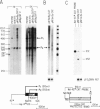
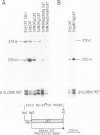
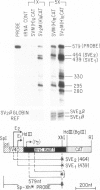
Abstract
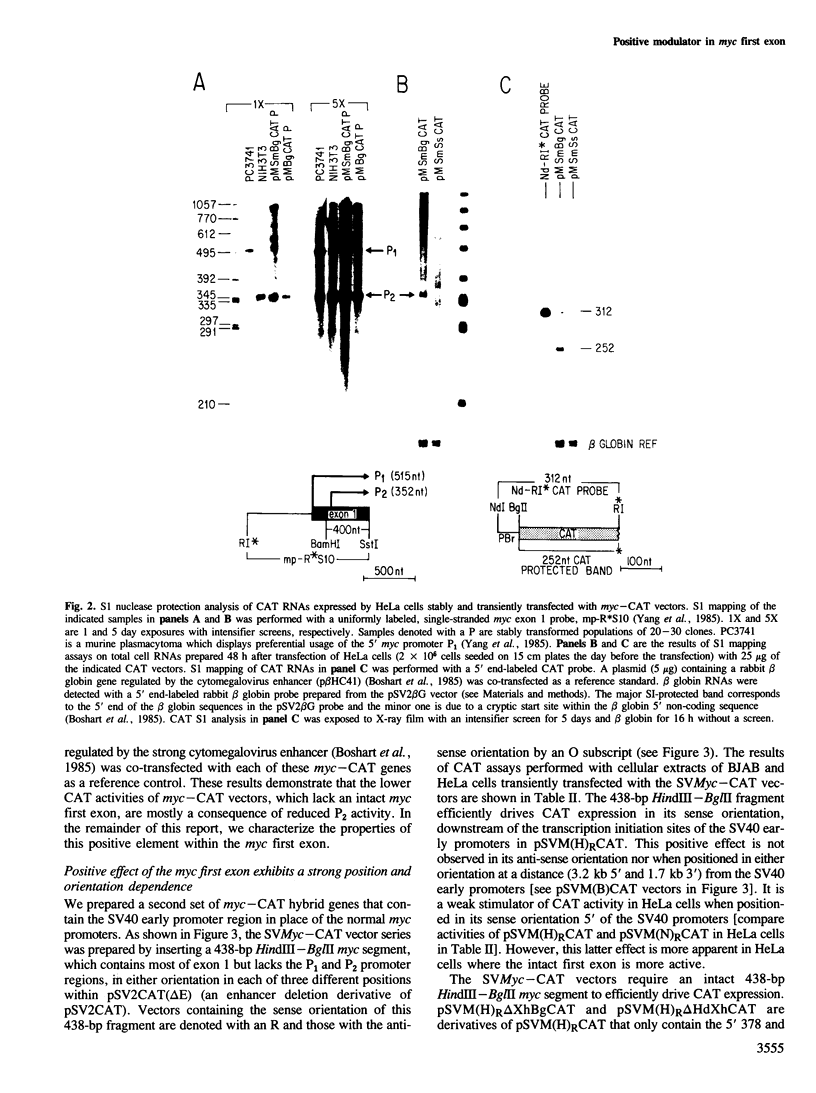
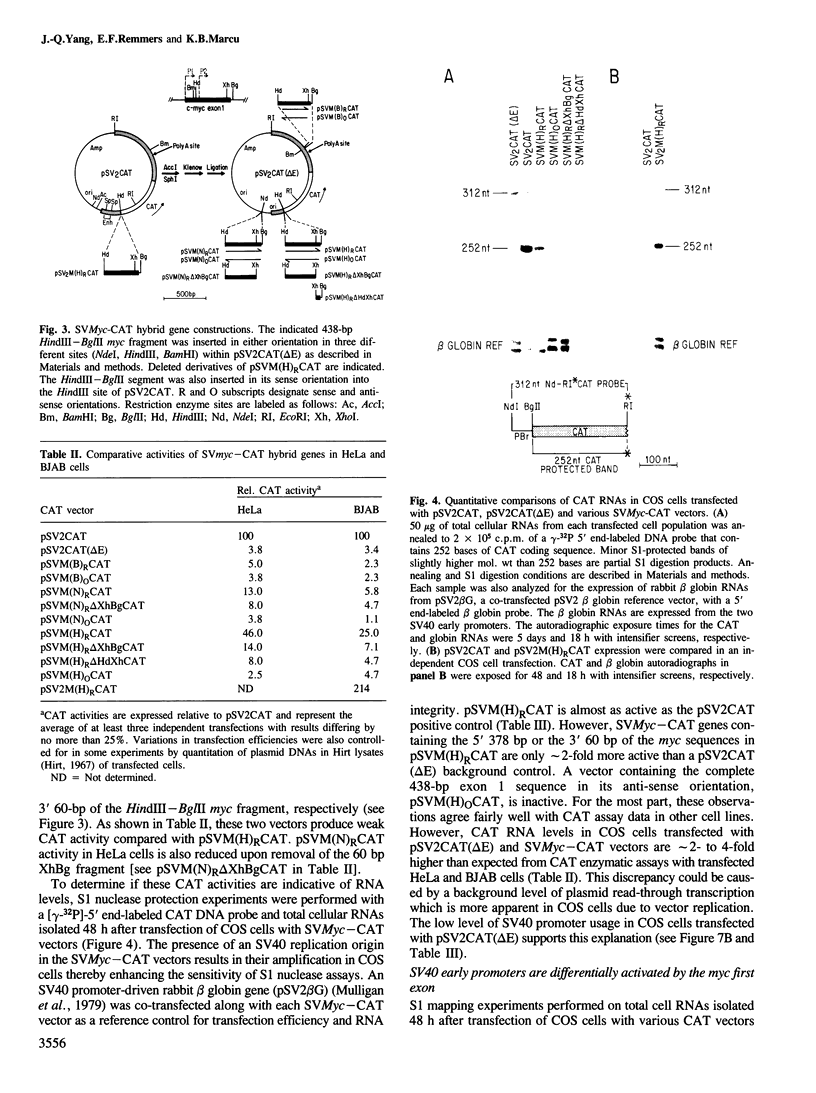
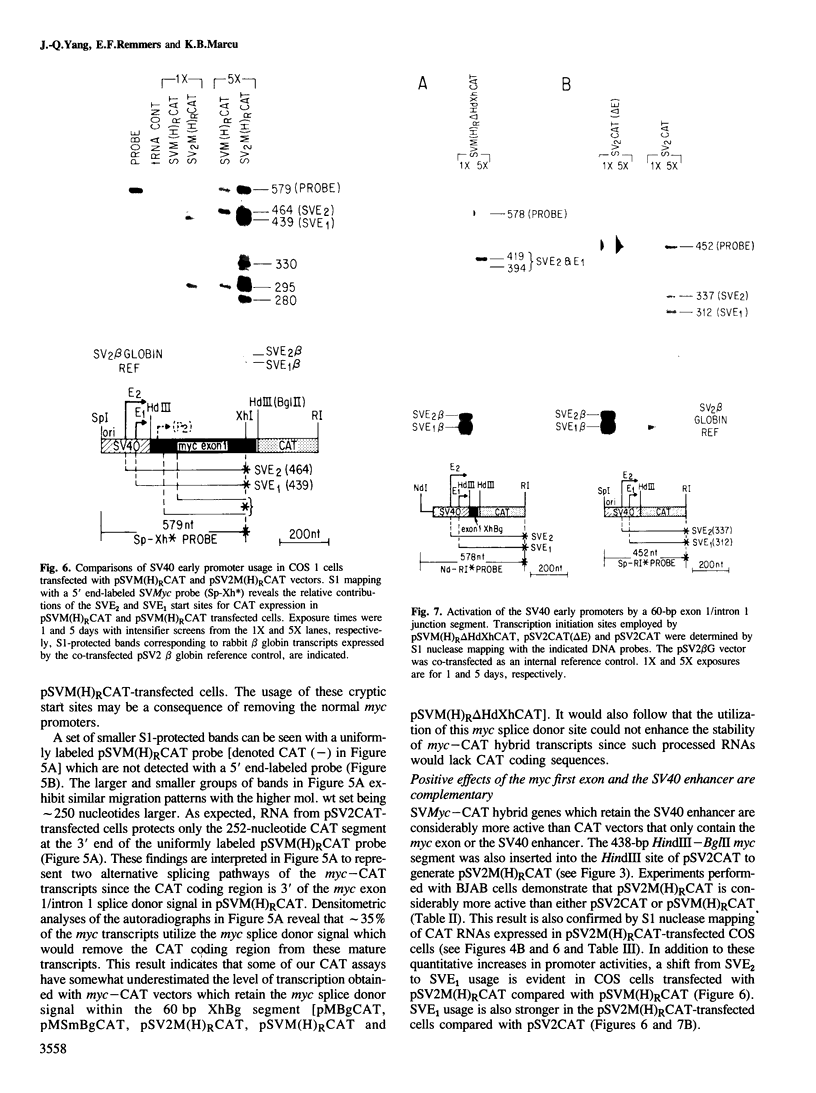
We have identified a positive modulator within the c-myc first exon downstream of the gene's transcription initiation sites, P1 and P2. We introduced myc-CAT (chloramphenicol acetyltransferase) hybrid genes into three cell lines (BJAB, COS and HeLa) and measured their expression by either CAT enzymatic activity, S1 nuclease protection or by a nuclear 'run-on' transcription assay. Removal of 46 bp from the 3' end of the first exon results in a decrease of myc-CAT expression and P2 activity. A 438-bp exon 1 segment, lacking the normal myc promoters, efficiently drives the expression of SV40 early promoters. We find that this first exon segment efficiently functions as a positive modulator only in its sense orientation, 3' of a nearby promoter. The positive effects of the myc first exon and the SV40 enhancer are complementary.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Armelin H. A., Armelin M. C., Kelly K., Stewart T., Leder P., Cochran B. H., Stiles C. D. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984 Aug 23;310(5979):655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bechade C., Calothy G., Pessac B., Martin P., Coll J., Denhez F., Saule S., Ghysdael J., Stéhelin D. Induction of proliferation or transformation of neuroretina cells by the mil and myc viral oncogenes. Nature. 1985 Aug 8;316(6028):559–562. doi: 10.1038/316559a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bernard O., Cory S., Gerondakis S., Webb E., Adams J. M. Sequence of the murine and human cellular myc oncogenes and two modes of myc transcription resulting from chromosome translocation in B lymphoid tumours. EMBO J. 1983;2(12):2375–2383. doi: 10.1002/j.1460-2075.1983.tb01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. M. Cellular oncogenes and retroviruses. Annu Rev Biochem. 1983;52:301–354. doi: 10.1146/annurev.bi.52.070183.001505. [DOI] [PubMed] [Google Scholar]
- Blanchard J. M., Piechaczyk M., Dani C., Chambard J. C., Franchi A., Pouyssegur J., Jeanteur P. c-myc gene is transcribed at high rate in G0-arrested fibroblasts and is post-transcriptionally regulated in response to growth factors. Nature. 1985 Oct 3;317(6036):443–445. doi: 10.1038/317443a0. [DOI] [PubMed] [Google Scholar]
- Boshart M., Weber F., Jahn G., Dorsch-Häsler K., Fleckenstein B., Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985 Jun;41(2):521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Fromm M., Berg P. Complex regulation of simian virus 40 early-region transcription from different overlapping promoters. Mol Cell Biol. 1984 Sep;4(9):1900–1914. doi: 10.1128/mcb.4.9.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butnick N. Z., Miyamoto C., Chizzonite R., Cullen B. R., Ju G., Skalka A. M. Regulation of the human c-myc gene: 5' noncoding sequences do not affect translation. Mol Cell Biol. 1985 Nov;5(11):3009–3016. doi: 10.1128/mcb.5.11.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calame K. L. Mechanisms that regulate immunoglobulin gene expression. Annu Rev Immunol. 1985;3:159–195. doi: 10.1146/annurev.iy.03.040185.001111. [DOI] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Charnay P., Treisman R., Mellon P., Chao M., Axel R., Maniatis T. Differences in human alpha- and beta-globin gene expression in mouse erythroleukemia cells: the role of intragenic sequences. Cell. 1984 Aug;38(1):251–263. doi: 10.1016/0092-8674(84)90547-6. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Colby W. W., Chen E. Y., Smith D. H., Levinson A. D. Identification and nucleotide sequence of a human locus homologous to the v-myc oncogene of avian myelocytomatosis virus MC29. Nature. 1983 Feb 24;301(5902):722–725. doi: 10.1038/301722a0. [DOI] [PubMed] [Google Scholar]
- Coughlin S. R., Lee W. M., Williams P. W., Giels G. M., Williams L. T. c-myc gene expression is stimulated by agents that activate protein kinase C and does not account for the mitogenic effect of PDGF. Cell. 1985 Nov;43(1):243–251. doi: 10.1016/0092-8674(85)90029-7. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Mechti N., Piechaczyk M., Lebleu B., Jeanteur P., Blanchard J. M. Increased rate of degradation of c-myc mRNA in interferon-treated Daudi cells. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4896–4899. doi: 10.1073/pnas.82.15.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau A., Pelletier J., Sonenberg N. Differential efficiencies of in vitro translation of mouse c-myc transcripts differing in the 5' untranslated region. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2315–2319. doi: 10.1073/pnas.82.8.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Campisi J. c-myc regulation during retinoic acid-induced differentiation of F9 cells is posttranscriptional and associated with growth arrest. Mol Cell Biol. 1986 Feb;6(2):518–524. doi: 10.1128/mcb.6.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Ran W., Kindy M. S., Sonenshein G. E., Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986 Jul 15;261(20):9161–9166. [PubMed] [Google Scholar]
- Denny C. T., Hollis G. F., Magrath I. T., Kirsch I. R. Burkitt lymphoma cell line carrying a variant translocation creates new DNA at the breakpoint and violates the hierarchy of immunoglobulin gene rearrangement. Mol Cell Biol. 1985 Nov;5(11):3199–3207. doi: 10.1128/mcb.5.11.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Casey J. W. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science. 1986 Mar 21;231(4744):1437–1440. doi: 10.1126/science.3006241. [DOI] [PubMed] [Google Scholar]
- Dony C., Kessel M., Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985 Oct 17;317(6038):636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Shell B. E., Dery C. DNA sequences near the site of reciprocal recombination between a c-myc oncogene and an immunoglobulin switch region. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7269–7273. doi: 10.1073/pnas.80.23.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Eick D., Piechaczyk M., Henglein B., Blanchard J. M., Traub B., Kofler E., Wiest S., Lenoir G. M., Bornkamm G. W. Aberrant c-myc RNAs of Burkitt's lymphoma cells have longer half-lives. EMBO J. 1985 Dec 30;4(13B):3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrlander P. D., Piechaczyk M., Marcu K. B. Chromatin structure of the murine c-myc locus: implications for the regulation of normal and chromosomally translocated genes. EMBO J. 1985 Dec 1;4(12):3195–3202. doi: 10.1002/j.1460-2075.1985.tb04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazin C., Dupont de Dinechin S., Hampe A., Masson J. M., Martin P., Stehelin D., Galibert F. Nucleotide sequence of the human c-myc locus: provocative open reading frame within the first exon. EMBO J. 1984 Feb;3(2):383–387. doi: 10.1002/j.1460-2075.1984.tb01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P. Simian virus 40 early mRNA's contain multiple 5' termini upstream and downstream from a Hogness-Goldberg sequence; a shift in 5' termini during the lytic cycle is mediated by large T antigen. J Virol. 1981 Oct;40(1):224–240. doi: 10.1128/jvi.40.1.224-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidoni D., Kadonaga J. T., Barrera-Saldaña H., Takahashi K., Chambon P., Tjian R. Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science. 1985 Nov 1;230(4725):511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
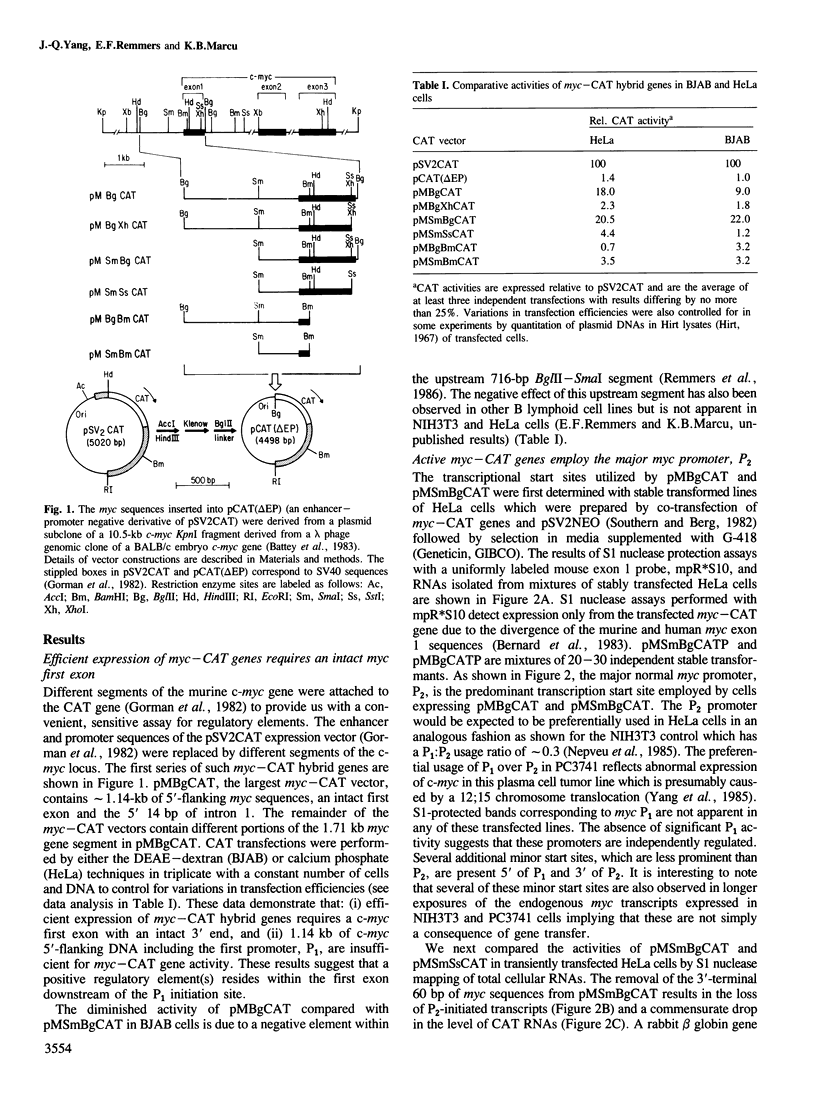
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Hermanowski A. L., Ziff E. B. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol Cell Biol. 1986 Apr;6(4):1050–1057. doi: 10.1128/mcb.6.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985 Jul;41(3):885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- Grosso L. E., Pitot H. C. Transcriptional regulation of c-myc during chemically induced differentiation of HL-60 cultures. Cancer Res. 1985 Feb;45(2):847–850. [PubMed] [Google Scholar]
- Hann S. R., Thompson C. B., Eisenman R. N. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. 1985 Mar 28-Apr 3Nature. 314(6009):366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Jonak G. J., Knight E., Jr Selective reduction of c-myc mRNA in Daudi cells by human beta interferon. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1747–1750. doi: 10.1073/pnas.81.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek L., Hyland J. K., Watt R., Rosenberg M., Baserga R. Microinjected c-myc as a competence factor. Science. 1985 Jun 14;228(4705):1313–1315. doi: 10.1126/science.4001943. [DOI] [PubMed] [Google Scholar]
- Keath E. J., Caimi P. G., Cole M. D. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984 Dec;39(2 Pt 1):339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Knight E., Jr, Anton E. D., Fahey D., Friedland B. K., Jonak G. J. Interferon regulates c-myc gene expression in Daudi cells at the post-transcriptional level. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1151–1154. doi: 10.1073/pnas.82.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman H. M., Hatton K. S., Skoultchi A. I., Schildkraut C. L. c-myc mRNA levels in the cell cycle change in mouse erythroleukemia cells following inducer treatment. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5323–5327. doi: 10.1073/pnas.82.16.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Lee W. M., Schwab M., Westaway D., Varmus H. E. Augmented expression of normal c-myc is sufficient for cotransformation of rat embryo cells with a mutant ras gene. Mol Cell Biol. 1985 Dec;5(12):3345–3356. doi: 10.1128/mcb.5.12.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. L., Grodzicker T., Tjian R. Downstream sequences affect transcription initiation from the adenovirus major late promoter. Mol Cell Biol. 1986 Jul;6(7):2684–2694. doi: 10.1128/mcb.6.7.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCormack J. E., Pepe V. H., Kent R. B., Dean M., Marshak-Rothstein A., Sonenshein G. E. Specific regulation of c-myc oncogene expression in a murine B-cell lymphoma. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5546–5550. doi: 10.1073/pnas.81.17.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill G. F., Hauschka S. D., McKnight S. L. tk Enzyme expression in differentiating muscle cells is regulated through an internal segment of the cellular tk gene. Mol Cell Biol. 1984 Sep;4(9):1777–1784. doi: 10.1128/mcb.4.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosthaf L., Pawlita M., Gruss P. A viral enhancer element specifically active in human haematopoietic cells. Nature. 1985 Jun 13;315(6020):597–600. doi: 10.1038/315597a0. [DOI] [PubMed] [Google Scholar]
- Mougneau E., Lemieux L., Rassoulzadegan M., Cuzin F. Biological activities of v-myc and rearranged c-myc oncogenes in rat fibroblast cells in culture. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5758–5762. doi: 10.1073/pnas.81.18.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan R. C., Howard B. H., Berg P. Synthesis of rabbit beta-globin in cultured monkey kidney cells following infection with a SV40 beta-globin recombinant genome. Nature. 1979 Jan 11;277(5692):108–114. doi: 10.1038/277108a0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Fahrlander P. D., Yang J. Q., Marcu K. B. Amplification and altered expression of the c-myc oncogene in A-MuLV-transformed fibroblasts. Nature. 1985 Oct 3;317(6036):440–443. doi: 10.1038/317440a0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., ar-Rushdi A., Erikson J., Watt R., Rovera G., Croce C. M. Differential expression of the normal and of the translocated human c-myc oncogenes in B cells. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4822–4826. doi: 10.1073/pnas.80.15.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H., Gray H. E., Godeau F. Growth-dependent synthesis of c-myc-encoded proteins: early stimulation by serum factors in synchronized mouse 3T3 cells. Mol Cell Biol. 1985 Nov;5(11):2903–2912. doi: 10.1128/mcb.5.11.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczyk M., Bonnieu A., Eick D., Remmers E., Yang J. Q., Marcu K., Jeanteur P., Blanchard J. M. Altered c-myc RNA metabolism in Burkitt's lymphomas and mouse plasmacytomas. Curr Top Microbiol Immunol. 1986;132:331–338. doi: 10.1007/978-3-642-71562-4_49. [DOI] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Rabbitts P. H., Forster A., Stinson M. A., Rabbitts T. H. Truncation of exon 1 from the c-myc gene results in prolonged c-myc mRNa stability. EMBO J. 1985 Dec 30;4(13B):3727–3733. doi: 10.1002/j.1460-2075.1985.tb04141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts T. H., Forster A., Hamlyn P., Baer R. Effect of somatic mutation within translocated c-myc genes in Burkitt's lymphoma. Nature. 1984 Jun 14;309(5969):592–597. doi: 10.1038/309592a0. [DOI] [PubMed] [Google Scholar]
- Rapp U. R., Cleveland J. L., Brightman K., Scott A., Ihle J. N. Abrogation of IL-3 and IL-2 dependence by recombinant murine retroviruses expressing v-myc oncogenes. Nature. 1985 Oct 3;317(6036):434–438. doi: 10.1038/317434a0. [DOI] [PubMed] [Google Scholar]
- Reed J. C., Nowell P. C., Hoover R. G. Regulation of c-myc mRNA levels in normal human lymphocytes by modulators of cell proliferation. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4221–4224. doi: 10.1073/pnas.82.12.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma P. H., Rothberg P. G., Astrin S. M., Trial J., Bar-Shavit Z., Hall A., Teitelbaum S. L., Kahn A. J. Regulation of myc gene expression in HL-60 leukaemia cells by a vitamin D metabolite. Nature. 1983 Dec 1;306(5942):492–494. doi: 10.1038/306492a0. [DOI] [PubMed] [Google Scholar]
- Remmers E. F., Yang J. Q., Marcu K. B. A negative transcriptional control element located upstream of the murine c-myc gene. EMBO J. 1986 May;5(5):899–904. doi: 10.1002/j.1460-2075.1986.tb04301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Haseltine W. A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985 Jul;41(3):813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- Saito H., Hayday A. C., Wiman K., Hayward W. S., Tonegawa S. Activation of the c-myc gene by translocation: a model for translational control. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7476–7480. doi: 10.1073/pnas.80.24.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Hagenbüchle O., Wellauer P. K., Pittet A. C. Two promoters of different strengths control the transcription of the mouse alpha-amylase gene Amy-1a in the parotid gland and the liver. Cell. 1983 Jun;33(2):501–508. doi: 10.1016/0092-8674(83)90431-2. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Stewart T. A., Bellvé A. R., Leder P. Transcription and promoter usage of the myc gene in normal somatic and spermatogenic cells. Science. 1984 Nov 9;226(4675):707–710. doi: 10.1126/science.6494906. [DOI] [PubMed] [Google Scholar]
- Stewart T. A., Pattengale P. K., Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984 Oct;38(3):627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Thompson C. B., Challoner P. B., Neiman P. E., Groudine M. Levels of c-myc oncogene mRNA are invariant throughout the cell cycle. 1985 Mar 28-Apr 3Nature. 314(6009):363–366. doi: 10.1038/314363a0. [DOI] [PubMed] [Google Scholar]
- Vennström B., Kahn P., Adkins B., Enrietto P., Hayman M. J., Graf T., Luciw P. Transformation of mammalian fibroblasts and macrophages in vitro by a murine retrovirus encoding an avian v-myc oncogene. EMBO J. 1984 Dec 20;3(13):3223–3229. doi: 10.1002/j.1460-2075.1984.tb02282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Stanton L. W., Marcu K. B., Gallo R. C., Croce C. M., Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983 Jun 23;303(5919):725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- Yang J. Q., Bauer S. R., Mushinski J. F., Marcu K. B. Chromosome translocations clustered 5' of the murine c-myc gene qualitatively affect promoter usage: implications for the site of normal c-myc regulation. EMBO J. 1985 Jun;4(6):1441–1447. doi: 10.1002/j.1460-2075.1985.tb03800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Q., Mushinski J. F., Stanton L. W., Fahrlander P. D., Tesser P. C., Marcu K. B. Modes of c-myc oncogene activation in murine plasmacytomas. Curr Top Microbiol Immunol. 1984;113:146–153. doi: 10.1007/978-3-642-69860-6_25. [DOI] [PubMed] [Google Scholar]
- Zech L., Haglund U., Nilsson K., Klein G. Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int J Cancer. 1976 Jan 15;17(1):47–56. doi: 10.1002/ijc.2910170108. [DOI] [PubMed] [Google Scholar]
- Zerlin M., Julius M. A., Cerni C., Marcu K. B. Biological effects of high level c-myc expression in FR3T3 fibroblasts. Curr Top Microbiol Immunol. 1986;132:320–326. doi: 10.1007/978-3-642-71562-4_47. [DOI] [PubMed] [Google Scholar]
- Zullo J. N., Cochran B. H., Huang A. S., Stiles C. D. Platelet-derived growth factor and double-stranded ribonucleic acids stimulate expression of the same genes in 3T3 cells. Cell. 1985 Dec;43(3 Pt 2):793–800. doi: 10.1016/0092-8674(85)90252-1. [DOI] [PubMed] [Google Scholar]