Abstract
Electrochemical biosensors are gaining attention as powerful tools in cancer diagnosis, particularly in liquid biopsy, due to their high efficiency, rapid response, exceptional sensitivity, and specificity. However, the complexity of intra- and intertumor heterogeneity, with variations in genetic and protein expression profiles and epigenetic modifications, makes electrochemical biosensors susceptible to false-positive or false-negative diagnostic outcomes. To address this challenge, there is growing interest in simultaneously analyzing multiple biomarkers to reveal molecular characteristics of tumor heterogeneity for precise cancer diagnosis. In this Perspective, we highlight recent advancements in utilizing electrochemical biosensors for cancer diagnosis, with a specific emphasis on the multitarget analysis of cancer biomarkers including tumor-associated nucleic acids, tumor protein markers, extracellular vesicles, and tumor cells. These biosensors hold significant promise for improving precision in early cancer diagnosis and monitoring, as well as potentially offering new insights into personalized cancer management.
Keywords: electrochemical biosensor, cancer biomarker, multitarget analysis, tumor heterogeneity, cancer diagnosis
Electrochemical biosensors incorporating biological recognition elements and electronic transducers can serve as robust analytical tools for a wide range of analytes, encompassing small molecules, nucleic acids, proteins, and even cells.1 Depending on the electroactive or nonelectroactive properties of analytes, electrochemical biosensors transduce biorecognition events into measurable signals that are subsequently interrogated using appropriate electrochemical techniques. For example, variations in the current generated by electrochemical oxidation or reduction reactions are monitored via amperometric and voltammetric techniques, while the resistance and capacitance characteristics of an electrochemical cell is measured using impedance techniques to establish a quantitative relationship with the analyte concentration. Compared with other conventional biosensors, electrochemical biosensors possess several distinct advantages, such as low cost, high sensitivity, simple operation, small analyte volume requirement, rapid readout, and ease of portability and miniaturization, which renders them applicable for point-of-care testing to acquire physiological and biochemical information.2−4
Among widespread applications of electrochemical biosensors, the analysis of cancer biomarkers, such as tumor-associated nucleic acids, tumor protein markers, extracellular vesicles (EVs), and tumor cells, is of particular interest, highlighting their potential for indicating the occurrence of cancer and monitoring the disease progression.5−7 Unlike biological species (e.g., glucose and lactate) commonly used in health monitoring, clinical biomarkers for cancer diagnosis present a low abundance in the body fluids.8−10 For instance, in the blood, the concentration of tumor-associated nucleic acids (such as microRNAs) is typically at the fM or pM level, tumor protein markers are often found in concentrations ranging from fg/mL to pg/mL or even lower, while the quantity of circulating tumor cells (CTCs) generally falls below ten per milliliter. And although EVs possess a relatively high abundance (approximately109 to 1010 particles/mL), the proportion of EVs derived from tumors is extremely limited. The low abundance along with the coexistence of a wealth of nontarget components brings about considerable challenges in sensitivity and specificity during the analysis of these cancer biomarkers.11 To address the challenges, recent researches on electrochemical biosensors have primarily focused on the advancement in biorecognition systems and associated signal amplification strategies. Emerging recognition elements such as aptamers, molecular imprinting polymers, and affinity peptides have been exploited and utilized as valuable complements to antibodies for meeting the specific recognition and analysis requirements of diverse cancer biomarkers.12−14 Meanwhile, a variety of signal amplification strategies have been devised based on nanomaterials or molecular assembly processes and applied in electrochemical biosensors to realize highly sensitive analysis of cancer biomarkers.15−18
Despite the improved sensitivity and specificity, the clinical practice of electrochemical biosensors in cancer diagnosis remains hindered, particularly attributed to the high tumor heterogeneity. As one of the main drivers of resistance to cancer therapies, tumor heterogeneity encompasses the variability in genetic and protein expression profiles as well as epigenetic modifications across different tumors (intertumor heterogeneity) or within the same tumor (intratumor heterogeneity), which is also evident in distinct disease lesions (spatial heterogeneity) or at different disease stages (temporal heterogeneity).19−21 Due to tumor heterogeneity, relying on a single cancer biomarker is not sufficient for accurate tumor identification or monitoring of tumor progression, resulting in limited accuracy and efficacy in both cancer diagnosis and treatment. Conversely, the utilization of multiple biomarkers holds great promise for addressing the challenges posed by tumor heterogeneity, thereby facilitating improved diagnostic insights and enhancing the therapeutic outcomes for cancer patients.22−25 In this regard, there is a growing interest in the development of electrochemical biosensors that enable multitarget analysis of coexisting cancer biomarkers, as these biosensors are anticipated to create a more comprehensive tumor molecular profile to reflect tumor heterogeneity. For instance, electrochemical biosensors designed for the multitarget analysis of nucleic acid biomarkers (such as tumor-associated DNA and RNA) can provide gene expression profiles that help differentiate among distinct cancer subgroups. Similarly, electrochemical biosensors designed for the multitarget analysis of protein biomarkers can reveal the diversity of protein expression and modification present in different malignant tumors. Moreover, through the multitarget analysis of different biomarkers on the surface and inside of EVs or intact tumor cells, electrochemical biosensors can more effectively uncover the molecular characteristics of the originating tumors, which allows for better monitoring of disease progression at various stages.26,27
To date, several reviews concerning the multitarget analysis of biomarkers have been published.28−31 Nevertheless, these reviews mainly concentrate on either the integration of various analytical techniques (not limited to electrochemical approaches) with micro/nanomaterials or the combined detection of multiple biomarker species (such as nucleic acids and proteins) across a range of diseases. Therefore, there remains a lack of reviews focusing on the applications of electrochemical biosensors in resolving tumor heterogeneity, although a large number of papers and groundbreaking studies have emerged in the past. In this context, we herein aim to provide an up-to-date overview of the process in this fast-moving field, presenting the latest achievements of electrochemical biosensors in the multitarget analysis of various cancer biomarkers, such as tumor-associated nucleic acids, tumor protein markers, EVs, and tumor cells (Figure 1), with a specific emphasis on the work of the past five years. Meanwhile, we are willing to share our prospects on the future research directions of this filed and hope that this perspective can contribute to promoting the clinical transformation of electrochemical biosensors in precision cancer diagnosis.
Figure 1.
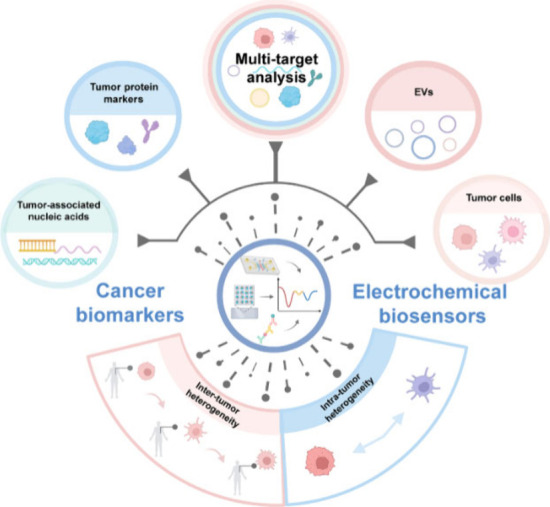
Schematic illustration of using electrochemical biosensors to address tumor heterogeneity challenges for cancer diagnosis, mainly focusing on multitarget analysis of tumor-associated nucleic acids, tumor protein markers, EVs and tumor cells. Created with BioRender.com.
Multitarget Analysis of Tumor-Associated Nucleic Acids
Tumor heterogeneity plays a crucial role in shaping the unique genetic profiles observed in cancer patients, thereby highlighting the significance of tumor-associated nucleic acids as biomarkers for cancer diagnosis, prognosis, and prediction. Specifically, cell-free DNA (cfDNA) from tumors, such as circulating tumor DNA (ctDNA), can accurately present the mutations existing in cancers.32 This makes the assessment of the mutation burden in cfDNA highly beneficial for diagnosing cancer patients. Moreover, the molecular testing of circulating RNA biomarkers, especially mRNAs (mRNAs), microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs), have emerged as another promising approach.33 Nevertheless, the precise identification of tumor-associated nucleic acids has always faced with the challenges posed by their low abundance and the extensive coexistence of nucleic acids with sequence similarities. Hence, an increasing number of studies have been dedicated to developing electrochemical biosensors for high-sensitivity and multitarget analysis to enable the molecular profiling of tumor-associated nucleic acids.
Multitarget Analysis of Tumor-Associated DNA Biomarkers
As a substitute for tissue-based genomic profiling, plasma ctDNA analysis can identify mutations in specific genes and reflect tumor heterogeneity.34 To overcome the difficulties presented by low-frequency site mutations in ctDNA, DNA-based amplification strategies are often integrated into electrochemical biosensors to enable sensitive analysis of tumor-specific mutations. For example, DNAzymes, specific DNA sequences with excellent catalytic activity comparable to protein enzymes, are utilized to enhance the accumulation of electroactive signals by modulating conformational changes of DNA probes;35 DNA polymerases that facilitate elongation of DNA strands are used to catalyze isothermal DNA amplification reactions for enhanced electrochemical signals;36 CRISPR/Cas systems, known for their exceptional precision in inducing site-specific cleavage, are engineered to be activated upon binding to the target ctDNA for unleashing inherent trans-cleavage capability;37 and functional nanoparticles are widely applied to preparing electroactive signaling probes, thereby amplifying the electrochemical detection signal of ctDNA mutation.38
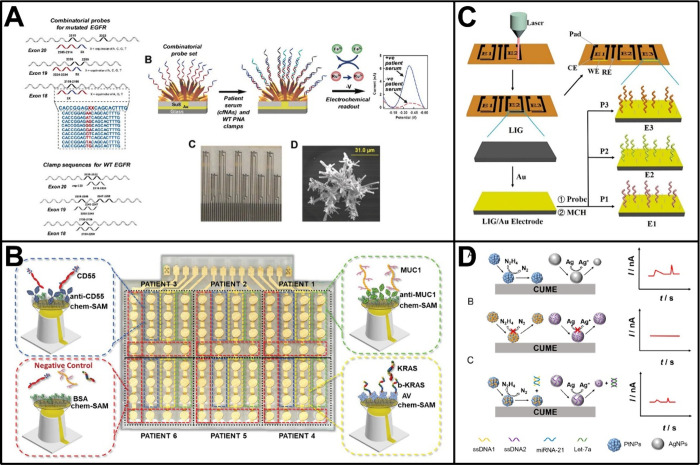
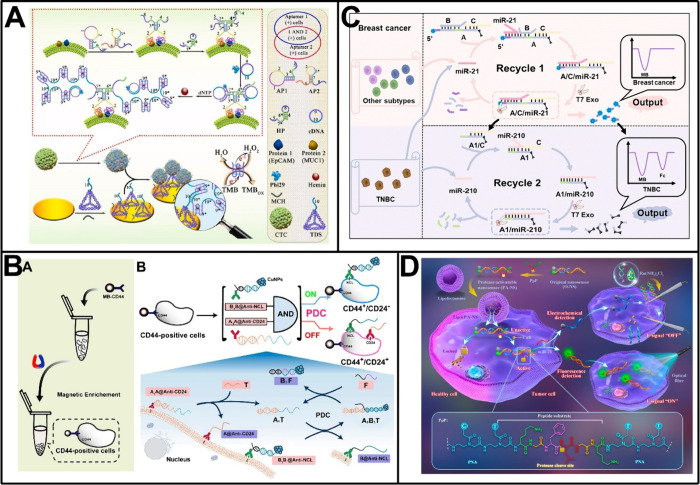
Combining these signal amplification strategies with spatially separated electrodes, such as electrode arrays, a number of electrochemical biosensors have been developed for spatial multiplexed analyses of ctDNAs.39−43 For instance, Kelley and co-workers developed an electrochemical biosensor to analyze mutated circulating ctDNA with a specific focus on the KRAS gene.39 The biosensor made use of DNA clutch probes to prevent denatured ctDNA from reassociating, thereby ensuring that only mutated ctDNA bound immobilized peptide nucleic acid (PNA) probes to generate an electrochemical signal. Based on this design and the further use of a microelectrode array, the biosensor was challenged with PCR products of wild-type KRAS and its seven mutant alleles and achieved analysis and discrimination of homozygous and heterozygous mutants even in samples collected from cancer patients. With a similar spatial multiplexing design, Kelley and co-workers also proposed an electrochemical biosensor to identify mutations of the epidermal growth factor receptor (EGFR) gene, which had dozens of different alterations that included deletions, insertions, and point mutations (Figure 2A).40 Combinatorial PNA probes with variable positions at the N-terminal region were utilized in the biosensor to effectively analyze serum samples from patients diagnosed with nonsmall cell lung cancer. Through the use of a multiplexed chip, the biosensor was demonstrated applicable in the analysis of 40 clinically relevant alterations within the EGFR gene, including 26 deletion mutations within exon 19, 6 insertion mutations within exon 20, and 3 point mutations within exon 18. Recently, Genco et al. reported another interesting work, in which a bioelectronic single-molecule sensing array was fabricated for the simultaneous analysis of both ctDNA and tumor protein markers (Figure 2B).43 The sensing array relied on the use of single-molecule-with-a-large-transistor technology and arranged in an 8 × 12 enzyme-linked immunosorbent assay-like layout. By using the reader based on a custom Si-IC chip to collect signals, analyses of KRAS mutation, mucin 1 (MUC1), and CD55 in 47 patients’ fluids were multiplexed at the single-molecule limit-of-identification, resulting in at least 96% sensitivity and 100% specificity for the diagnosis of pancreatic cancer.
Figure 2.
Electrochemical biosensors for multitarget analysis of tumor-associated nucleic acids. (A) Electrochemical biosensor based on a multiplexed chip and combinatorial PNA probes to identify mutations of the EGFR gene. Reproduced with permission from ref (40). Copyright 2018 Wiley-VCH. (B) Bioelectronic single-molecule sensing array for the simultaneous analysis of both ctDNA and tumor protein markers. Reproduced from ref (43). Available under a CC-BY 4.0 license. Copyright 2023 The authors. (C) Laser-induced graphene electrode array with microfluidic channels for high-throughput analysis of miRNAs. Reproduced with permission from ref (60). Copyright 2024 Elsevier. (D) Dual analysis of miRNA-21 and Let-7a in a single run based on SPEC. Reproduced from ref (65). Copyright 2023 American Chemical Society.
Multitarget Analysis of Tumor-Associated RNA Biomarkers
mRNAs, serving as the direct carriers of genetic information and the templates for encoding proteins, play an essential role in biological processes. In cancer patients, alterations of mRNAs can influence various facets of tumor development, and the discovery of mRNAs in the plasma has been shown to facilitate the development of noninvasive diagnostic and prognostic tools.44,45 The analysis of tumor-associated mRNAs is capable of reflecting the abnormalities in specific gene expression and making valuable diagnostic contributions beyond DNA analyses. Meanwhile, starting with miRNAs, many efforts have been made to explore the biological roles of linear noncoding RNAs, which also include lncRNAs.46 These linear noncoding RNAs are found to participate in various cellular processes by interfering with post-transcriptional and translational regulations and are increasingly recognized as potential RNA biomarkers in liquid biopsy due to their high abundance and inherent stability in the blood.47−49 In addition, unlike linear RNA biomarkers, circRNAs having a covalently closed loop structure confer increased stability and resistance to RNase degradation, which renders them as promising noncoding RNA biomarkers for cancer diagnosis.50 Notably, circRNAs contain unique back-splice junction sequences that are formed via cyclization. This distinctive sequence can serve as a recognition domain, facilitating the incorporation of circRNAs into nucleic acids-based analysis strategies, such as the activation of CRISPR-Cas systems and the initiation of DNA amplification reactions.51,52
Compared to traditional RT-PCR techniques, electrochemical biosensors are generally easy to operate and thus have found impressive success in multiple analyses of tumor-associated RNA biomarkers, which may enhance the accuracy of cancer diagnosis. One popular way to develop such biosensors is with the help of the electrode array, similar to that employed in multiple analyses of ctDNAs. In a groundbreaking work, Xie et al. fabricated an 8 × 8 sensor array by evaporating a titanium adhesion layer on a glass slide and employed it to electrochemically detect the expression of breast cancer susceptibility genes at the mRNA level, i.e., tumor protein p53 (TP53), heat-shock protein 90 (HSP90), breast cancer gene 1 (BRCA1), and Histone H4 (His4).53 In human breast tissues, the sensor array allowed the direct quantification of the target genes from mRNA extracts with high sensitivity and specificity. The estimated lowest detectable amount of a specific gene was approximately 800 copies in 1.5 ng of mRNA. Since then, a succession of array-based multitarget analyses for tumor-associated RNA biomarkers have been accomplished.54−60 For instance, Kelley and co-workers devised multiplexed sensor arrays that integrated novel nanostructured microelectrodes (NMEs) with rapid electrocatalytic readout for profiling mRNAs and miRNAs.54,55 These arrays enabled the direct and amplification-free analysis of multiple RNAs from cell extracts and clinical tumor samples, using only nanograms of total RNA, and thereby were capable of identifying gene fusions related to aggressive prostate cancer and miRNA sequences overexpressed in human head and neck cancer cells. Later, they extended the NMEs-based sensor array to the specific detection of cancer-related mutations in cell-free nucleic acids, encompassing not only cfDNAs but also cfRNAs.56 Through the introduction of a clamp cocktail, the biosensor exhibited remarkable practicality in unprocessed serum specimens from patients with lung cancer and melanoma, and yielded outcomes in accordance with the PCR. This marked the first successful analysis of cell-free nucleic acids in serum without the necessity of enzymatic amplification. Likewise, with the array design, Torrente-Rodríguez et al. developed a magnetobiosensor for the simultaneous analysis of two miRNAs (miRNA-21 and miRNA-205) related to breast cancer.57 By employing the p19 RNA binding protein as a capture receptor and the H2O2/hydroquinone system to generate amperometric signals, the sensor exhibited desirable detection limits of 6 × 105 fM for both target miRNAs, and demonstrated good usability in metastatic breast cancer cells and tissues. Bruch et al. utilized the dry film photoresist technology to manufacture a CRISPR/Cas13a-powered electrochemical microfluidic biosensor for miRNA detection.58 Based on the cleavage of the reporter RNA by target miRNA-activated CRISPR/Cas13a, the biosensor allowed the sensitive analysis of target miRNA (e.g., miRNA-19b and miRNA-20a) down to 1 × 104 fM and had the capacity for simultaneous analysis of up to eight miRNA biomarkers. Very recently, Liu et al. integrated a laser-induced graphene electrode array with microfluidic channels to construct a high-throughput electrochemical biosensor for miRNA analysis (Figure 2C).60 Thanks to the signal amplification mediated by λ-exonuclease, the biosensor was able to perform multiplexed analysis of miR-21, miR-1246, and miR-155 with detection limits of 140 fM, 240 fM, and 110 fM, respectively. The high potential of the biosensor in cancer diagnosis was also validated by assessing the expression levels of these three miRNAs in exosomes and clinical serum samples, showing high accuracy in distinguishing breast cancer patients from healthy donors.
In addition to using electrode arrays, other strategies have also been attempted for the development of electrochemical biosensors to achieve multiplex analysis of cancer-associated RNA biomarkers.61−67 For example, Labib et al. developed an electrochemical biosensor to detect ultralow levels of miRNAs, which possessed three detection modalities based on hybridization, p19 protein binding, and protein displacement.61 By adopting voltammetric and impedance techniques (such as square wave voltammetry and electrochemical impedance spectroscopy) to acquire signals, this three-mode sensor was capable of identifying target miRNA as low as 5 × 10–3 fM without amplification, and was applicable for sequential analysis of multiple miRNAs on a single electrode. Meanwhile, this three-mode sensor achieved success in directly profiling three endogenous miRNAs in human serum, including hsa-miRNA-21, has-miRNA-32, and hsa-miRNA-122, with quantitative results validated by qPCR. Also using a sequential multiplexing strategy, Sheng et al. presented a sensitive biosensor for detecting a set of nonsmall cell lung cancer-associated RNA biomarkers.62 In the biosensor, the sequential multiplex was realized through the regeneration of electrode interface induced by the combined enzymatic cleavage by Uracil-DNA glycosylase and endonuclease IV. As a result, this biosensor maintained high stability for up to 37 sequential RNA measurements and was able to analyze multiple RNA biomarkers, including miRNA-17, miRNA-155, TTF-1 mRNA, miRNA-19b, miRNA-210 and EGFR mRNA. In another approach, Qiu et al. employed a different but interesting strategy, single particle electrochemical collision (SPEC), to realize multitarget analysis of RNAs in a single run (Figure 2D).65 In their design, the complementary base pairing between target miRNAs and single-stranded DNAs at the surface of nanoparticles accelerated the removal of DNA coating, thereby restoring the SPEC of the electrocatalysis of Pt nanoparticles and the oxidation of Ag nanoparticles. By monitoring the dual signals, two target miRNAs, miRNA-21 and Let-7a, were detected simultaneously with limits of 5.3 × 105 fM and 8.8 × 105 fM, respectively. Recently, Ye et al. developed a novel “on–off-on” electrochemiluminescence-based biosensor with dual targets of breast cancer-related miRNA-21 and miRNA-105.67 Two different DNA assembly circuits, namely catalytic triple hairpin assembly and toehold-mediated strand displacement reactions, were introduced in this biosensor, facilitating the ultrasensitive quantitation of the two miRNAs with limits of 2.03 × 10–3 and 8.04 × 10–2 fM, respectively. Validation in cellular samples further demonstrated that this sensor could effectively distinguish breast cancer cells from nonbreast cancer cells and accurately identify metastatic breast cancer cells, such as MDA-MB-231 cells.
In view of the intricate nature of tumor heterogeneity at the molecular level, there is a growing interest in the multidimensional analysis of tumor-associated RNAs in combination with other cancer biomarkers.68−70 In this regard, Zuo and co-workers developed a DNA-framework-based molecular classifier by using programmable atom-like nanoparticles (PANs).71 As designed, the PANs were encoded with multivalence reporters and thus allowed the conversion of heterogeneous molecular binding events into unified electrochemical sensing signals, enabling the acquisition of comprehensive molecular information in clinical samples. Benefiting from these valence-encoded PANs, the molecular classifier demonstrated effectiveness in analyzing a panel of six prostate cancer-associated biomarkers, including miRNA-153, miRNA-183, ROR2 mRNA, MEIS2 mRNA, prostate-specific antigen (PSA), and sarcosine. Furthermore, the molecular classifier displayed a high diagnostic power for prostate cancer with an area under the curve of 100% based on optimized weight sets for the biomarker panel, showing greatly enhanced accuracy compared to that using a single biomarker (miRNA or mRNA).
Multitarget Analysis of Tumor Protein Markers
The accessibility of blood as a readily available sample renders circulating cancer biomarkers as potential tools for cancer diagnosis and physical examination. Specifically, circulating tumor protein markers, which originate from tumor-associated tissues in response to cancer and related pathological states, have been clinically utilized for screening and diagnosing various cancers, evaluating responses to cancer treatment, and monitoring prognosis during the disease progression.72−74 In recent years, electrochemical biosensors have made remarkable achievements in the detection of tumor protein markers. However, it is evident that current biosensors are typically confronted with two challenges: one is the challenge of sensitivity due to the low abundance of the target protein, and the other is the challenge of specificity due to nonspecific adsorption from clinical samples at the electrode interface. Furthermore, tumor protein markers encompass a range of proteins from diverse sources, including glycoproteins, hormones, enzymes, antigens, and other varieties. However, extensive clinical data suggests that no single tumor protein marker is universally applicable to all patients, even those with the same type of cancer.75,76 Therefore, many efforts have been and are being made to improve the performance of electrochemical biosensors and to achieve the combined analysis of tumor protein markers, which would be essential for enhancing diagnostic accuracy while facilitating personalized risk assessment for cancer management.
Immunosensors for Multitarget Analysis of Tumor Protein Markers
Since the initial discovery of tumor protein markers, such as alpha-fetoprotein (AFP), carcinoembryonic antigen (CEA), and PSA, immunosensors have become a fundamental tool for their analysis, relying on the high-affinity and noncovalent interaction between the target and the antibody.77−80 For instance, our group engineered an Escherichia coli biocarrier harboring redox indicators and antibodies and developed an electrochemical immunosensor for analyzing human epidermal growth factor receptor-2 (HER-2), a key protein biomarker in breast cancer subtyping.77 Because of the great signal amplification capability of Escherichia coli biocarrier arising from its superior loading aptitude, as low as 35 pg/mL HER-2 was sensitively detected using our immunosensor. In another representative work, Vargas et al. reported on a novel design using hybrid cell membranes derived from human macrophages and red blood cells to construct the electrochemical immunosensor.78 Taking tumor necrosis factor-alpha (TNF-α) as a model target, the immunosensor displayed a remarkable limit of detection of 150 pM (approximately 2.55 × 103 pg/mL), demonstrating antifouling abilities of the hybrid membrane in preventing nonspecific adsorption.
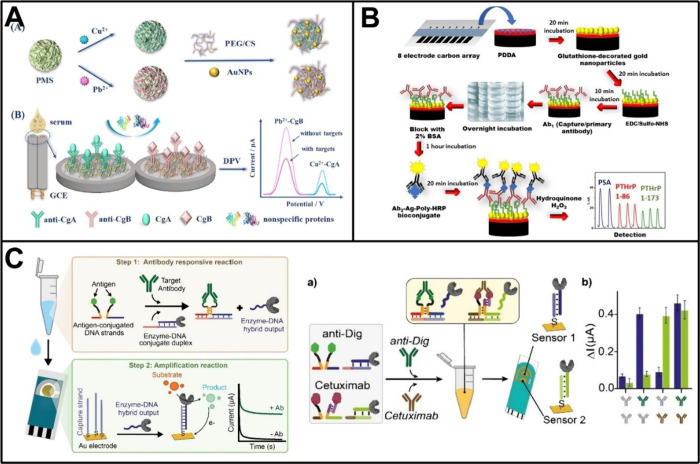
Given subtle changes at the protein level in the initiation of cancer that may not be readily detectable through single protein assays, developing electrochemical immunosensors for the identification of multiple tumor protein markers is believed to enhance predictive precision in cancer diagnosis. The utilization of differentiated electrochemical signal labeling is one popular approach in multitarget analysis. In a pioneering work, Hayes et al. employed metal ions (such as bismuth and indium ions) as electrochemical tracers to develop an immunosensor capable of simultaneously detecting two proteins.81 Thereafter, Liu et al. further enhanced the design of multiplex analysis through the utilization of inorganic metal nanoparticles for electrochemical coding.82 Since then, the sensing designs integrating different metal-based labels have been frequently employed for the multitarget analysis of tumor protein markers. For example, Kong et al. fabricated CdS/DNA and PbS/DNA nanochains via in situ growth approach and tethered them to relevant antibodies to serve as distinguishable nanolabels in an immunosensor.83 Based on the electrochemical stripping analysis of Cd2+ and Pb2+ dissolved from these nanolabels, simultaneous detection of CEA and AFP was achieved in a single run. The detect limit for CEA reached 3.3 pg/mL while for AFP reached 7.8 pg/mL. Liu et al. reported an electrochemical immunosensor for analyzing two neuroendocrine tumor-associated proteins, chromogranin A (CgA) and chromogranin B (CgB), employing independent signals derived from Cu2+ and Pb2+-deposited porous magnesium silicate (Figure 3A).84 Under optimal conditions, this immunosensor was feasible to detect CgA and CgB down to 5.3 × 10–3 pg/mL and 2.1 × 10–3 pg/mL, and exhibited satisfactory accuracy in real serum samples from neuroendocrine tumor patients. Recently, Zhang et al. developed an electrochemical homogeneous platform based on metal ions/SiO2NPs/magnetic beads for multitarget immunoassay of tumor protein markers.85 Utilizing the distinct oxidation peak potentials and current signals of different metal ions (Cd2+, Cu2+, Pb2+, and Hg2+), this platform enabled the simultaneous detection of four standard biomarkers for the clinical molecular typing of breast cancer (i.e., HER-2, estrogen receptor [ER], cell proliferation biomarker Ki67, and progesterone receptor [PR]), with detection limits of 2 pg/mL, 1.8 pg/mL, 10.36 pg/mL, and 1.33 pg/mL, respectively. Apart from metal-based labels, redox molecules and enzymes have also been employed in immunosensors to achieve multitarget analysis of tumor protein markers.86−88 In one such work, Zhao et al. developed a sandwich-type immunosensor for multiplexed analysis of three tumor protein markers using ferrocene-carboxylic acid, anthraquinone-2-carboxylic acid, and acetylsalicylic acid as signal sources.86 Distinguishable electrochemical signals were characterized by cyclic voltammetry and differential pulse voltammetry, demonstrating satisfactory performance for simultaneous determinations of CEA, AFP, and PSA. Similarly, Xie et al. proposed an immobilization-free dual-target electrochemical biosensor that combined distinguishable magnetic signal reporters.87 This biosensor was capable of converting the quantities of target tumor protein markers (i.e., CEA and AFP) into the signals of potential-resolved methylene blue (MB) and 6-(ferrocenyl)hexanethiol, and thereby achieved delightful detection performance with detection limits of 3.334 × 10–2 pg/mL and 1.702 × 10–2 pg/mL for the two proteins, respectively.
Figure 3.
Electrochemical immunosensors for tumor protein marker analysis. (A) Electrochemical immunosensor utilizing metal ions-deposited porous magnesium silicate for the simultaneous analysis of CgA and CgB. Reproduced with permission from ref (84). Copyright 2021 Elsevier. (B) A semiautonomous electrochemical microfluidic platform for analyzing PTHrP along with its fragments. Reproduced from ref (104). Copyright 2022 American Chemical Society. (C) ELIDIS for ultrasensitive detection of antibodies. Reproduced with permission from ref (107). Copyright 2023 Wiley-VCH.
Electrode arrays and other forms of spatially separated electrodes have also been extensively explored for multitarget analysis of tumor protein markers over a long period.89−94 One of the early efforts was to position multiple gold electrodes on a single microporous membrane, with each electrode being functionalized by specific capture antibodies for independently detecting two protein markers, PSA and human chorionic gonadotropin.89 In another early attempt, Wilson patterned two iridium oxide electrodes on a glass substrate and developed an alkaline phosphatase-based amperometric immunosensor.91 The spatial separation among the electrodes empowered the sensor to concurrently measure AFP and CEA with high precision and accuracy comparable to single-analyte ELISAs. Likewise, La Belle et al. presented the first report on an impedimetric biosensor for the detection of glycoproteins bearing distinct glycan moieties.92 This sensor was dependent on the functionalization of printed circuit board electrodes with either peanut agglutinin or Sambucus nigra agglutinin, which preferentially bound to the tumor-associated Galα1–3GalNAc and α2,6-linked sialic acid, respectively. In this field, Rusling and colleagues have also been at the forefront of research, making many significant contributions. For instance, they utilized a nanostructured electrode based on single-wall carbon nanotube forest to develop an amperometric immunosensor with a detection limit as low as pg/mL.95 By integrating it with a simple four-electrode array, the sensor was expanded to detect four tumor protein markers, including PSA, prostate specific membrane antigen (PSMA), platelet factor-4, and interleukin-6 (IL-6), with the necessary high sensitivity for detecting these markers at physiological levels.96 Concurrently, they developed another sensitive amperometric immunosensor by combining a densely packed gold nanoparticle platform with massively labeled magnetic beads.97 Through integration with a microfluidic chip featuring eight sensing elements, this sensor could simultaneously measure several tumor protein markers related to head and neck squamous cell carcinoma, i.e., IL-6, IL-8, vascular endothelial growth factor [VEGF], and VEGF-C, while achieving high sensitivity with subfemtomolar detection limits (5 × 10–3 pg/mL for IL-6, 1 × 10–2 pg/mL for VEGF and IL-8, and 5 × 10–2 pg/mL for VEGF-C).98 Subsequently, they further enhanced the performance of amperometric immunosensors in multitarget analysis of tumor protein markers by increasing array throughput, improving protein capture procedure, or employing Fe3O4@graphene oxide nanozymes instead of massively labeled magnetic beads.99−101 Recently, Rusling and co-workers have concentrated on prostate cancer diagnosis and developed several immunosensors with unprecedented sensitivity. For example, by using a screen-printed electrode array coated with a dense layer of gold nanoparticles, they achieved subzeptomole detection of PSA, VEGF-D, ETS-related gene protein, and insulin-like growth factor-1 down to 1.3 × 10–4, 8.8 × 10–5, 6.3 × 10–5, and 1.3 × 10–5 pg/mL.102 With the construction of an optimized microfluidic immunoarray with subfg/mL detection limits, they also accomplished simultaneous analysis of an 8-protein panel while identifying a subset comprising four proteins that demonstrated a much better accuracy in predicting prostate cancer than using PSA alone.103 Additionally, they developed a semiautonomous microfluidic platform without magnetic beads for the amperometric analysis of parathyroid hormone-related peptide (PTHrP) along with its fragments (Figure 3B).104 This sensor not only achieved an ultralow detection limit of 3 × 10–4 pg/mL but also highlighted the value of PTHrP in diagnosing aggressive prostate cancer.
Integrating new sensing designs with spatially separated electrodes has also presented opportunities for the multitarget analysis of tumor protein markers.105−108 For instance, Zhong et al. developed a photoelectrochemical immunosensor for analyzing two breast cancer-associated protein markers, CA15–3 and CEA, in which metal–organic framework incorporating Ag2S was adopted to yield robust and stable dual photocurrent signals.105 In a recent work, Díaz-Fernández et al. designed an antigen-DNA conjugates-based electrochemical biosensor for antibody detection, which was termed as Enzyme-Linked DNA Displacement (ELIDIS).107 The interaction of antigen-DNA conjugate and the target antibody facilitated the release of a GOx-linked DNA strand, which subsequently hybridized with probe DNA to generate a measurable signal upon the catalysis reaction. With the use of a gold screen printed disposable electrode that presented two working interfaces, the ELIDIS allowed simultaneous analysis of different antibodies including the clinically relevant one, Cetuximab, an FDA-approved chimeric monoclonal antibody that specifically targeted EGFR to inhibit tumor growth (Figure 3C).
Aptamer-Based Biosensors for Multitarget Analysis of Tumor Protein Markers
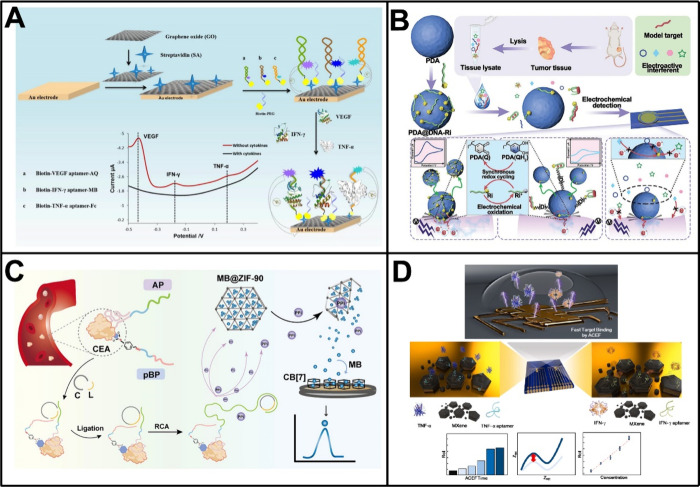
Aptamers, a kind of synthetic single-stranded nucleic acid ligands generated through SELEX (systematic evolution of ligands by exponential enrichment), demonstrate remarkable affinity and specificity in binding to a variety of biomolecules and thus can serve as substitutes for antibodies in the development of electrochemical biosensors.109,110 In an early landmark study, aptamers were combined with the encoding and amplification capabilities of quantum dots, achieving electrochemical simultaneous detection of thrombin and lysozyme.111 Despite the fact that the biosensor merely employed model proteins as targets, it initiated the application of aptamers in the multiplex analysis of proteins. Since then, by adopting appropriate sensing designs, particularly through employing unique three-dimensional structures upon target binding via conformational changes or strand competitions, aptamers have obtained considerable success in the multitarget analysis of proteins, especially tumor protein markers.112,113 In one such work, Shen et al. designed an electrochemical biosensor utilizing redox probes-tagged molecular beacon aptamers for simultaneous detection of multiple proteins.112 As shown in Figure 4A, these molecular beacon aptamers labeled with anthraquinone, MB, and ferrocene underwent conformational changes upon binding to target proteins, leading to departures of the tagged redox probes from the electrode surface and resulting in obvious alterations in electrochemical response. By employing three breast cancer-associated cytokines (i.e., VEGF, interferon-γ [IFN-γ], and TNF-α) as models, the biosensor demonstrated sensitive quantification of target proteins as low as 5 pg/mL while showed desirable recoveries in human serum samples.
Figure 4.
Aptamer-based electrochemical biosensors for multitarget analysis of tumor protein markers. (A) Electrochemical biosensor utilizing redox probes-tagged molecular beacon aptamers for simultaneous detection of multiple proteins. Reproduced with permission from ref (112). Copyright 2021 Elsevier. (B) Homogeneous electrochemical biosensor employing Ri-labeled aptamers for quantitative analysis of VEGF, miRNA-21, and ATP. Reproduced with permission from ref (115). Copyright 2023 Wiley-VCH. (C) Versatile electrochemical biosensor for analyzing tumor-associated glycoprotein biomarkers. Reproduced from ref (116). Copyright 2023 American Chemical Society. (D) Dual-target electrochemical biosensor based on an Au microgap electrode coated with aptamer/MXene nanosheets. Reproduced with permission from ref (119). Copyright 2022 Elsevier.
Moreover, the noncovalent interaction between single-stranded aptamers and various nanomaterials offers new opportunities for the advancement of electrochemical biosensors. In a representative work, a dual-signal biosensor was developed by adopting the aptamer-coating Sb quantum dots@ZIF-67 nanocomposites with the encapsulation of electroactive molecules, such as MB and 3,3′,5,5′-tetramethylbenzidine (TMB).114 Following the incubation with target proteins, positively charged MB and TMB were released from the nanocomposites after the removal of aptamer coating, achieving the sensitive and accurate analysis of HER-2 and estrogen receptor in a label-free manner. Similarly, Zhang and co-workers devised a homogeneous electrochemical biosensor by employing electron transfer cascades on the electrode-nanoparticle interface (Figure 4B).115 In their design, redox indicator (Ri)-labeled aptamers or complementary single-stranded DNA probes were initially adsorbed on the surface of polydopamine nanoparticles (PDA). In this case, the collision of PDA on the electrode triggered a cascade redox cycling between the Ri and the nanoparticle through synchronous electron transfer, producing an amplified signal output. However, the presence of targets, not only tumor protein markers but also tumor-associated nucleic acids or small molecule metabolites, caused the Ri-labeled aptamers or DNA probes to detach from the surface of PDA, thus disrupting the cascade redox cycling and resulting in significantly reduced signal output. Based on this design, the biosensor was successfully used for quantitative analysis of VEGF, miRNA-21, and ATP, three upregulated cancer biomarkers, with wide linear ranges and low limits of detection (32 fM [approximately 0.608 pg/mL], 3.86 × 105 fM, and 2.8 pM, respectively).
Many other interesting schemes have also been incorporated in the construction of aptamer-based electrochemical biosensors.116−119 For instance, our group reported a universal electrochemical biosensor for the analysis of tumor-associated glycoprotein biomarkers by simultaneously using an aptamer probe and a phenylboronic acid-modified DNA probe.116 The binding of the two probes to a single glycoprotein resulted in a spatial proximity effect that facilitated the formation of a circular template for rolling circle amplification (RCA). Subsequently, the amplification reaction produced a large number of byproducts, pyrophosphate, which accelerated the breakdown of ZIF-90 to release MB for electrochemical measurements. The biosensor effectively avoided false positive results associated with the single recognition, enabling precise determination of a variety of tumor-associated glycoprotein biomarkers, including CEA, AFP, and MUC1 (Figure 4C). Recently, Noh et al. prepared an Au microgap electrode coated with aptamer/MXene nanosheets to determine TNF-α and IFN-γ levels using a single biosensor.119 Depending on the alternating current electrothermal flow, both targets could be detected within a short time (<10 min) and with desirable limits of detection of 0.25 pg/mL and 0.26 pg/mL, respectively (Figure 4D).
Multitarget Analysis of EVs
According to the definition established by the International Society for Extracellular Vesicles (ISEV), EVs are the nano- or microsized particles secreted by cells, which are delimited by a lipid bilayer and incapable of self-replication.120 EVs, including exosomes and microvesicles, generally contain a large amount of biocomponents from the parental cells, such as proteins, nucleic acids, lipids, and metabolites. They are involved in intercellular communication and contribute to cancer progression by promoting tumor growth, angiogenesis, immune suppression, and distant metastasis.121,122 Therefore, EVs have become appealing circulating biomarkers in liquid biopsy for early cancer detection and disease progression monitoring, as well as for indicating drug resistance.123 So far, electrochemical biosensors have been utilized to determinate a series of EV biomarkers, such as surface proteins and endogenous RNAs, indicating their potential for diagnostic applications. Moreover, the simultaneous targeting of two or more EV biomarkers has been shown to overcome the stochastic expression effect of a single biomarker on or within an individual vesicle, thereby enhancing the utility of EV analysis in addressing tumor heterogeneity for precise cancer diagnosis. Typically, the analysis of EVs demands an initial step of isolating them from diverse sources (e.g., blood and urine). A variety of techniques, for instance, ultracentrifugation, size-exclusion chromatography, polymer-based precipitation, and affinity approaches based on molecular recognition, have been developed to fulfill this research requirement.120 These techniques leverage the biophysical or biochemical characteristics of EVs, like size, density, charge, surface composition (such as specific surface proteins or lipids), and their advancement provides increasing possibilities for multitarget analysis of EVs. Nonetheless, distinct isolation techniques exhibit notable variations in terms of efficiency, specificity, cost-effectiveness, and their impact on the composition of isolated EVs. Hence, careful consideration should be given to the choice of isolation techniques during the development of electrochemical biosensors for EV analysis, which might directly influence both the accuracy and diagnostic value of final analytical outcomes.
Multitarget Analysis of EV Surface Proteins
EVs carry a wide variety of surface proteins mirroring parental cells, which play a crucial role in their identification and analysis. The sandwich-type model has been extensively used to illustrate the interaction between the recognition element, the surface protein of EVs, and the signaling probe to produce electrochemical signals. Unlike tumor protein markers, EVs display a greater surface area and more binding sites for the attachment of recognition and signaling probes, which may help to enhance the detection sensitivity.
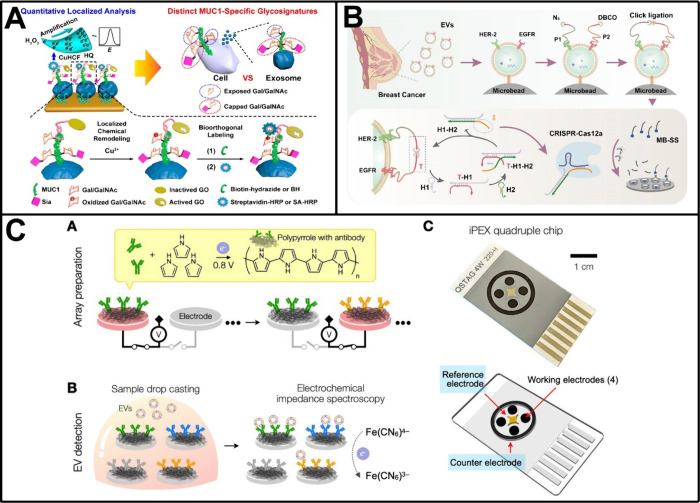
Based on the simultaneous targeting of dual surface proteins, electrochemical biosensors have been widely developed for analyzing EVs that expressed either CD9 and epithelial cell adhesion molecule (EpCAM),124 MUC1 and EpCAM,125 CD63 and nucleolin,126 CD63 and PD-L1,127,128 HER-2 and EGFR,129 CD49f and EpCAM,130 tyrosine kinase-like 7 (PTK7) and PSMA,131 or CD63 and MUC1.132,133 In this aspect, Ding and co-workers reported a representative work.125 As shown in Figure 5A, an electrode-confined aptamer was designed to target EpCAM for capturing EVs derived from the breast cancer cell line MCF-7, and an inactivated galactose oxidase (GO)-coupled aptamer was utilized to bind to the glycoprotein MUC1 on the EV surface. Upon Cu2+-mediated activation of the GO, localized remodeling of terminal galactose/N-acetylgalactosamine was performed on the MUC1 surface, enabling the bioorthogonal labeling with biotin-hydrazide and the subsequent labeling with streptavidin-horseradish peroxidase for electrochemical signal output. In combination with sialic acid cleavage manipulation, the method, named quantitative localized analysis, revealed distinct MUC1-specific sialylation capping ratios in EVs derived from MCF-7 and MDA-MB-231 cells, providing new insights into the use of EVs as noninvasive indicators for breast cancer subtyping. Not long ago, Ge et al. constructed a double-hook type aptamer-based electrochemical biosensor to facilitate accurate early diagnosis of oral cancer based on EV analysis.126 This sensor utilized CD63 aptamer as the first hook to capture EVs, while another aptamer to target specific oral cancer-associated EV surface protein, such as nucleolin. Through the adoption of electrochemical impedance spectroscopy, this sensor confirmed excellent linearity within the EV concentration range of 3.1 × 104 to 3.1 × 109 particles/mL, with a detection limit as low as 1.2 × 104 particles/mL. Our group also proposed some interesting biosensors for the analysis of EVs through the recognition of coexisting surface proteins.128,129 For example, we recently developed a proximity-guaranteed DNA machine-based electrochemical biosensor for analyzing breast cancer EVs expressing both HER-2 and EGFR (Figure 5B).129 In the biosensor, target EVs were initially captured by phosphatidylserine-targeting peptide-functionalized microbeads and then were selectively labeled by two proximity probes that bound to HER-2 and EGFR respectively, triggering a proximity-guaranteed copper-free click ligation. As a result, a ligated intact activator was generated to manipulate the DNA machine, which eventually activated the trans-cleavage activity of CRISPR-Cas12a to arouse amplified signals. Challenging with EVs derived from the breast cancer cell line BT474 and circulating EVs found in plasma samples, the biosensor demonstrated high sensitivity with an impressive detection limit of 985 particles/mL and also good clinical practicability, which may have potential use in the subtype-based diagnosis of breast cancer.
Figure 5.
Electrochemical biosensors for multitarget analysis of EV surface proteins. (A) Electrochemical biosensor for EV surface protein-specific glycoform analysis based on simultaneous targeting of MUC1 and EpCAM. Reproduced from ref (125). Copyright 2020 American Chemical Society. (B) Proximity-guaranteed DNA machine-based electrochemical biosensor for analyzing breast cancer EVs expressing both HER-2 and EGFR. Reproduced from ref (129). Copyright 2024 American Chemical Society. (C) Impedance profiling of EV surface proteins based on a quadruple sensor chip. Reproduced from ref (137). Copyright 2022 American Chemical Society.
Despite the progress made in EV analysis using electrochemical biosensors based on simultaneous targeting of surface proteins, which has enhanced our comprehension of the molecular characteristics of tumors, these biosensors are unlikely to be effective for all cancer patients, even those with the same cancer due to the highly heterogeneous nature of tumors. Instead, efforts have been devoted to developing electrochemical biosensors for protein profiling of EVs.134−141 These biosensors can analyze a panel of EV surface proteins, thereby providing a more comprehensive picture of the surface molecular composition of EVs, which is of great significance for accurately diagnosing cancers.142,143 One of the early milestones in this aspect was accomplished by Lee and co-workers, who developed an integrated magneto–electrochemical sensor for multiplexed EV analyses.134 This sensor combined two orthogonal modalities, magnetic isolation and electrochemical measurement, and adopted the chronoamperometry approach for signal collection. By employing a portable, eight-channel device, this sensor allowed for the simultaneous profiling of multiple EV surface proteins, such as the three typical tetraspanin proteins (CD63, CD9, CD81) or a combination of six representative tumor markers (i.e., EpCAM, CD24, CA125, HER-2, MUC18, and EGFR), showing superior sensitivity and speed compared to conventional methods. In another work, Lee and co-workers presented an improved version of the integrated magneto-electrochemical biosensor, known as HiMEX, for the rapid protein profiling of EVs.135 The HiMEX streamlined EV protein profiling by combining the immune-magnetic enrichment of EVs and the probe labeling for signal generation in a single workflow. Based on integrating a compact reader to carry out 96 parallel measurements, the HiMEX assessed expression levels of a panel of EV surface proteins, including EpCAM, EGFR, CD133, GPA33, CD24, and CD63, in less than 1 h, empowering the classification of plasma samples from 102 colorectal cancer patients and 40 controls with an accuracy of exceeded 96%. Recently, Kilic et al. prepared a quadruple sensor chip to perform impedance profiling of EV surface proteins (Figure 5C).137 The sensor chip was custom-designed with four antibody-functionalized working electrodes, which facilitated the parallel analysis of multiple proteins based on changes in electrochemical impedance spectroscopy. Through one-step electropolymerization with different antibodies, a glioblastoma (GBM) chip was successfully tailored to simultaneously identify three known GBM biomarkers (EGFR, EGFRvIII, PDGFRα) and showed potential clinical utility by directly analyzing plasma samples from GBM patients. Also using a sensor chip with multiple antibody-functionalized working electrodes, Zhang et al. constructed an integrated electrochemical liquid biopsy platform based on ZIF-90-ZnO-MoS2 nanohybrids for the direct profiling of EVs in blood.139 In a cohort of 24 participants (n = 12 for healthy individuals and n = 12 for lung cancer patients), the platform revealed distinct expression profiles of EV surface proteins, such as CD9, CD63, and CD81, and demonstrated effectiveness in differentiating cancerous groups from healthy controls.
Multitarget Analysis of EV RNAs
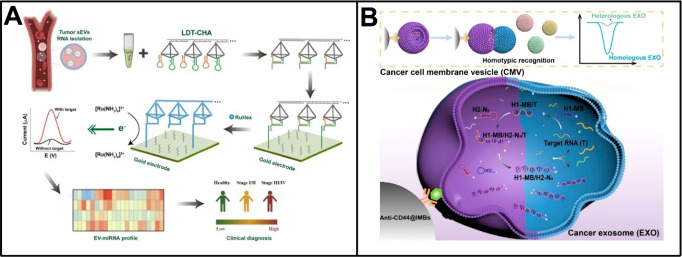
There is an abundance of RNA species inside EVs with specialized functions, such as miRNAs, mRNAs, and lncRNAs.144 Due to the protection of the outer lipid membrane, EV RNAs exhibit much higher stability compared to free RNAs in biofluids and are regarded as reliable biomarkers in the noninvasive diagnosis of cancers.145,146 Increasing evidence suggests that EVs from different cell types and status have distinct RNA profiles. Consequently, in recent years, electrochemical biosensors have been put forward for the multitarget analysis of EV RNAs. For example, Yang et al. developed a disposable paper-based electrochemical biosensor for simultaneous detection of EV miRNAs (such as miRNA-155 and miRNA-21).147 To achieve this, two electroactive dyes-loaded functional MOFs were prepared with hairpin DNA probe caps and were subsequently used in conjunction with gold nanoparticles and graphene walls stacked carbon fiber papers. The presence of target EV miRNAs was designed to trigger strand displacement reactions that produced obvious signals from the electroactive dyes, giving the biosensor ideal performance, even in clinical plasma samples. Furthermore, Zhang et al. proposed an electrochemical biosensor utilizing localized DNA tetrahedrons assisted catalytic hairpin assembly (LDT-CHA) for sensitive profiling of EV miRNAs (Figure 6A).148 The LDT-CHA leveraged an efficient localized reaction that reduced the distance between reaction units and increased substrate concentration within a specific region, contributing to the enhanced sensitivity in the analysis of miRNA targets. Based on the LDT-CHA, a panel of four EV miRNAs (i.e., miRNA-1246, miRNA-21, miRNA-183–5P, and miRNA-142–5P) were quantitatively detected down to 2.5 × 10–2 fM in 30 min, demonstrating improved accuracy for diagnosing gastric cancer and monitoring treatment effects compared to that using a single miRNA biomarker. Very recently, these researchers also reported a sensitive electrochemical analysis of multiple EV-associated circRNAs through the integration of DT-CHA and tetrahedron-Dox-AuNPs (TDA) tags.149 The dual signal amplification provided by DT-CHA and TDA enabled this biosensor to achieve an exceptional detection limit for target circRNAs, reaching 0.1531 fM. By profiling four specific circRNAs (circNRIP1, circRANGAP1, circCORO1C, and circSHKBP1), this biosensor further demonstrated remarkable efficacy in distinguishing early gastric cancer patients from healthy donors.
Figure 6.
Electrochemical biosensors for multitarget analysis of EV RNAs. (A) LDT-CHA-based electrochemical profiling of EV RNAs. Reproduced from ref (149). Available under a CC-BY 4.0 license. Copyright 2022 The authors. (B) Electrochemical biosensor based on homotypic recognition for analyzing EVs and their carried RNAs. Reproduced from ref (150). Copyright 2022 American Chemical Society.
Electrochemical biosensors are undoubtedly valuable for the analysis of EV RNAs. However, current sensors either rely on recognizing a single surface biomarker for EV enrichment, leaving them susceptible to the expression variability of the marker, or they analyze endogenous RNA through EV disintegration, making it challenging to avoid interference from free RNAs. To address these issues, our group recently introduced an electrochemical biosensor based on homotypic recognition for analyzing EVs and their carried RNAs.150 As shown in Figure 6B, a biomimetic vesicle was prepared by camouflaging a catalytic DNA machine with a breast cancer cell membrane, which could identify and fuse with breast cancer EVs sharing similar subtype characteristics. Following the fusion, EV RNAs, such as miRNA-375 and PD-L1 mRNA, acted as endogenous triggers to manipulate the catalytic DNA machine, producing numerous clickable double-stranded DNA products, which were finally enriched onto electrodes via click chemistry for electrochemical determination. Leveraging the homotypic recognition, our biosensor exhibited satisfactory performance in evaluating the molecular features of breast cancer EVs and demonstrated high reliability for subtype-based diagnosis and stage-specific monitoring of breast cancer patients.
Multitarget Analysis of Tumor Cells
Tumor cells, characterized by uncontrolled proliferation, migration, and invasive capabilities, comprise the main components of neoplastic tissues. Understanding their behaviors and characteristics is essential for uncovering the mechanisms behind cancers. This knowledge can offer invaluable insights into fundamental processes that drive tumorigenesis and can help identify potential therapeutic targets. Of particular interest in liquid biopsy are CTCs that disseminate from the primary tumor into the bloodstream.151,152 They can provide abundant information about disease progression, treatment evaluation, and prediction of recurrence and distant metastasis. As intricate biological entities, tumor cells carry a variety of genetic, epigenetic, and molecular information and can be exploited to elucidate tumor heterogeneity.153 However, the analysis of tumor cells is primarily impeded by their low abundance in the bloodstream, particularly during the early stage of cancer, as well as by the concurrent presence of numerous interfering cells. In this sense, electrochemical biosensors capable of multitarget analysis are essential for analyzing tumor cells because of their inherent sensitivity and the comprehensive information they provide.
Multitarget Analysis of Tumor Cell Surface Proteins
Remarkable advancements have been witnessed in the coidentification of multiple cell surface proteins by electrochemical biosensors, which would aid in the accurate targeting and analysis of tumor cells.154−160 In an earlier study, we developed a sensitive electrochemical immunosensor for the detection of breast cancer cells by simultaneously recognizing coexpressed cell surface proteins, MUC1 and CEA.154 This effectively enhanced the detection accuracy and facilitated the classification of cancer cells. In another work, Wan et al. reported a quite different strategy for the electrochemical identification of cancer cells.155 Their strategy relied on the use of a family of aptamer- or antibody-modified metal nanoparticles (including Cu nanoparticles, Ag nanoparticles, and Pd nanoparticles), which were capable of generating well-resolved and distinguishable voltammetric responses corresponding to their respective target surface proteins. With the aid of these nanoparticle labels, their strategy permitted simultaneous analysis of multiple proteins on the surface of cancer cells, such as HER-2, PSMA, and MUC1, yielding results comparable to those obtained from the gold standard immunostaining. Figure 7A shows another work reported by our group, in which an electrochemical biosensor was proposed based on the simultaneous binding of dual aptamer probes to cell surface-expressing proteins, EpCAM and MUC1.156 The hybridization of dual probes with each other at the same breast cancer cell triggered a dimer-like RCA reaction, resulting in the generation of long DNA products with repeated G-quadruplex-forming sequences. Subsequently, a significant current signal upon catalyzing the oxidation of TMB was obtained and allowed for the sensitive detection of target breast cancer cells down to 3 cells/mL. In a recent work, Zhang et al. proposed an electrochemical and fluorescent dual-mode biosensor based on three kinds of aptamer-functionalized Co–Fe-MOF nanomaterials.159 These nanomaterials adsorbed aptamers for specific cell surface proteins, and displayed nanozyme activity after the binding of aptamers to their targets, leading to self-catalytic disintegration in the presence of hydrogen peroxide. As a result, a large amount of internally loaded dyes (MB, TMB or NR) were released to generate corresponding electrochemical and fluorescent signals. Using surface proteins such as PTK7, EpCAM, and MUC1 as targets, this dual-mode biosensor realized accurate typing of different cancer cells and normal cells by simultaneously analyzing the expression of these proteins in one step.
Figure 7.
Electrochemical biosensors for multitarget analysis of tumor cells. (A) Dual-recognition-controlled electrochemical biosensor for the accurate analysis of specific tumor cells. Reproduced with permission from ref (156). Copyright 2022 Elsevier. (B) In situ PDC-based electrochemical biosensor for the precise differentiation of the stem-like subpopulation in breast cancer cells. Reproduced from ref (161). Copyright 2021 American Chemical Society. (C) Endogenous miRNA discriminator utilizing enzyme-powered strand displacement reactions for the subtype-specific diagnosis of breast cancer. Reproduced from ref (163). Available under a CC BY-NC 3.0 license. Copyright 2023 The authors. (D) PNA-peptide-DNA copolymers-guided nanosensor for the AND-gated and dual-model sensing of cathepsin B and miRNA-21. Reproduced from ref (165). Copyright 2023 American Chemical Society.
Phenotypic characteristics of tumor cells are closely associated with their biological properties. Recently, our group has focused on the phenotypic characterization of a specific subpopulation of breast cancer cells known as breast cancer stem cells (BCSCs), which could be identified based on the positive expression of surface protein CD44 and the negative/low expression of another surface protein CD24. As shown in Figure 7B, we first developed an electrochemical biosensor for the precise differentiation of the stem-like subpopulation in breast cancer cells, in which an in situ programmable DNA circuit (PDC) was engineered following the cell enrichment on anti-CD44-functionalized magnetic beads.161 Specifically, two capture probes (A1A@Anti-CD24 and B1B@Antinucleolin) bound to respective surface proteins via antibody parts, while their nucleic acid parts served as toeholds to operate the in situ PDC, eventually eliminating signal labels from CD24+ breast cancer cells. In this way, an “always-ON” electrochemical sensor was established for BCSC identification, which demonstrated comparable accuracy to the “gold-standard” flow cytometry especially when used in breast tumor-bearing mice. Very recently, we further constructed a phenotype-directed DNA nanomachine for in situ multidemand analysis of the stem-like subpopulation in breast tumors.162 Two antibody-nucleic acid recognition probes (I@Anti-CD44 and I’@anti-CD24) were able to modulate the strand migration pathway with varying displacement rates when interacting with a three-stranded signal probe C/N/T. As a result, the assembly product of C/T/I was exclusively anchored at BCSCs, enabling the signal strand C to perform diverse functions. The integration of electroactive silver nanoclusters with the strand C allowed for the quantitative determination of BCSCs, while the integration of biotin enabled the “one-step” magnetic isolation of this subpopulation. This method not only demonstrated comparable accuracy to flow cytometry but also showed advantages of simple operation and low cost in discriminating specific cell subgroups, thereby advancing BCSC-targeted diagnosis, treatment, and related mechanism researches.
Multitarget Analysis of Tumor Cell Endogenous Biomarkers
Endogenous biomarkers within tumor cells, including but not limited to RNAs, enzymes, lipids, thiols, and carbohydrates, are closely associated with tumor progression. Accordingly, emerging electrochemical biosensors are designed for their multitarget analysis, which help to understand the biological processes of cancer development and drug screening.163−165 In one such work, our group constructed an endogenous miRNA discriminator utilizing enzyme-powered strand displacement reactions to differentiate breast cancer cells from normal cells and identify subtype-specific characteristics among them (Figure 7C).163 As designed, miRNA-21 was employed as a universal biomarker of breast cancer cells to initiate the first round of T7 exonuclease-powered strand displacement reactions, while miRNA-210 was utilized to identify features specific to the triple-negative subtype by initiating the second round of the T7 exonuclease-powered strand displacement reactions. Consequently, our discriminator demonstrated high sensitivity to determine the two endogenous miRNAs at the femtomolar level and facilitated the precise discrimination of the subtype feature among different breast cancer cells. In another interesting work, Xi et al. proposed a PNA-peptide-DNA copolymers-guided nanosensor for the AND-gated and dual-model sensing of cathepsin B and miRNA-21 (Figure 7D).165 The nanosensor was constructed by hybridizing PNA-peptide-PNA triblock complexes with FAM-DNA/BHQ-DNA duplexes and was designed to run a miRNA-21-assisted strand displacement reaction upon cathepsin B-catalyzed activation, leading to an obvious fluorescence recovery. In the meanwhile, the released BHQ-DNA initiated another strand displacement reaction at the inserted gold nanoelectrode, resulting in a ∼ 30% decrease in electrochemical signals at the single-cell level.
Conclusion and Perspectives
In this perspective, we provide an overview of recent advancements in electrochemical biosensors, particularly for the simultaneous analysis of multiple cancer biomarkers, including tumor-associated nucleic acids, tumor protein markers, EVs, and tumor cells. Electrochemical biosensors have demonstrated superior characteristics of inherent simplicity, wide measurement range, and low detection limit, significantly contributing to early cancer diagnosis and screening. More importantly, the electrochemical biosensors capable of multitarget analysis of cancer biomarkers are advantageous in addressing tumor heterogeneity. They can overcome the relatively limited specificity and sensitivity of single-target analysis in cancer diagnosis and offer more profound details concerning the molecular characteristics of malignant tumors from multiple perspectives, potentially providing more valuable tools for cancer management.
Despite the promising performance in the identification and analysis of multiple cancer biomarkers, the integration of electrochemical biosensors into clinical diagnostic practice still confronts many challenges. Thus, ongoing efforts are required to develop advanced electrochemical biosensors with superior clinical practicality. For instance: (1) The diversity of cancer biomarkers within a specific tumor is dictated by unique molecular characteristics and varying abundances in biological systems, including the potential presence of certain biomarkers at extremely low levels. Consequently, there remains a desire for recognition molecules with enhanced specificity and affinity or signal amplification units with higher efficiency, in order to improve detection sensitivity. (2) The risk of signal cross-interference in multitarget analysis may give rise to false-negative or false-positive results, ultimately affecting the diagnostic accuracy. Thus, it is necessary to explore combinations of electrochemical reporters with greater discriminatory power or novel designs of microchannels and electrode arrays that offer more rational spatial distributions. (3) The complex biological composition of clinical samples renders the electrochemical sensing interface highly vulnerable to nonspecific adsorption. As such, integrating more effective antifouling strategies is crucial for enhancing the stability and reproducibility of electrochemical biosensors when analyzing actual samples. (4) Multitarget analysis utilizing electrochemical biosensors can yield large quantities of current, potential, or resistance data; this presents difficulties in extracting relevant molecular information applicable to cancer diagnosis. In this context, leveraging artificial intelligence—particularly machine learning algorithms—is expected to streamline data analysis processes while reducing signal noise interference and improving both accuracy and depth in multidiagnostic information extraction.
Overall, it is anticipated that the progression of future electrochemical biosensors will be closely associated with the enhancement of multimolecular recognition and clinical utility, which would provide a more reliable tool for the precise detection of cancers and the monitoring of treatment efficacy.
Acknowledgments
This work was supported by the Natural Science Foundation of Shanghai (Grant No. 23ZR1421400) and the National Natural Science Foundation of China (Grant No. 81972799).
Author Contributions
The manuscript was written through contributions of all authors. CRediT: Ya Cao funding acquisition, writing - original draft, writing - review & editing; Jianan Xia writing - original draft; Lijuan Li writing - original draft; Yujing Zeng writing - original draft; Jing Zhao funding acquisition, supervision, writing - review & editing; Genxi Li supervision, writing - review & editing.
The authors declare no competing financial interest.
References
- Wu J.; Liu H.; Chen W.; Ma B.; Ju H. Device Integration of Electrochemical Biosensors. Nat. Rev. Bioeng. 2023, 1, 346–360. 10.1038/s44222-023-00032-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn C. D.; Chang D.; Mahmud A.; Yousefi H.; Das J.; Riordan K. T.; Sargent E. H.; Kelley S. O. Biomolecular Sensors for Advanced Physiological Monitoring. Nat. Rev. Bioeng. 2023, 1, 560–575. 10.1038/s44222-023-00067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford A.; Das J.; Yousefi H.; Mahmud A.; Chen J. B.; Kelley S. O. Strategies for Biomolecular Analysis and Continuous Physiological Monitoring. J. Am. Chem. Soc. 2021, 143, 5281–5294. 10.1021/jacs.0c13138. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Jin K.; Li J.; Sheng K.; Huang W.; Liu Y. Flexible and Stretchable Electrochemical Sensors for Biological Monitoring. Adv. Mater. 2023, 2305917. 10.1002/adma.202305917. [DOI] [PubMed] [Google Scholar]
- Natalia A.; Zhang L.; Sundah N. R.; Zhang Y.; Shao H. Analytical Device Miniaturization for the Detection of Circulating Biomarkers. Nat. Rev. Bioeng. 2023, 1, 481–498. 10.1038/s44222-023-00050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Sen P.; Adhikari B. R.; Li Y.; Soleymani L. Development of Nucleic-Acid-Based Electrochemical Biosensors for Clinical Applications. Angew. Chem., Int. Ed. 2022, 61, e202212496 10.1002/anie.202212496. [DOI] [PubMed] [Google Scholar]
- Dai Y.; Liu C. C. Recent Advances on Electrochemical Biosensing Strategies Toward Universal Point-of-Care Systems. Angew. Chem., Int. Ed. 2019, 58, 12355–12368. 10.1002/anie.201901879. [DOI] [PubMed] [Google Scholar]
- Li S.; Zhang H.; Zhu M.; Kuang Z.; Li X.; Xu F.; Miao S.; Zhang Z.; Lou X.; Li H.; et al. Electrochemical Biosensors for Whole Blood Analysis: Recent Progress, Challenges, and Future Perspectives. Chem. Rev. 2023, 123, 7953–8039. 10.1021/acs.chemrev.1c00759. [DOI] [PubMed] [Google Scholar]
- Rayamajhi S.; Sipes J.; Tetlow A. L.; Saha S.; Bansal A.; Godwin A. K. Extracellular Vesicles as Liquid Biopsy Biomarkers across the Cancer Journey: From Early Detection to Recurrence. Clin. Chem. 2024, 70, 206–219. 10.1093/clinchem/hvad176. [DOI] [PubMed] [Google Scholar]
- Wu L.; Wang Y.; Xu X.; Liu Y.; Lin B.; Zhang M.; Zhang J.; Wan S.; Yang C.; Tan W. Aptamer-Based Detection of Circulating Targets for Precision Medicine. Chem. Rev. 2021, 121, 12035–12105. 10.1021/acs.chemrev.0c01140. [DOI] [PubMed] [Google Scholar]
- Crosby D.; Bhatia S.; Brindle K. M.; Coussens L. M.; Dive C.; Emberton M.; Esener S.; Fitzgerald R.; Gambhir S. S.; Kuhn P.; et al. Early Detection of Cancer. Science 2022, 375, eaay9040 10.1126/science.aay9040. [DOI] [PubMed] [Google Scholar]
- Wang M.; Li L.; Zhang L.; Zhao J.; Jiang Z.; Wang W. Peptide-Derived Biosensors and Their Applications in Tumor Immunology-Related Detection. Anal. Chem. 2022, 94, 431–441. 10.1021/acs.analchem.1c04461. [DOI] [PubMed] [Google Scholar]
- Xu J.; Miao H.; Wang J.; Pan G. Molecularly Imprinted Synthetic Antibodies: From Chemical Design to Biomedical Applications. Small 2020, 16, 1906644. 10.1002/smll.201906644. [DOI] [PubMed] [Google Scholar]
- Campuzano S.; Pingarrón J. M. Electrochemical Affinity Biosensors: Pervasive Devices with Exciting Alliances and Horizons Ahead. ACS Sens. 2023, 8, 3276–3293. 10.1021/acssensors.3c01172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissanaprasit A.; Key C. M.; Pontula S.; LaBean T. H. Self-Assembling Nucleic Acid Nanostructures Functionalized with Aptamers. Chem. Rev. 2021, 121, 13797–13868. 10.1021/acs.chemrev.0c01332. [DOI] [PubMed] [Google Scholar]
- Wang B.; Zhao J.; Zhang J.; Wei T.; Han K.; Gao T. Electrochemical Biosensing Interfaced with Cell-free Synthetic Biology. TrAC-Trend. Anal. Chem. 2024, 176, 117756. 10.1016/j.trac.2024.117756. [DOI] [Google Scholar]
- Yang S.; Zhu R.; Wang S.; Xiong Y.; Zhou G.; Cao Y.; Zhao J. Recent Advances in DNA-Based Molecular Devices and Their Applications in Cancer Diagnosis. Coordin. Chem. Rev. 2023, 493, 215331. 10.1016/j.ccr.2023.215331. [DOI] [Google Scholar]
- Reddy K. K.; Bandal H.; Satyanarayana M.; Goud K. Y.; Gobi K. V.; Jayaramudu T.; Amalraj J.; Kim H. Recent Trends in Electrochemical Sensors for Vital Biomedical Markers Using Hybrid Nanostructured Materials. Adv. Sci. 2020, 7, 1902980. 10.1002/advs.201902980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham C. E.; Morrison S. J. Tumour Heterogeneity and Cancer Cell Plasticity. Nature 2013, 501, 328–337. 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagogo-Jack L.; Shaw A. T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- Hausser J.; Alon U. Tumour Heterogeneity and the Evolutionary Trade-offs of Cancer. Nat. Rev. Cancer 2020, 20, 247–257. 10.1038/s41568-020-0241-6. [DOI] [PubMed] [Google Scholar]
- Liu X.; Wu W.; Cui D.; Chen X.; Li W. Functional Micro-/Nanomaterials for Multiplexed Biodetection. Adv. Mater. 2021, 33, 2004734. 10.1002/adma.202004734. [DOI] [PubMed] [Google Scholar]
- Cai S.; Pataillot-Meakin T.; Shibakawa A.; Ren R.; Bevan C. L.; Ladame S.; Ivanov A. P.; Edel J. B. Single-Molecule Amplification-Free Multiplexed Detection of Circulating MicroRNA Cancer Biomarkers from Serum. Nat. Commun. 2021, 12, 3515. 10.1038/s41467-021-23497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez-Gabriel M.; Heymann M.; Heymann D. Circulating Tumor Cells as a Tool for Assessing Tumor Heterogeneity. Theranostics 2019, 9, 4580–4594. 10.7150/thno.34337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu V.; Kalluri R. Exosomes as a Multicomponent Biomarker Platform in Cancer. Trend. Cancer 2020, 6, 767–774. 10.1016/j.trecan.2020.03.007. [DOI] [PubMed] [Google Scholar]
- Keller L.; Pantel K. Unravelling Tumour Heterogeneity by Single-Cell Profiling of Circulating Tumour Cells. Nat. Rev. Cancer 2019, 19, 553–567. 10.1038/s41568-019-0180-2. [DOI] [PubMed] [Google Scholar]
- Bordanaba-Florit G.; Royo F.; Kruglik S. G.; Falcón-Pérez J. M. Using Single-Vesicle Technologies to Unravel the Heterogeneity of Extracellular Vesicles. Nat. Protoc. 2021, 16, 3163–3185. 10.1038/s41596-021-00551-z. [DOI] [PubMed] [Google Scholar]
- Klebes A.; Ates H. C.; Verboket R. D.; Urban G. A.; von Stetten F.; Dincer C.; Früh S. M. Emerging Multianalyte Biosensors for the Simultaneous Detection of Protein and Nucleic Acid Biomarkers. Biosens. Bioelectron. 2024, 244, 115800. 10.1016/j.bios.2023.115800. [DOI] [PubMed] [Google Scholar]
- Li J.; Wuethrich A.; Dey S.; Lane R. E.; Sina A. A. I.; Wang J.; Wang Y.; Puttick S.; Koo K. M.; Trau M. The Growing Impact of Micro/Nanomaterial-Based Systems in Precision Oncology: Translating “Multiomics” Technologies. Adv. Funct. Mater. 2020, 30, 1909306. 10.1002/adfm.202070248. [DOI] [Google Scholar]
- Sharafeldin M.; Rusling J. F. Multiplexed Electrochemical Assays for Clinical Applications. Curr. Opin. Electrochem. 2023, 39, 101256. 10.1016/j.coelec.2023.101256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.; Dhanapala L.; Kankanamage R. N. T.; Kumar C. V.; Rusling J. F. Multiplexed Immunosensors and Immunoarrays. Anal. Chem. 2020, 92, 345–362. 10.1021/acs.analchem.9b05080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.; Zeng Q.; Wang Z.; Li C.; Xu Y.; Cui P.; Zhu X.; Lu H.; Wang G.; Cai S.; et al. Circulating Cell-Free DNA for Cancer Early Detection. Innovation 2022, 4, 100259. 10.1016/j.xinn.2022.100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J.; Wickramasinghe V. O. RNA in Cancer. Nat. Rev. Cancer 2021, 21, 22–36. 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- Cheng M.; Pectasides E.; Hanna G. J.; Parsons H. A.; Choudhury A. D.; Oxnard G. R. Circulating Tumor DNA in Advanced Solid Tumors: Clinical Relevance and Future Directions. CA Cancer J. Clin. 2021, 71, 176–190. 10.3322/caac.21650. [DOI] [PubMed] [Google Scholar]
- Miao P.; Chai H.; Tang Y. DNA Hairpins and Dumbbell-Wheel Transitions Amplified Walking Nanomachine for Ultrasensitive Nucleic Acid Detection. ACS Nano 2022, 16, 4726–4733. 10.1021/acsnano.1c11582. [DOI] [PubMed] [Google Scholar]
- Park B. C.; Soh J. O.; Choi H.-J.; Park H. S.; Lee S. M.; Fu H. E.; Kim M. S.; Ko M. J.; Koo T. M.; Lee J.-Y.; Kim Y. K.; Lee J. H. Ultrasensitive and Rapid Circulating Tumor DNA Liquid Biopsy Using Surface-Confined Gene Amplification on Dispersible Magnetic Nano-Electrodes. ACS Nano 2024, 18, 12781–12794. 10.1021/acsnano.3c12266. [DOI] [PubMed] [Google Scholar]
- Dong J.; Li X.; Hou H.; Hou J.; Huo D. A Novel CRISPR/Cas12a-Mediated Ratiometric Dual-Signal Electrochemical Biosensor for Ultrasensitive and Reliable Detection of Circulating Tumor Deoxyribonucleic Acid. Anal. Chem. 2024, 96, 6930–6939. 10.1021/acs.analchem.3c05700. [DOI] [PubMed] [Google Scholar]
- Fu Y.; Li J.; Niu D.; Yang G.; Yang M.; Shen H.; Li L. Synergistic Signal Amplification of a 3D Dual-Core DNA Nanomachine and PCNs@AuPdCe Hybrid Nanozymes for Ultrasensitive Electrochemical Detection of Cell-free DNA. Chem. Eng. J. 2023, 475, 146323. 10.1016/j.cej.2023.146323. [DOI] [Google Scholar]
- Das J.; Ivanov I.; Sargent E. H.; Kelley S. O. DNA Clutch Probes for Circulating Tumor DNA Analysis. J. Am. Chem. Soc. 2016, 138, 11009–11016. 10.1021/jacs.6b05679. [DOI] [PubMed] [Google Scholar]
- Das J.; Ivanov I.; Safaei T. S.; Sargent E. H.; Kelley S. O. Combinatorial Probes for High-Throughput Electrochemical Analysis of Circulating Nucleic Acids in Clinical Samples. Angew. Chem., Int. Ed. 2018, 57, 3711–3716. 10.1002/anie.201800455. [DOI] [PubMed] [Google Scholar]
- Koo K. M.; Trau M. Direct Enhanced Detection of Multiple Circulating Tumor DNA Variants in Unprocessed Plasma by Magnetic-Assisted Bioelectrocatalytic Cycling. ACS Sens. 2020, 5, 3217–3225. 10.1021/acssensors.0c01512. [DOI] [PubMed] [Google Scholar]
- Cai Q.; Li H.; Wang B.; Jie G. A Spatial-Potential-Resolved Electrochemiluminescence Biosensor for Simultaneous Detection of BRCA1 and BRCA2 Based on a Novel Self-Luminescent Metal-Organic Framework. Chem. Eng. J. 2023, 476, 146799. 10.1016/j.cej.2023.146799. [DOI] [Google Scholar]
- Genco E.; Modena F.; Sarcina L.; Björkström K.; Brunetti C.; Caironi M.; Caputo M.; Demartis V. M.; Di Franco C. D.; Frusconi G.; et al. A Single-Molecule Bioelectronic Portable Array for Early Diagnosis of Pancreatic Cancer Precursors. Adv. Mater. 2023, 35, 2304102. 10.1002/adma.202304102. [DOI] [PubMed] [Google Scholar]
- García V.; García J. M.; Peña C.; Silva J.; Domínguez G.; Lorenzo Y.; Diaz R.; Espinosa P.; de Sola J. G.; Cantos B.; Bonilla F. Free Circulating mRNA in Plasma from Breast Cancer Patients and Clinical Outcome. Cancer Lett. 2008, 263, 312–320. 10.1016/j.canlet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Luo Y.; Chen X.; Li H.; Huang B.; Zhou B.; Zhu L.; Kang X.; Geng W. The Role of mRNA in the Development, Diagnosis, Treatment and Prognosis of Neural Tumors. Mol. Cancer 2021, 20, 49. 10.1186/s12943-021-01341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth K.; Bayraktar R.; Ferracin M.; Calin G. A. Non-Coding RNAs in Disease: From Mechanisms to Therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. 10.1038/s41576-023-00662-1. [DOI] [PubMed] [Google Scholar]
- Slack F. J.; Chinnaiyan A. M. The Role of Non-Coding RNAs in Oncology. Cell 2019, 179, 1033–1055. 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Yin F.; Song L.; Mao X.; Li F.; Fan C.; Zuo X.; Xia Q. Nucleic Acid Tests for Clinical Translation. Chem. Rev. 2021, 121, 10469–10558. 10.1021/acs.chemrev.1c00241. [DOI] [PubMed] [Google Scholar]
- Moro G.; Fratte C. D.; Normanno N.; Polo F.; Cinti S. Point-of-Care Testing for the Detection of MicroRNAs: Towards Liquid Biopsy on a Chip. Angew. Chem., Int. Ed. 2023, 62, e202309135 10.1002/anie.202309135. [DOI] [PubMed] [Google Scholar]
- Wang S.; Zhang K.; Tan S.; Xin J.; Yuan Q.; Xu H.; Xu X.; Liang Q.; Christiani D. C.; Wang M.; et al. Circular RNAs in Body Fluids as Cancer Biomarkers: The New frontier of Liquid Biopsies. Mol. Cancer 2021, 20, 13. 10.1186/s12943-020-01298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broto M.; Kaminski M. M.; Adrianus C.; Kim N.; Greensmith R.; Dissanayake-Perera S.; Schubert A. J.; Tan X.; Kim H.; Dighe A. S.; Collins J. J.; Stevens M. M. Nanozyme-Catalysed CRISPR Assay for Preamplification-Free Detection of Non-Coding RNAs. Nat. Nanotechnol. 2022, 17, 1120–1126. 10.1038/s41565-022-01179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N.-N.; Li F.-Z.; Zhang X.; Liu M.; Cao H.; Zhang C.-Y. Construction of Genetically Encoded Light-Up RNA Aptamers for Label-free and Ultrasensitive Detection of CircRNAs in Cancer Cells and Tissues. Anal. Chem. 2023, 95, 8728–8734. 10.1021/acs.analchem.3c01624. [DOI] [PubMed] [Google Scholar]
- Xie H.; Yu Y. H.; Xie F.; Lao Y. Z.; Gao Z. Breast Cancer Susceptibility Gene mRNAs Quantified by Microarrays with Electrochemical Detection. Clin. Chem. 2004, 50, 1231–1233. 10.1373/clinchem.2004.031559. [DOI] [PubMed] [Google Scholar]
- Fang Z.; Soleymani L.; Pampalakis G.; Yoshimoto M.; Squire J. A.; Sargent E. H.; Kelley S. O. Direct Profiling of Cancer Biomarkers in Tumor Tissue Using a Multiplexed Nanostructured Microelectrode Integrated Circuit. ACS Nano 2009, 3, 3207–3213. 10.1021/nn900733d. [DOI] [PubMed] [Google Scholar]
- Yang H.; Hui A.; Pampalakis G.; Soleymani L.; Liu F.-F.; Sargent E. H.; Kelley S. O. Direct, Electronic MicroRNA Detection for the Rapid Determination of Differential Expression Profiles. Angew. Chem., Int. Ed. 2009, 48, 8461–8464. 10.1002/anie.200902577. [DOI] [PubMed] [Google Scholar]
- Das J.; Ivanov I.; Montermini L.; Rak J.; Sargent E. H.; Kelley S. O. An Electrochemical Clamp Assay for Direct, Rapid Analysis of Circulating Nucleic Acids in Serum. Nat. Chem. 2015, 7, 569–575. 10.1038/nchem.2270. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez R. M.; Campuzano S.; López-Hernández E.; Montiel R. R.-V.; Barderas R.; Granados R.; Sánchez-Puelles J. M.; Pingarron J. M. Simultaneous Detection of Two Breast Cancer-Related miRNAs in Tumor Tissues Using p19-Based Disposable Amperometric Magnetobiosensing Platforms. Biosens. Bioelectron. 2015, 66, 385–391. 10.1016/j.bios.2014.11.047. [DOI] [PubMed] [Google Scholar]
- Bruch R.; Baaske J.; Chatelle C.; Meirich M.; Madlener S.; Weber W.; Dincer C.; Urban G. A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, 1905311. 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- Povedano E.; Ruiz-Valdepenas Montiel V. R.; Sebuyoya R.; Torrente-Rodríguez R. M.; Garranzo-Asensio M.; Montero-Calle A.; Pingarrón J. M.; Barderas R.; Bartosik M.; Campuzano S. Bringing to Light the Importance of the miRNA Methylome in Colorectal Cancer Prognosis Through Electrochemical Bioplatforms. Anal. Chem. 2024, 96, 4580–4588. 10.1021/acs.analchem.3c05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Wang Y.; Du Y.; Zhang J.; Wang Y.; Xue Y.; Zhao J.; Ge L.; Yang L.; Li F. Laser-Induced Graphene (LIG)-Based Electrochemical Microfluidic Chip for Simultaneous Analysis of Multiplex microRNAs. Chem. Eng. J. 2024, 486, 150233. 10.1016/j.cej.2024.150233. [DOI] [Google Scholar]
- Labib M.; Khan N.; Ghobadloo S. M.; Cheng J.; Pezacki J. P.; Berezovski M. V. Three-Mode Electrochemical Sensing of Ultralow MicroRNA Levels. J. Am. Chem. Soc. 2013, 135, 3027–3038. 10.1021/ja308216z. [DOI] [PubMed] [Google Scholar]
- Sheng Y.; Zhang T.; Zhang S.; Johnston M.; Zheng X.; Shan Y.; Liu T.; Huang Z.; Qian F.; Xie Z.; Ai Y.; Zhong H.; Kuang T.; Dincer C.; Urban G. A.; Hu J. A CRISPR/Cas13a-Powered Catalytic Electrochemical Biosensor for Successive and Highly Sensitive RNA Diagnostics. Biosens. Bioelectron. 2021, 178, 113027. 10.1016/j.bios.2021.113027. [DOI] [PubMed] [Google Scholar]
- Bruch R.; Johnston M.; Kling A.; Mattmüller T.; Baaske J.; Partel S.; Madlener S.; Weber W.; Urban G. A.; Dincer C. CRISPR-Powered Electrochemical Microfluidic Multiplexed Biosensor for Target Amplification-Free miRNA Diagnostics. Biosens. Bioelectron. 2021, 177, 112887. 10.1016/j.bios.2020.112887. [DOI] [PubMed] [Google Scholar]
- Xu J.; Liu Y.; Li Y.; Liu Y.; Huang K.-J. Smartphone-Assisted Flexible Electrochemical Sensor Platform by a Homology DNA Nanomanager Tailored for Multiple Cancer Markers Field Inspection. Anal. Chem. 2023, 95, 13305–13312. 10.1021/acs.analchem.3c02481. [DOI] [PubMed] [Google Scholar]
- Qiu X.; Dai Q.; Tang H.; Li Y. Multiplex Assays of MicroRNAs by Using Single Particle Electrochemical Collision in a Single Run. Anal. Chem. 2023, 95, 13376–13384. 10.1021/acs.analchem.3c02892. [DOI] [PubMed] [Google Scholar]
- Wang F.; Cai R.; Tan W. Self-Powered Biosensor for a Highly Efficient and Ultrasensitive Dual-Biomarker Assay. Anal. Chem. 2023, 95, 6046–6052. 10.1021/acs.analchem.3c00097. [DOI] [PubMed] [Google Scholar]
- Ye Z.; Ma M.; Chen Y.; Liu R.; Zhang Y.; Ma P.; Song D. Dual-microRNA-Controlled Electrochemiluminescence Biosensor for Breast Cancer Diagnosis and Supplemental Identification of Breast Cancer Metastasis. Anal. Chem. 2024, 96, 3636–3644. 10.1021/acs.analchem.3c05766. [DOI] [PubMed] [Google Scholar]
- Xu T.; Song Y.; Gao W.; Wu T.; Xu L.; Zhang X.; Wang S. Superwettable Electrochemical Biosensor toward Detection of Cancer Biomarkers. ACS Sens. 2018, 3, 72–78. 10.1021/acssensors.7b00868. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez R. M.; Campuzano S.; Ruiz-Valdepeñas Montiel V.; Gamella M.; Pingarrón J. M. Electrochemical Bioplatforms for the Simultaneous Determination of Interleukin (IL)-8 mRNA and IL-8 Protein Oral Cancer Biomarkers in Raw Saliva. Biosens. Bioelectron. 2016, 77, 543–548. 10.1016/j.bios.2015.10.016. [DOI] [PubMed] [Google Scholar]
- Kong L.; Han Z.; Zhao M.; Zhang X.; Zhuo Y.; Chai Y.; Li Z.; Yuan R. Versatile Electrochemical Biosensor Based on the Target-Controlled Capture and Release of DNA Nanotubes for the Ultrasensitive Detection of Multiplexed Biomarkers. Anal. Chem. 2022, 94, 11416–11424. 10.1021/acs.analchem.2c02541. [DOI] [PubMed] [Google Scholar]
- Yin F.; Zhao H.; Lu S.; Shen J.; Li M.; Mao X.; Li F.; Shi J.; Li J.; Dong B.; Xue W.; Zuo X.; Yang X.; Fan C. DNA-Framework-Based Multidimensional Molecular Classifiers for Cancer Diagnosis. Nat. Nanotechnol. 2023, 18, 677–686. 10.1038/s41565-023-01348-9. [DOI] [PubMed] [Google Scholar]
- Rifai N.; Gillette M. A.; Carr S. A. Protein Biomarker Discovery and Validation: The Long and Uncertain Path to Clinical Utility. Nat. Biotechnol. 2006, 24, 971–983. 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- Landegren U.; Hammond M. Cancer Diagnostics Based on Plasma Protein Biomarkers: Hard Times but Great Expectations. Mol. Oncol. 2021, 15, 1715–1726. 10.1002/1878-0261.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou M. P.; Diamandis E. P.; Blasutig M. L. The Long Journey of Cancer Biomarkers from the Bench to the Clinic. Clin. Chem. 2013, 59, 147–157. 10.1373/clinchem.2012.184614. [DOI] [PubMed] [Google Scholar]
- Borrebaeck C. A. K. Precision Diagnostics: Moving Towards Protein Biomarker Signatures of Clinical Utility in Cancer. Nat. Rev. Cancer 2017, 17, 199–204. 10.1038/nrc.2016.153. [DOI] [PubMed] [Google Scholar]
- Srinivas P. R.; Kramer S. B.; Srivastava S. Trends in Biomarker Research for Cancer Detection. Lancet Oncol. 2001, 2, 698–704. 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- Zeng Y.; Mu Z.; Nie B.; Qu X.; Zhang Y.; Li C.; Sun L.; Li G. Engineered Escherichia coli as a Controlled-Release Biocarrier for Electrochemical Immunoassay. Nano Lett. 2023, 23, 2854–2861. 10.1021/acs.nanolett.3c00184. [DOI] [PubMed] [Google Scholar]
- Vargas E.; Zhang F.; Ben Hassine A.; Ruiz-Valdepenas Montiel V.; Mundaca-Uribe R.; Nandhakumar P.; He P.; Guo Z.; Zhou Z.; Fang R. H.; et al. Using Cell Membranes as Recognition Layers to Construct Ultrasensitive and Selective Bioelectronic Affinity Sensors. J. Am. Chem. Soc. 2022, 144, 17700–17708. 10.1021/jacs.2c07956. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Liu M.; Zhou F.; Yan H.; Zhou Y. Homogeneous Electrochemical Immunoassay Using an Aggregation-Collision Strategy for Alpha-Fetoprotein Detection. Anal. Chem. 2023, 95, 3045–3053. 10.1021/acs.analchem.2c05193. [DOI] [PubMed] [Google Scholar]
- Traynor S. M.; Wang G. A.; Pandey R.; Li F.; Soleymani L. Dynamic Bio-Barcode Assay Enables Electrochemical Detection of a Cancer Biomarker in Undiluted Human Plasma: A Sample-In-Answer-Out Approach. Angew. Chem., Int. Ed. 2020, 59, 22617–22622. 10.1002/anie.202009664. [DOI] [PubMed] [Google Scholar]
- Hayes F. J.; Halsall H. B.; Heineman W. R. Simultaneous Immunoassay Using Electrochemical Detection of Metal Ion Labels. Anal. Chem. 1994, 66, 1860–1865. 10.1021/ac00083a014. [DOI] [PubMed] [Google Scholar]
- Liu G.; Wang J.; Kim J.; Jan M. R.; Collins G. E. Electrochemical Coding for Multiplexed Immunoassays of Proteins. Anal. Chem. 2004, 76, 7126–7130. 10.1021/ac049107l. [DOI] [PubMed] [Google Scholar]
- Kong F.-Y.; Xu B.-Y.; Xu J.-J.; Chen H.-Y. Simultaneous Electrochemical Immunoassay Using CdS/DNA and PbS/DNA Nanochains as Labels. Biosens. Bioelectron. 2013, 39, 177–182. 10.1016/j.bios.2012.07.023. [DOI] [PubMed] [Google Scholar]
- Liu X.; Li Y.; He L.; Feng Y.; Tan H.; Chen X.; Yang W. Simultaneous Detection of Multiple Neuroendocrine Tumor Markers in Patient Serum with an Ultrasensitive and Antifouling Electrochemical Immunosensor. Biosens. Bioelectron. 2021, 194, 113603. 10.1016/j.bios.2021.113603. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Ma W.; Li N.; Yang M.; Hou C.; Huo D. A Clinically Feasible Diagnostic Typing of Breast Cancer Built on a Homogeneous Electrochemical Biosensor for Simultaneous Multiplex Detection. Anal. Chem. 2024, 96, 13870–13878. 10.1021/acs.analchem.4c01877. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Wang J.; Chen H.; Xu H.; Bai L.; Wang W.; Yang H.; Wei D.; Yuan B. A Multiple Signal Amplification Based on PEI and rGO Nanocomposite for Simultaneous Multiple Electrochemical Immunoassay. Sensor Actuat. B-Chem. 2019, 301, 127071. 10.1016/j.snb.2019.127071. [DOI] [Google Scholar]
- Xie Y.-R.; Pan H.-J.; Zhang Z.-H.; Jia L.-P.; Zhang W.; Shang L.; Li X.-J.; Xue Q.-W.; Wang H.-S.; Ma R.-N. Distinguishable Magnetic Reporter Coordination with Buoyancy-Magnetism Separation for Immobilization-Free Dual-Target Electrochemical Immunosensing. Anal. Chem. 2024, 96, 8365–8372. 10.1021/acs.analchem.3c05391. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Liu X.; Xu Y.; Wang Q.; Hou J.; Hou C.; Huo D. A Clinically Feasible Diagnostic Electrochemical Micronano Motors Biosensor Built on Miniature Swimmer for Multiplex Detection and Grading of Breast Cancer Biomarkers. Anal. Chem. 2024, 96, 12316–12322. 10.1021/acs.analchem.4c01385. [DOI] [PubMed] [Google Scholar]
- Meyerhoff M. E.; Duan C.; Meusel M. Novel Nonseparation Sandwich-Type Electrochemical Enzyme Immunoassay System for Detecting Marker Proteins in Undiluted Blood. Clin. Chem. 1995, 41, 1378–1384. 10.1093/clinchem/41.9.1378. [DOI] [PubMed] [Google Scholar]
- Kojima K.; Hiratsuka A.; Suzuki H.; Yano K.; Ikebukuro K.; Karube I. Electrochemical Protein Chip with Arrayed Immunosensors with Antibodies Immobilized in a Plasma-Polymerized Film. Anal. Chem. 2003, 75, 1116–1122. 10.1021/ac0257391. [DOI] [PubMed] [Google Scholar]
- Wilson M. S. Electrochemical Immunosensors for the Simultaneous Detection of Two Tumor Markers. Anal. Chem. 2005, 77, 1496–1502. 10.1021/ac0485278. [DOI] [PubMed] [Google Scholar]
- La Belle J. T.; Gerlach J. Q.; Svarovsky S.; Joshi L. Label-Free Impedimetric Detection of Glycan-Lectin Interactions. Anal. Chem. 2007, 79, 6959–6964. 10.1021/ac070651e. [DOI] [PubMed] [Google Scholar]
- Li J.; Lee S. H.; Yoo D. O.; Woo H. C.; Jhung S. H.; Jovíc M.; Girault H.; Lee H. J. A Spatially Multiplexed Voltammetric Magneto-Sandwich Assay Involving Fe3O4/Fe-Based Metal-Organic Framework for Dual Liver Cancer Biomarkers. Sensor Actuat. B-Chem. 2023, 380, 133313. 10.1016/j.snb.2023.133313. [DOI] [Google Scholar]
- Duan L.; Yobas L. Label-Free Multiplexed Electrical Detection of Cancer Markers on a Microchip Featuring an Integrated Fluidic Diode Nanopore Array. ACS Nano 2018, 12, 7892–7900. 10.1021/acsnano.8b02260. [DOI] [PubMed] [Google Scholar]
- Yu X.; Munge B.; Patel V.; Jensen G.; Bhirde A.; Gong J. D.; Kim S. N.; Gillespie J.; Gutkind J. S.; Papadimitrakopoulos F.; Rusling J. F. Carbon Nanotube Amplification Strategies for Highly Sensitive Immunodetection of Cancer Biomarkers. J. Am. Chem. Soc. 2006, 128, 11199–11205. 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikkaveeraiah B. V.; Bhirde A.; Malhotra R.; Patel V.; Gutkind J. S.; Rusling J. F. Single-Wall Carbon Nanotube Forest Arrays for Immunoelectrochemical Measurement of Four Protein Biomarkers for Prostate Cancer. Anal. Chem. 2009, 81, 9129–9134. 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani V.; Chikkaveeraiah B. V.; Patel V.; Gutkind J. S.; Rusling J. F. Ultrasensitive Immunosensor for Cancer Biomarker Proteins Using Gold Nanoparticle Film Electrodes and Multienzyme-Particle Amplification. ACS Nano 2009, 3, 585–594. 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra R.; Patel V.; Chikkaveeraiah B. V.; Munge B. S.; Cheong S. C.; Zain R. B.; Abraham M. T.; Dey D. K.; Gutkind J. S.; Rusling J. F. Ultrasensitive Detection of Cancer Biomarkers in the Clinic by Use of a Nanostructured Microfluidic Array. Anal. Chem. 2012, 84, 6249–6255. 10.1021/ac301392g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. K.; Vaze A.; Shen M.; Rusling J. F. High-Throughput Electrochemical Microfluidic Immunoarray for Multiplexed Detection of Cancer Biomarker Proteins. ACS Sens. 2016, 1, 1036–1043. 10.1021/acssensors.6b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otieno B. A.; Krause C. E.; Latus A.; Chikkaveeraiah B. V.; Faria R. C.; Rusling J. F. On-line Protein Capture on Magnetic Beads for Ultrasensitive Microfluidic Immunoassays of Cancer Biomarkers. Biosens. Bioelectron. 2014, 53, 268–274. 10.1016/j.bios.2013.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharafeldin M.; Bishop G. W.; Bhakta S.; El-Sawy A.; Suib S. L.; Rusling J. F. Fe3O4 Nanoparticles on Graphene Oxide Sheets for Isolation and Ultrasensitive Amperometric Detection of Cancer Biomarker Proteins. Biosens. Bioelectron. 2017, 91, 359–366. 10.1016/j.bios.2016.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanapala L.; Jones A. L.; Czarnecki P.; Rusling J. F. Sub-zeptomole Detection of Biomarker Proteins Using a Microfluidic Immunoarray with Nanostructured Sensors. Anal. Chem. 2020, 92, 8021–8025. 10.1021/acs.analchem.0c01507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. L.; Dhanapala L.; Baldo T. A.; Sharafeldin M.; Krause C. E.; Shen M.; Moghaddam S.; Faria R. C.; Dey D. K.; Watson R. W.; et al. Prostate Cancer Diagnosis in the Clinic Using an 8-Protein Biomarker Panel. Anal. Chem. 2021, 93, 1059–1067. 10.1021/acs.analchem.0c04034. [DOI] [PubMed] [Google Scholar]
- Dhanapala L.; Joseph S.; Jones A. L.; Moghaddam S.; Lee N.; Kremer R. B.; Rusling J. F. Immunoarray Measurements of Parathyroid Hormone-Related Peptides Combined with Other Biomarkers to Diagnose Aggressive Prostate Cancer. Anal. Chem. 2022, 94, 12788–12797. 10.1021/acs.analchem.2c02648. [DOI] [PubMed] [Google Scholar]
- Zhong Z.; Ding L.; Man Z.; Zeng Y.; Pan B.; Zhu J.-J.; Zhang M.; Cheng F. Versatile Metal-Organic Framework Incorporating Ag2S for Constructing a Photoelectrochemical Immunosensor for Two Breast Cancer Markers. Anal. Chem. 2024, 96, 8837–8843. 10.1021/acs.analchem.4c02091. [DOI] [PubMed] [Google Scholar]
- Das J.; Gomis S.; Chen J. B.; Yousefi H.; Ahmed S.; Mahmud A.; Zhou W.; Sargent E. H.; Kelley S. O. Reagentless Biomolecular Analysis Using a Molecular Pendulum. Nat. Chem. 2021, 13, 428–434. 10.1038/s41557-021-00644-y. [DOI] [PubMed] [Google Scholar]
- Díaz-Fernández A.; Ranallo S.; Ricci F. Enzyme-Linked DNA Displacement (ELIDIS) Assay for Ultrasensitive Electrochemical Detection of Antibodies. Angew. Chem., Int. Ed. 2024, 63, e202314818 10.1002/anie.202314818. [DOI] [PubMed] [Google Scholar]
- Garranzo-Asensio M.; Guzmán-Aránguez A.; Povedano E.; Ruiz-Valdepeñas Montiel V. R. V.; Poves C.; Fernandez-Aceñero M. J.; Montero-Calle A.; Solís-Fernández G.; Fernandez-Diez S.; Camps J.; et al. Multiplexed Monitoring of a Novel Autoantibody Diagnostic Signature of Colorectal Cancer Using HaloTag Technology-Based Electrochemical Immunosensing Platform. Theranostics 2020, 10, 3022–3034. 10.7150/thno.42507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Yang W.; Zhao B.; Yang S.; Tang Q.; Chen X.; Dai H.; Liu P. Ultrasensitive DNA-Biomacromolecule Sensor for the Detection Application of Clinical Cancer Samples. Adv. Sci. 2022, 9, 2102804. 10.1002/advs.202102804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Liang X.; Zhong R.; Liu M.; Liu X.; Yan H.; Zhou Y.-G. Clinically Applicable Homogeneous Assay for Serological Diagnosis of Alpha-Fetoprotein by Impact Electrochemistry. ACS Sens. 2022, 7, 3216–3222. 10.1021/acssensors.2c01887. [DOI] [PubMed] [Google Scholar]
- Hansen J. A.; Wang J.; Kawde A.-N.; Xiang Y.; Gothelf K. V.; Collins G. Quantum-Dot/Aptamer-Based Ultrasensitive Multi-Analyte Electrochemical Biosensor. J. Am. Chem. Soc. 2006, 128, 2228–2229. 10.1021/ja060005h. [DOI] [PubMed] [Google Scholar]
- Shen Z.; Ni S.; Yang W.; Sun W.; Yang G.; Liu G. Redox Probes Tagged Electrochemical Aptasensing Device for Simultaneous Detection of Multiple Cytokines in Real Time. Sensor Actuat. B-Chem. 2021, 336, 129747. 10.1016/j.snb.2021.129747. [DOI] [Google Scholar]
- Yan R.; Lu N.; Han S. P.; Lu Z.; Xiao Y.; Zhao Z.; Zhang M. Simultaneous Detection of Dual Biomarkers Using Hierarchical MoS2 Nanostructuring and Nano-Signal Amplification-Based Electrochemical Aptasensor Toward Accurate Diagnosis of Prostate Cancer. Biosens. Bioelectron. 2022, 197, 113797. 10.1016/j.bios.2021.113797. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Li N.; Xu Y.; Lu P.; Qi N.; Yang M.; Hou C.; Huo D. An Ultrasensitive Dual-Signal Aptasensor Based on Functionalized Sb@ZIF-67 Nanocomposites for Simultaneously Detect Multiple Biomarkers. Biosens. Bioelectron. 2022, 214, 114508. 10.1016/j.bios.2022.114508. [DOI] [PubMed] [Google Scholar]
- Xie X.; Jin K.; Wang Z.; Wang S.; Zhu J.; Huang J.; Tang S.; Cai K.; Zhang J. Constraint Coupling of Redox Cascade and Electron Transfer Synchronization on Electrode-Nanosensor Interface for Repeatable Detection of Tumor Biomarkers. Small Methods 2024, 8, 2301330. 10.1002/smtd.202301330. [DOI] [PubMed] [Google Scholar]
- Yu X.; Cao Y.; Zhao Y.; Xia J.; Yang J.; Xu Y.; Zhao J. Proximity Amplification-Enabled Electrochemical Analysis of Tumor-Associated Glycoprotein Biomarkers. Anal. Chem. 2023, 95, 15900–15907. 10.1021/acs.analchem.3c02266. [DOI] [PubMed] [Google Scholar]
- Díaz-Fernández A.; Miranda-Castro R.; de-los-Santos-Alvarez N.; Lobo-Castañón M. J.; Estrela P. Impedimetric Aptamer-Based Glycan PSA Score for Discrimination of Prostate Cancer from Other Prostate Diseases. Biosens. Bioelectron. 2021, 175, 112872. 10.1016/j.bios.2020.112872. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Liu Q.; Qin Y.; Cao Y.; Zhao J.; Zhang K.; Cao Y. Ordered Labeling-Facilitated Electrochemical Assay of Alpha-Fetoprotein-L3 Ratio for Diagnosing Hepatocellular Carcinoma. ACS Appl. Mater. Interfaces 2023, 15, 6411–6419. 10.1021/acsami.2c19231. [DOI] [PubMed] [Google Scholar]
- Noh S.; Lee H.; Kim J.; Jang H.; An J.; Park C.; Lee M.-H.; Lee T. Rapid Electrochemical Dual-Target Biosensor Composed of an Aptamer/MXene Hybrid on Au Microgap Electrodes for Cytokines Detection. Biosens. Bioelectron. 2022, 207, 114159. 10.1016/j.bios.2022.114159. [DOI] [PubMed] [Google Scholar]
- Welsh J. A.; Goberdhan D. C. I.; O’Driscoll L.; Buzas E. I.; Blenkiron C.; Bussolati B.; Cai H.; Di Vizio D.; Driedonks T. A. P.; Erdbrügger U.; et al. Minimal Information for Studies of Extracellular Vesicles (MISEV2023): From Basic to Advanced Approaches. J. Extracell. Vesicles 2024, 13, e12404 10.1002/jev2.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R.; LeBleu V. S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R.; McAndrews K. M. The Role of Extracellular Vesicles in Cancer. Cell 2023, 186, 1610–1626. 10.1016/j.cell.2023.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller A.; Lobb R. J. The Evolving Translational Potential of Small Extracellular Vesicles in Cancer. Nat. Rev. Cancer 2020, 20, 697–709. 10.1038/s41568-020-00299-w. [DOI] [PubMed] [Google Scholar]
- Sabate del Rio J.; Woo H.-K.; Park J.; Ha H. K.; Kim J.-R.; Cho Y.-K. SEEDING to Enable Sensitive Electrochemical Detection of Biomarkers in Undiluted Biological Samples. Adv. Mater. 2022, 34, 2200981. 10.1002/adma.202200981. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Tao J.; Li Y.; Feng Y.; Ju H.; Wang Z.; Ding L. Quantitative Localized Analysis Reveals Distinct Exosomal Protein-Specific Glycosignatures: Implications in Cancer Cell Subtyping, Exosome Biogenesis, and Function. J. Am. Chem. Soc. 2020, 142, 7404–7412. 10.1021/jacs.9b12182. [DOI] [PubMed] [Google Scholar]
- Ge F.; Ding W.; Han C.; Zhang L.; Liu Q.; Zhao J.; Luo Z.; Jia C.; Qu P.; Zhang L. Electrochemical Sensor for the Detection and Accurate Early Diagnosis of Ovarian Cancer. ACS Sens. 2024, 9, 2897–2906. 10.1021/acssensors.3c02776. [DOI] [PubMed] [Google Scholar]
- Chang L.; Wu H.; Chen R.; Sun X.; Yang Y.; Huang C.; Ding S.; Liu C.; Cheng W. Microporous PdCuB Nanotag-Based Electrochemical Aptasensor with Au@CuCl2 Nanowires Interface for Ultrasensitive Detection of PD-L1-Positive Exosomes in the Serum of Lung Cancer Patients. J. Nanobiotechnol. 2023, 21, 86. 10.1186/s12951-023-01845-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Wang Y.; Yu X.; Jiang X.; Li G.; Zhao J. Identification of Programmed Death Ligand-1 Positive Exosomes in Breast Cancer Based on DNA Amplification-Responsive Metal-Organic Frameworks. Biosens. Bioelectron. 2020, 166, 112452. 10.1016/j.bios.2020.112452. [DOI] [PubMed] [Google Scholar]
- Yang S.; Zhou L.; Fang Z.; Wang Y.; Zhou G.; Jin X.; Cao Y.; Zhao J. Proximity-Guaranteed DNA Machine for Accurate Identification of Breast Cancer Extracellular Vesicles. ACS Sens. 2024, 9, 2194–2202. 10.1021/acssensors.4c00491. [DOI] [PubMed] [Google Scholar]
- Gurudatt N. G.; Gwak H.; Hyun K.-A.; Jeong S.-E.; Lee K.; Park S.; Chung M. J.; Kim S.-E.; Jo J. H.; Jung H.-I. Electrochemical Detection and Analysis of Tumor-Derived Extracellular Vesicles to Evaluate Malignancy of Pancreatic Cystic Neoplasm Using Integrated Microfluidic Device. Biosens. Bioelectron. 2023, 226, 115124. 10.1016/j.bios.2023.115124. [DOI] [PubMed] [Google Scholar]
- Yu Y.; Guo Q.; Jiang W.; Zhang H.; Cai C. Dual-Aptamer-Assisted AND Logic Gate for Cyclic Enzymatic Signal Amplification Electrochemical Detection of Tumor-Derived Small Extracellular Vesicles. Anal. Chem. 2021, 93, 11298–11304. 10.1021/acs.analchem.1c02489. [DOI] [PubMed] [Google Scholar]
- Yang L.; Yin X.; An B.; Li F. Precise Capture and Direct Quantification of Tumor Exosomes via a Highly Efficient Dual-Aptamer Recognition-Assisted Ratiometric Immobilization-Free Electrochemical Strategy. Anal. Chem. 2021, 93, 1709–1716. 10.1021/acs.analchem.0c04308. [DOI] [PubMed] [Google Scholar]
- Zhao Z.; Yang S.; Tang X.; Feng L.; Ding Z.; Chen Z.; Luo X.; Deng R. J.; Sheng J.; Xie S.; Chang K.; Chen M. DNA Four-way Junction-Driven Dual-Rolling Circle Amplification Sandwich-Type Aptasensor for Ultra-Sensitive and Specific Detection of Tumor-Derived Exosomes. Biosens. Bioelectron. 2024, 246, 115841. 10.1016/j.bios.2023.115841. [DOI] [PubMed] [Google Scholar]
- Jeong S.; Park J.; Pathania D.; Castro C. M.; Weissleder R.; Lee H. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano 2016, 10, 1802–1809. 10.1021/acsnano.5b07584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.; Park J. S.; Huang C.-H.; Jo A.; Cook K.; Wang R.; Lin H.-Y.; Van Deun J.; Li H.; Min J.; et al. An Integrated Magneto-Electrochemical Device for the Rapid Profiling of Tumour Extracellular Vesicles from Blood Plasma. Nat. Biomed. Eng. 2021, 5, 678–689. 10.1038/s41551-021-00752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y.; Li R.; Zhang F.; He P. Magneto-Mediated Electrochemical Sensor for Simultaneous Analysis of Breast Cancer Exosomal Proteins. Anal. Chem. 2020, 92, 5404–5410. 10.1021/acs.analchem.0c00106. [DOI] [PubMed] [Google Scholar]
- Kilic T.; Cho Y. K.; Jeong N.; Shin I. S.; Carter B. S.; Balaj L.; Weissleder R.; Lee H. Multielectrode Spectroscopy Enables Rapid and Sensitive Molecular Profiling of Extracellular Vesicles. ACS Cent. Sci. 2022, 8, 110–117. 10.1021/acscentsci.1c01193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Gui Y.; Liu W.; Li C.; Yang Y. Precise Molecular Profiling of Circulating Exosomes Using a Metal-Organic Framework-Based Sensing Interface and an Enzyme-Based Electrochemical Logic Platform. Anal. Chem. 2022, 94, 875–883. 10.1021/acs.analchem.1c03644. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Zhu H.; Ying Z.; Gao X.; Chen W.; Zhan Y.; Feng L.; Liu C. C.; Dai Y. Design and Application of Metal Organic Framework ZIF-90-ZnO-MoS2 Nanohybrid for an Integrated Electrochemical Liquid Biopsy. Nano Lett. 2022, 22, 6833–6840. 10.1021/acs.nanolett.2c01613. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Gao W.; Sun M.; Feng B.; Shen H.; Zhu J.; Chen X.; Yu S. A Filter-Electrochemical Microfluidic Chip for Multiple Surface Protein Analysis of Exosomes to Detect and Classify Breast Cancer. Biosens. Bioelectron. 2023, 239, 115590. 10.1016/j.bios.2023.115590. [DOI] [PubMed] [Google Scholar]
- Qin X.; Wei B.; Xiang Y.; Lu H.; Liu F.; Li X.; Yang F. Exosome-Tuned MOF Signal Amplifier Boosting Tumor Exosome Phenotyping with High-Affinity Nanostars. Biosens. Bioelectron. 2024, 245, 115828. 10.1016/j.bios.2023.115828. [DOI] [PubMed] [Google Scholar]
- Hallal S.; Tuzesi A.; Grau G. E.; Buckland M. E.; Alexander K. L. Understanding the Extracellular Vesicle Surface for Clinical Molecular Biology. J. Extracell. Vesicles 2022, 11, e12260 10.1002/jev2.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C.; Fu Y.; Liu G.; Shu B.; Davis J.; Tofaris G. K. Multiplexed Profiling of Extracellular Vesicles for Biomarker Development. Nano-Micro Lett. 2022, 14, 3. 10.1007/s40820-021-00753-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K.; Breyne K.; Ughetto S.; Laurent L. C.; Breakefield X. O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.; Liu C.; Bi Z.-Y.; Zhou Q.; Zhang H.; Li L.-L.; Zhang J.; Zhu W.; Song Y.-Y.-Y.; Zhang F.; et al. Comprehensive Landscape of Extracellular Vesicle-Derived RNAs in Cancer Initiation, Progression, Metastasis and Cancer Immunology. Mol. Cancer 2020, 19, 102. 10.1186/s12943-020-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Zhu X.; Huang S. Extracellular Vesicle Long Non-Coding RNAs and Circular RNAs: Biology, Functions and Applications in Cancer. Cancer Lett. 2020, 489, 111–120. 10.1016/j.canlet.2020.06.006. [DOI] [PubMed] [Google Scholar]
- Yang H.; Zhao J.; Dong J.; Wen L.; Hu Z.; He C.; Xu F.; Huo D.; Hou C. Simultaneous Detection of Exosomal MicroRNAs by Nucleic Acid Functionalized Disposable Paper-Based Sensors. Chem. Eng. J. 2022, 438, 135594. 10.1016/j.cej.2022.135594. [DOI] [Google Scholar]
- Zhang Y.; Li W.; Ji T.; Luo S.; Qiu J.; Situ B.; Li B.; Zhang X.; Zhang T.; Wang W.; et al. Localized DNA Tetrahedrons Assisted Catalytic Hairpin Assembly for the Rapid and Sensitive Profiling of Small Extracellular Vesicle-Associated MicroRNAs. J. Nanobiotechnol. 2022, 20, 503. 10.1186/s12951-022-01700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.; Lan F.; Yang C.; Ji T.; Lou Y.; Zhu Y.; Li W.; Chen S.; Gao Z.; Luo S.; Zhang Y. Sensitive Small Extracellular Vesicles Associated CircRNAs Analysis Combined with Machine Learning for Precision Identification of Gastric Cancer. Chem. Eng. J. 2024, 491, 152094. 10.1016/j.cej.2024.152094. [DOI] [Google Scholar]
- Cao Y.; Yu X.; Zeng T.; Fu Z.; Zhao Y.; Nie B.; Zhao J.; Yin Y.; Li G. Molecular Characterization of Exosomes for Subtype-Based Diagnosis of Breast Cancer. J. Am. Chem. Soc. 2022, 144, 13475–13486. 10.1021/jacs.2c00119. [DOI] [PubMed] [Google Scholar]
- Lawrence R.; Watters M.; Davies C. R.; Pantel K.; Lu Y.-J. Circulating Tumour Cells for Early Detection of Clinically Relevant Cancer. Nat. Rev. Clin. Oncol. 2023, 20, 487–500. 10.1038/s41571-023-00781-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.; Shen L.; Luo M.; Zhang K.; Li J.; Yang Q.; Zhu F.; Zhou D.; Zheng S.; Chen Y.; Zhu J.; et al. Circulating Tumor Cells: Biology and Clinical Significance. Sig. Transduct. Target Ther. 2021, 6, 404. 10.1038/s41392-021-00817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. G.; Metcalf R. L.; Carter L.; Brady G.; Blackhall F. H.; Dive C. Molecular Analysis of Circulating Tumour Cells—Biology and Biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- Li T.; Fan Q.; Liu T.; Zhu X.; Zhao J.; Li G. Detection of Breast Cancer Cells Specially and Accurately by an Electrochemical Method. Biosens. Bioelectron. 2010, 25, 2686–2689. 10.1016/j.bios.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Wan Y.; Zhou Y.-G.; Poudineh M.; Safaei T. S.; Mohamadi R. M.; Sargent E. H.; Kelley S. O. Highly Specific Electrochemical Analysis of Cancer Cells using Multi-Nanoparticle Labeling. Angew. Chem., Int. Ed. 2014, 53, 13145–13149. 10.1002/anie.201407982. [DOI] [PubMed] [Google Scholar]
- Peng Y.; Lu B.; Deng Y.; Yang N.; Li G. A Dual-Recognition-Controlled Electrochemical Biosensor for Accurate and Sensitive Detection of Specific Circulating Tumor Cells. Biosens. Bioelectron. 2022, 201, 113973. 10.1016/j.bios.2022.113973. [DOI] [PubMed] [Google Scholar]
- Han B.; Sha L.; Yu X.; Yang M.; Cao Y.; Zhao J. Identification of Dual Therapeutic Targets Assisted by in Situ Automatous DNA Assembly for Combined Therapy in Breast Cancer. Biosens. Bioelectron. 2021, 176, 112913. 10.1016/j.bios.2020.112913. [DOI] [PubMed] [Google Scholar]
- Tang Y.; Dai Y.; Huang X.; Li L.; Han B.; Cao Y.; Zhao J. Self-Assembling Peptide-Based Multifunctional Nanofibers for Electrochemical Identification of Breast Cancer Stem-like Cells. Anal. Chem. 2019, 91, 7531–7537. 10.1021/acs.analchem.8b05359. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Liu X.; Li N.; Xu Y.; Ma Y.; Huang Z.; Luo H.; Hou C.; Huo D. Typing of Cancer Cells by Microswimmer Based on Co-Fe-MOF for One-step Simultaneously Detect Multiple Biomarkers. Biosens. Bioelectron. 2023, 230, 115263. 10.1016/j.bios.2023.115263. [DOI] [PubMed] [Google Scholar]
- Wei X.; Xiong H.; Zhou Y.; Chen X.; Yang W. Tracking Epithelial-Mesenchymal Transition in Breast Cancer Cells Based on a Multiplex Electrochemical Immunosensor. Biosens. Bioelectron. 2024, 258, 116372. 10.1016/j.bios.2024.116372. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Yu X.; Han B.; Dong L.; Xu J.; Dai Y.; Li G.; Zhao J. In Situ Programmable DNA Circuit-Promoted Electrochemical Characterization of Stemlike Phenotype in Breast Cancer. J. Am. Chem. Soc. 2021, 143, 16078–16086. 10.1021/jacs.1c06436. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Mao H.; Sha L.; Liu Q.; Han B.; Zhao J.; Li G. Phenotype-Directed DNA Nanomachine for in Situ Analysis of Stem Cell-like Subpopulation in Breast Cancer. CCS Chem. 2024, 6, 1034–1046. 10.31635/ccschem.023.202303010. [DOI] [Google Scholar]
- Mao H.; Cao Y.; Zou Z.; Xia J.; Zhao J. An Enzyme-powered MicroRNA Discriminator for the Subtype-Specific Diagnosis of Breast Cancer. Chem. Sci. 2023, 14, 2097–2106. 10.1039/D3SC00090G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y.-L.; Li X.-Q.; Wang Z.-X.; Gao H.; Chen H.-Y.; Xu J.-J. Logic Signal Amplification System for Sensitive Electrochemiluminescence Detection and Subtype Identification of Cancer Cells. Anal. Chem. 2024, 96, 7172–7178. 10.1021/acs.analchem.4c00754. [DOI] [PubMed] [Google Scholar]
- Xi X.; Wu Z.; Zhang X.; Li Y.; Zhao Y.; Wen W.; Wang S. Endogenous Protease-Activatable Nanosensor Based on PNA-Peptide-DNA Engineering for AND-Gated and Dual-Model Detection of MicroRNA in Single Living Tumor Cells. ACS Appl. Mater. Interfaces 2023, 15, 21917–21928. 10.1021/acsami.3c02012. [DOI] [PubMed] [Google Scholar]