Abstract
Background
Smoking is a significant risk factor for pancreatic ductal adenocarcinoma (PDAC). This study aimed to investigate the effects of smoking on the pancreatic microbiome and metabolome in resectable and unresectable male PDAC patients.
Methods
The pancreatic tissue samples were collected from resectable PDACs via surgery and unresectable PDACs via endoscopic ultrasound fine needle aspiration (EUS-FNA). Surgical samples obtained from 10 smoking and 6 non-smoking PDACs were measured by 16S ribosomal RNA (16S rRNA) gene sequencing and liquid chromatography-mass spectrometry (LC/MS). Fine needle aspiration (FNA) samples obtained from 20 smoking and 14 non-smoking PDACs were measured by 16S rRNA gene sequencing.
Results
From resectable to unresectable patients, the dominant genus in the pancreas changed from Achromobacter to Delftia. Smoking further altered the abundance of specific bacteria, mainly manifested as an increase of Slackia in surgical tumor tissue of the smoking group, and an enrichment of Aggregatibacter and Peptococcus in FNA samples of the smoking group. In tumor tissue, smoking caused an enrichment of the cancer-promoting cAMP signaling pathway and L-lactic acid. In paracancerous tissue, smoking also induced a detrimental disturbance in the pancreatic microbiome and metabolome, including an enrichment of Veillonella, Novosphingobium, Deinococcus, and 3-hydroxybutanoic acid, and a reduction of linoleic acid. Besides, the cancer-promoting L-lactic acid was negatively correlated with Faecalibacterium in tumor tissue based on the correlation analysis.
Conclusion
There were differences in the pancreatic microbiome of PDAC patients at different stages, and smoking can further disrupt the pancreatic microbiome and metabolism in PDAC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03688-5.
Keywords: Pancreatic ductal adenocarcinoma, Smoking, Endoscopic ultrasound-guided fine needle aspiration, Microbiome, Metabolome
Introduction
Pancreatic cancer, mostly pancreatic ductal adenocarcinoma (PDAC), is a leading cause of cancer death worldwide [1]. About 80–85% of patients have advanced unresectable tumors [2]. In recent years, the incidence of PDAC has shown a rapidly increasing trend, which burdens families and society [3]. However, the etiology of PDAC is still unclear.
Microorganisms with complex functions have been found to be a potential component of tumor microenvironment (TME) in various tumors [4], and epidemiological studies have shown that PDAC was also related to microorganisms in different parts of the human body [5]. Tumors of different stages often have varied intracellular microbial compositions [6], which are related to tumor prognosis. However, there is little research to explore the microbial features of PDAC at different stages. Moreover, metabolic remodeling is considered as an essential hallmark of cancer [7], and the microbiome can disrupt the balance of metabolites, resulting in tumorigenesis [8]. Nevertheless, there are few studies focusing on the microbiome and metabolome of PDAC patients.
Smoking is the most certain and top risk factor of PDAC [9], and smoking patients also have a poorer prognosis and a higher risk of death [10]. There are many hypotheses about how smoking causes and exacerbates PDAC, including tobacco components causing the pathophysiological changes of pancreatic cells, disruption of pancreatic enzyme secretion, excessive production of pro-inflammatory cytokines leading to chronic inflammation, immune system dysfunction, pancreatic fibrosis, KRAS mutation, genomic instability [11], etc. These changes eventually lead to alterations in TME and carcinogenesis. Until now, whether smoking could affect PDAC through microbiome and metabolome remains inconclusive.
Solid pancreatic lesions suspected as PDAC are typically targets for endoscopic ultrasound fine needle aspiration (EUS-FNA). Metastases and primary lesion sampling are indicated when the lesion is unresectable [12]. Therefore, we used the surgical and fine needle aspiration (FNA) samples to represent two stages of PDAC patients, resectable and unresectable, respectively. In this study, we conducted the microbiome analysis, metabolomic analysis, and correlation analysis on surgical samples (tumor and paracancerous tissue) from the resectable male PDAC patients, and microbiome analysis on FNA samples (tumor tissue) from the unresectable male PDAC patients. Using different sampling methods, our study aimed to reveal the pancreatic microbial composition of PDAC at different stages under the influence of smoking and further explore the metabolic mechanism behind it.
Methods
Subjects, samples, and grouping
We performed an observational study of subjects aged > 18 years with suspected pancreatic lesions requiring surgery (n = 16) or EUS-FNA (n = 34) recruited from Shandong University Qilu Hospital, using the same protocols for biological sample collection, processing, and storage prior to any treatment. Due to the rarity of female smokers in China and the potential influence of gender as a confounding factor on microbiome and metabolome [13, 14], we included only male patients in this study. Additional exclusion criteria were as follows: (I) comorbidity with other cancers; (II) underwent ERCP, X-ray-guided puncture, chemotherapy, radiotherapy; and (III) use of antibiotics, probiotics, prebiotics, or synbiotics in the previous month. For patients who underwent EUS-FNA, those found to be in the resectable stage and subsequently underwent surgery were excluded. Institutional review board ethical approval (KYLL-202107-097, KYLL-202111-018-1) and written informed consent were obtained from the Ethics Review Committee of Shandong University Qilu Hospital and study participants, respectively. Patients’ basic information, including name, gender, age, carbohydrate antigen 19–9 (CA19-9), carcinoembryonic antigen (CEA), total cholesterol (CHO), free fatty acid (FFA), low-density lipoprotein cholesterol (LDL), body mass index (BMI), smoking history, etc., was collected through the electronic medical record system. The postoperative pathology of tumors (tumor type, cytology, etc.) in all patients was confirmed by the pathology query system.
Surgical samples of PDAC tumor and paracancerous tissue were collected during surgery. FNA samples were taken using a linear array echo endoscope (PENTAX EG-3870UTK and EG-3270UK, Tokyo, Japan). All samples were immediately flash-frozen in liquid nitrogen, transferred via temporary liquid nitrogen transport containers, and stored in lock-in freezer chambers at −80℃ until DNA extraction. The subjects were divided into smoking and non-smoking groups according to smoking history (surgical subjects: smoking PDAC group, n = 10; non-smoking PDAC group, n = 6. FNA subjects: smoking PDAC group, n = 20; non-smoking PDAC group, n = 14). In this study, the variable smoking was defined as patients who currently smoke any tobacco products [15]. Surgical samples were divided into two subgroups based on the sampling location: the tumor tissue group and the paracancerous tissue group.
DNA extraction, Illumina MiSeq 16S sequencing, and data analysis
All samples were thawed on ice and aliquoted, and microbial DNA was extracted using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.). The DNA extract was checked on 1% agarose gel and the NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA). The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with primer pairs 338 F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) by an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA). Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, USA).
The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp version 0.20.0 [16], and merged by FLASH version 1.2.7 [17]. Reads were truncated at any site receiving an average quality score of < 20, and the truncated reads shorter than 50 bp were discarded. Operational taxonomic units (OTUs) with a 97% similarity cut-off were clustered using UPARSE version 7.1 [18], and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 [19] against the 16S rRNA database (Silva v138) using a confidence threshold of 0.7. Bioinformatic analysis of microbiota was carried out using the Majorbio Cloud platform (https://cloud.majorbio.com).
Metabolomics analysis
A total of 50 mg of tissue samples were used to extract the metabolites. The instrument platform for liquid chromatography-mass spectrometry (LC/MS) analysis is the UHPLC-Q Exactive system of Thermo Fisher Scientific. The mobile phases consisted of 0.1% formic acid in water: acetonitrile (95:5, v/v) (solvent A) and 0.1% formic acid in acetonitrile: isopropanol: water (47.5:47.5:5, v/v) (solvent B). The solvent gradient changed according to the following conditions: from 0 to 0.1 min, 0% B to 5% B; from 0.1 to 2 min, 5% B to 25% B; from 2 to 9 min, 25% B to 100% B; from 9 to 13 min, 100% B to 100% B; from 13 to 13.1 min, 100% B to 0% B; from 13.1 to 16 min, 0% B to 0% B for equilibrating the systems. The sample injection volume was 2 µL, and the flow rate was 0.4 mL/min. The raw data of LC/MS was preprocessed by Progenesis QI (Waters Corporation, Milford, USA) software. The metabolites were searched and identified, and the primary databases were the HMDB (http://www.hmdb.ca/), Metlin (https://metlin.scripps.edu/), and Majorbio Database. After the database search, the data was uploaded to the Majorbio cloud platform (https://cloud.majorbio.com) for data analysis. The R package ropls (Version 1.6.2) performed orthogonal least partial squares discriminant analysis (OPLS-DA). The metabolites with variable importance in the projection (VIP) > 1 and P < 0.05 were significantly differential. The functions of these metabolites and metabolic pathways were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [20]. The correlation between the differential metabolites and differential bacteria was calculated according to the Pearson correlation analysis.
Results
Microbiome of tumor tissue from surgical samples of resectable PDAC patients
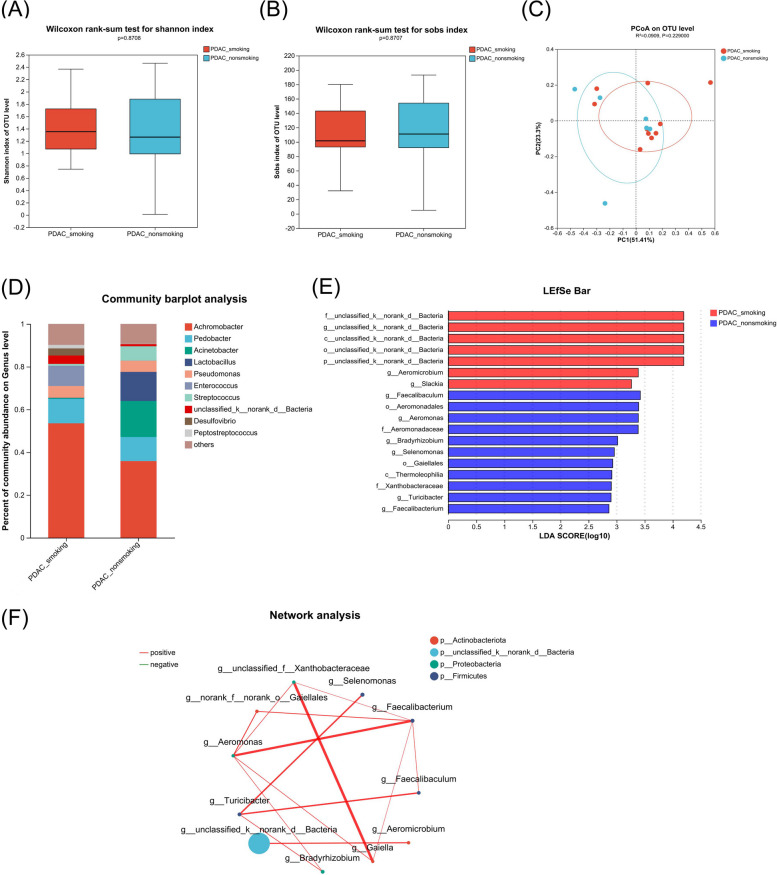
To explore the effects of smoking on pancreatic microbiome and metabolism in resectable PDAC patients, we divided the surgical PDAC samples into smoking and non-smoking PDAC groups. The baseline data of the subjects are shown in Table 1. There was no significant difference in alpha and beta diversity (Fig. 1A and C). At the class level, the abundance of Gammaproteobacteria was the highest in both groups (Fig. S1). At the genus level, the abundance of Achromobacter was the highest in both groups (Fig. 1D). Of note, at the genus level, Aeromicrobium and Slackia were enriched, while Faecalibaculum, Aeromonas, Bradyrhizobium, Selenomonas, Turicibacter, and Faecalibacterium were reduced in the smoking PDAC group compared with the non-smoking PDAC group according to LDA effect size (LEfSe) difference analysis (LDA threshold = 2; Fig. 1E and Table S1). The network analysis showed the differential genera with correlation coefficient > 0.5 and P < 0.05, which were generally positively correlated with each other (Fig. 1F).
Table 1.
Clinical data of the subjects in the smoking and non-smoking PDAC group of surgical samples
| Smoking PDAC (n = 10) | Non-smoking PDAC (n = 6) | P | |
|---|---|---|---|
| Age (ys, mean ± SD) | 64 ± 10.03 | 60 ± 4.56 | 0.377 |
| BMI (mean ± SD) | 21.72 ± 3.17 | 23.74 ± 2.16 | 0.199 |
| CA19-9 (U/ml, mean ± SD) | 295.43 ± 383.43 | 410.13 ± 472.22 | 0.638 |
| CEA (ng/ml, mean ± SD) | 4.08 ± 2.23 | 1.93 ± 0.58 | 0.044 |
| CHO (mmol/L, mean ± SD) | 5.54 ± 3.47 | 4.90 ± 0.90 | 0.701 |
| LDL (mmol/L, mean ± SD) | 2.53 ± 0.31 | 2.77 ± 0.34 | 0.259 |
| FFA (µmmol/dl, mean ± SD) | 61.29 ± 35.76 | 75.60 ± 21.69 | 0.441 |
| Diabetes | 0.889 | ||
| Yes | 3 | 2 | |
| No | 7 | 4 |
Fig. 1.
Comparison of the microbiome in tumor tissue of surgical samples from smoking vs. non-smoking PDAC. A, B Alpha diversity analysis of the smoking PDAC group (PDAC_smoking, n = 10) and non-smoking PDAC group (PDAC_non-smoking, n = 6). Box plots show the distribution of the Shannon and Sobs indices of alpha diversity scores. C Principal co-ordinates analysis (PCoA) quantified as weighted-unifrac dissimilarity. D Distinct bacterial composition at the genus level of subjects between the smoking PDAC group and non-smoking PDAC group. E Linear discriminant analysis (LDA) demonstrated distinct bacterial genera enriched in the smoking PDAC group and non-smoking PDAC group. Microorganisms with P < 0.05 and LDA score > 2 were considered significant. F Network analysis of the differential genera between the smoking PDAC group and non-smoking PDAC group. Only the genera with correlation coefficient > 0.5 and P < 0.05 were illustrated in the graph. The red line indicates positive correlation, while the green line indicates negative correlation. The size of the node indicates the abundance of the genus, and the thickness of the lines between nodes indicates the magnitude of the correlation
Metabolome of tumor tissue from surgical samples of resectable PDAC patients
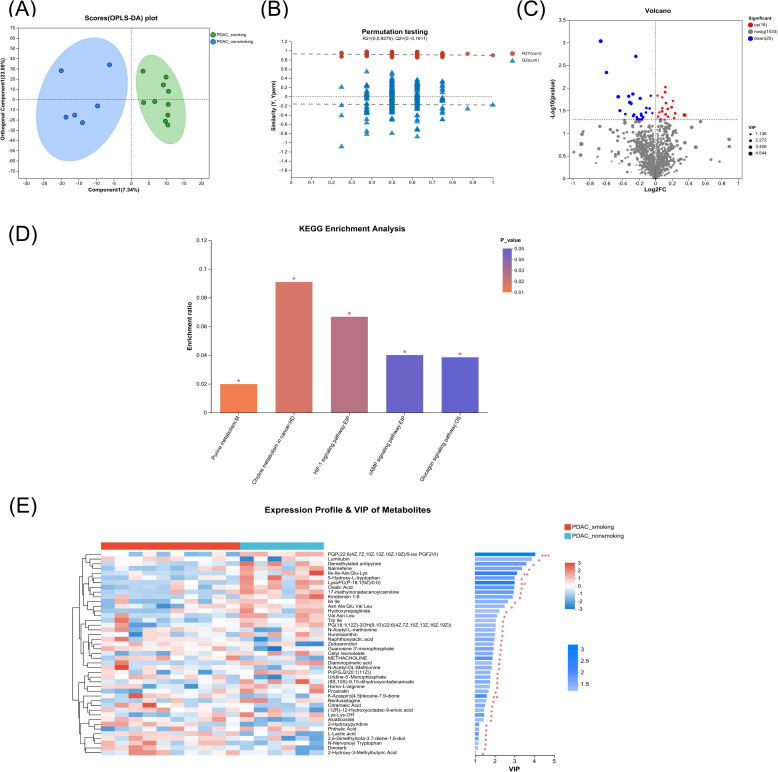
We determined alterations in pancreatic metabolites by LC/MS analysis in the above tumor tissue samples obtained from the surgery. It was found that the pancreatic metabolic profile in the smoking PDAC group was significantly differed from the non-smoking PDAC group in the tumor tissue (R2Y = 0.898 and R2X = 0.303) (Fig. 2A). The test for the OPLS-DA model illustrated that the R2 value was larger than the Q2 value and that the Q2 regression line had a negative intercept (R2 = [0.0, 0.9279], Q2 = [0.0, −0.1611]), indicating that the OPLS-DA model for this study was valid (Fig. 2B). Total of 43 metabolites (P < 0.05, VIP of OPLS-DA > 1 and fold-change > 1) altered in the tumor tissue of the smoking PDAC group compared with the non-smoking PDAC group, including 18 upregulated metabolites and 25 downregulated metabolites (Fig. 2C and Table S2), though not significantly different after multiple comparison adjustment. The altered metabolites were enriched or decreased in different metabolomic signaling pathways (Fig. 2D and Table S3). The purine metabolism pathway was the top altered pathway in the smoking PDAC group compared with the non-smoking PDAC group. In addition, choline metabolism in cancer, HIF-1 signaling pathway, cAMP signaling pathway, and glucagon signaling pathway were also significantly altered in the smoking PDAC group. The abundance of L-lactic acid and 2-hydroxy-3-methylbutyric acid was increased significantly in the smoking PDAC group. Some metabolites, including diaminopimelic acid and 5-hydroxy-L-tryptophan, were decreased in the smoking PDAC group (Fig. 2E and Fig. S2).
Fig. 2.
Comparison of the metabolome in tumor tissue of surgical samples from smoking vs. non-smoking PDAC. A, B Pancreatic metabolic profile significantly differed between smoking PDAC and non-smoking PDAC group by OPLS-DA method. Test for the OPLS-DA model showed that the model for this study was valid. C Metabolites with significant differences between groups were presented in the volcano plot. Different metabolites were depicted by setting the fold-change > 1 and P < 0.05 as the threshold. The red points represented the upregulated metabolites (n = 18), and the blue points represented the downregulated metabolites (n = 25). D The altered signaling pathways in the smoking PDAC group compared with the non-smoking PDAC group by KEGG enrichment analysis, marked as * for P < 0.05. E Heatmap representative differential metabolites between the smoking PDAC group and non-smoking PDAC group. The color from blue to red indicates that the expression differential abundance of metabolites is from low to high. The right side shows the VIP bar chart of the metabolite, and the length of the bar represents the contribution value of the metabolite to the difference between the two groups. The color of the bar indicates the significance of the difference between the two groups of samples, P < 0.001 is marked as ***, P < 0.01 is marked as **, P < 0.05 is marked as *
Microbiome of paracancerous tissue from surgical samples of resectable PDAC patients
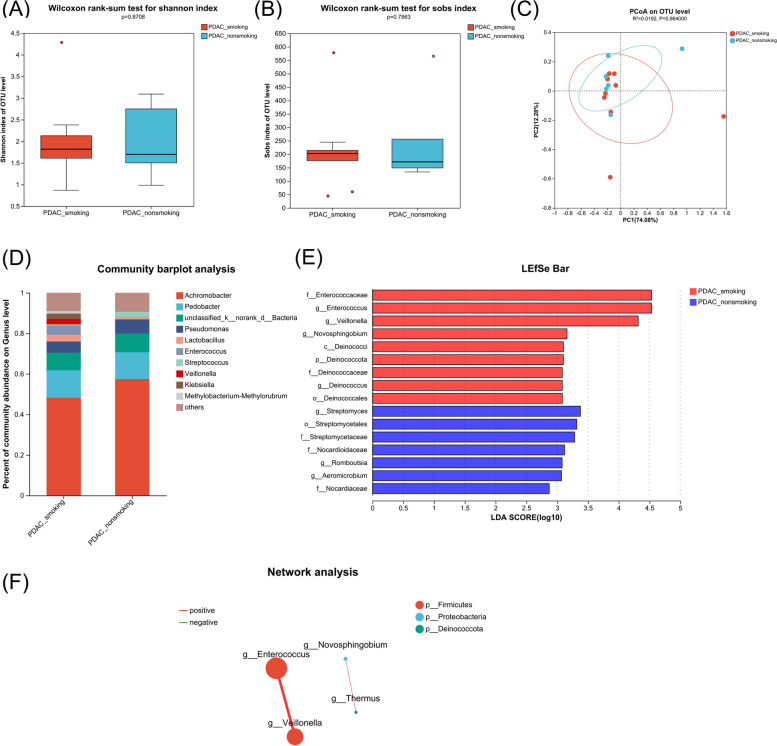
Similarly, we divided the subjects into the smoking and non-smoking groups to investigate the impact of smoking on normal paracancerous pancreatic tissue. Consistent with the tumor tissue, there was no significant difference in alpha and beta diversity (Fig. 3A and C). In addition, the abundance of the Gammaproteobacteria class (Fig. S3) and Achromobacter genus was still the highest in both groups (Fig. 3D). Of note, at the genus level, Enterococcus, Veillonella, Novosphingobium, and Deinococcus were enriched, whereas Streptomyces, Romboutsia, and Aeromicrobium were reduced in paracancerous tissue of the smoking group compared with the non-smoking group based on LEfSe (Fig. 3E and Table S4). The network analysis showed that the abundance of Enterococcus and Veillonella was significantly positively correlated, as was the abundance of Novosphingobium and Thermus (correlation coefficient > 0.5 and P < 0.05, Fig. 3F).
Fig. 3.
Comparison of the microbiome in paracancerous tissue of surgical samples from smoking vs. non-smoking PDAC. A, B, Alpha diversity analysis of the smoking PDAC group (PDAC_smoking, n = 10) and non-smoking PDAC group (PDAC_non-smoking, n = 6). Box plots show the distribution of the Shannon and Sobs indices of alpha diversity scores. C PCoA quantified as weighted-unifrac dissimilarity. D Distinct bacterial composition at the genus level of subjects between the smoking PDAC group and non-smoking PDAC group. E LDA demonstrated distinct bacterial genera enriched in the smoking PDAC group and non-smoking PDAC group. Microorganisms with P < 0.05 and LDA score > 2 were considered significant. F Network analysis of the differential genera between the smoking PDAC group and non-smoking PDAC group. Only the genera with correlation coefficient > 0.5 and P < 0.05 were illustrated in the graph. The red line indicates positive correlation, while the green line indicates negative correlation. The size of the node indicates the abundance of the genus, and the thickness of the lines between nodes indicates the magnitude of the correlation
Metabolome of paracancerous tissue from surgical samples of resectable PDAC patients
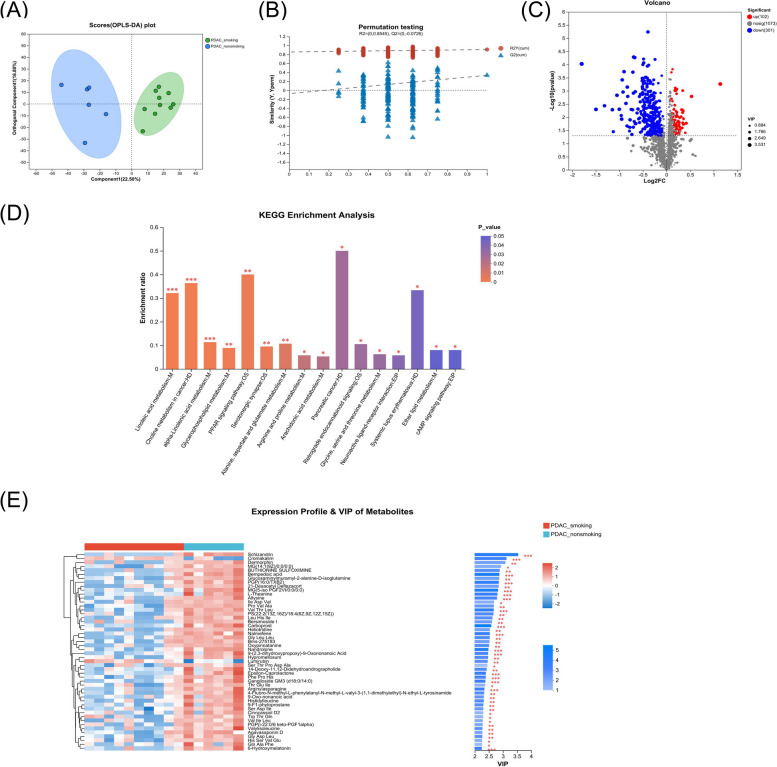
We also evaluated alterations in pancreatic metabolites by LC/MS analysis in the above paracancerous tissue samples. The results showed that the pancreatic metabolic profile in paracancerous tissue of the smoking group was significantly different from the non-smoking group (R2Y = 0.910 and R2X = 0.394) (Fig. 4A). The test for the OPLS-DA model illustrated that the R2 value was larger than the Q2 value and that the Q2 regression line had a negative intercept (R2 = [0.0, 0.8545], Q2 = [0.0, −0.0726]), indicating that the OPLS-DA model for this study was valid (Fig. 4B). Total of 403 metabolites (P < 0.05, VIP of OPLS-DA > 1 and fold-change > 1) altered in the paracancerous tissue of the smoking group compared with the non-smoking group, including 102 upregulated metabolites and 301 downregulated metabolites (Fig. 4C and Table S5). Of them, 129 metabolites were still significantly different after multiple comparison adjustment. The altered metabolites participated in different metabolomic signaling pathways (Fig. 4D and Table S6). The linoleic acid metabolism pathway was the top altered pathway in the paracancerous tissue of the smoking group compared with the non-smoking group. Besides, the choline metabolism in cancer, alpha-linolenic acid metabolism, glycerophospholipid metabolism, PPAR signaling pathway, etc., were significantly altered in the smoking group. Moreover, the abundance of 3-hydroxybutanoic acid and 2-hydroxy-3-methylbutyric acid was significantly increased in the smoking group. Some metabolites, including linoleic acid, elaidic acid, L-theanine, sinomenine, glucosamine, 14-deoxy-11,12-didehydroandrographolid (14-DDA), prostaglandin A2, etc., were significantly decreased in the smoking group (Fig. 4E and Fig. S4).
Fig. 4.
Comparison of the metabolome in paracancerous tissue of surgical samples from smoking vs. non-smoking PDAC. A, B, Pancreatic metabolic profile significantly differed between smoking PDAC and non-smoking PDAC group by orthogonal least partial squares discriminant analysis (OPLS-DA) method. Test for the OPLS-DA model showed that the model for this study was valid. C Metabolites with significant differences between groups were presented in the volcano plot. Different metabolites were depicted by setting the fold-change > 1 and P < 0.05 as the threshold. The red points represented the upregulated metabolites (n = 102) and the blue points represented the downregulated metabolites (n = 301). D The altered signaling pathways in the smoking PDAC group compared with the non-smoking PDAC group by KEGG enrichment analysis, marked as *** for P < 0.001, ** for P < 0.01, and * for P < 0.05. E Heatmap representative differential metabolites between the smoking PDAC group and non-smoking PDAC group. The color from blue to red indicates that the expression differential abundance of metabolites is from low to high. The right side shows the VIP bar chart of the metabolite, and the length of the bar represents the contribution value of the metabolite to the difference between the two groups. The color of the bar indicates the significance of the difference between the two groups of samples, P < 0.001 is marked as ***, P < 0.01 is marked as **, P < 0.05 is marked as *
Correlation analysis of microbiome and metabolome from surgical samples of resectable PDAC patients
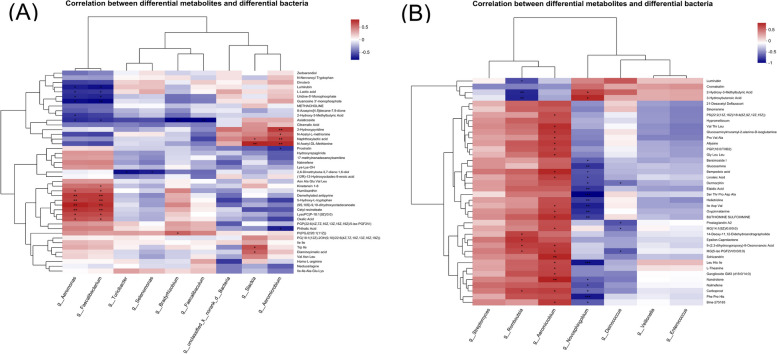
To further study the interaction between microbiome and metabolism, we conducted a correlation analysis of differential bacteria and metabolites in both tumor and paracancerous tissue. In the tumor tissue, the abundance of 5-hydroxy-L-tryptophan, which was decreased in the smoking group, was significantly positively correlated with Faecalibacterium, which was reduced in the smoking group. Whereas L-lactic acid, which was increased in the smoking group, was significantly negatively correlated with Faecalibacterium (Fig. 5A).
Fig. 5.
Correlation analysis between differential bacteria and metabolites of surgical samples from smoking vs. non-smoking PDAC. A tumor tissue; B, paracancerous tissue. The abscissa represents differential bacteria, and the ordinate represents differential metabolites. P < 0.001 is marked as ***, P < 0.01 is marked as **, P < 0.05 is marked as *
In the paracancerous tissue, metabolites significantly upregulated in the smoking group, such as 3-hydroxybutanoic acid and 2-hydroxy-3-methylbutyric acid, were positively correlated with Novosphingobium, which was enriched in the smoking group. In contrast, metabolites significantly downregulated in the smoking group, such as linoleic acid, were negatively correlated with Novosphingobium (Fig. 5B).
Microbiome of FNA samples from unresectable PDAC patients
We further explored the impact of smoking on the pancreatic microbiome of late-stage unresectable PDAC patients using FNA samples of the tumor tissue. The baseline data of the subjects are shown in Table 2. Consistent with the results of the surgical samples, it was found that there was no significant difference in the alpha and beta diversity between the two groups (Fig. 6A-C). The most abundant class in both groups was still Gammaproteobacteria (Fig.S5). However, the most abundant genus was Delftia (Fig. 6D). At the genus level, the LEfSe analysis showed that Aggregatibacter, Soonwooa, Eubacterium brachy group, etc. increased while Simiduia, Jatrophihabitans, Polynucleobacter, etc. decreased in the smoking PDAC group compared with the non-smoking PDAC group (LDA threshold = 2; Fig. 6E and Table S7). Similar to surgical samples, network analysis showed the differential genera with correlation coefficient > 0.5 and P < 0.05, which were generally positively correlated with each other (Fig. 6F).
Table 2.
Clinical data of the subjects in the smoking and non-smoking PDAC group of FNA samples
| Smoking PDAC (n = 20) | Non-smoking PDAC (n = 14) | P | |
|---|---|---|---|
| Age (ys, mean ± SD) | 56.75 ± 9.80 | 59 ± 13.96 | 0.584 |
| BMI (mean ± SD) | 21.45 ± 2.84 | 22.64 ± 3.50 | 0.440 |
| CA19-9 (U/ml, mean ± SD) | 328.19 ± 283.26 | 174.22 ± 255.16 | 0.253 |
| CEA (ng/ml, mean ± SD) | 6.10 ± 5.45 | 9.89 ± 15.32 | 0.492 |
| CHO (mmol/L, mean ± SD) | 5.93 ± 4.07 | 4.61 ± 1.16 | 0.330 |
| LDL (mmol/L, mean ± SD) | 4.09 ± 3.76 | 2.89 ± 1.07 | 0.341 |
| FFA (µmmol/dl, mean ± SD) | 73.29 ± 35.85 | 74.52 ± 34.71 | 0.935 |
| Diabetes | 0.493 | ||
| Yes | 8 | 4 | |
| No | 12 | 10 |
Fig. 6.
Comparison of the microbiome in FNA samples from smoking vs. non-smoking PDAC. A, B Alpha diversity analysis of the smoking PDAC group (PDAC_smoking, n = 20) and non-smoking PDAC group (PDAC_non-smoking, n = 14). Box plots show the distribution of the Shannon and Sobs indices of alpha diversity scores. C PCoA quantified as weighted-unifrac dissimilarity. D, Distinct bacterial composition at the genus level of subjects between the smoking PDAC group and non-smoking PDAC group. E LDA demonstrated distinct bacterial genera enriched in the smoking PDAC group and non-smoking PDAC group. Microorganisms with P < 0.05 and LDA score > 2 were considered significant. F Network analysis of the differential genera between the smoking PDAC group and non-smoking PDAC group. Only the genera with correlation coefficient > 0.5 and P < 0.05 were illustrated in the graph. The red line indicates positive correlation, while the green line indicates negative correlation. The size of the node indicates the abundance of the genus, and the thickness of the lines between nodes indicates the magnitude of the correlation
Discussion
In this study, we used surgical and FNA pancreatic samples to investigate the differences in pancreatic microbiome between resectable and unresectable male PDAC patients, and focused on the effects of smoking on the pancreatic microbiome and metabolomics of PDAC. We found that from the resectable stage to the unresectable stage, the dominant genus in the pancreas changed from Achromobacter to Delftia, and smoking further altered the abundance of specific bacteria. In the tumor tissue of resectable patients, the cAMP signaling pathway and L-lactic acid were enriched under the influence of smoking. In addition, smoking also caused a detrimental disturbance in the microbiome and metabolism in paracancerous tissue, such as the enrichment of linoleic acid metabolism and decrease of linoleic acid.
Smoking can alter the pancreatic microbiome of resectable PDAC patients. In surgical samples, whether in tumor or paracancerous tissue, the dominant class and genus of all groups were Gammaproteobacteria and Achromobacter. Gammaproteobacteria is the most abundant bacterium in the tumor tissue of PDAC, which has been confirmed in multiple previous studies, and may be related to the resistance of PDAC to gemcitabine [21]. Consistent with the results of the previous research [22], there was no statistical difference in alpha and beta diversity between the smoking and non-smoking groups, indicating that smoking may have little impact on the overall distribution of pancreatic microbiome in the early-stage. However, smoking caused changes in the abundance of specific bacteria in tumor and paracancerous tissues. In tumor tissue, Slackia was significantly increased in the smoking group compared with the non-smoking group. Slackia was also demonstrated to be continuously enriched from superficial gastritis to gastric cancer [23]. However, Faecalibaculum, Selenomonas, Faecalibacterium, and Turicibacter were significantly decreased in the smoking group. Faecalibaculum, Selenomonas, and Faecalibacterium can produce short-chain fatty acids (SCFAs), which could inhibit oncogenic pathways, such as TGF-β signaling [24]. Turicibacter is a probiotic [25] and a prominent member of the mammalian gut microbiome, involved in modifying bile acids and host lipids [26]. In paracancerous tissue, Veillonella, Novosphingobium and Deinococcus were enriched in the smoking group compared with the non-smoking group. Veillonella was associated with an early recurrence of hepatocellular carcinoma (HCC) [27]. Novosphingobium was found to increase in the biliary tract of extrahepatic cholangiocarcinoma [28]. Deinococcus was more abundant in the duodenal mucosa of PDAC patients compared with healthy controls [29]. In contrast, Streptomyces was reduced in the smoking group. An oxygenated anthrabenzoxocinone compound derived from Streptomyces was demonstrated to show an inhibitory activity against non-small cell lung cancer [30]. In brief, the increase of these harmful bacteria and the decrease of the beneficial bacteria suggest that smoking also has adverse effects on normal paracancerous tissue. Moreover, the identified differential bacteria in this study were inconsistent with previous reports [22], possibly due to differences in microbial sequencing methods and races of the subjects.
In unresectable PDAC patients, smoking further disturbed the pancreatic microbiome. In FNA samples, the dominant class of all groups was still Gammaproteobacteria, but the dominant genus was Delftia. The Delftia genus is enriched in the duodenal mucosa of PDAC patients and colorectal cancer mice [31]. From resectable to unresectable PDAC tissue, the dominant genus changed from Achromobacter to Delftia, but the mechanism and significance of this change are not yet clear. Furthermore, smoking still caused significant changes in the abundance of specific bacteria, but the types of bacteria differed from those of resectable patients. Aggregatibacter and Peptococcus were enriched in the smoking group compared with the non-smoking group. Aggregatibacter was enriched in tumor samples of oral squamous cell carcinoma (OSCC) patients [32]. Similarly, Peptococcus was also confirmed to be significantly enriched in OSCC samples [33]. However, the specific roles of the aforementioned bacteria in PDAC remain to be investigated.
Smoking also has great impacts on the pancreatic metabolism of PDAC. In tumor tissue, purine metabolism, cAMP signaling, and HIF-1 signaling pathways were significantly altered in the smoking group. Purine metabolism pathway was the top altered among these. Impaired purine metabolism is associated with the progression of cancer [34], and may participate in the development of PDAC [35, 36]. cAMP signaling pathway is closely related to oncogenesis and represents a potential therapeutic target [37]. Previous in vitro studies have shown that nicotine and tobacco-specific carcinogenic nitrosamine 4-methylnitrosamino(3-pyridyl)−1-butanone (NNK) can promote the proliferation and migration of human PDAC cells by increasing cAMP downstream of beta-adrenergic receptors [38], which may provide clues to the role of smoking. Moreover, smoking can activate HIF-1α and promote the Warburg effect in lung cancer [39]. Despite many studies reporting the critical role of the HIF-1 signaling pathway in PDAC [40], it remains unclear whether such smoking-induced HIF-1 activation also exists in PDAC.L-lactic acid, a common component of the cAMP signaling pathway and HIF-1 signaling pathway, was also significantly increased in the smoking group. L-lactic acid regulates the energy metabolism in cancer and establishes a favorable acidic microenvironment for tumorigenesis [41]. A study found that smoking episodes increase the lactic acid appearance rate in the blood [42], indicating that smoking may worsen PDAC by further accumulating lactic acid. These evidences suggest a possible causal chain between smoking, metabolites, and PDAC. In contrast, diaminopimelic acid was significantly decreased in the smoking group. Nod1, a cytosolic protein that senses meso-diaminopimelic acid-containing ligands, plays a role in the host’s control of tumor growth. In estrogen-dependent tumors, the absence of Nod1 correlates with tumor growth and cell proliferation [43]. However, it is currently unclear whether Nod1 has a similar effect in other tumors, including PDAC. Further research is needed to validate the impact of diaminopimelic acid on PDAC.
The pancreatic metabolism was also disrupted by smoking in the paracancerous tissue. Same as tumor tissue, cAMP signaling pathway and choline metabolism in cancer altered similarly in the smoking group. Linoleic acid metabolism was the top altered pathway in the smoking group, with its vital component linoleic acid significantly decreased. Linoleic acid was decreased in HCC patients and can inhibit the proliferation of cancer cells [44]. Besides, linoleic acid could induce cancer dormancy and quiescence in colorectal cancer [45], and was a major positive regulator of CD8 + T cell activity and an adoptive T cell therapy potentiator in tumor therapy [46]. Further investigation is needed to confirm the impact of linoleic acid on PDAC. Besides, 3-hydroxybutanoic acid was also significantly enriched in the smoking group. 3-hydroxybutanoic acid, also known as beta-hydroxybutyrate (betaOHB), is an alternative cell-intrinsic or systemic fuel to promote PDAC growth and progression [47]. In addition, some other anticancer metabolites, including elaidic acid [48], L-theanine [49], sinomenine [50], glucosamine [51], 14-deoxy-11,12-didehydroandrographolid (14-DDA) [52], and prostaglandin A2 [53], were significantly decreased in the smoking group. It needs to be clarified whether the above metabolites and pathways indeed have an inhibitory effect on PDAC and whether smoking can suppress this trend. At least we can confirm that the paracancerous tissue is not truly “normal” tissue even in the early-stage resectable PDAC, but it has many similarities with the tumor tissue. Some precancerous metabolic and microbial changes may also have occurred in the paracancerous tissue, and smoking can exacerbate these changes.
Correlation analysis indicated that there were crosstalk and interactions between microbiome and metabolism in the pancreas under the influence of smoking. In this study, the abundance of 5-hydroxy-L-tryptophan was significantly positively correlated with the cancer-inhibiting Faecalibacterium. 5-hydroxy-L-tryptophan can promote antitumor immunity by inhibiting PD-L1 inducible expression [54]. On the contrary, the cancer-promoting L-lactic acid was enriched in the smoking group, and significantly negatively correlated with Faecalibacterium. In the paracancerous tissue, 3-hydroxybutanoic acid and 2-hydroxy-3-methylbutyric acid were upregulated in the smoking group, and positively correlated with Novosphingobium. As previously mentioned, 3-hydroxybutanoic acid and Novosphingobium have potential cancer-promoting effects [28, 47]. Additionally, 2-hydroxy-3-methylbutyric acid was reported to be associated with the risk of HCC and PDAC [55]. Our findings suggest that there may be a causal chain from smoking, through bacteria, and then to metabolites, but the specific mechanism needs further investigation.
This study has some limitations. First, this study included only male PDAC patients due to the rarity of female smokers in China and the potential impact of gender on the microbiome and metabolism. Second, the number of surgical samples was not large enough, as 80–85% of PDAC patients are late-stage and unresectable [2]. Lastly, we only performed microbiome analysis on FNA samples from unresectable patients, without metabolomics. However, previous studies have found that the total tumor burden has little influence on the metabolic signature of PDAC [56]. In the future, studies with larger sample sizes that involve female patients and multiple FNA sampling of the same patient are needed.
In conclusion, we found variations in the pancreatic microbiome of male PDAC patients at different stages, and smoking could further lead to significant changes in the abundance of specific bacteria. Smoking can also cause pancreatic metabolic disorders in PDAC, and partial of the differential metabolites were significantly associated with certain differential bacteria. More research is warranted to further explore the interaction mechanisms among smoking, bacteria, and metabolites, as well as their specific roles in PDAC.
Supplementary Information
Acknowledgements
We would like to thank all our participants and investigators as well as the participating staff for their contributions to the study.
Abbreviations
- PDAC
Pancreatic ductal adenocarcinoma
- EUS-FNA
Endoscopic ultrasound-guided fine needle aspiration
- FNA
Fine needle aspiration
- 16S rRNA
16S ribosomal RNA
- LC/MS
Liquid chromatography mass spectrometry
- TME
Tumor microenvironment
- CA19-9
Carbohydrate antigen 19–9
- CEA
Carcinoembryonic antigen
- CHO
Total cholesterol
- LDL-C
Low-density lipoprotein cholesterol
- FFA
Free fatty acid
- BMI
Body mass index
- OTU
Operational taxonomic unit
- OPLS-DA
Orthogonal least partial squares discriminant analysis
- LDA
Linear discriminant analysis
- LEfSe
LDA effect size
- VIP
Variable importance in the projection
- 14-DDA
14-deoxy-11,12-didehydroandrographolid
- SCFAs
Short-chain fatty acids
- HCC
Hepatocellular carcinoma
- OSCC
Oral squamous cell carcinoma
- betaOHB
beta-hydroxybutyrate)
Authors' contributions
Z.L., LX.L., and N.Z. contributed to the study concept and design and are the corresponding authors of this paper. M.D., GM.Z., HY.T., CX.C., LM.W., P.W., YF.W., RG.M., and XY.C. enrolled subjects for the study and contributed clinical data. X.L., YQ.Z., YQ.B., J.W., and GX.Y. managed and analyzed the data. The manuscript was drafted by X.L. with considerable input from YQ.Z. and reviewed by all authors. X.L. and YQ.Z. contributed equally to this paper. The work reported in the paper has been performed by the authors, unless clearly specified in the text.
Funding
This study is supported by the Key R&D Program of Shandong Province, China (Major Scientific and Technological Innovation Project) (NO. 2021CXGC010506), the National Natural Science Foundation of China (No. 82261160396), the Taishan Scholars Program of Shandong Province (No. tsqn202306344), and the ECCM Program of Clinical Research Center of Shandong University (No. 2021SDUCRCB004).
Data availability
The raw sequencing data from this study have been deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database and is publicly accessible under the accession number: PRJNA1188451.
Declarations
Ethics approval and consent to participate
The samples and clinical information used in this study were obtained under conditions of informed consent and with approval of the Ethics Committee of the Qilu Hospital of Shandong University (KYLL-202107-097).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiao Liang and Yiqing Zhu have contributed equally to this work and share first authorship.
Contributor Information
Ning Zhong, zhongning@email.sdu.edu.cn.
Lixiang Li, Email: lilixiang@sdu.edu.cn.
Zhen Li, Email: qilulizhen@sdu.edu.cn.
References
- 1.Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. 2021;18:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, et al. Pancreat cancer Lancet. 2011;378:607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 4.Xie Y, Xie F, Zhou X, et al. Microbiota in tumors: from understanding to application. Adv Sci (Weinh). 2022;9:e2200470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kartal E, Schmidt TSB, Molina-Montes E, et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut. 2022;71:1359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968–76. [DOI] [PubMed] [Google Scholar]
- 7.Luo X, Liu J, Wang H, et al. Metabolomics identified new biomarkers for the precise diagnosis of pancreatic cancer and associated tissue metastasis. Pharmacol Res. 2020;156:104805. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Misra BB, Liang L, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9:4101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres PJ, Fletcher EM, Gibbons SM, et al. Characterization of the salivary microbiome in patients with pancreatic cancer. Peerj. 2015;3:e1373. [DOI] [PMC free article] [PubMed]
- 10.Hu JX, Zhao CF, Chen WB, et al. Pancreatic cancer: a review of epidemiology, trend, and risk factors. World J Gastroenterol. 2021;27:4298–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olakowski M, Buldak L. Modifiable and non-modifiable risk factors for the development of non-hereditary pancreatic Cancer. Medicina-Lithuania. 2022;58:978. [DOI] [PMC free article] [PubMed]
- 12.Jenssen C, Hocke M, Fusaroli P, et al. EFSUMB guidelines on Interventional Ultrasound (INVUS), part IV - EUS-guided interventions: General aspects and EUS-guided sampling (Long Version). Ultraschall Med. 2016;37:E33–76. [DOI] [PubMed] [Google Scholar]
- 13.Chakladar J, Kuo SZ, Castaneda G, et al. The pancreatic microbiome is Associated with Carcinogenesis and worse prognosis in males and smokers. Cancers (Basel) 2020;12:2672. [DOI] [PMC free article] [PubMed]
- 14.Mauvais-Jarvis F. Sex differences in energy metabolism: natural selection, mechanisms and consequences. Nat Rev Nephrol. 2024;20:56–69. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization. Noncommunicable D, Mental Health Cluster. Surveillance T. STEPS instruments for NCD risk factors (core and expanded version 1.4) : the WHO STEPwise approach to Surveillance of noncommunicable diseases (STEPS). Geneva: World Health Organization; 2001.
- 16.Chen S, Zhou Y, Chen Y, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Garrity GM, Tiedje JM, et al. Naïve bayesian classifier for Rapid assignment of rRNA sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren Y, Yu G, Shi C, et al. Majorbio Cloud: a one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta. 2022;1:e12. [DOI] [PMC free article] [PubMed]
- 21.Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakladar J, Kuo SAZ, Castaneda G, et al. The pancreatic microbiome is Associated with Carcinogenesis and worse prognosis in males and smokers. Cancers. 2020;12:2672. [DOI] [PMC free article] [PubMed]
- 23.Zhang X, Li C, Cao W, et al. Alterations of gastric microbiota in gastric Cancer and precancerous stages. Front Cell Infect Microbiol. 2021;11:559148. [DOI] [PMC free article] [PubMed]
- 24.Singh V, Lee G, Koh H, et al. Butyrate producers,the Sentinel of Gut: their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front Microbiol. 2023;13:1103836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q, Liang Q, Balakrishnan B, et al. Role of Dietary nutrients in the modulation of gut microbiota: a narrative review. Nutrients. 2020;12:381. [DOI] [PMC free article] [PubMed]
- 26.Lynch JB, Gonzalez EL, Choy K, et al. Gut microbiota Turicibacter strains differentially modify bile acids and host lipids. Nat Commun. 2023;14:3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng C, Lu F, Chen B, et al. Gut microbiome as a biomarker for predicting early recurrence of HBV-related hepatocellular carcinoma. Cancer Sci. 2023;114:4717–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avilés-Jiménez F, Guitron A, Segura-López FK, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infection: Official Publication Eur Soc Clin Microbiol Infect Dis. 2016;22(2):e17811–22. [DOI] [PubMed] [Google Scholar]
- 29.Mei Q, Huang C, Luo S, et al. Characterization of the duodenal bacterial microbiota in patients with pancreatic head cancer vs. healthy controls. Pancreatology: Official J Int Assoc Pancreatology (IAP). 2018;18(4):438–45. [DOI] [PubMed] [Google Scholar]
- 30.Li XQ, Cheng XJ, Wu J, et al. Targeted inhibition of the PI3K/AKT/mTOR pathway by (+)-anthrabenzoxocinone induces cell cycle arrest, apoptosis, and autophagy in non-small cell lung cancer. Cell Mol Biol Lett. 2024;29:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitali F, Tortora K, Di Paola M, et al. Intestinal microbiota profiles in a genetic model of colon tumorigenesis correlates with colon cancer biomarkers. Sci Rep. 2022;12:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nie F, Wang L, Huang Y, et al. Characteristics of Microbial distribution in different oral niches of oral squamous cell carcinoma. Front Cell Infect Microbiol. 2022;12:905653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Chu M, Huang Z, et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017;7:11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin J, Ren W, Huang X, et al. Potential mechanisms connecting Purine Metabolism and Cancer Therapy. Front Immunol. 2018;9:1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolzenberg-Solomon R, Derkach A, Moore S, et al. Associations between metabolites and pancreatic cancer risk in a large prospective epidemiological study. Gut. 2020;69:2008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Q, Qin Y, Ji S, et al. MTAP Deficiency-Induced metabolic reprogramming creates a vulnerability to Cotargeting De Novo Purine Synthesis and Glycolysis in Pancreatic Cancer. Cancer Res. 2021;81:4964–80. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Liu Y, Liu J, et al. cAMP-PKA/EPAC signaling and cancer: the interplay in tumor microenvironment. J Hematol Oncol. 2024;17:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Zhang W, Liu R, et al. NNK from tobacco smoking enhances pancreatic cancer cell stemness and chemoresistance by creating a β2AR-Akt feedback loop that activates autophagy. Mol Oncol. 2022;16:2881–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang HK, Lin PC, Huang TT, et al. Nicotine activates HIF-1α and regulates acid extruders through the nicotinic acetylcholine receptor to promote the Warburg effect in non-small cell lung cancer cells. Eur J Pharmacol. 2023;950:175778. [DOI] [PubMed] [Google Scholar]
- 40.Shukla SK, Purohit V, Mehla K, et al. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell. 2017;32:71–e877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: still emerging. Cell Metab. 2022;34:355–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huie MJ, Casazza GA, Horning MA, et al. Smoking increases conversion of lactate to glucose during submaximal exercise. J Appl Physiol (1985). 1996;80:1554–9. [DOI] [PubMed] [Google Scholar]
- 43.da Silva Correia J, Miranda Y, Austin-Brown N, et al. Nod1-dependent control of tumor growth. Proc Natl Acad Sci U S A. 2006;103:1840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Geng W, Sun H, et al. Integrative metabolomic characterisation identifies altered portal vein serum metabolome contributing to human hepatocellular carcinoma. Gut. 2022;71:1203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogata R, Mori S, Kishi S, et al. Linoleic acid Upregulates Microrna-494 to induce quiescence in Colorectal Cancer. Int J Mol Sci. 2021;23:225. [DOI] [PMC free article] [PubMed]
- 46.Nava Lauson CB, Tiberti S, Corsetto PA, et al. Linoleic acid potentiates CD8(+) T cell metabolic fitness and antitumor immunity. Cell Metab. 2023;35:633–e6509. [DOI] [PubMed] [Google Scholar]
- 47.Gouirand V, Gicquel T, Lien EC, et al. Ketogenic HMG-CoA lyase and its product β-hydroxybutyrate promote pancreatic cancer progression. Embo j. 2022;41:e110466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai Y, Gao Y, Lin J, et al. Dietary elaidic acid boosts tumoral antigen presentation and cancer immunity via ACSL5. Cell Metab. 2024;36:822–e8388. [DOI] [PubMed] [Google Scholar]
- 49.Fan X, Xiao X, Mao X, et al. Tea bioactive components prevent carcinogenesis via anti-pathogen, anti-inflammation, and cell survival pathways. IUBMB Life. 2021;73:328–40. [DOI] [PubMed] [Google Scholar]
- 50.Yan J, Yang J, Shen H, et al. Sinomenine regulates circTRPM7-related pathway to inhibit gastric cancer cell growth and metastasis. Chem Biol Drug Des. 2023;102:870–81. [DOI] [PubMed] [Google Scholar]
- 51.Zahedipour F, Dalirfardouei R, Karimi G, et al. Molecular mechanisms of anticancer effects of glucosamine. Biomed Pharmacother. 2017;95:1051–8. [DOI] [PubMed] [Google Scholar]
- 52.Tan ML, Tan HK, Oon CE, et al. Identification of genes involved in the regulation of 14-deoxy-11,12-didehydroandrographolide-induced toxicity in T-47D mammary cells. Food Chem Toxicol. 2012;50:431–44. [DOI] [PubMed] [Google Scholar]
- 53.Lee SB, Lee S, Park JY, et al. Induction of p53-Dependent apoptosis by prostaglandin A(2). Biomolecules. 2020;10:492. [DOI] [PMC free article] [PubMed]
- 54.Huang J, Wang X, Li B, et al. L-5-hydroxytryptophan promotes antitumor immunity by inhibiting PD-L1 inducible expression. J Immunother Cancer. 2022;10:e003957. [DOI] [PMC free article] [PubMed]
- 55.Loftfield E, Stepien M, Viallon V, et al. Novel biomarkers of Habitual Alcohol Intake and associations with Risk of Pancreatic and Liver cancers and Liver Disease Mortality. J Natl Cancer Inst. 2021;113:1542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayerle J, Kalthoff H, Reszka R, et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut. 2018;67:128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data from this study have been deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database and is publicly accessible under the accession number: PRJNA1188451.