Abstract
Background
Ribosome engineering is a semi-empirical technique used to select antibiotic-resistant mutants that exhibit altered secondary metabolism. This method has been demonstrated to effectively select mutants with enhanced synthesis of natural products in many bacterial species, including actinomycetes. Myxobacteria are recognized as fascinating producers of natural active products. However, it remains uncertain whether this technique is similarly effective in myxobacteria, especially for the heterologous production of epothilones in Myxococcus xanthus.
Results
Antibiotics that target the ribosome and RNA polymerase (RNAP) were evaluated for ribosome engineering of the epothilone-producing strain M. xanthus ZE9. The production of epothilone was dramatically altered in different resistant mutants. We screened the mutants resistant to neomycin and rifampicin and found that the yield of epothilones in the resistant mutant ZE9N-R22 was improved by sixfold compared to that of ZE9. Our findings indicate that the improved growth of the mutants, the upregulation of epothilone biosynthetic genes, and specific mutations identified through genome re-sequencing may collectively contribute to the yield improvement. Ultimately, the total titer of epothilones achieved in a 10 L bioreactor reached 93.4 mg/L.
Conclusions
Ribosome engineering is an efficient approach to obtain M. xanthus strains with enhanced production of epothilones through various interference mechanisms. Here, we discuss the potential mechanisms of the semi-empirical method.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-024-02627-3.
Keywords: Ribosome engineering, Myxococcus xanthus, Secondary metabolism, Production of epothilones, Metabolic interference
Background
Ribosome engineering (RE) is a semi-empirical method employed to select mutants resistant to ribosome- or RNAP-associated antibiotics, and the mutations in ribosomal proteins or RNAP may stimulate the synthesis of secondary metabolites [1–3]. Hosaka et al. reported that 43% of the tested non-producing Streptomyces strains commenced antibacterial production upon acquisition of spontaneous mutations that conferred resistance to rifamycin (Rif) or streptomycin (Str) [1]. Mutations in the rpsL gene encoding the ribosomal protein S12 [4, 5] or the rsmG gene encoding 16S rRNA methyltransferase [6], were identified in the mutant strains resistant to Str, whereas mutants obtained by selection with Rif typically harbored mutations in the RNAP subunit (specifically in the rpoB gene) [7]. These RE mutations result in the structural and functional alterations in the ribosome or RNAP, thereby enhancing the production of natural active products (NAPs). Over the past two decades, many other ribosome-associated antibiotics, such as paromomycin (Par), gentamicin (Gen), and neomycin (Neo), have been successfully applied to select mutants with elevated production of NAPs, predominantly in Streptomyces genus [2, 3, 8, 9]. The combination of various drug-resistant mutations can further augment the bacterial productivity. For example, the introduction of octuple drug-resistant mutations in Streptomyces coelicolor resulted in a 180-fold increase in actinorhodin production [10]. Although RE has been extensively applied in Actinomyces to improve NAP production or to activate novel secondary metabolites, it has not, to our knowledge, been described in myxobacteria.
Myxobacteria, initially recognized for their fascinating social behaviors, are characterized by their abundant NAPs and are considered important drug-producing microorganisms, following actinomycetes and fungi [11, 12]. Epothilones are famous antitumor compounds isolated from some Sorangium cellulosum strains, from which two kinds of anticancer drugs have been developed and approved for the treatment of advanced breast cancer [13, 14]. Owing to the challenges associated with the low yield and the difficulties in the genetic manipulation of S. cellulosum, the biosynthetic gene cluster (BGC) of epothilone has been expressed in various heterologous hosts; however, the yields obtained remain inferior to those in native producers [15]. Myxococcus xanthus, as a fermentation-friendly and genetically amenable host that is phylogenetically related to S. cellulosum, presented the highest initial yields of epothilones and exhibits considerable potential for application [15].
In previous research, we successfully expressed the epothilone BGC in M. xanthus DZ2 and obtained a high-yielding strain designated M. xanthus ZE9, in which the epothilone BGC was integrated into the DZ2 genome [16]. The production of epothilone in ZE9 was optimized at the transcriptional level through various strategies, including the modification of promoters [17], transcriptional regulators [18], and the application of CRISPR-dCas9-mediated transcriptional activation [19, 20]. In this study, we performed the RE in M. xanthus strain ZE9, and investigated its capacity to select mutants with stimulated production of secondary metabolites in M. xanthus.
Materials and methods
Strains and culture conditions
The strains used in this study are listed in Table S1. M. xanthus ZE9 and mutant strains were cultivated at 30 °C in CYE medium [5 g/L yeast extract, 10 g/L casitone, 10 mM 3-(N-morpholino) propanesulfonic acid (MOPS), and 4 mM MgSO4, pH 7.4. For solid medium, 15 g/L agar was added]. CMO medium [CYE medium added with 7 mL/L of methyl oleate and 2% XAD-16 resin (Merck, Germany)] was used for fermentation. The medium was supplemented with the following antibiotics if required: apramycin [Apra, 30 μg/mL]; rifampicin [Rif, 2 μg/mL]; neomycin [Neo, 150 μg/mL]; paromomycin [Par, 200 μg/mL]; tetracycline [Tet, 10 μg/mL].
Determination of MICs for antibiotics against M. xanthus ZE9
The minimum inhibitor concentrations (MICs) for different antibiotics were determined using microdilution in 96-well microplates. The antibiotics with a concentration of 256, 128, 64, 32, 16, 8, 4, 2, and 1 µg/mL were added to the microplate containing 150 µL of test microorganisms, which were diluted from the bacterial suspension cultured overnight (OD600 is about 1) by 200 times. The microplates were incubated at 30 °C for 36 h, and the minimum antibiotic concentrations at which no visible growth was observed were considered as the MIC values.
Ribosome engineering of M. xanthus ZE9
ZE9 was cultured overnight in 50 mL of CYE liquid medium. Then, 1 OD of bacteria was mixed with soft agar and spread on CYE plates containing antibiotics (Rif, 2 µg/mL; Neo, 150 µg/mL; Par, 200 µg/mL; Tet, 10 µg/mL) and cultured at 30 °C. The drug-resistant colonies appeared after 6–7 days were transferred to new plates containing the same antibiotics and regrown to reconfirm resistance.
For combined selection with multiple antibiotics, the ZE9-N18 was incubated using the same method, and the mutant strains resistant to two antibiotics were selected on plates containing Neo plus Rif or plates containing Neo plus Par. The ZE9N-R14, ZE9N-R22, ZE9N-R25, ZE9N-R27, and ZE9N-R28 strains were cultured similarly and mixed together before being screened on plates containing Neo, Rif, and Tet.
Genetic characteristics of mutant strains
Chromosomal DNAs of the M. xanthus mutants were extracted and purified with Bacteria Genomic DNA kit (TIANGEN, China). The potentially mutated genes, rpoB and rpsL, were amplified from Rif-resistant and Par-resistant mutants, respectively, and then sequenced at Tsingke Biotech Co., Ltd. (Beijing, China). Primers used are listed in Table S2.
The ZE9 genome was sequenced with PacBio Sequel II platform at Frasergen Genetic Information Co. Ltd. (Wuhan, China). The genome was assembled using Microbial Assembly (smrtlink8) and annotated with Glimmer (v3.02) [21], tRNAscan-SE (v2.0) [22], and RNAmmer (v1.2) [23]. Genome re-sequencing was conducted with Illumina NovaSeq X Plus platform at Novogene Technology Co. Ltd. (Beijing, China). Sequence data was mapped with the ZE9 reference genome using BWA (parameters: mem -t 4 -k 32 -M) [24]. The SNPs and InDels were detected and annotated with GATK (parameters: HaplotypeCaller-pair-hmm-gap-continuation-penalty 10-genotyping-mode DISCOVERY-stand-call-conf 30) [25] and ANNOVAR [26]. The sequencing data was uploaded into the NCBI database as BioProject PRJNA1184456.
Detection of the epothilone production in flasks
The ZE9 and resistant mutants were cultured in 50 mL of CYE medium overnight and then inoculated into 50 mL of CMO medium at a ratio of 1:50. After 7 days of incubation, the resin was collected into the 15 mL centrifuge tubes in which 8 mL of methanol was added to extract the epothilones overnight. The leaching liquor was filtered with 0.22 μm filter and analyzed by high-performance liquid chromatography (HPLC) (SHIMADZU, Japan) as we previously described [17]. Briefly, 20 µL of filtered sample was used for the chromatographic separation with a C18 column (4.6 mm × 250 mm, 4.60 μm, Thermo Scientific) at a flow rate of 1.0 mL/min, with mobile phase of 40% of H2O and 60% of methanol. According to the standard curve of epothilones, the yields of epothilones were quantified according to the formula Y = (A × 1.1 × 10^−7–3 × 10^−5) × 160 (Y represents the yield of epothilone, and A presents the peak area of epothilone in the UV chromatogram).
LC–MS analysis of secondary metabolites
The LC–MS analysis was performed on a UHPLC system (Thermo Scientific Vanquish, USA) coupled with a Q Exactive HFX Hybrid Quadrupole-Orbitrap mass spectrometer (Q Exactive HFX, Thermo Scientific, USA) equipped with an ESI source. The chromatographic separation was done using a C18 column (4.6 mm × 250 mm, 4.60 μm, Thermo Scientific, USA). The elution solvent system was water and methanol, with a linear gradient from 5 to 100% methanol in water in 0−30 min, at a constant flow rate of 0.8 mL/min. ESI source parameters were used as follows: heater temperature 425 °C, sheath gas flow rate 50 arb, Aux gas flow rate 13 arb, Spray voltage ± 3500 V, and normalized collision energy 20, 40, 60 eV.
Transcriptional analysis of epothilone genes
The transcription of epothilone genes were determined by real-time quantitative PCR (RT-qPCR) as we previously reported [17]. Briefly, cells were collected at 48 h during fermentation, followed with RNA extraction with BIOZOL kits (Total RNA Extraction Reagent, BioFlux, China). The total RNA was reverse-transcribed into cDNA with the PrimeScript™ Regent Kit with DNase (Takara, Japan) for the RT-qPCR analysis on LightCycler® 480 (Roche, Switzerland) with ChamQ Universal SYBR qPCR Master Mix (Vazyme, China). The gapA gene (glyceraldehyde-3-phosphate dehydrogenase gene, MXAN_2815) was chosen as the reference gene for normalization. All the primers used are listed in Table S2.
Detection of intracellular protein concentration
The strains were cultured in 50 mL CYE medium overnight and then inoculated into 50 mL CMO medium with initial 0.04 OD. 2 mL of suspension was harvested at 24 h and 48 h of incubation, washed with cold PBS buffer (KH2PO4 0.27 g/L, Na2HPO4 1.42 g/L, NaCl 8 g/L, KCl 0.2 g/L, pH 7.4) three times and then disrupted with ultrasonic processor (5 s of sonication with 10-s interval was performed 4 times). After centrifugation of 12,000 rpm, 25 min at 4 °C, 2 µL of the protein supernatant was used for automated detection with NanoDrop2000C (Thermo Scientific, USA) in the protein-A280 mode.
Fermentation of ZE9N-R22 in bioreactors
The ZE9N-R22 was incubated in CYE medium containing Neo (64 µg/ml) and Rif (2 µg/ml) overnight at 30 °C, and then inoculated into 3 L fermenter (BioFlo 310, Eppendorf NBS, Germany) with 2 L of CMO medium. The fermentation was performed at 30 °C for 7 days with dissolved oxygen (DO) controlled at 50%, while pH not controlled. For fed-batch fermentation in 10 L bioreactor (BioFlo 310, Eppendorf NBS, Germany) containing 5 L of CMO medium, nitrogen nutrition (casitone 150 g/L and yeast extract 75 g/L) and methyl oleate were added at feed rated of 140 ml /L and 6 ml/L, respectively. The feeds were started after 4 days of incubation, and delivered 5 times as a single shot every 48 h. Cultivation pH was maintained at 7.4 by automated addition of 2.5 M H2SO4, and the DO was controlled at 50%. The cells were allowed to grow for 18 days before being harvest together with resin, followed with extraction with 800 mL of methanol. The epothilone products were detected as mentioned above.
Results
Screening of antibiotics applicable for the RE of M. xanthus ZE9
Seven antibiotics commonly used in RE were evaluated for their MICs against M. xanthus ZE9 (Table 1). Mutations in the rpsL gene [5] or the rsmG gene [6] have been documented in streptomycin-resistant mutants, with analogous mutations in rpsL reported in mutants selected with Par [27]. In Rif-resistant mutants, mutations are localized within the rpoB gene [7]. Mutations in the ribosome influence the protein translation process, while mutations in rpoB may result in transcriptional-level regulation. The mutational mechanisms associated with Tet, Neo, Gen, and spectinomycin (Spe) remain to be elucidated.
Table 1.
Antibiotics used for ribosome engineering of M. xanthus ZE9
| Antibiotics | Targets | Mutant gene | MIC (µg/mL) | Screening concentration (µg/mL) | Mutant rate |
|---|---|---|---|---|---|
| Streptomycin | 30S | rpsL or rsmG | > 256 | – | – |
| Gentamicin | 30S | rplF | > 256 | – | – |
| Spectinomycin | 30S | – | > 256 | – | – |
| Paromomycin | 30S | rpsL | 64 | 200 | 6 × 10–7 |
| Rifampicin | β subunit of RNA polymerase | rpoB | 1 | 2 | 3 × 10–8 |
| Neomycin | 30S | – | 64 | 150 | 1 × 10–8 |
| Tetracycline | 30S | – | 8 | 10 | 1 × 10–7 |
As shown in Table 1, ZE9 was tolerant to Str, Gen, and Spe, with MIC values exceeding 256 µg/mL. The remaining four antibiotics, of which the Rif exhibited the strongest inhibitory activity, were used for subsequent experiments.
Disruption of epothilone production in single antibiotic-resistant mutants
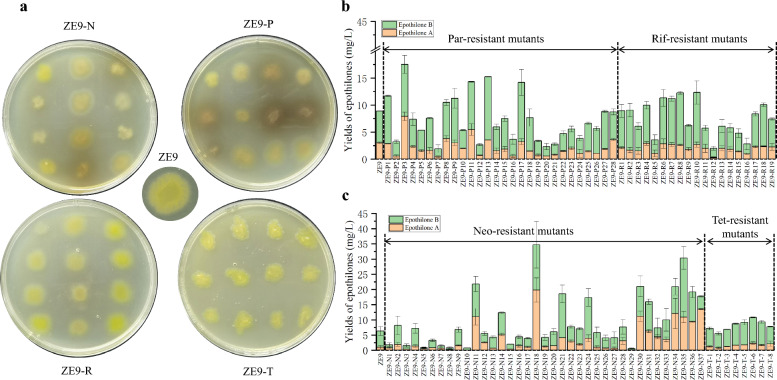
Resistant mutants were selected against Par, Rif, Neo, or Tet at concentrations higher than their MICs. Variations in mutant rate were observed during selection with individual antibiotic (Table 1). Ultimately, we obtained 19 Par-resistant mutants, 19 Rif-resistant mutants, 37 Neo-resistant mutants, and 8 Tet-resistant mutants. Notable phenotypic alterations were observed in certain mutant strains, including a change in colony color from bright yellow to buff, gray, or orange (Fig. 1a), which may be associated with disruptions in the synthesis of endogenous pigments.
Fig. 1.
Colony growth and epothilone yields of mutant strains selected by single antibiotic. a Growth of mutant colonies. ZE9-N, mutants resistant to neomycin; ZE9-P, mutants resistant to paromomycin; ZE9-R, mutants resistant to rifampicin; ZE9-T, mutants resistant to tetracycline. b Detection of epothilone yields in ZE9-P and ZE9-R mutants. The error bars represent the standard deviation of three independent experiments. c Detection of epothilone yields in ZE9-N and ZE9-T mutants. The error bars represent the standard deviation of three independent experiments
All resistant mutant strains were fermented and subsequently evaluated for epothilone production. Among the Par-resistant mutants, 60% exhibited increased epothilone yields, while 25% demonstrated decreased yields, with a maximum variation of 70% (Fig. 1b). Similarly, approximately 50% of the Rif-resistant mutants produced less epothilones compared to ZE9, with the lowest yield of ZE9-R12 being only 23% of that of ZE9 (Fig. 1b).
Improved production of epothilones was observed in 22% of the Rif-resistant mutants, with a maximum increase of 40%. Therefore, a significant proportion of the mutations selected by Rif and Par resulted in detrimental effects on epothilone biosynthesis. In relation to the Neo-resistant mutants, 27% of the mutants exhibited reduced epothilone production, while a greater proportion (approximately 40%) presented increased titers (Fig. 1c). Among these mutants, ZE9-N29 showed the lowest epothilone yield, which was 91% lower than that in ZE9. Conversely, the yield of ZE9-N18 was increased by 400%, reaching 34.7 mg/L. Although only 8 Tet-resistant mutants were identified, 4 of these mutants were found to be more productive strains (Fig. 1c), suggesting that mutations selected by Tet may have a more positive effect on epothilone synthesis, thereby generating more high-yielding mutants. This study demonstrated, for the first time, the differences observed in mutants of M. xanthus ZE9 selected with different antibiotics, and their influences on epothilone production. These findings undoubtedly serve as a reference for the future application of RE technique in myxobacteria.
Selection of multiple antibiotics for mutants exhibiting improved epothilone production
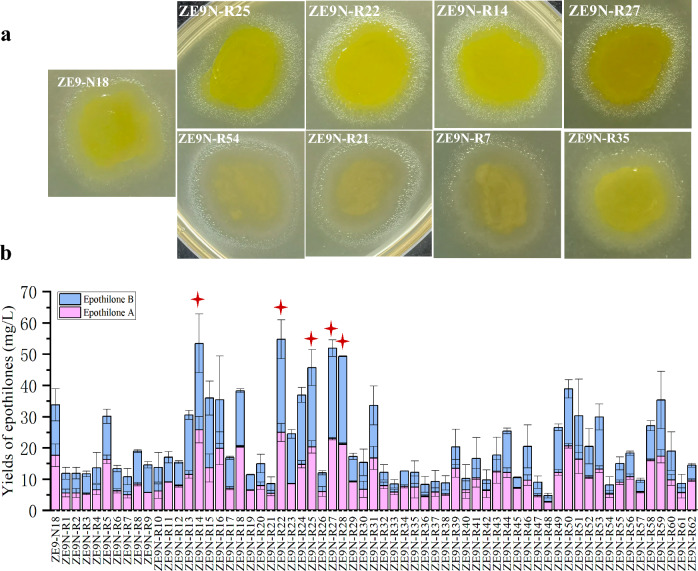
With RE, disparate mutations may be selected by different antibiotics. Consequently, the desired mutations that stimulate the synthesis of target products can be integrated by successive selection through multiple antibiotics. To further screen high-yielding mutants, the Neo-resistant mutant ZE9-N18, which exhibited the highest epothilone yield, was chosen for sequential selection with Rif. Although selection with Par gave 60% of mutants with increased production, Par was un-adopted in combined selection due to the resistance of the ZE9-N18 strain to Par (MIC > 256 µg/mL). 62 double-resistant strains designated ZE9N-Rs were obtained; these strains displayed similar growth characteristics with minor color variations (Fig. 2a) but different epothilone productivities (Fig. 2b). In comparison to ZE9-N18, most ZE9N-R strains (73%) presented reduced epothilone yields, which was consistent with the observed effect of Rif on ZE9. Among the five mutants with enhanced epothilone production (marked with red stars in Fig. 2b), the highest yield was improved by 62%, rising from 33.7 mg/L in ZE9-N18 to 54.8 mg/L in ZE9N-R22.
Fig. 2.
Mutant strains induced by neomycin plus rifampicin. a Growth of mutant colonies. b Detection of epothilones in mutants. The error bars represent the standard deviation of three independent experiments. The high-yielding mutants selected for further mutagenesis were marked with red asterisk
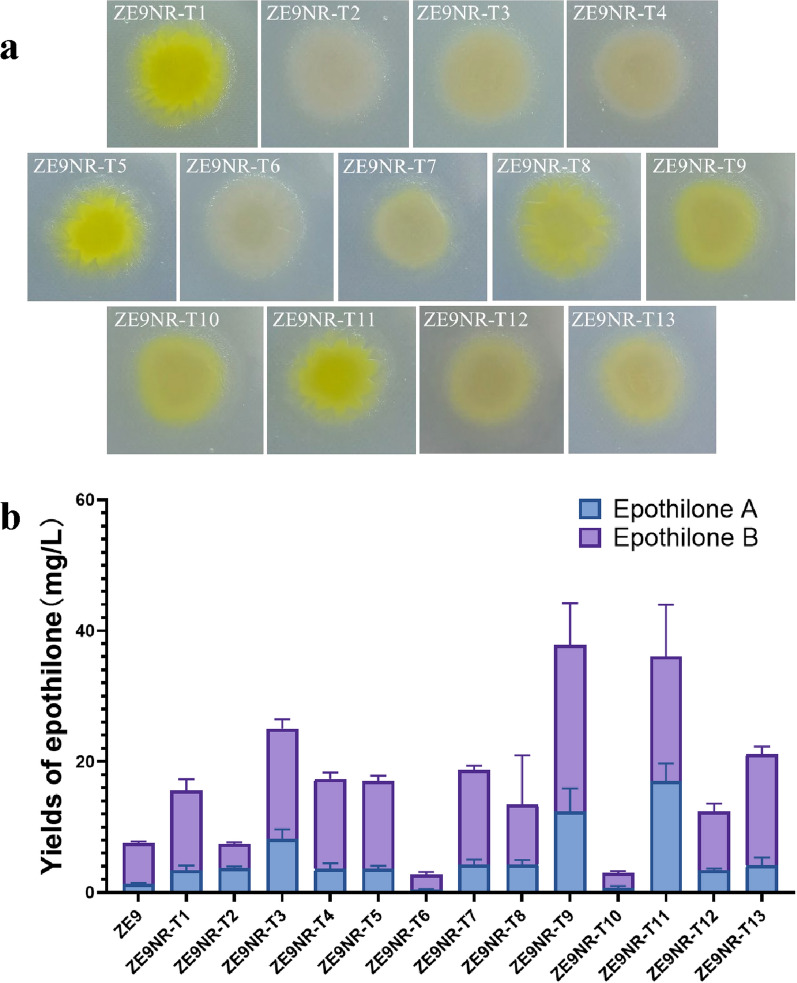
The five high-yielding dual-resistant strains, including ZE9N-R14, ZE9N-R22, ZE9N-R25, ZE9N-R27, and ZE9N-R28, were found to produce epothilones without any significant difference (t-test, p > 0.05). The optimal strain for subsequent screening aimed at achieving mutants with higher yield of epothilones remains indeterminate. These five strains were consequently mixed together for the selection of tri-resistant mutants with Tet. Thirteen mutants, named as ZE9NR-T strains, were isolated and further analyzed. As shown in Fig. 3, the mutants presented different colors, with the bright yellow stains demonstrating higher yields of epothilones compared to the off-white ones. However, the acquisition of the third resistance to Tet did not further promote the synthesis of epothilones among the tested mutants.
Fig. 3.
Colony growth and epothilone yields of mutant strains selected by neomycin, rifampicin, plus tetracycline. a Growth of mutant colonies. b Detection of epothilones in mutants. The error bars represent the standard deviation of three independent experiments
Genotype identification of high-yielding mutants
Mutations in ribosomes or RNAP selected by RE may affect the global transcription and translation activities, consequently influencing the synthesis of natural products in mutant strains. This is considered the primary mechanism of RE [28]. Therefore, we investigated how the mutations surviving antibiotic selection influence epothilone production in high-yield strains, in terms of genotype, growth, transcription, and product synthesis.
Among the four antibiotics used in this study, Rif and Par have been reported to select mutations in rpoB and rpsL, respectively. The target genes associated with Rif-resistant and Par-resistant mutants were amplified and sequenced. A total of three distinct single-base mutations in the rpoB gene were identified in 17 out of 19 Rif-resistant mutants (Table S1), including H548Y (11 strains), D538V (2 strains) and V152F (4 strains). Notably, the production of epothilones was unaffected in the mutants harboring the D538V mutation (ZE9-R1 and ZE9-R2), whereas it was significantly impacted in the mutants containing the V152F or H548Y mutation (Fig. 1b). Among the strains carrying the H548Y mutation, variations in epothilone yields were observed, suggesting that the Rif-associated mutation in rpoB may not the primary determinant of epothilone yield variability. As to the Par-resistant mutants, decreased epothilone yields were observed in all 13 strains containing various mutations in the rpsL gene, with the exception of ZE9-P27 and ZE9-P28 (Fig. 1b, Table S1). This further demonstrates that the identified mutations do not contribute to enhance the epothilone production.
High-yielding mutants, including ZE9N-18 and the dual-resistant mutants ZE9N-R14, ZE9N-R22, ZE9N-R25, and ZE9N-R27, were analyzed by genome resequencing, which revealed only a few mutations (Table S3). In ZE9N-18, a single nucleotide polymorphism (SNP) mutation was found in orf05968, which encodes the CarD-like family RNAP-interacting protein CdnL [29]. This SNP resulted in the loss of the termination codon, leading to extension of the C-terminus with 32 amino acids. Notably, this alteration did not disrupt the primary function of CdnL, as it is indispensable essential in M. xanthus. The other SNP was found upstream of the coding region of the 50S ribosomal protein L10 (rpsJ), which might contribute to the acquisition of resistance to Neo. In addition, an insertion-deletion (InDel) mutation was detected in a hypothetical protein-encoding gene (Table S3). For the dual-resistant mutants, the typical mutations in rpoB of the four tested mutants were two novel substitutions (Q535L and H548D) not present in the ZE9-R mutant strains. In conclusion, mutant strains acquired few mutations during the RE progression, and the combined effect of mutations resulted in changes in strain resistance, phenotype, and activated production of epothilones.
Characteristic identification of high-yielding mutants
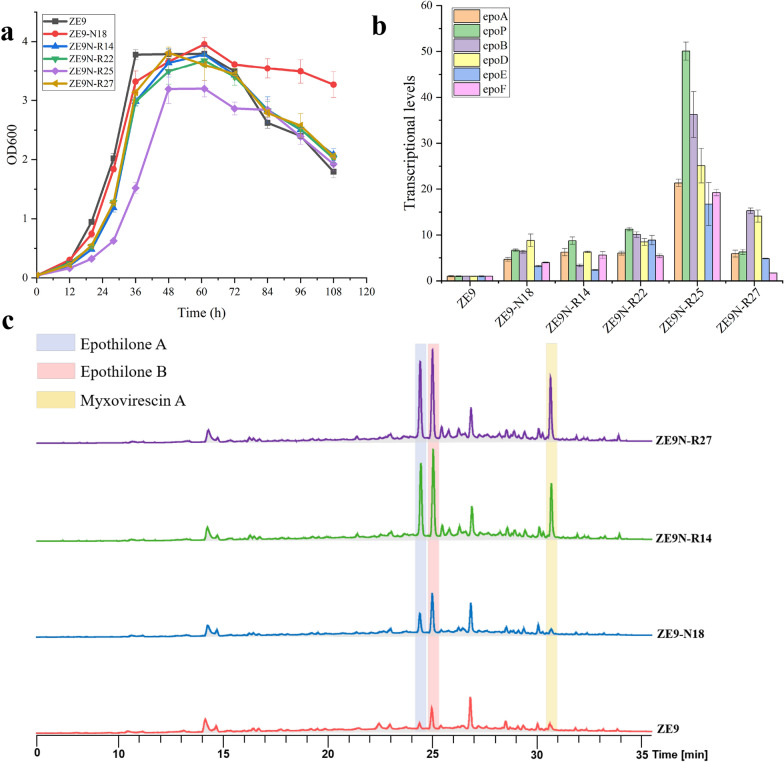
As shown in Fig. 4a, ZE9-N18 exhibited slightly delayed exponential growth phase, accompanied by an extended stationary phase, which is theoretically conducive to the biosynthesis of secondary metabolites, such as epothilones. The combined mutations selected with Rif led to further delayed exponential growth in high-yielding ZE9N-R strains. For instance, when cultured to the midpoint of the exponential growth phase (36 h), the biomass of ZE9-N18 was found to be 20% lower than that of ZE9. In comparison, the biomass reduction in the ZE9N-R strains reached 30% in ZE9N-R14, ZE9N-R22, and ZE9N-R27, and escalating to 65% in ZE9N-R25 and ZE9N-R27. In addition, the maximum biomass observed during the stationary phase in the high-yielding ZE9N-R mutants did not exceed that of ZE9, and the subsequent trends of apoptosis were similar to those in ZE9. This indicates the presence of additional, unidentified factors, beyond enhanced growth, that may play a role in the efficient synthesis of epothilone in these mutants.
Fig. 4.
Characteristic identification of high-yielding mutants. a Growth curves of ZE9 and mutants. b Transcriptional analysis of the epothilone genes in ZE9 and different mutants. c HPLC analysis of the secondary metabolites in ZE9 and different mutants
The transcriptional levels of epothilone biosynthetic genes were analyzed with RT-qPCR. The transcription of epothilone genes in all representative high-yielding strains exhibited a significant increase (Fig. 4b). For example, the transcription levels of epothilone-encoding genes in ZE9-N18 were found to be 2.2-fold (epoE) to 7.8-fold (epoD) higher than those in ZE9. Compared with those in ZE9-N18, most epothilone-encoding genes were further activated in the ZE9N-R strains, especially in ZE9N-R25. While the elevated transcription of biosynthetic genes is a crucial factor contributing to epothilone production, it is not the sole determinant. Although the transcription levels of the epothilone-encoding genes in ZE9N-R25 were 1.8 to 2.5 times greater than those in ZE9N-R22 (Fig. 4b), no significant difference in epothilone yields was detected between these two mutants. This might be attributed to the delayed growth of ZE9N-R25 compared to ZE9N-R22 (Fig. 4a). It has been indicated that mutant ribosomes may exhibit enhanced stability and facilitate the synthesis of intracellular proteins during stationary phases [1, 30]. We determined the intracellular protein content of ZE9 and its mutants; however, no significant differences were observed (Figure S1).
At the metabolite level, the fermentation products of representative strains were analyzed with LC–MS, which revealed that, in addition to epothilones, the synthesis of endogenous secondary metabolites was significantly affected. Four NAPs of M. xanthus were identified, including three hybrid polyketide-nonribosomal peptide (PK−NRP, myxovirescin A [31], DKxanthene [32], and myxalamid [33]), as well as one ribosomally synthesized and posttranslationally modified peptide (RiPP, cittilin [34]). These compunds consumes malonyl-CoA, methylmalonyl-CoA, and amino acids as precursors, akin to the biosynthesis of epothilones [35] (Table S4). Compared with that in ZE9, the production of myxovirescin A in the tested mutants was significantly enhanced (Fig. 4c), while the synthesis of cittilin and DKxanthene was inhibited, and the production of myxalamid remained unaffected (Figure S2). The varying trends in product synthesis observed in this study further demonstrated the complex effects of RE mutagenesis on intracellular secondary metabolism. Although no new products were identified in the high-yield mutants tested, the potential for discovering activated unknown products in other myxobacteria through RE, as demonstrated in Streptomyces [1], merits further investigation.
Scaled-up fermentation of high-yielding mutant ZE9N-R22 in bioreactors
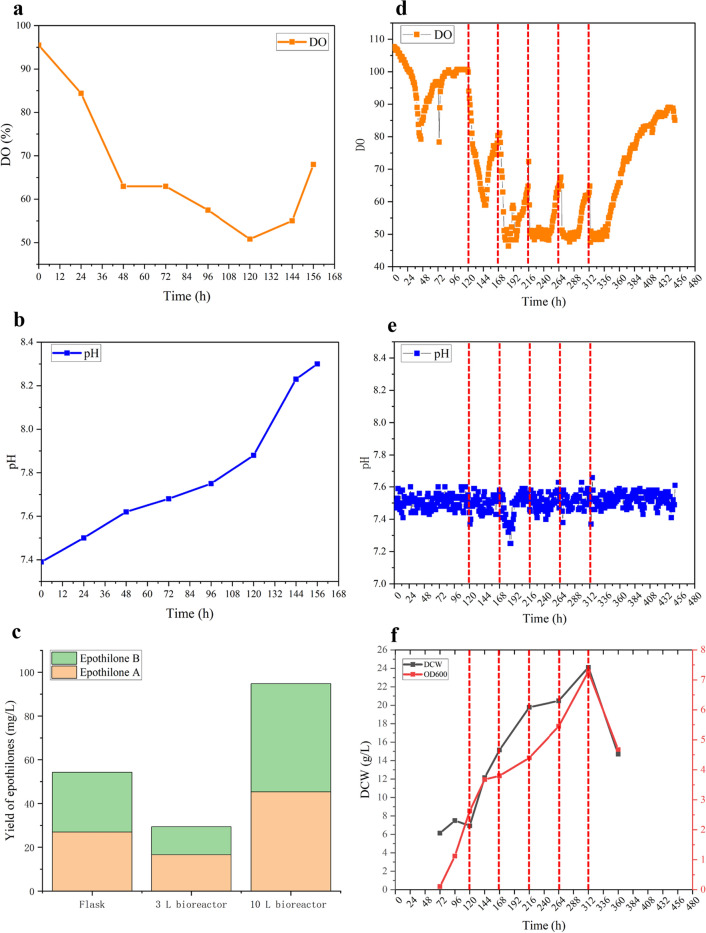
The improved epothilone titer in ZR9N-R22 was demonstrated through fermentation experiment in flasks mentioned above. To further validate its productivity, bioreactor fermentation was performed using both 3 L and 10 L fermenters. A 7 day batch fermentation of ZR9N-R22 was conducted in a 3 L bioreactor. After incubation with 10% of seed solution, the DO showed a gradual decline corresponding with the increase in bacterial growth (Fig. 5a). The pH level increased from 7.4 to 8.3 after six days of incubation in the 3 L bioreactor without any supplementation (Fig. 5b). This rise in pH might be attributed to the release of NH3 during the metabolic processes of abundant amino acids in the CMO medium. The epothilone titer achieved was 29.4 mg/L, which was 46% lower than that in flasks (Fig. 5c).
Fig. 5.
Fermentation of ZE9N-R22 in bioreactors. a DO curve in 3 L fermenter. b pH curve in 3 L fermenter. c Yields of epothilones in flasks and fermenters. d DO curve in 10 L fermenter. e pH curve in 10 L fermenter. f Biomass detection in 10 L fermenter. The red dotted line indicated the feeding time nodes
A fed-batch fermentation lasting 18 days was subsequently conducted in a 10-L bioreactor. As illustrated in Fig. 5d and e, nitrogen nutrition and methyl oleate were added in five batches after four days of fermentation (feed nodes were indicated by red dashed lines). Each feeding effectively promoted cell growth, resulting in an instantaneous decrease in DO abundance, while the pH remained relatively stable. After 5 feedings, the maximum biomass attained was 12.5 g/L (Fig. 5f). The OD600 value and dry cell weight (DCW) exhibited a decline upon the cessation of feeding. Ultimately, after 18 days of cultivation, the titers of epothilone A and epothilone B were 44.8 mg/L and 48.6 mg/L, respectively, culminating in a total yield of 93.4 mg/L (Fig. 5c). This is the highest epothilone heterologous expression yield reported in the literature [15].
Discussion
Since the BGC of epothilone was heterologously expressed in M. xanthus, various genetic manipulation strategies have been applied to optimize the yield of epothilones. Based on the characterization of the promoter within the epothilone BGC [36], the original promoter was replaced or arranged in series, resulting in a 1.2-fold enhancement in epothilone production [17]. Deletion of the transcriptional inhibitor upstream the epothilone BGC led to a 4.1-fold increase in the production of epothilones [18]. Besides, the CRISPR-dCas9 system was introduced into ZE9 to activate either single [20] or multiple promoters [19] within the epothilone BGC, and promoted the epothilone synthesis by about 2 times. Compared with the above genetic manipulations, RE generates mutants exhibiting unpredictable yield of epothilones. This non-directional characteristic indicates that it may be necessary to screen a substantial number of mutants in order to identify high-yielding strains. However, RE is independent of the genetic flexibility of the strain, which makes the method particularly advantageous for application in non-model bacteria.
Although RE has been demonstrated to be effective in activating the synthesis of secondary metabolites, the mechanism of such activation is still unclear. A mutation in rpsL was identified in Str-resistant Streptomyces mutants with activated production of NAPs [1], which has been explained by the maintained translational accuracy [37] or increased stability of ribosomes [3]. Certain rpsL mutations reportedly increase protein synthesis during later stages of growth, which coincides with NAP production in most microorganisms [38]. However, some studies have refuted the causal relationship between simple mutations in ribosomal proteins and increased product yield. For example, the direct introduction of mutations in rpsL or rsmG resulted in resistance to Str, but did not improve the production of tiancimycins [39]. In our study, mutants harboring the same mutation exhibited different epothilone production. This also demonstrates the intricate nature of the activation on the production of NAPs by RE.
Mutations in rpoB during the Rif selection have been shown to alter the affinity of RNAP for specific promoters, effectively recognizing and transcribing silenced BGCs and thereby promoting product synthesis [1]. The mutations confering resistance to Rif have been well reviewed [40]. Most (> 95%) of the mutations are clustered within four distinct sites within the rpoB gene, the so-called Rif cluster N, I, II and III (Figure S3). We compared the Rif cluster in M. xanthus with the sequences summarized in the literature [40, 41], and marked the mutations identified in this study (Red mark in Figure S3). No novel mutation was found in the M. xanthus mutants we obtained. Although certain mutations, such as D427V, S433P, S433L, H437Y, H437R, have been identified as particularly effective in activating the production of actinorhodin and undecylprodigiosin in Streptomyces, their applicability may not extend to other organisms. Apparently, enhanced production of epothilone was not observed in the ZE9-R1/ZE9-R2 strain containing the D538V mutation (corresponding to the D427V in Streptomyces), nor in the ZE9-R3/ZE9-R4 harboring the H548Y (corresponding to the H437Y in Streptomyces).
Among the mutants derived from RE that exhibit enhanced production of NAPs, certain strains demonstrated significant alterations in growth characteristics, such as the delayed sporulation in Streptomyces diastatochromogenes [42] and slender mycelium in Micromonospora yangpuensis [43]. The extended stationary phase in M. xanthus mutants has never been reported in other strains. Since the secondary metabolites are usually produced during the stationary phase of fermentation, we speculate that the extended stationary phase may contribute to the yield improvement of epothilones in the mutants identified in this study. We observed significantly increased transcription of epothilone genes in mutants carrying mutations in rpoB and producing more epothilones (Fig. 4b), but this could not explain why some strains carrying the same mutation presented decreased epothilone yields. Although enhanced protein synthesis has only been reported in mutants selected by Str, which was not used in this study, the total intracellular proteins in ZE9 and the mutants producing more epothilones were cursorily analyzed, and the results revealed no significant differences (Figure S1).
Recently, RE has been used in combination with other random mutagenesis strategies, such as UV/ARTP mutagenesis, to activate secondary metabolism, and the mechanism of activation has been explored using genome resequencing, metabolomic analysis, transcriptomic analysis and other omics methods, revealing that many mutations unrelated to resistance to antibiotics, along with transcriptional changes in genes involved in primary metabolism, might influence the precursors and energy supply, and thus affect the synthesis of NAPs [43–45]. We re-sequenced the genomes of representative mutant strains, and detected some mutations in ribosomal proteins, RNAP, CarD-like RNAP-interacting proteins, etc. The CdnL brought to our attention because it has been demonstrated to bind to β subunit of the RNAP, stabilizing the formation of transcriptionally competent, open complexes by the primary σA-RNAP holoenzyme at an rRNA promoter in vitro [46]. The C-terminal region of CdnL is critical for its function, as mutations in this region lead to severe growth defects without affecting its interaction with endogenous RNAP. In our study, a mutation was identified in the CdnL of ZE9-N18, resulting in elongated C-terminal region. However, the relationship between this change and the acquisition of resistance to Rif, as well as the improvement of epothilone yield, requires further verification. We speculated that the combined effects of these mutations promoted the synthesis of epothilones.
During the biosynthesis of epothilones, epothilone A or epothilone B is produced when the AT domain in the third module of polyketide synthase uses a malonyl-CoA or methylmalonyl-CoA as the extended unit, respectively [35, 47]. S. cellulosum So0157-2 produced more epothilone A than epothilone B, while the B/A ratio ranged from 2 to 10 in M. xanthus host, possibly because more methylmalonyl-CoA was present in M. xanthus substrate pool [16]. However, reduced B/A ratios were observed in most high-yielding RE mutants (Figure S4). ZE9N-R22, derived from ZE9-N18 (B/A ratio of 0.75), produced equal amounts of epothilone A and epothilone B or even less epothilone B in fermenters (Fig. 5c), suggesting potential changes in the precursor supply, which can be rationally modified by subsequent metabolic engineering for the tendentious synthesis of epothilone B.
Conclusions and outlook
The ribosome engineering strategy was successfully employed to enhance the synthesis of NAPs in M. xanthus. A variety of resistant mutants were obtained through selection against single or multiple antibiotics, leading to the selection of high-yielding mutant strains that were tolerant to neomycin and rifampicin. The production of epothilones, along with other secondary metabolites such as myxovirescin A, was dramatically improved, reaching 54.8 mg/L in flasks and 93.4 mg/L in bioreactors. In combination with the transcriptional regulation strategies previously described, further increases in epothilone yield are anticipated. In addition, it is theoretically plausible to augment the synthesis of methylmalonyl-coenzyme A through metabolic engineering to further increase the production of epothilone B. Although no new products were activated in the mutant strains examined in this study, additional strain screening, especially in non-model myxobacteria that lack genetic manipulation methods, is expected to excavate new NAPs.
Supplementary Information
Acknowledgements
We thank Professor Jun-qiang Fang from National Glycoengineering Research Center (Shandong University), and Cheng-jia Zhang and Nan-nan Dong from Core Facilities for Life and Environmental Sciences (State Key Laboratory of Microbial Technology, Shandong University) for assistance in microbial fermentation. We also thank Zhi-feng Li, Jing Zhu, Jing-yao Qu, and Guan-nan Lin from the State Key Laboratory of Microbial Technology (Shandong University) for help in HPLC-MS analysis.
Abbreviations
- RE
Ribosome engineering
- Rif
Rifamycin
- Str
Streptomycin
- Par
Paromomycin
- Gen
Gentamicin
- Neo
Neomycin
- Tet
Tetracycline
- Spe
Spectinomycin
- NAPs
Natural active products
- DO
Dissolved oxygen
- DCW
Dry cell weight
Author contributions
XJY, XK, and YZL designed the researches; XK, XRY and CXW performed researches; XJY, XK, and XRY analyzed the data; XJY wrote the main manuscript text; JRW, JNZ, ZPY, QKF, WH, CSW and YZL revised the manuscript. All authors reviewed the manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (2021YFC2101000), and the National Natural Science Foundation of China (32301220).
Data availability
Sequence data that support the findings of this study have been deposited in the European Nucleotide Archive with the primary accession code PRJNA1184456.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol. 2009;27:462–4. [DOI] [PubMed] [Google Scholar]
- 2.Ochi K. Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects. J Antibiot. 2017;70:25–40. [DOI] [PubMed] [Google Scholar]
- 3.Zhu SB, Duan YW, Huang Y. The application of ribosome engineering to natural product discovery and yield improvement in Streptomyces. Antibiot Basel. 2019;8(3):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol. 1996;178:7276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koshla O, Lopatniuk M, Borys O, Misaki Y, Kravets V, Ostash I, Shemediuk A, Ochi K, Luzhetskyy A, Fedorenko V, Ostash B. Genetically engineered merodiploidy impacts secondary metabolism and antibiotic resistance in Streptomyces. World J Microb Biot. 2021;37:1–9. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura K, Hosaka T, Tokuyama S, Okamoto S, Ochi K. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J Bacteriol. 2007;189:3876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H, Kim SH, Ying YH, Kim HJ, Koh YH, Kim CJ, Lee SH, Cha CY, Kook YH, Kim BJ. Mechanism of natural rifampin resistance of Streptomyces spp. Syst Appl Microbiol. 2005;28:398–404. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka Y, Kasahara K, Hirose Y, Murakami K, Kugimiya R, Ochi K. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J Bacteriol. 2013;195:2959–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopatniuk M, Myronovskyi M, Nottebrock A, Busche T, Kalinowski J, Ostash B, Fedorenko V, Luzhetskyy A. Effect of “ribosome engineering” on the transcription level and production of S. albus indigenous secondary metabolites. Appl Microbiol Biot. 2019;103:7097–110. [DOI] [PubMed] [Google Scholar]
- 10.Wang GJ, Hosaka T, Ochi K. Dramatic activation of antibiotic production in Actinomadura sp. by cumulative drug resistance mutations. Appl Environ Microb. 2008;74:2834–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CY, Hu JQ, Wang DG, Li YZ, Wu CS. Recent advances in discovery and biosynthesis of natural products from myxobacteria: an overview from 2017 to 2023. Nat Prod Rep. 2024. 10.1039/D3NP00062A. [DOI] [PubMed] [Google Scholar]
- 12.Saggu SK, Nath A, Kumar S. Myxobacteria: biology and bioactive secondary metabolites. Res Microbiol. 2023. 10.1016/j.resmic.2023.104079. [DOI] [PubMed] [Google Scholar]
- 13.Peterson JK, Tucker C, Favours E, Cheshire PJ, Creech J, Billups CA, Smykla R, Lee FYF, Houghton PJ. In vivo evaluation of ixabepilone (BMS247550), a novel epothilone B derivative, against pediatric cancer models. Clin Cancer Res. 2005;11:6950–8. [DOI] [PubMed] [Google Scholar]
- 14.Xu B, Sun T, Zhang Q, Zhang P, Yuan Z, Jiang Z, Wang X, Cui S, Teng Y, Hu XC, et al. Efficacy of utidelone plus capecitabine versus capecitabine for heavily pretreated, anthracycline- and taxane-refractory metastatic breast cancer: final analysis of overall survival in a phase III randomised controlled trial. Ann Oncol. 2021;32:218–28. [DOI] [PubMed] [Google Scholar]
- 15.Yue XJ, Sheng DH, Zhuo L, Li YZ. Genetic manipulation and tools in myxobacteria for the exploitation of secondary metabolism. Engin Microbiol. 2023. 10.1016/j.engmic.2023.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu LP, Yue XJ, Han K, Li ZF, Zheng LS, Yi XN, Wang HL, Zhang YM, Li YZ. Allopatric integrations selectively change host transcriptomes, leading to varied expression efficiencies of exotic genes in Myxococcus xanthus. Microb Cell Fact. 2015;14:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue XJ, Cui XW, Zhang Z, Hu WF, Li ZF, Zhang YM, Li YZ. Effects of transcriptional mode on promoter substitution and tandem engineering for the production of epothilones in Myxococcus xanthus. Appl Microbiol Biotechnol. 2018;102:5599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue XJ, Cui XW, Zhang Z, Peng R, Zhang P, Li ZF, Li YZ. A bacterial negative transcription regulator binding on an inverted repeat in the promoter for epothilone biosynthesis. Microb Cell Fact. 2017;16:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Yue XJ, Yuan SF, Hong Y, Hu WF, Li YZ. Internal promoters and their effects on the transcription of operon genes for epothilone production in Myxococcus xanthus. Front Bioeng Biotech. 2021;9: 758561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng R, Wang Y, Feng WW, Yue XJ, Chen JH, Hu XZ, Li ZF, Sheng DH, Zhang YM, Li YZ. CRISPR/dCas9-mediated transcriptional improvement of the biosynthetic gene cluster for the epothilone production in Myxococcus xanthus. Microb Cell Fact. 2018;17:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics. 2007;23:673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acid Res. 1997;25:955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagesen K, Hallin P, Rodland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acid Res. 2007;35:3100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011. 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li MY, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl Acid Res. 2010. 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto-Hosoya Y, Sato TA, Ochi K. Resistance to paromomycin is conferred by mutations, accompanied by an enhanced antibiotic production in Streptomyces coelicolor A3(2). J Antibiot. 2000;53:1424–7. [DOI] [PubMed] [Google Scholar]
- 28.Ochi K, Okamoto S, Tozawa Y, Inaoka T, Hosaka T, Xu J, Kurosawa K. Ribosome engineering and secondary metabolite production. Adv Appl Microbiol. 2004. 10.1016/S0065-2164(04)56005-7. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Moreno D, Abellon-Ruiz J, Garcia-Heras F, Murillo FJ, Padmanabhan S, Elias-Arnanz M. CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucl Acid Res. 2010;38:4586–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamehiro N, Hosaka T, Xu J, Hu HF, Otake N, Ochi K. Innovative approach for improvement of an antibiotic- overproducing industrial strain of Streptomyces albus. Appl Environ Microb. 2003;69:6412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simunovic V, Zapp J, Rachid S, Krug D, Meiser P, Müller R. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. ChemBioChem. 2006;7:1206–20. [DOI] [PubMed] [Google Scholar]
- 32.Meiser P, Weissman KJ, Bode HB, Krug D, Dickschat JS, Sandmann A, Müller R. DKxanthene biosynthesis-understanding the basis for diversity-oriented secondary metabolism. Chem Biol. 2008;15:771–81. [DOI] [PubMed] [Google Scholar]
- 33.Silakowski B, Nordsiek G, Kunze B, Blöcker H, Müller R. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15. Chem Biol. 2001;8:59–69. [DOI] [PubMed] [Google Scholar]
- 34.Hug JJ, Dastbaz J, Adam S, Revermann O, Koehnke J, Krug D, Müller R. Biosynthesis of cittilins, unusual ribosomally synthesized and post-translationally modified peptides from Myxococcus xanthus. Acs Chem Biol. 2020;15:2221–31. [DOI] [PubMed] [Google Scholar]
- 35.Gerth K, Steinmetz H, Höfle G, Reichenbach H. Studies on the biosynthesis of epothilones: the biosynthetic origin of the carbon skeleton. J Antibiot. 2000;53:1373–7. [DOI] [PubMed] [Google Scholar]
- 36.Zhu LP, Li ZF, Sun X, Li SG, Li YZ. Characteristics and activity analysis of epothilone operon promoters from Sorangium cellulosum strains in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:6857–66. [DOI] [PubMed] [Google Scholar]
- 37.Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol. 2007;374:1065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang GJ, Hosaka T, Ochi K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microb. 2008;74:2834–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuang ZK, Kong WP, Wen ZQ, Tong N, Lin J, Zhang F, Fan ZY, Yi LW, Huang Y, Duan YW, et al. Combinatorial metabolic engineering of Streptomyces sp. CB03234-S for the enhanced production of anthraquinone-fused enediyne tiancimycins. Microbial Cell Fact. 2024. 10.1186/s12934-024-02399-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alifano P, Palumbo C, Pasanisi D, Talà A. Rifampicin-resistance, rpoB polymorphism and RNA polymerase genetic engineering. J Biotechnol. 2015;202:60–77. [DOI] [PubMed] [Google Scholar]
- 41.Achour W, Guenni F, Fines M, Leclercq R, Ben Hassen A. mutations in Streptococcus mitis clinical isolates resistant to rifampin. Antimicrob Agent Ch. 2004;48:2757–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Z, Luo S, Xu XH, Bechthold A, Yu XP. Characterization of representative gene mutations leading to a significant change in toyocamycin production of Streptomyces diastatochromogenes 1628. J Ind Microbiol Biot. 2016;43:463–71. [DOI] [PubMed] [Google Scholar]
- 43.Wang ZL, Sun RZ, Li M, Liu L, Duan YW, Huang Y. Yield improvement of enediyne yangpumicins in Micromonospora yangpuensis through ribosome engineering and fermentation optimization. Biotechnol J. 2021. 10.1002/biot.202100250. [DOI] [PubMed] [Google Scholar]
- 44.Zhao XL, Hussain MH, Mohsin A, Liu ZB, Xu ZX, Li ZX, Guo WQ, Guo MJ. Mechanistic insight for improving butenyl-spinosyn production through combined ARTP/UV mutagenesis and ribosome engineering in Saccharopolyspora pogona. Front Bioeng Biotech. 2024. 10.3389/fbioe.2023.1329859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaikh AA, Nothias LF, Srivastava SK, Dorrestein PC, Tahlan K. Specialized metabolites from ribosome engineered strains of Streptomyces clavuligerus. Metabolites. 2021. 10.3390/metabo11040239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallego-García A, Mirassou Y, García-Moreno D, Elías-Arnanz M, Jiménez MA, Padmanabhan S. Structural insights into RNA polymerase recognition and essential function of Myxococcus xanthus CdnL. PLoS ONE. 2014. 10.1371/journal.pone.0108946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang HM, Liang JH, Yue QW, Li L, Shi Y, Chen GS, Li YZ, Bian XY, Zhang YM, Zhao GP, Ding XM. Engineering the acyltransferase domain of epothilone polyketide synthase to alter the substrate specificity. Microbial Cell Fact. 2021. 10.1186/s12934-021-01578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the European Nucleotide Archive with the primary accession code PRJNA1184456.