C served as the negative control. 5 μl of each PCR amplicon was used for agarose gel for electrophoresis. The 400bp PCR amplicons are shown
BLAST analysis on the selected positive samples revealed high homology to sequences retrieved from the NCBI online repository. Multiple sequence analyses of all five ITS-2 nucleotide sequences were obtained from this study, and six separate nucleotide sequences were retrieved from the NCBI online repository, with accession codes: LC430924 (NGR), LC430923 (NGR), LC366992 (NGR), MG669349 (India), KU870651 (India), LS997564 (UK) was carried out. Substitution A217T, A226G, and G240A distinguishes H. contortus from H. placei (Fig. 3). Multiple sequence analysis of the five ETS nucleotide sequences from this study and five individual sequences retrieved from the NCBI online repository with accession codes: AF343971 (USA), MH581148 (India), LS997564 (UK), KF266762 (Ireland) L04152; KM259915 (Pakistan) was as shown in Fig. 4, nucleotide substitutions A988C, C1030G and C1058T distinguishes H. contortus from H. placei. Also, in the ETS gene, substitutions A988G, AG to GC at positions 1026–1027, C1030T, C1058A, and GTGC to CCAT at positions 1059 to 1062 differentiate H. similis from H. placei and H. contortus.
Fig. 3:
Multiple sequence alignment of the nucleotide sequences of partial ITS-2 genes in Haemonchus of Nigerian origin and corresponding gene sequences retrieved from the NCBI online repository.
Fig. 4:
Multiple sequence alignment of the nucleotide sequences of partial ETS genes in Haemonchus of Nigerian origin and corresponding gene sequences retrieved from the NCBI online repository
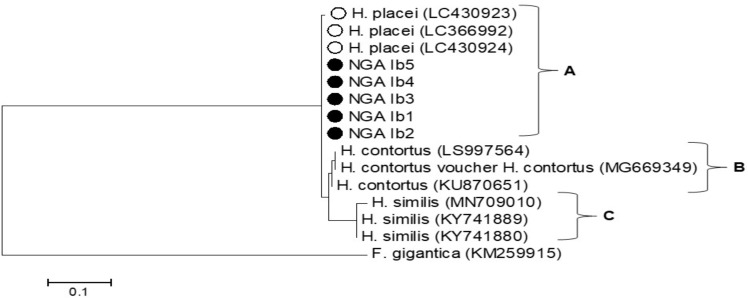
The phylogenetic tree was computed from the multiple sequence alignments of ITS-2 and ETS genes from H. placei of Nigerian origin and other Haemonchus species from GenBank sequences. For the ITS-2 gene, the five nucleotide sequences from this study clustered with H. placei (clade A) indicating that they are H. placei whereas H. contortus clustered separately as clade B (Fig. 5). There are three clades in the ETS phylogenetic tree, the five sequences from this study and three Nigerian sequences in the GenBank clustered in clade A, H. contortus sequences clustered in clade B and H. similis in clade C (Fig. 5).
Fig. 5:
Phylogenetic analyses of H. placei based on partial ITS-2 and ETS genes. The phylogenetic tree was computed using the multiple alignments of the nucleotide sequence of the ITS-2 gene from Nigeria H. placei and other Haemonchus spp. from GenBank sequences. Fasciola gigantica gene was used as the out-group. The tree was computed by the maximum likelihood method with bootstrapping at 1000 replicates. The Haemonchus sequences from this study are labeled with black circles while H. placei sequences retrieved from the online repository are labeled with white circles
The ITS-2 sequences are represented by the codes NGA_Ib1, NGA_Ib 2, NGA_Ib3, NGA_Ib4, NGA_Ib5; H. placei (LC430924), H. placei (LC430923), H. placei (LC366992), H. contortus (LS997564), H. contortus voucher H. contortus (MG669349), H. contortus (KU870651). The sites of interspecies mutations of the Haemonchus ITS-2 gene in the Nigerian strains are at positions 217, 226, and 240 in the highlighted boxes
The ETS gene sequences are represented by the codes NGA_Ib1, NGA_Ib 2, NGA_Ib3, NGA_Ib4, NGA_Ib5; H. placei (AF343971), H. placei (MH581148), H. contortus (LS997564), H. contortus (KF266762), H. similis (L04152). The sites of mutations of the Haemonchus ETS gene in the Nigerian strains are at positions 1026–1027, 1030, and 1058–1062 in the highlighted boxes
Discussion
Genetic information plays a crucial role in managing parasitic diseases, enabling the easy identification of the specific parasites infecting the host (26). Molecular data serves as valuable markers for species description and determining genetic populations. For example, the use of ITS genes, known for their variability, is particularly important for evolutionary studies and the discrimination of nematode species (23). This study highlights the significance of comparative sequence analysis in inferring the functionality of ETS and ITS-2 as novel functional genes for identifying H. placei. Additionally, the use of ITS gene is notably important for quickly distinguishing between known species, due to lower levels of intra-specific polymorphism (27).
Adult worm counts done at necropsy give a better picture of the extent of parasitic infestation than fecal egg counts (23). In other words, adult worm count allows for precise Morphological identification of the worms present, while also giving a direct means of assessing the total worm burden. Although considered the most accurate means of diagnosing helminthosis, postmortem worm count is rather expensive. The differentiation of Haemonchus spp. ova from those of other gastrointestinal worms requires expertise. Owing to Morphological similarities among nematode ova, it is often challenging to distinguish them microscopically, particularly up to the species level (28).
Although larva culture offers a better alternative to fecal floatation methods, it is a time-consuming technique that requires certain expertise to identify subtleties in the Morphological appearance of the L3 larvae produced; the diagnosis from larva culture can therefore be subject to the expertise and experience of the microscopist (29).
To address this issue, the use of PCR in diagnosing bovine hemonchosis by identifying and assessing the genetic diversity among the parasite population studied is a viable alternative. The use of primers specific to the Haemonchus genus removes the need for PCR-positive controls (30). In the present study, the ITS-2 region is sufficient for differentiating H. placei from H. contortus. An ETS PCR has been employed to identify animals infested with H. similis (31). Although, H. similis has been isolated from cattle, in a study done in Brazil, there have been no reports of its presence in sheep, even in sheep grazed on the same pasture as H. similis-infected cattle (17, 32). This can be explained by a high predilection for cattle. Conversely, there has been a report of H. similis in wild ruminants in Brazil (33).
Amplification of the rDNA ITS-2 and ETS regions of the adult worm samples aids the determination of H. species, using species-specific primers which produced (ITS-2) 320 bp and (ETS) 400 bp bands, respectively on agarose gel electrophoresis. H. placei has also been characterized in Brazilian cattle using these genes (34,2).
In conclusion, this study revealed that H. placei is a distinct and major Haemonchus species in Nigerian cattle found in Ibadan southwestern Nigeria. The study animals were infected only with H. placei. This aligns with the species delineation of Haemonchus in another study (31). Genetically identifying Haemonchus spp. isolates and phylogenetic analysis will help monitor the spread of Haemonchus species within and outside Nigeria as well as provide epidemiological data on Haemonchus spp. in Nigeria.
Identifying the specific species is crucial for understanding the parasites that infect animal hosts (35). The evolutionary presence of Haemonchus is linked to genetic drift, mutation, and changes in its morphological characteristics, which have been observed to influence the development of drug resistance. Employing molecular markers for identification can offer insights into the genetic variations of the species, potentially enhancing our understanding of treatment procedures, including drug discovery.
Conclusion
We advocate for further research into parasite intensity and the development of strategies to control Haemonchus species in Nigeria. This will give a perspective on the possibility of using genetic, genomic, and bioinformatic tools to better understand these parasites, anthelminthic resistance, vaccine immunoprophylaxis, and control of parasitic diseases. This study confirmed that the study samples are H. placei with distinct subtypes and delineated species nomenclature from other sequences subtypes belonging to other Haemonchus species.
Acknowledgements
This study received no financial aid and was self-funded.
Footnotes
Conflict of interest
All authors hereby declare no financial, academic, commercial, or personal conflict of interest.
References
- 1.Gibbs HC, Herd RP. Nematodiasis in cattle. Importance, species involved, immunity, and resistance. Vet Clin North Am Food Anim Pract. 1986; 2(2): 211–224. [PubMed] [Google Scholar]
- 2.Rudolphi CA. New Observations on Intestinal Worms. Arch Zool Comp Anat. 1803; 3: 1–32. [Google Scholar]
- 3.Place FE. Anaemic diarrhoea in young cattle. Vet Rec. 1893; 5: 589. [Google Scholar]
- 4.Travassos LP. Trichostrongylideos brazileiros (III preliminary note). Brazil-Med. 1914; 28(34): 325–327. [Google Scholar]
- 5.Achi YL, Zinsstag J, Yao K, Yeo N, Dorchies P, Jacquiet P. Host specificity of Haemonchus spp. for domestic ruminants in the savanna in northern Ivory Coast. Vet Parasitol. 2003; 116(2): 151–158. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenfels JR, Pilitt PA, Hoberg EP. New morphological characters for identifying individual specimens of Haemonchus spp. (Nematoda: Trichostrongyloidea) and a key to species in ruminants of North America. J Parasitol. 1994; 80(1): 107–119. [PubMed] [Google Scholar]
- 7.Alessandro F, Amarante T. Why is it important to correctly identify Haemonchus species? (Colégio Brasileiro de Parasitologia Veterinaria). Rev Bras Parasitol Vet. 2011; 20(4): 263–268. [DOI] [PubMed] [Google Scholar]
- 8.Lichtenfels JR, Pilitt PA, Le Jambre LF. Spicule lengths of the ruminant stomach nematodes Haemonchus contortus, Haemonchus placei, and their hybrids. Proc Helminthol Soc Wash. 1988; 55: 97–100. [Google Scholar]
- 9.Ali Q, Rashid MI, Ashraf K, Zahid MN, Ashraf S, Chaudhry U. Genetic variation in the rDNA ITS-2 sequence of Haemonchus placei from cattle host. J Inf Mol Biol. 2015; 3(1): 13–18. [Google Scholar]
- 10.Jeremiah OT, Banwo OG. Haematologic profile and prevalence survey of haemonchosis in various breeds of slaughtered cattle in Ibadan, Nigeria. Nig J Anim Prod. 2019; 46(3): 151–162. [Google Scholar]
- 11.Odeniran PO, Jegede HO, Adewoga TOS. Prevalence and risk perception of adult-stage parasites in slaughtered food animals (cattle, sheep and goat) among local meat personnel in Ipata abattoir, Ilorin, Nigeria. Vet Med Anim Sci. 2016; 4:1. [Google Scholar]
- 12.Almeida FA. Multiple resistance to anthelmintics by Haemonchus contortus and Trichostrongylus colubriformis in sheep in Brazil. Parasitol Int. 2010; 59(4): 622–625. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland IA, Leathwick DM. Anthelmintic resistance in nematode parasites of cattle: a global issue? Trends Parasitol.2011;27(4):176–181. [DOI] [PubMed] [Google Scholar]
- 14.Hoberg EP, Lichtenfels JR, Gibbons L. Phylogeny for species of the genus Haemonchus (Nematoda: Trichostrongylo-idea): considerations of their evolutionary history and global biogeography among Camelidae and Pecora (Artiodactyla). J Parasitol. 2004; 90(5): 1085–1102. [DOI] [PubMed] [Google Scholar]
- 15.Almeida JL. Note on the species of the genus Haemonchus Cobb, 1898 (Nematoda: Trichostrongyloidea). Proceedings of the Society of Biology and Its Branches. 1935; 114: 960–961. [Google Scholar]
- 16.Gibbons LM. Revision of the genus Haemonchus Cobb, 1898 (Nematoda: Trichostronglidae). Syst Parasitol. 1979; 1: 3–24. [Google Scholar]
- 17.Amarante AF, Bagnola J, Amarante MR, Barbosa MA. Host specificity of sheep and cattle nematodes in Sao Paulo state, Brazil. Vet Parasitol. 1997; 73(1–2): 89–104. [DOI] [PubMed] [Google Scholar]
- 18.Jacquiet P, Cabaret J, Thiam E, Cheikh D. Host range and the maintenance of Haemonchus spp. in an adverse arid climate. Int J Parasitol. 1998; 28(2): 253–261. [DOI] [PubMed] [Google Scholar]
- 19.Le Jambre LF. Hybridization studies of Haemonchus contortus (Rudolphi. 1803) and H. placei (Place, 1893) (Nematoda: Trichostrongylidae). Int J Parasitol. 1979;9: 455–463. [DOI] [PubMed] [Google Scholar]
- 20.Stevenson LA, Chilton NB, Gasser RB. Differentiation of Haemonchus placei from H. contortus (Nematoda: Trichostrongylidae) by the ribosomal DNA second internal transcribed spacer. Int J Parasitol. 1995; 25(4): 483–488. [DOI] [PubMed] [Google Scholar]
- 21.Lasisi OT, Ojo NA, Otesile EB. Estimation of age of cattle in Nigeria using rostral dentition. Trop Vet. 2002; 20: 204–208. [Google Scholar]
- 22.Hansen J, Perry B. The Epidemiology, Diagnosis and Control of Helminthes Parasites of Ruminants. A Handbook, ILRAD. 1994.
- 23.Cerutti MC, Citterio CV, Bazzocchi C., et al. Genetic variability of Haemonchus contortus (Nematoda: Trichostrongyloidea) in alpine ruminant host species. J Helminthol. 2010; 84(3): 276–283. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990; 215(3): 403–410. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximumlikelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28(10): 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palevich N, Britton C, Kamenetzky L, Mitreva M, de Moraes MM, Bennuru S, et al. Tackling hypotheticals in helminth genomes. Trends Parasitol. 2018; 34: 179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ndosi BA, Lee D, Bia MM, et al. Morphometry and Molecular Identification of Haemonchus Cobb, 1898 (Trichostrongylidae: Nematoda) Isolates from Small Ruminants in Tanzania Based on Mitochondrial cox 1 and rRNA-ITS genes. J Parasitol Res. 2023; 2023:1923804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mochizuki R, Endoh D, Onuma M, Fukumoto S. PCR-based species-specific amplification of ITS of Mecistocirrus digitatus and its application in identification of GI nematode eggs in bovine faeces. J Vet Med Sci. 2006; 68(4): 345–351. [DOI] [PubMed] [Google Scholar]
- 29.Food and Agricultural Organization (FAO) . “Distribution and impact of helminth diseases of livestock in developing countries,” in FAO Corporate Document Repository—Agriculture and Consumer Protection. 2000.
- 30.Zarlenga DS, Chute MB, Martin A, Kapel CMO. A multiplex PCR for unequivocal differentiation of all encapsulated and non-encapsulated genotypes of Trichinella. Int J Parasitol. 1999; 29(11): 1859–1867. [DOI] [PubMed] [Google Scholar]
- 31.Ademola IO, Krücken J, Ramünke S, Demeler J, Von G, Himmelstjerna S. Absence of detectable benzimidazole resistance associated alleles in Haemonchus placei in cattle in Nigeria revealed by pyrosequencing of β-tubulin isotype 1. Parasitol Res. 2015;114(5):1997–2001. [DOI] [PubMed] [Google Scholar]
- 32.Rocha RA, Rocha R, Bresciani K, et al. Sheep and cattle grazing alternately: Nematode parasitism and pasture decontamination. Small Rumin Res. 2008;75(2–3):135–143. [Google Scholar]
- 33.Nascimento AA, Bonuti MR, Mapeli EB, et al. Natural infections in cervids (Mammalia: Cervidae) from the states of Mato Grosso do Sul and São Paulo by Trichostrongyloidea nematodes. Braz J Vet Res Anim Sci. 1927;37(2):153–158. [Google Scholar]
- 34.Zarlenga DS, Stringfellow F, Nobary M, Lichtenfels JR. Cloning and characterization of ribosomal RNA genes from three species of Haemonchus (Nematoda: Trichostrongyloidea) and identification of PCR primers for rapid differentiation. Exp Parasitol. 1994; 78(1): 28–36. [DOI] [PubMed] [Google Scholar]
- 35.Palevich N, Maclean PH, Choi YJ, Mitreva M. Characterization of the complete mitochondrial genomes of two sibling species of parasitic roundworms, Haemonchus contortus and Teladorsagia circumcincta. Front Genet. 2020;11: 573395. [DOI] [PMC free article] [PubMed] [Google Scholar]