Abstract
Background
Annotations of completely sequenced genomes reveal that nearly half of the genes identified are of unknown function, and that some belong to uncharacterized gene families. To help resolve such issues, information can be obtained from the comparative analysis of homologous genes in model organisms.
Results
While characterizing genes from the retinitis pigmentosa locus RP26 at 2q31-q33, we have identified a new gene, ORMDL1, that belongs to a novel gene family comprising three genes in humans (ORMDL1, ORMDL2 and ORMDL3), and homologs in yeast, microsporidia, plants, Drosophila, urochordates and vertebrates. The human genes are expressed ubiquitously in adult and fetal tissues. The Drosophila ORMDL homolog is also expressed throughout embryonic and larval stages, particularly in ectodermally derived tissues. The ORMDL genes encode transmembrane proteins anchored in the endoplasmic reticulum (ER). Double knockout of the two Saccharomyces cerevisiae homologs leads to decreased growth rate and greater sensitivity to tunicamycin and dithiothreitol. Yeast mutants can be rescued by human ORMDL homologs.
Conclusions
From protein sequence comparisons we have defined a novel gene family, not previously recognized because of the absence of a characterized functional signature. The sequence conservation of this family from yeast to vertebrates, the maintenance of duplicate copies in different lineages, the ubiquitous pattern of expression in human and Drosophila, the partial functional redundancy of the yeast homologs and phenotypic rescue by the human homologs, strongly support functional conservation. Subcellular localization and the response of yeast mutants to specific agents point to the involvement of ORMDL in protein folding in the ER.
Background
The human genome project has generated raw information on an increasing number of novel genes and gene families whose function is still unknown. Positional cloning and large-scale genome analysis allow preliminary functional assignment of human genes on the basis of linkage to genetic diseases and reported information from model organisms. Although the available computational tools may fail to provide clear functional clues, they are still of great value in defining structural domains, pinpointing intra- and interspecific sequence homologies and establishing new gene families. In the human genome, a mutational approach to characterizing genes functionally is limited to patients that carry well characterized disease alleles. On the other hand, the availability of the mouse genome sequence is providing new tools for systematic functional characterization. This approach has already been used in yeast by the European Functional Analysis Network (EUROFAN) and has provided functional insights on evolutionarily conserved genes.
We previously reported linkage of autosomal recessive retinitis pigmentosa (RP26) to chromosome 2q31-33, between markers D2S148 and D2S117 [1]. Subsequent analyses narrowed the locus to the D2S350-D2S161 interval. This chromosomal region, (approximately 8 million base pairs (Mbp)) is characterized by a low percentage of G+C - basically made of L1+L2 isochores - and by sparse gene content [2]. As no obvious positional candidates were found among known genes, an expressed sequence tag (EST) database search was undertaken to identify sequences expressed in the retina. One of the identified ESTs was homologous with two yeast open reading frames (ORFs), two predicted genes in Arabidopsis and one in Drosophila and other human genomic and EST sequences. After full cDNA analyses and characterization of the corresponding genomic regions, a functional approach was undertaken. We report here a new evolutionarily conserved gene family, named ORMDL, for ORM1 (Saccharomyces cerevisiae)-like genes, following the HUGO Gene Nomenclature Committee guidelines, and present what could be called a 'vertical' genomics approach in several model organisms. This comprises subcellular localization of the encoded proteins, expression analyses on human tissues and Drosophila embryos, and single and double yeast knockouts.
Results
Characterization of the full-length human ORMDL1 cDNA
A human retinal cDNA library was screened using a 647 base pair (bp) probe containing the WI-18706 STS (located at the RP26 locus, see Materials and methods). A total of 13 positive clones were isolated, subcloned in pBluescript II KS(+), and sequenced (Figure 1). Eight of the clones contained an apparently complete ORF, and the other five were truncated. The 5' and 3' ends of the messages were verified by rapid amplification of cDNA ends (RACE) using placental RNA as template. In the 5' experiment, two extended products were detected with the same 5' end but a differentially spliced 110 bp non-coding exon. The longer RACE product started 175 bp upstream of a putative initiation codon and this 5' untranslated region (5'-UTR) contained two in-frame stop codons. The shorter RACE product did not contain an in-frame stop codon. In the 3' experiment, a single extension product was detected which contained a polyadenylation signal (ATTAAA) situated 24 nucleotides 5' of the poly(A) tail. Some of the cDNA clones had an extended 3'-UTR which could be the result of the use of different polyadenylation signals further downstream. The full-length cDNA (1,092 bp) contained an ORF consisting of 462 bp, from nucleotides 176 to 637. The deduced protein chain consisted of 153 amino acids with an estimated molecular mass of 17.4 kDa.
Figure 1.

Nucleotide sequence of the ORMDL1 cDNA. The translation is shown below. Intron positions are marked with black triangles. The exon shown between square brackets in the 5'-UTR is alternatively spliced. Underlines mark the positions of the primers used for the RACE experiments.
Characterization of ORMDL1 homologs cDNAs
When searching the nucleotide databases with the full-length human ORMDL1 cDNA, human homologous EST sequences were identified which belonged to two separate UniGene clusters (Hs.13144 and Hs.293711). Corresponding IMAGE cDNA clones were obtained and sequenced. The deduced ORFs (denoted ORMDL2 and ORMDL3) had the same size as ORMDL1. Comparison of the proteins showed between 80% and 84% positional identities (Table 1), and 116 out of 153 amino-acid residues were conserved between the three sequences. Moreover, in 26 of the 37 remaining positions the substitutions are conservative.
Table 1.
Percentage identity between members of the ORMDL family
| HsapORMDL1 | - | ||||||||||||
| MmusORMDL1 | 99 | - | |||||||||||
| HsapORMDL2 | 83 | 82 | - | ||||||||||
| MmusORMDL2 | 83 | 82 | 97 | - | |||||||||
| HsapORMDL3 | 84 | 83 | 80 | 82 | - | ||||||||
| MmusORMDL3 | 84 | 84 | 81 | 83 | 96 | - | |||||||
| DmelORMDL | 48 | 48 | 50 | 50 | 50 | 50 | - | ||||||
| AthaORMDLa | 40 | 41 | 39 | 39 | 41 | 41 | 35 | - | |||||
| AthaORMDLb | 39 | 39 | 38 | 38 | 39 | 39 | 37 | 81 | - | ||||
| ScerORM1 | 32 | 32 | 32 | 32 | 34 | 34 | 32 | 31 | 32 | - | |||
| SmonORM1 | 33 | 33 | 34 | 33 | 36 | 36 | 33 | 32 | 32 | 92 | - | ||
| ScerORM2 | 31 | 31 | 33 | 34 | 34 | 34 | 31 | 29 | 30 | 68 | 68 | - | |
| SpomORM | 39 | 39 | 39 | 40 | 41 | 41 | 35 | 31 | 34 | 48 | 49 | 46 | - |
| HsapORMDL1 | MmusORMDL1 | HsapORMDL2 | MmusORMDL2 | HsapORMDL3 | MmusORMDL3 | DmelORMDL | AthaORMDLa | AthaORMDLb | ScerORM1 | SmonORM1 | ScerORM2 | SpomORM |
The calculations are based on the alignment shown in Figure 2.
No homologous sequences were identified in Drosophila EST databases. Screening of an adult Drosophila melanogaster cDNA library using the human ORMDL1 cDNA as a heterologous probe was not successful either. We then designed a homologous probe based on the Drosophila genomic high-throughput sequence. Five positive clones were isolated and sequenced. Although none of the clones was full-length, one covered more than 80% of the ORF. The complete ORF could then be deduced by overlapping this sequence to the genomic data. The conceptual translated sequence was one amino acid longer at the amino terminus than the human forms and shared between 48% and 50% residue identities. Subsequently, in the completed Drosophila Genome Project, this sequence has appeared annotated as predicted gene CG14577 [3].
ORMDL1 belongs to an evolutionarily conserved gene family
The ORMDL1 ORF was used for database sequence comparisons using the National Center for Biotechnology Information (NCBI) BLAST server [4,5] (tBLASTN against nucleotide databases, and BLASTP, PSI-BLAST and PHI-BLAST against the non-redundant protein database). Homologous protein sequences were found in yeast, microsporidia (Encephalitozoon cuniculi, an opportunistic pathogen in AIDS), plants (including Arabidopsis), invertebrates (Drosophila), urochordates (Ciona intestinalis) and vertebrates. The search for distant relatives using PSI-BLAST and PHI-BLAST programs and for short, nearly exact, matches (BLASTP) did not yield any additional homologs. In vertebrates, three different genes were distinguished, corresponding to ORMDL1, ORMDL2 and ORMDL3. S. cerevisiae and A. thaliana showed two copies each, while a single copy was found in C. intestinalis, D. melanogaster, Saccharomyces monacensis, Schizosaccharomyces pombe and E. cuniculi. Table 2 lists most of the genes, with their corresponding annotations in the public databases, if available, and summarizes other relevant features.
Table 2.
Summary of experimental and in silico data on the ORMDL gene family members
| Organism | Gene name | cDNA (bp) | Protein (amino acids) | Annotation (from RefSeq, GenBank, TAIR, FlyBase, SGD, YPD, PombePD) | Chromosomal location | Structural domains* | Expression pattern† | Subcellular localization† |
| H. sapiens | ORMDL1 | 1,092† | 153 | LOC51240 [39] | 2q32.2 | 4 TM | Ubiquitous in adult and fetal tissues | Endoplasmic reticulum |
| ORMDL2 | 934† | 153 | HSPC160[39] | 12q13.2 | 3 TM | Ubiquitous in adult and fetal tissues | Endoplasmic reticulum | |
| ORMDL3 | 869† | 153 | None | 17q21.1 | 4 TM | Ubiquitous in adult and fetal tissues | Endoplasmic reticulum | |
|
|
(pseudogene) | None | 10p14 | - | - | - | ||
|
|
(pseudogene) | None | 8q22.1 | - | - | - | ||
| M. musculus | ORMDL1 | 1,815 | 153 | None | 1‡ | 4 TM | NT | NT |
| ORMDL2 | 1,053 | 153 | 0610012C09 gene [12] | 10‡ | 4 TM | NT | NT | |
| ORMDL3 | 2,024 | 153 | 2810011N17 gene [12] | 11‡ | 4 TM | NT | NT | |
| T. rubripes | ORMDL1 | - | 153 | None | 4 TM | NT | NT | |
| ORMDL2 | - | 153 | None | 4 TM | NT | NT | ||
| ORMDL3 | - | 153 | None | 4 TM | NT | NT | ||
| C. intestinalis | ORMDL | - | 153 | None | 4 TM | NT | NT | |
| D. melanogaster | ORMDL | 388†, partial cds | 154 | CG14577 [3] | 3L 78E6 | 3 TM | Ubiquitous at early embryonic stages and in ectodermal derived tissues at later stages | NT |
| A. thaliana | ORMDLa | 795 | 157 | F6F3.4; At1g01230§ | 1 | 2 TM | NT | NT |
| ORMDLb | none | 154 | MJC20.10; At5g42000¶ | 5 | 4 TM | NT | NT | |
| S. cerevisiae | ORM1¥ | none | 222 | ORM1 [40]; G4089; YGR038w | VII | 4 TM | NT | NT |
| ORM2# | none | 216 | YLR350w [41]; L8300.1 | XII | 4 TM | NT | NT | |
| S. monacensis | ORM1 | None | 222 | ORM1 [40] | 4 TM | NT | NT | |
| S. pombe | ORM | 1167, partial cds | 186 | SPBC119.09c [42] | 2 | 3 TM | NT | NT |
| E. cuniculi | ORMDL | - | 147 | ECU11_1150 [43] | XI | 4 TM | NT | NT |
*Potential transmembrane segments were predicted using the HMMTOP 2.0 program [26]. †Experimental data obtained in the present study. ‡Mapping of mouse ORMDL genes was inferred from syntenic regions between human and mouse chromosomes. § GenBank accession no. AF360237. ¶GenBank accession no. NC_003076. ¥The null mutant constructed in the present study is viable; the double null mutant with ORM2 is viable but shows an impaired growth rate†; the ORM1 mRNA is more abundant in MATα cells than in MATa cells, and at 39°C than at 30°C [44]; ORM1 is coregulated with other 117 genes under 26 cell-damaging conditions [45] and is induced during sporulation [46]; Orm1 protein is produced in mid-log cells [47]. #The null mutant constructed in the present study is viable; the double null mutant with YGR038w (ORM1) is viable but shows an impaired growth rate†; Orm2 interacts with Slt2p in a systematic two-hybrid assay [21]; ORM2 is coregulated with other 121 genes under 26 cell-damaging conditions [45] and is induced by a 30 min and 90 min treatment with 1 M NaCl [48]. TM, transmembrane segments; NT, not tested.
When all the protein sequences were aligned using CLUSTALW, several conserved domains became evident (Figure 2). In addition, all yeast proteins were longer, at both the amino- and carboxy-terminal ends. Pairwise comparisons revealed a very high level of identity (Table 1). Searches for characterized functional domains did not reveal any significant homology. However, four putative transmembrane domains were detected when considering the protein alignment of all known members of the family. Similar results were obtained when analyzing the ORMDL1 sequence on its own (Figure 2).
Figure 2.

Alignment of deduced ORMDL amino-acid sequences. Highly conserved positions (≥ 95%) are shown against a black background, whereas those with conservative exchanges are shown against a grey background. Alignment was performed using the CLUSTALW program [23]. Potential transmembrane segments (TM1 to TM4) are marked with bars above the alignment: Upper bars according to HMMTOP [25] and lower bars following TMAP [26]. Species abbreviations are as follows: Hsap, human; Mmus, mouse; Rnor, rat; Sscr, pig; Btau, cow; Ggal, chicken; Xlae, Xenopus laevis; Stro, Silurana tropicalis; Frub, Takifugu rubripes (pufferfish); Drer, Danio rerio (zebrafish); Cint, Ciona intestinalis; Dmel, Drosophila melanogaster; Atha, Arabidopsis thaliana; Hvul, Hordeum vulgare (barley); Sbic, Sorghum vulgare; Sreb, Stevia rebaudiana; Lesc, Lycopersicon esculentum (tomato); Gmax, Glycine max (soybean); Mtru, Medicago truncatula; Lpen, Lycopersicon pennellii; Zmay, Zea mays (maize); Scer, Saccharomyces cerevisiae; Smon, Saccharomyces monacensis; Spom, Schizosaccharomyces pombe; Ecun, Encephalitozoon cuniculi.
A phylogenetic tree using all the available protein sequences was drawn with CLUSTALW and the bootstrap values were also calculated (Figure 3). According to this tree, each human ORMDL gene has an extremely conserved counterpart in all vertebrates analyzed. Vertebrate sequences cluster together in a separate branch. Similarly, yeast, microsporidian and plant ORMDLs group in well differentiated lineages.
Figure 3.
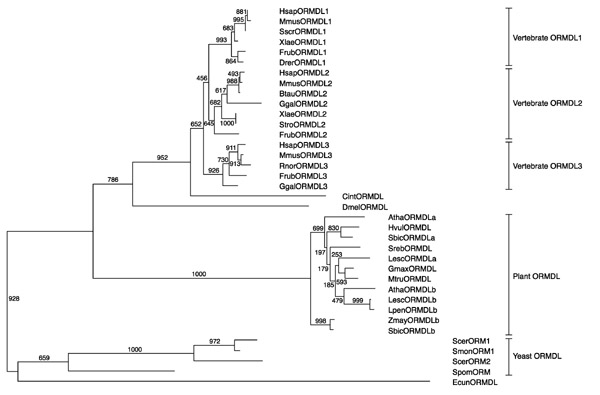
Relationships between the ORMDL amino-acid sequences shown as an unrooted phylogenetic tree. The tree was obtained with the program CLUSTALW [23], with distances corrected for multiple substitutions, and positions with gaps excluded. Numbers show results from bootstrap analysis [4]. The branches are labeled in the same manner as for the alignment (Figure 2).
Genomic structure of the ORMDL sequences
The full-length human ORMDL1 cDNA clone was compared to non-redundant DNA databases and the 5' end was found to be homologous to the promoter region of the PMS1 gene [6]. The P1 genomic clone 1670, containing this region, was analyzed by restriction mapping and Southern blot with the ORMDL1 cDNA as a probe. Positive fragments were sub-cloned and sequenced. The nucleotide sequence contained 4.8 kilobases (kb) of the upstream region, all of the five exons, and the four introns (Figure 4). The third intron was not fully sequenced and the size was determined by restriction mapping after amplification by polymerase chain reaction (PCR). All introns were flanked by the consensus donor and acceptor splice sites. No putative TATA box was found on the upstream genomic sequence, although several Sp1-binding sites were detected using the TESS server [7]. Using the EMBL CpG Islands service [8] a presumptive 0.9 kb CpG island containing 0.4 kb of the upstream region, the first exon of the 5'-UTR, and parts of the first intron was detected.
Figure 4.
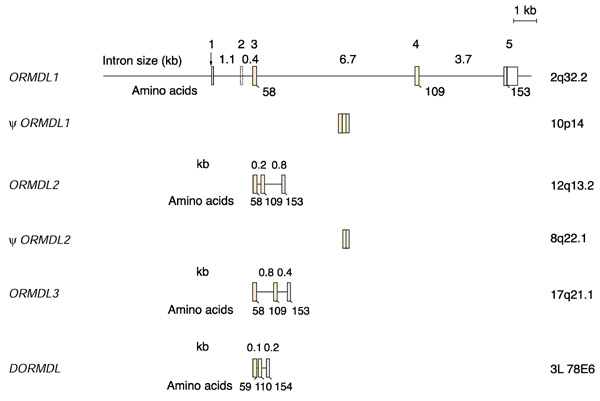
Gene organization of human ORMDL1, ORMDL2, ORMDL3, 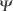 ORMDL1,
ORMDL1, 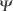 ORMDL2, and Drosophila ORMDL. Exon numbers of ORMDL1 are shown above the bars. Coding exons are shown in color and their sizes (in nucleotides) are: exon 3, 181 (orange); exon 4, 152 (yellow); exon 5, 133 (green). The intron sizes in kilobases are shown in-between the bars. The numbers below the bars denote the amino-acid positions for the exon-intron boundaries, and for the end of the ORF. The dotted appearance of the bar representing exon 2 of ORMDL1 indicates that it is alternatively spliced. The structure of the two human pseudogenes is also shown. The chromosomal location of each gene or pseudogene is indicated to the right.
ORMDL2, and Drosophila ORMDL. Exon numbers of ORMDL1 are shown above the bars. Coding exons are shown in color and their sizes (in nucleotides) are: exon 3, 181 (orange); exon 4, 152 (yellow); exon 5, 133 (green). The intron sizes in kilobases are shown in-between the bars. The numbers below the bars denote the amino-acid positions for the exon-intron boundaries, and for the end of the ORF. The dotted appearance of the bar representing exon 2 of ORMDL1 indicates that it is alternatively spliced. The structure of the two human pseudogenes is also shown. The chromosomal location of each gene or pseudogene is indicated to the right.
Database searches against releases of the public Human Genome Project [4] confirmed the genomic ORMDL1 structure. Furthermore, the genomic structures of the coding region of the two human paralogs ORMDL2 and ORMDL3 and the D. melanogaster homolog were deduced after alignment of the characterized human and Drosophila cDNAs with genomic sequence databases. For the A. thaliana ORMDL homolog, tBLASTN searches were carried out with the human ORMDL proteins against the Arabidopsis genome. The data obtained were contrasted with gene predictions from the Arabidopsis Genome Initiative (actually, one of these predictions has already been confirmed; GenBank accession no. AF360237). In all these genes a genomic structure of three coding exons is totally conserved, including the positions of the exon-intron boundaries ( 4). In contrast, the yeast homologs were each encoded in one single continuous ORF. Interestingly, the introns of human ORMDL2 and ORMDL3 genes were found to be much smaller than those of ORMDL1. The human ORMDL homologs mapped to different chromosomes from ORMDL1 (ORMDL2 to 12q13.2 and ORMDL3 to 17q21.1). No further human homologs were detected in the public databanks [4] or in the Celera database [9], except for two putative processed pseudogenes. One of these showed 86% positional identity to ORMDL1 and mapped to chromosome 10p14, and the other showed 84% identity to ORMDL2 and mapped to 8q22.1. Both 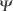 ORMDL1 and
ORMDL1 and  ORMDL2 lacked introns and appeared to have acquired several indels (insertions/deletions). In addition,
ORMDL2 lacked introns and appeared to have acquired several indels (insertions/deletions). In addition,  ORMDL2 lacked the first coding exon of ORMDL2.
ORMDL2 lacked the first coding exon of ORMDL2.
Expression analysis of human ORMDL genes
To study the expression pattern of human ORMDL genes, RT-PCR and northern analyses were performed. RT-PCR expression analysis on sixteen adult and fetal tissue samples showed a ubiquitous expression pattern for all three human genes (Figure 5a). Northern analysis using full-length as well as 3'-UTR ORMDL1 cDNA probes identified three transcripts of 1.4, 2.5 and 3.3 kb in most human adult tissues (Figure 5b). The expression of ORMDL1 was moderately high in pancreas, placenta and brain but low in skeletal muscle and lung.
Figure 5.
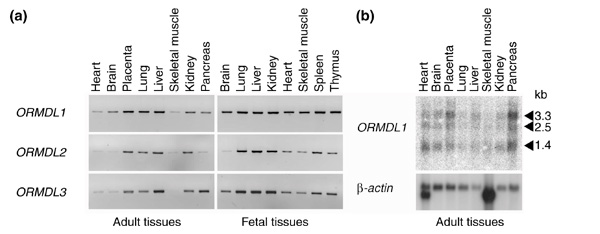
RT-PCR analyses of expression patterns of ORMDL1, ORMDL2, and ORMDL3. (a) Pattern of expression in a normalized panel of adult and fetal human tissues. (b) Northern blot hybridization using a ORMDL1 probe (containing the whole coding region) against the mRNA of human adult tissues. The size of the three detected transcripts is shown. The same pattern was obtained using a probe containing the ORMDL1 3'-UTR (data not shown).
Subcellular localization of human ORMDL proteins: expression of ORMDL-EGFP fusion proteins in COS-7 cells
To determine the subcellular localization of the ORMDL proteins, the cDNAs corresponding to the human ORMDL genes were cloned into pEGFP-derived vectors and transiently expressed in transfected COS-7 cells. Twenty-four hours after transfection, fusion proteins of enhanced green fluorescent protein and ORMDL (EGFP-ORMDL) appeared predominantly in the perinuclear endoplasmic reticulum (ER) and, to a lesser extent, throughout the extended ER network (Figure 6a). Confocal scanning microscopy using an antibody against protein-disulfide isomerase (PDI), an ER-marker protein, showed an overlapping signal in the endoplasmic reticulum (Figure 6a,6b,6c). In contrast, double-label fluorescence with biotin-labeled concanavalin A (which shows affinity for terminal α-D-mannosyl and α-D-glucosyl residues, and thus binds to the plasma membrane in non-permeabilized cells), detected with Texas Red-conjugated streptavidin, showed no overlapping signal with EGFP-ORMDL at the plasmatic membrane (Figure 6d,6e,6f). The subcellular patterns observed for EGFP-ORMDL1, EGFP-ORMDL2 and EGFP-ORMDL3 fusion proteins were similar, in either fixed (Figure 6g,6h,6i, respectively) or in vivo cells (Figure 6j,6k), co-localizing with an in vivo ER marker. Similar results were obtained from COS-7 cells transfected with either pEGFP-N2- or pEGFP-C2-derived constructs. In contrast, EGFP alone was uniformly distributed throughout the cell, including the nucleus (Figure 6l), in all experiments. Overall, these data show that the ORMDL proteins locate at the ER membrane in agreement with their predicted transmembrane topology.
Figure 6.
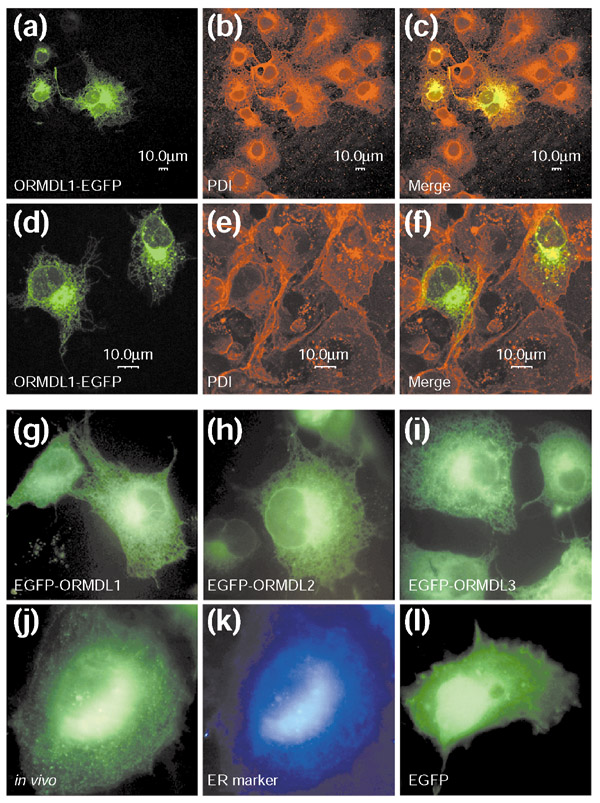
ORMDL fused to EGFP at the amino or carboxyl terminus localizes in the perinuclear ER and throughout the ER network. (a-c) Confocal scanning microscopy of COS-7 cells transfected with ORMDL1-EGFP (a) co-localize with protein-disulfide isomerase (PDI) (b), an ER-marker protein, showing an overlapping signal in the endoplasmic reticulum (c). (d-f) Double-label fluorescence with biotin-labeled concanavalin A showed no overlapping signal with ORMDL1-EGFP at the plasma membrane. (g-l) COS-7 cells transfected with EGFP-ORMDL1, EGFP-ORMDL2 and EGFP-ORMDL3 fusion proteins were similar either in fixed (g, h, and i, respectively) and in vivo (j) cells, co-localizing with an in vivo ER marker (k). EGFP alone was uniformly distributed throughout the cell, including the nucleus (l).
In situ hybridization in Drosophila embryos and larval imaginal discs
We analyzed the expression pattern of the Drosophila ORMDL homolog (DORMDL) during different developmental stages. To this end, a 391 bp antisense riboprobe containing most of the coding sequence was used for in situ hybridization on wild-type Canton S D. melanogaster 0 h to 24 h embryos (stages 1 to 17). DORMDL was ubiquitously and homogeneously expressed in the syncytial blastoderm and during the cellularization stage (Figure 7a,7b). In stages 11-12, the germ-band layer (which later gives rise to all somatic cell lines, including ectoderm, mesoderm and endoderm) gave a positive hybridization signal (Figure 7c,7d). In later stages (stage 14), the signal was mainly detected in the ectodermal tissues (Figure 7e). The DORMDL riboprobe was also used to analyze the expression pattern in imaginal discs in third-instar larvae. Uniform and homogeneous expression was observed in eye-antenna, leg, wing, (Figure 7g,7h,7i) and haltere (data not shown) imaginal discs, which was consistent with the former ectodermal expression detected in late-stage embryos.
Figure 7.
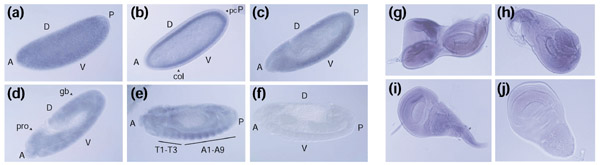
Whole-mount in situ hybridization to detect DORMDL expression in Drosophila embryos and imaginal discs. DORMDL is ubiquitously expressed in the ectodermal tissues. (a) Syncytial blastoderm (stage 3/4, approximately 2 h); (b) cellular blastoderm focused in the plane where polar cells are visible (stage 5, 3 h-3 h 15 min); (c) lateral view of an stage 11 embryo with the fully stretched germ band; (d) lateral view of an stage 11-12 embryo when the germ band begins to recede; (e) lateral view of a stage 14 embryo; (f) sense probe (stage 13). A, anterior; P, posterior; D, dorsal; V, ventral; pc, polar cells; cel, cellularization; pro, procephalon; gb, germ band; T1-T3, thoracic segments; A1-A9, abdominal segments. Timings of developmental stages are approximate (development at 25°C). (g) Eye-antenna imaginal disc; (h) leg imaginal disc; (i) wing imaginal disc; (j) negative control - wing imaginal disc hybridized with the sense probe.
Yeast single and double knockouts: analysis of deletion strains
The S. cerevisiae genome contained two members of this family: ORM1 (ORF YGR038w) at chromosome VII and ORM2 (ORF YLR350w) at chromosome XII. To provide functional clues for the human homologs, we generated and analyzed single and double knockouts. We used the W303-1A and W303-1B yeast wild-type strains (see genotypes in Materials and methods) and the kanMX4 cassette for gene disruption [10]. Single gene targetings were assessed by designing specific PCR reactions. The double knockout mutant was obtained by mating the two single knockout strains, subsequent sporulation, tetrad dissection and analysis of single spores. Verification of ORM1 and ORM2 gene integrity and/or deletion for the clones derived from each spore was carried out by independent PCRs with specific primers (Table 3; Figure 8). Overall, 42 independent clones were analyzed, from which 23.8% were wild-type for ORM1 and ORM2 (and thus did not grow on G418-supplemented medium), 28.57% single ORM1 deletants, 28.57% single ORM2 deletants, 14.28% double knockouts and 4.76% diploids. The two single and double knockouts were viable and no morphological differences were detected between them and the wild-type cells. However, growth rates on rich media (YPD) differed: the double mutant grew much slower than the wild-type and single-knockout strains (data not shown).
Table 3.
Primers used for the generation and verification of yeast single knockouts
| Oligonucleotides for the synthesis of kanMX4 cassettes | |
| Primer | Sequence 5' to 3' |
| YGR038w F | AGG AAC TGC TGA GGC GGC TTC TAC CTC GTA TAG TCG AAA TCA AAC CGT ACG CTG CAG GTC GAC |
| YGR038w R | TGT TCA ACT AAT TTG GGC GCG ACC TGT GAT ACC TGG GAT AGA AAT ATC GAT GAA TTC GAG CTC G |
| YLR350w F | ATC ATG ATT GAC CGC ACT AAA AAC GAA TCT CCA GCT TTT GCG TAC GCT GCA GGT CGA C |
| YLR350w R | CTA ACT AAT TTG AGC ACG GCC CGT AAT ACC AGG GAT GGA TAT CGA TGA ATT CGA GCT CG |
| Oligonucleotides for gene integrity and gene deletion verification | |
| Verification of ORM1 gene integrity | |
| A1 (F) | GTT TAG GAA CGG ATT ATA GA |
| A2 (R) | CTG GTC ATT TTC ATC TTC AA |
| Verification of ORM1 substitution | |
| A1 (F) | GTT TAG GAA CGG ATT ATA GA |
| KAN5 (R) | GTT CGG ATG TGA TGT GAG |
| Verification of ORM2 gene integrity | |
| YLR350w up400 (F) | GCG TAT TTT GAT TGC TCA TA |
| YLR350w ver (R) | TGG TTT CAG GTT AGA CAC AT |
| Verification of ORM2 substitution | |
| YLR350w up400 (F) | GCG TAT TTT GAT TGC TCA TA |
| KAN5 (R) | GTT CGG ATG TGA TGT GAG |
Oligonucleotides in bold are 18-19 nucleotide homology regions to pFA6a MCS sequences.
Figure 8.
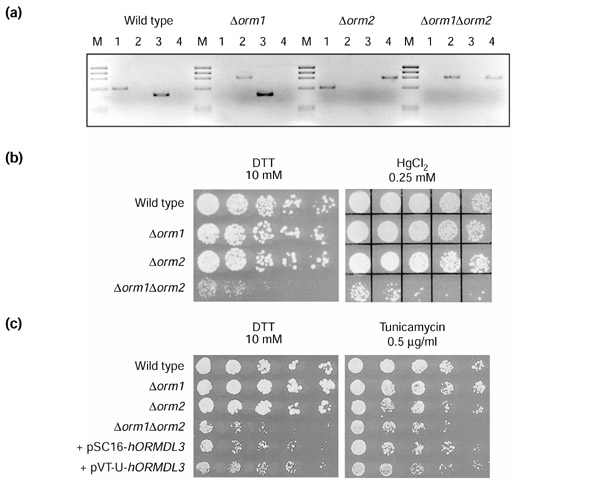
Qualitative phenotypic analysis of yeast knockout strains and functional complementation of human ORMDL3. (a) Genotype PCR analyses of spores derived from tetrad dissection. The deduced genotype for each clone is indicated above. Four different PCR reactions were performed to assess the following: lane 1, ORM1 integrity; lane 2, ORM1 deletion; lane 3, ORM2 integrity; lane 4, ORM2 deletion. (b) Dropout plate showing the growth of haploid wild-type, single-knockout and double-knockout strains. (c) Growth of wild-type, single-knockout and double-knockout strains as well as functional complementation of double knockouts transformed with human ORMDL3. pSC16-hORMDL3 and pVT-U-hORMDL3 denote, respectively, yeast centromeric and non-centromeric plasmid constructs used in the complementation assay. Five sequential dilutions (1:5 each) were plated, left to right, in YPAD medium supplemented with the toxic agents at the indicated concentrations.
A battery of tests was carried out to characterize the yeast mutants on the basis of their sensitivity or resistance to toxic compounds. The YPAD medium was supplemented with one of the following: dithiothreitol (DTT), tunicamycin, HgCl2, cycloheximide, CaCl2, KCl, caffeine, EGTA or SDS. In all cases the double-knockout strain was more sensitive to the toxic agent. In contrast, the growth of the single knockouts did not differ from that of the wild-type strain. This result was particularly evident in the plates supplemented with DTT, HgCl2 or tunicamycin (Figure 8b,8c). We also carried out complementation analyses with ORMDL3, the human counterpart with the greatest sequence similarity to yeast ORM1 and ORM2. To this end, double-knockout yeast cells were transformed with either multicopy or centromeric expression vectors bearing ORMDL3 under the control of a constitutive promoter, and dropout-plated in media supplemented with DTT, HgCl2 or tunicamycin. In all cases, some degree of phenotypic rescue was clearly observed, further arguing in favor of functional conservation in the ORMDL family (Figure 8c).
Discussion
We report here the characterization of a novel gene family highly conserved in eukaryotes from yeast to mammals. Most of the species studied have more than one member of this gene family: S. cerevisiae and A. thaliana each contain two ORMDL genes, whereas three copies are present in vertebrates. On the other hand, Encephalitozoon, Drosophila and Ciona appear to contain a single ORMDL gene and no homologous sequence was found in Caenorhabditis elegans.
While this work was being carried out, many of the family members became incorporated in the genomic databases of the corresponding organisms, some only as predicted genes. However, even after a significant number of sequences had been annotated, the family had not been formally defined. This omission most probably reflects the fact that present annotations of gene families rely on previously characterized functional signatures or domains. In their absence, protein sequence comparisons remain as the only valuable tool.
A phylogenetic tree was constructed to study the evolutionary relationships between the ORMDL members (Figure 3). Several gene duplication events have taken place independently in different lineages, and have given rise to multiple copies in most of the species analyzed. The three human paralogs (ORMDL1, ORMDL2 and ORMDL3) showed amino-acid identities of around 80%, whereas pairwise comparisons revealed more than 95% identity between human and mouse orthologs (Table 1). Recently, genomic data derived from the complete genome sequence of the pufferfish Takifugu rubripes [11], cDNA annotation data in Mus musculus [12], and EST data collected from several vertebrate EST projects [4] support the evidence that the presence of three ORMDL genes - as characterized in humans - is common to all vertebrates. In addition, a single-copy ORMDL has been identified in the pre-vertebrate urochordate C. intestinalis. Overall, these data support the evidence that vertebrate ORMDL genes are true orthologs, probably arising by the postulated large-scale duplications, either genome doublings or extensive chromosomal duplications, in the vertebrate ancestry ([13] and references therein). In the human genome, the three ORMDL genes map within the large paralogous regions defined at 2q, 12q and 17q, which also include segments of chromosomes 7 and 3 [13,14]. These regions contain the HOX clusters as well as two to four members of other families: ORMDL1 and HOXD at 2q31-32; ORMDL2 and HOXC at 12q13, and ORMDL3 and HOXB at 17q21. Moreover, the HOX clusters in mammals are located within chromosomal regions that have maintained extensive chromosomal synteny throughout the vertebrates [15]. No further ORMDL sequences were found next to HOXA at 7p15 but, in this context, loss of a fourth copy is a feasible assumption.
In invertebrates, one ORMDL gene has been characterized in Drosophila. Although no sequence with amino-acid identities above 30% has been found in Caenorhabditis elegans, it is possible that a homolog will be identified as the worm genome assembly is completed. Alternatively, it could be argued that ORMDL may have been lost in the phylogenetic lineage that leads to the nematode, as has happened with some genes involved in developmental pathways, which are broadly represented in animal phylogeny but missing in C. elegans [16,17].
In plants, different lineages show duplicated copies of ORMDL genes: the Solanaceae Lycopersicon esculentum (tomato) and Solarium tuberosum (potato; data not shown), the grasses Sorghum bicolor (sorghum) and Zea mays (maize; data not shown), and the brassica Arabidopsis. The two gene sequences found in A. thaliana and yeast are consistent either with the postulated genome duplications or large-scale segmental duplications in the Arabidopsis and Saccharomyces lineages [18,19,20]. Interestingly, the yeast ORM1 and ORM2 are located within duplicated chromosomal blocks in chromosomes VII and XII, respectively, which include 15 other paralogous gene pairs [19].
Protein alignment showed extended amino-terminal segments of around 20-60 amino acid residues for the yeast ORM proteins in comparison to the plant and animal forms. The conserved residues were mostly clustered in the middle segment of the protein chain. Transmembrane (TM) segment prediction yielded from two to four TM domains, (Table 2 and Figure 2). When using aligned sequences, four hydrophobic domains were apparent, and in most programs were recognized as putative TM sequences. Interestingly, the TM domains are conserved in their relative position but are much less conserved at the residue level than their flanking segments. In fact, the most conserved amino-acid segments are flanked by the two central hydrophobic domains, and probably indicate the location of an important functional domain. Whether this region is relevant to the reported association of yeast ORM2 with SLT2, a serine/threonine protein kinase of the MAP kinase family, remains to be proved [21]. Unfortunately, the reported interaction, from a comprehensive yeast proteome-wide two-hybrid analysis, has not been substantiated with further experimental data. In accordance with the TM prediction, subcellular localization using GFP fusion proteins showed ER anchorage of the human ORMDL proteins. Although co-localization with PDI was used to verify the ER-staining pattern, one cannot discard the possibility that overexpression could influence protein folding and trafficking. Further experiments to characterize protein topology are underway. Searches for signal peptides, nuclear-localization signals, protease-cleavage sites and conserved protein domains were also carried out but did not yield any significant similarities.
Human ORMDL homologs were ubiquitously expressed in adult and fetal samples with some minor tissue-specific differences. There was no evidence of alternative splicing affecting the coding region. However, the northern-blot analysis of ORMDL1 revealed three transcripts of different size, which may be explained by the use of either different promoters or polyadenylation signals, and/or by alternative splicing affecting only the 5'- or 3'-UTRs. In fact, an alternative exon in the 5'-UTR of ORMDL1 is reported here.
Functional characterization of gene homologs in model organisms is an invaluable reference for investigating human genes with unknown function. In this respect, Drosophila is particularly informative when mutant strains for a specific gene are available. Unfortunately, neither P-element insertions next to DORMDL nor information on gene expression were available. Our data from in situ hybridization analysis in different developmental stages and imaginal discs showed ubiquitous expression at early embryonic stages, although at late stages the signal was stronger in the ectodermally derived tissues. Overall, these data support a basic cellular function for this gene family.
The search for additional functional clues prompted us to construct and analyze single and double yeast knockouts. Partial functional redundancy of the two yeast genes was deduced, as single mutants were viable and showed similar responses to the wild-type strain in sensitivity/resistance assays to toxic compounds. Deletion of both genes was not lethal, although yeast-cell growth and ability to respond to chemical aggression were partially affected in double knockouts. It is well known that DTT, a reducing agent that impairs disulfide bond formation, or tunicamycin, a glycosylation inhibitor that blocks the first step in the biosynthesis of N-linked oligosaccharides, induce the accumulation of unfolded proteins in the ER lumen. The cellular response to this type of stress (the unfolded protein response (UPR) pathway) eventually leads to the activation of several ER-resident protein chaperons and chaperon cofactors [22]. Given the ER-membrane localization of the ORMDL proteins and the higher sensitivity of the double knockouts to both tunicamycin and DTT, it is reasonable to postulate that these proteins participate in correct protein folding and/or trafficking in the ER, or are involved in the cellular UPR. Interestingly, phenotypic rescue of the double-knockout mutants was achieved by functional complementation with a human homolog, thus clearly supporting functional as well as structural evolutionary conservation of this gene family.
In summary, this study defines a novel eukaryotic gene family whose function is not yet clear. Several questions should be addressed in the future. They mainly concern protein topology, the cellular function of the family members and their association, if any, to human disease.
Materials and methods
Cloning of human ORMDL1 cDNAs
A PCR-derived probe of 647 bp was obtained from an IMAGE clone (220362, Research Genetics) using a primer pair (5'-GGGAACAACTGGACTATG-3' and 5'-ACTTGTATAGAGAGGATTCC-3') designed after the retinal EST H86933. This EST contained the STS WI-18706, which mapped to the RP26 locus. The probe was labeled with [α-32P]dCTP by random priming (Roche Molecular Biochemicals) and was used to screen 106 recombinant phages from a commercial λ gt10 retinal cDNA library (Clontech). Hybridization was carried out using standard protocols under highly stringent conditions [23]. Positive λ phages were isolated through several rounds of rescreening. Purified phage DNA was EcoRI-digested and inserts were subcloned into pBluescript II KS(+) (Stratagene). Sequencing was carried out using a ThermoSequenase II sequencing kit (Amersham Biosciences) and an ABI377 automatic sequencer (Applied Biosystems).
Rapid amplification of ORMDL1 cDNA ends
The 5' and 3' transcript ends were characterized using the SMART RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions. Total RNA from human placenta was used for first-strand cDNA synthesis using SuperScript II RNase H- reverse transcriptase (Invitrogen Life Technologies). The primers used were: 5'-AGGAGCCTTGCTTTACCCTGGTCAG-3' for the 5'-RACE and 5'-CAATGGCTGGTCCTTCAAGTGCTGT-3' for the 3'-RACE. The RACE products were subcloned using the SureClone ligation kit (Amersham Biosciences) and sequenced.
Characterization of human ORMDL1 homologs
BLAST searches [4,5] against the EST section of the GenBank/EMBL databases identified sequences homologous to ORMDL1, which belonged to the two UniGene clusters Hs.13144 and Hs.293711. The IMAGE clones 212500, 221470, 1407036, 1407667, and 2364281 were obtained from the UK HGMP Resource Centre and sequenced.
Computer sequence analysis
A search for homologous protein sequences was carried out with tBLASTN, BLASTP, PSI-BLAST and PHI-BLAST programs at the NCBI BLAST server [4,5]. Translated sequences were aligned using the program CLUSTALW [24], and visualized using the program Boxshade (created by Kay Hofmann and Michael D. Baron). Phylogenetic trees were constructed with the program CLUSTALW, excluding gap positions and correcting for multiple substitutions. Bootstrap analysis [25] was used for confidence evaluation. Potential transmembrane segments were predicted using the HMMTOP, TMAP and TMPRED programs [26,27,28]. Conserved protein domains and signal peptide searches were performed with reverse position-specific BLAST at the NCBI Conserved Domain Database, including Pfam and Smart databases [4,5,29,30,31]. Analysis of putative binding sites for transcription factors was carried out using the TESS server [7]. CpG islands were detected using the EMBL CpG Islands service [8]. The human ORMDL sequences were mapped using the BLAT program against the Human Genome Working Draft (December 22, 2001 assembly) at UCSC [32,33].
Accession numbers of sequences used are as follows. This work: translations from AF395704 (HsapORMDL1), AF395705 (HsapORMDL2), and AF395708 (HsapORMDL3). From data banks: AAH02146 (MmusORMDL2), BAB26397 (MmusORMDL3), AAF51758 (DmelORMDL), AAK25947 (AthaORMDLa), BAB08433 (AthaORMDLb), CAA97026 (ScerORM1), AAB67252 (ScerORM2), CAA17924 (SpomORM), and translations from BE625123 (MmusORMDL1), AC098491 (RnorORMDL3), BI182456 (SscrORMDL1), BE588447 (BtauORMDL2), BM486897 (GgalORMDL2), BM489594 (GgalORMDL3), BJ037340 (XlaeORMDL1), BJ067072 (XlaeORMDL2), AL633210 (StroORMDL2), fugu clone T008329 (FrubORMDL1), fugu clone T005201 (FrubORMDL2), fugu clone T008602 (FrubORMDL3), AL591593 (DrerORMDL1), AL669202+AV885287 (CintORMDL), BE421632 (HvulORMDL), AW747032 (SbicORMDLa), AW678224 (SbicORMDLb), BG522023 (SrebORMDL), AI772256 (LescORMDLa), AI899675 (LescORMDLb), BM093316 (GmaxORMDL), BE202511 (MtruORMDL), BG140432 (LpenORMDLb), BE238653 (ZmayORMDLb), Y08688 (SmonORM1), and AL590450 (EcunORMDL).
Sequence data for this article were deposited with the GenBank data library under accession nos. AF395701 (ORMDL1 exons 1, 2, and 3); AF395702 (ORMDL1 exons 4 and 5); AF395704 (ORMDL1 mRNA); AF395705, AF395706, AF395707 (ORMDL2 mRNA); AF395708 (ORMDL3 mRNA); AF395703 (DORMDL mRNA).
Characterization of the genomic region of ORMDL1
The human P1 genomic clone 1670, mapped to 2q31-2q33 [6], was obtained from Incyte Genomics. Plasmid DNA was obtained according to the supplier's instructions. Restriction fragments containing the ORMDL1 exons were detected by Southern blot analysis using a full-length ORMDL1 cDNA probe. Positive fragments were subcloned into pBluescript II KS(+), and sequenced. The size of intron 3 was determined by restriction mapping after PCR-amplification.
RT-PCR and northern blot analysis
For RT-PCR expression analysis on 16 samples from adult and fetal tissues (Multiple Tissue cDNA Panels, Clontech), a pair of primers was designed for each human ORMDL gene to amplify the complete ORF, as follows: ORMDL1 forward: 5'-CATGAACGTTGGAGTTGCC-3', ORMDL1 reverse: 5'-GTAAAATTTTTTTTCAGTTTCAAA-3', ORMDL2 forward: GAAGAATGAATGTGGGGGTG-3', ORMDL2 reverse: 5'-CATGGAGCTGTCCCAAAAC-3', ORMDL3 forward: 5'-GCAGGATGAATGTGGGCACA-3', ORMDL3 reverse: 5'-GGCAGGGGAAGGGGCTGCA-3'.
PCR amplifications were carried out in 25 μl reactions containing 2.5 μl template cDNA, 0.2 mM dNTPs, 5 μM each primer, 20 mM Tris-HCl, 50 mM KCl, 3 mM MgCl2, and 1 U Taq Platinum (Invitrogen Life Technologies). After an initial 3 min step at 94°C, reactions were subjected to 35 cycles of a denaturation step of 30 sec at 94°C, and an annealing-extension step of 45 sec at either 59°C (ORMDL1), 62°C (ORMDL2), or 65°C (ORMDL3). A final extension step was performed at 72°C for 5 min.
For northern blot analysis, radiolabeled full-length and 3'-UTR ORMDL1 cDNA probes were hybridized against a commercial human Multiple Tissue Northern blot (Clontech) using standard highly stringent conditions. A human actin probe was used as a control for RNA loading.
Cell culture, transient expression of ORMDL1, ORMDL2 and ORMDL3 fused to EGFP, immunolocalization, and confocal laser scanning microscopy
COS-7 (African green monkey kidney) cells were routinely grown in DMEM containing 10% fetal calf serum, 2 mM L-glutamine, 100 U/ml of penicillin and 100 μg/ml of streptomycin (Invitrogen Life Technologies). ORMDL1, ORMDL2 and ORMDL3 cDNAs were cloned in-frame either upstream or downstream of the gene for GFP using the pEGFP-N2 and pEGFP-C2 vectors (Clontech), respectively. All constructs were verified by sequencing. Transient transfections on COS-7 cells grown on coverslips were performed using the FuGENE reagent (Roche Molecular Biochemicals) following the manufacturer's protocol (the ratio was 1.5 μg DNA to 7.5 μl FuGENE). Empty pEGFP-N2 and pEGFP-C2 vectors were used as controls. Twenty-four hours after transfection, cells were rinsed with 100 mM PBS and fixed in 3% paraformaldehyde and 2% sucrose in 0.1 M phosphate buffer at 4°C for 45 min, then washed and permeabilized (if required) with 0.1% Triton X-100/20 mM glycine/10 mM PBS for 10 min. Following permeabilization, cells were rinsed and blocked in 0.5% BSA/20 mM glycine/10 mM PBS. Cells were incubated either with a specific anti-PDI primary antibody (permeabilized cells) or with biotin-labeled concanavalin A (Sigma) (non-permeabilized cells) at 37°C for 1 h. Upon washing, cells were incubated either with Rhodamine Red™-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories) (permeabilized cells) or with Texas Red-streptavidin (Amersham Biosciences) (non-permeabilized cells) at 37°C for 45 min. In vivo co-localization with the ER marker (ER-Tracker Blue-White DPX, from Molecular Probes) was performed 24 h after transfection by rinsing coverslips twice with PBS followed by 30 min incubation at 37°C in a 4 μM ER-Tracker solution. All preparations were embedded in Vectashield mounting medium (Vector Laboratories). Fluorescence was analyzed with a Zeiss Axiophot epifluorescence microscope. Confocal laser scanning microscopy was performed with an Olympus IX-70 inverted laser scanning microscope.
Cloning of D. melanogaster cDNA
A 529-bp probe was amplified by PCR from genomic D. melanogaster DNA. The primers used (5'-TCAGTGCC-CTTCGTTAGC-3' and 5'-AGTGTTCCGTGTTGTTTC-3') were designed after the high-throughput genomic sequences (GenBank) that were identified in a BLAST search using the human ORMDL1 cDNA sequence. The radiolabeled probe was used to screen 8 × 105 recombinant phages from an adult D. melanogaster cDNA library (Stratagene) constructed in Lambda ZAP II. Hybridization, washes, and autoradiography were carried out in standard conditions. After several rescreening rounds, pBluescript SK(+) plasmids containing the cDNA insert were excised from the isolated phages and sequenced.
In situ hybridization on Drosophila embryos and larval imaginal discs
Digoxigenin (DIG)-labeled antisense and sense riboprobes covering 84% of the Drosophila ORMDL ORF were synthesized using the DIG RNA labeling kit (Roche Molecular Biochemicals) according to the manufacturer's instructions. Drosophila embryos at different developmental stages were collected from a 24-h lay, dechorionated, fixed in 2% formaldehyde and 0.5 M EGTA in PBS, prehybridized for 1 h, and hybridized overnight at 55°C. After removing the hybridization solution and washing, embryos were incubated with 1/2000 anti-DIG antibody conjugated to alkaline phosphatase (Roche Molecular Biochemicals), preabsorbed against fixed embryos. The signal was detected with NBT/BCIP (Roche Molecular Biochemicals). Embryos were mounted on Permount, and examined and photographed under a Zeiss Axiophot microscope.
Wild-type D. melanogaster Canton S third-instar larvae were dissected in PBS and fixed in 4% paraformaldehyde in PBS for 20 min. After rinsing in PBS, a second round of fixation was performed in 4% paraformaldehyde in PBS with addition of 0.1% Triton X-100 and 0.1% deoxycholate. After further rinsing with PBS, the imaginal discs were dehydrated through a graded ethanol series. Further protocol steps before the hybridization were carried out as described [34] except that proteinase K digestion was omitted. After 1 h prehybridization at 60°C, the riboprobe was added hybridized overnight at 65°C. Thereafter, the discs were washed for 20 min each in a series of decreasing concentrations of hybridization solution in PBT at 65°C. Two final washes were done with PBT at room temperature. After 1 h of blocking with 2% BSA in PBT, the discs were incubated with preabsorbed anti-DIG, washed and detected as described above. Dissected discs were then mounted in glycerol, examined and photographed under a Zeiss Axiophot microscope.
Yeast strains and culture media
S. cerevisiae W303-1A (MATa; ura3-1, ade2-1, leu2-3,112; his3-11,15; trp1- ; cam-100) and W303-1B (MATα; ura3-1, ade2-1, leu2-3,112; his3-11,15; trp1-
; cam-100) and W303-1B (MATα; ura3-1, ade2-1, leu2-3,112; his3-11,15; trp1- 2; can1-100) strains were grown on yeast extract peptone dextrose (YPD) media. G418-resistant strains were grown on YPD plates containing 200 mg/l of G418 sulphate (geneticin, Invitrogen Life Technologies).
2; can1-100) strains were grown on yeast extract peptone dextrose (YPD) media. G418-resistant strains were grown on YPD plates containing 200 mg/l of G418 sulphate (geneticin, Invitrogen Life Technologies).
Generation of yeast single knockouts and selection
To disrupt YGR038w and YLR350W ORFs, kanMX4 substitution cassettes were PCR-amplified on plasmid pFA6a-kanMX4 [10] using two sets of primers with 18-19 nucleotide homology to the pFA6a MCS plus 45 nucleotide homology to the flanking regions of either YGR038w (ORM1) or YLR350W (ORM2) ORFs (Table 3). One microgram of each gel-purified PCR product was used to transform yeast W303-1A and W303-1B strains by the lithium acetate method [35]. Transformed yeast cells were recovered by G418 selection. Homologous integration of the kanMX4 cassette was verified by colony PCR. Two sets of primers were used for each gene to test for gene disruption and gene integrity, respectively (Table 3). Primer design and PCR conditions essentially followed the EUROFAN guidelines [36].
Generation of yeast double knockouts
The W303-1A Δorm1 (MATa) and W303-1B Δorm2 (MATα) deletant strains were crossed to obtain the double heterozygous diploid strain. The reciprocal cross was also performed. The double heterozygous diploids were plated on sporulation medium (1% potassium acetate, 0.1% yeast extract, 0.05% dextrose, 2% agar) to induce meiosis [37]. Tetrad dissection and analysis were carried out as described [37]. Clones from spores were re-streaked on YPD plates containing 200 mg/l G418, and G418-resistant transformants were verified by colony PCR to assess single and double knockouts.
Qualitative phenotypic analysis of knockout strains
To test growth differences and sensitivity/resistance of the mutant strains against toxic compounds, single and double knockouts as well as wild-type clones were grown on liquid yeast extract peptone adenine dextrose (YPAD) medium for 14 h at 30°C, diluted to OD600 0.1 and grown to OD600 0.6-0.8. They were then sequentially diluted (5 dilutions, 1:5 each) with YPAD into the wells of microtiter plates and dropout-plated onto the corresponding test media (0.25 mM and 0.4 mM HgCl2; 10 mM and 15 mM DTT; 0.5 μg/ml and 1 μg/ml tunicamycin; 0.7 M CaCl2; 0.1% caffeine; 0.18 mg/ml and decreasing concentrations of cycloheximide; 3 mM EGTA; 1.5 M KCl; 0.01% SDS, all in YPAD). To test heat sensitivity, cultures were incubated in a water bath at 52°C for 35 min, sequentially diluted and plated on YPAD. We analyzed five independent clones for each mutant strain and three for each wild type, respectively.
Functional complementation assay with human ORMDL3 in yeast double knockouts
A yeast centromeric plasmid, pSC16 [38] (kindly provided by S. Cervantes), and a non-centromeric plasmid (pVT-U) containing the ADH1 promoter and termination expression cassettes were used to clone human ORMDL3 ORF. Each construct (100 ng) was used to transform yeast W303 Δorm1 Δorm2 strain by the lithium acetate method. Transformed yeast cells were recovered by selection on minimal synthetic dropout (SD) medium without leucine (centromeric plasmid) or without uracil (non-centromeric plasmid). Transformants were grown on liquid YPAD, sequentially diluted and dropout-plated onto the corresponding test media (0.25 mM and 0.4 mM HgCl2; 10 mM and 15 mM DTT; 0.5 μg/ml and 1 μg/ml tunicamycin, all in YPAD).
Acknowledgments
Acknowledgements
We thank Florenci Serras and Sergio González-Crespo for support on Drosophila in situ hybridizations, Ricardo Casaroli for advice on immunolocalization, and Maite Rodríguez-Manzaneque for technical advice on spore dissection. We are grateful to Charles Brenner for critically reviewing the manuscript and suggesting the use of tunicamycin and DTT tests in the yeast mutants. We also thank Robin Rycroft for revising the English. The Serveis Científico-Tècnics (Universitat de Barcelona) provided DNA sequencing and confocal laser scanning microscopy facilities. This study was funded by MCyT PM99-0168, CIRIT 1999SGR 00027, and Fundaluce to R.G.-D., and Magn. Bergvall Foundation to L.H. L.H. was in receipt of a postdoctoral fellowship from the Swedish Foundation for International Cooperation in Research and Higher Education (STINT) and a travel grant from the Swedish Society of Medicine. M.T. is in receipt of a predoctoral fellowship from the CIRIT (Generalitat de Catalunya).
References
- Bayés M, Goldaracena B, Martínez-Mir A, Iragui-Madoz MI, Solans T, Chivelet P, Bussaglia E, Ramos-Arroyo MA, Baiget M, Vilageliu L, et al. A new autosomal recessive retinitis pigmentosa locus maps on chromosome 2q31-q33. J Med Genet. 1998;35:141–145. doi: 10.1136/jmg.35.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- NCBI BLAST Server http://www.ncbi.nlm.nih.gov/BLAST/
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programmes. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Transcription Element Search System (TESS) Server http://www.cbil.upenn.edu/tess/index.html
- CpGPlot/CpGReport/Isochore http://www.ebi.ac.uk/emboss/cpgplot/
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- JGI Fugu Genome Project http://www.jgi.doe.gov/fugu/index.html
- The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Sidow A. Gen(om)e duplications in the evolution of early vertebrates. Curr Opin Genet Dev. 1996;6:715–722. doi: 10.1016/s0959-437x(96)80026-8. [DOI] [PubMed] [Google Scholar]
- Lundin LG. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics. 1993;16:1–19. doi: 10.1006/geno.1993.1133. [DOI] [PubMed] [Google Scholar]
- Popovici C, Leveugle M, Birnbaum D, Coulier F. Homeobox gene clusters and the human paralogy map. FEBS Lett. 2001;491:237–242. doi: 10.1016/s0014-5793(01)02187-1. [DOI] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Development. 1994;Suppl:125–133. [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH. Updated map of duplicated regions in the yeast genome. Gene. 1999;238:253–261. doi: 10.1016/s0378-1119(99)00319-4. [DOI] [PubMed] [Google Scholar]
- Llorente B, Malpertuy A, Neuveglise C, de Montigny J, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, et al. Genomic exploration of the Hemiascomycetous yeasts: 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 2000;487:101–112. doi: 10.1016/s0014-5793(00)02289-4. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol. 2001;13:349–356. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel D. Molecular Cloning: A Laboratory Manual Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 2000.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- TMpred Server http://www.ch.embnet.org/software/TMPRED_form.html
- NCBI Conserved Domain Database http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam Protein Families Database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signalling domains. Proc Natl Acad SciUSA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UCSC Human Genome Working Draft http://genome.cse.ucsc.edu/
- Kent WJ, Haussler D. Assembly of the working draft of the human genome with GigAssembler. Genome Res. 2001;11:1541–1548. doi: 10.1101/gr.183201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. Drosophila cdc2 homologs: a functional homolog is coexpressed with a cognate variant. EMBO J. 1990;9:3573–3581. doi: 10.1002/j.1460-2075.1990.tb07568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. High efficiency transformation with lithium acetate. In Molecular Genetics of Yeast: A Practical Approach Edited by Johnston JR Oxford: IRL Press; 1994. pp. 121–134.
- Yeast EUROFAN Projects Web Page http://www.mips.biochem.mpg.de/proj/eurofan/
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1994.
- Cervantes S, Gonzàlez-Duarte R, Marfany G. Homodimerization of presenilin N-terminal fragments is affected by mutations linked to Alzheimer's disease. FEBS Lett. 2001;505:81–86. doi: 10.1016/s0014-5793(01)02785-5. [DOI] [PubMed] [Google Scholar]
- Zhang QH, Ye M, Wu XY, Ren SX, Zhao M, Zhao CJ, Fu G, Shen Y, Fan HY, Lu G, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Børsting C, Hummel R, Schultz ER, Rose TM, Pedersen MB, Knudsen J, Kristiansen K. Saccharomyces carlsbergensis contains two functional genes encoding the acyl-CoA binding protein, one similar to the ACB1 gene from S. cerevisiae and one identical to the ACB1 gene from S. monacensis. Yeast. 1997;13:1409–1421. doi: 10.1002/(SICI)1097-0061(199712)13:15<1409::AID-YEA188>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Yoshioka S, Kato K, Nakai K, Okayama H, Nojima H. Identification of open reading frames in Schizosaccharomyces pombe cDNAs. DNA Res. 1997;4:363–369. doi: 10.1093/dnares/4.6.363. [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Yale J, Bohnert HJ. Transcript expression in Saccharomyces cerevisiae at high salinity. J Biol Chem. 2001;276:15996–16007. doi: 10.1074/jbc.M008209200. [DOI] [PubMed] [Google Scholar]


